Abstract
This study aimed to describe a Bacillus subtilis expression system based on genetically modified B. subtilis. Abaecin, an antimicrobial peptide obtained from Apis mellifera, can enhance the effect of pore-forming peptides from other species on the inhibition of bacterial growth. For the exogenous expression, the abaecin gene was fused with a tobacco etch virus protease cleavage site, a promoter Pglv, and a mature beta-glucanase signal peptide. Also, a B. subtilis expression system was constructed. The recombinant abaecin gene was expressed and purified as a recombinant protein in the culture supernatant. The purified abaecin did not inhibit the growth of Escherichia coli strain K88. Cecropin A and hymenoptaecin exhibited potent bactericidal activities at concentrations of 1 and 1.5 μM. Combinatorial assays revealed that cecropin A and hymenoptaecin had sublethal concentrations of 0.3 and 0.5 μM. This potentiating functional interaction represents a promising therapeutic strategy. It provides an opportunity to address the rising threat of multidrug-resistant pathogens that are recalcitrant to conventional antibiotics.
Keywords: Abaecin, Antimicrobial peptides, Bacillus subtilis, Expression
Introduction
Antimicrobial peptides (AMPs) are important components of the innate immune defense against microbial pathogens in a wide range of organisms.1 Abaecin is a major AMP found in the honeybees. It was originally isolated from the insect Apis mellifera.2 The bumblebee long-chain proline-rich peptide abaecin can reduce the minimal inhibitory concentrations.3 However, the expression of abaecin is extremely limited in honeybees. Therefore, a highly efficient expression system is needed for abaecin to allow its commercial application.
Abaecin consists of a single polypeptide chain of common amino acids and is well suited for economical production through the application of recombinant DNA technology or peptide synthesis.4 It can potentiate the activity of pore-forming AMPs from other species, for example, cecropin A and stomoxyn.5 This potentiating functional interaction represents a promising therapeutic strategy because the potency and range of combinations of AMPs are likely to extend well beyond the capabilities of individual peptides.6 It provides an opportunity to address the rising threat of multidrug-resistant pathogens that are recalcitrant to conventional antibiotics.7
The expression of foreign genes in Escherichia coli often leads to the formation of densely packed denatured peptide molecules in the form of insoluble particles called inclusion bodies.8 Inclusion body proteins are devoid of biological activity. An elaborate process involving solubilization, refolding, and purification is needed to partially recover a functionally active product.9 A yeast expression system, such as Pichia pastoris, may solve the problem of post-translational modifications.10 It needs significant investment during the later period of batch cultivation, rendering it uneconomical.11
The Bacillus subtilis expression system does not have the disadvantages of the two aforementioned expression systems and can potentially serve as an efficient expression host,12 especially for the secretion of heterologous proteins.13 Moreover, the secreted foreign proteins usually remain in biologically active forms,14 and the downstream purification is greatly simplified.15 Also, it has other advantages, such as theoretically higher yield, no aggregation of the product, and the possibility for continuous cultivation and production.16
This study aimed to describe a B. subtilis expression system based on B. subtilis cells genetically modified with a small tobacco etch virus (TEV) protease and two-cistron expression vector gene of abaecin fused to TEV.17
Materials and methods
Enzymes, chemicals, and antibiotics
T4 DNA ligase, Taq DNA polymerase, and all restriction enzymes were purchased from Promega (WI, USA). The protein marker was obtained from TaKaRa Biotechnology (Shiga, Japan). Antibiotics were purchased from Sigma (MO, USA). All the other chemicals used were of the highest grade commercially available.
Bacterial strains and plasmids
The gene encoding the antibacterial peptide abaecin, with the previously reported sequence FVPYNPPRPGQSKPFPSFPGHGPFNPK IQWPYPLPNPGH,18 was synthesized using a TEV protease cleavage locus gene, a beta-glucanase mature peptide, and signal peptide gene (gmp and gsp) as a full-length nucleotide by standard solid-phase methods at the AUGCT Biotechnology Company (Beijing, China). The recombinant plasmid named ABA containing the abaecin gene, a TEV protease cleavage locus gene, and a beta-glucanase mature peptide (gmp) with the signal peptide (gsp) were supplied by the AUGCT Biotechnology Company (Beijing, China). The E. coli DH5α, shuttle vector pGJ103, and promoter Pglv were obtained from National Feed Engineering Technology Research Center (Beijing, China). Preparation of plasmid DNA from E. coli cells and transformation of B. subtilis were carried out using standard procedures.19
Plasmid construction and expression
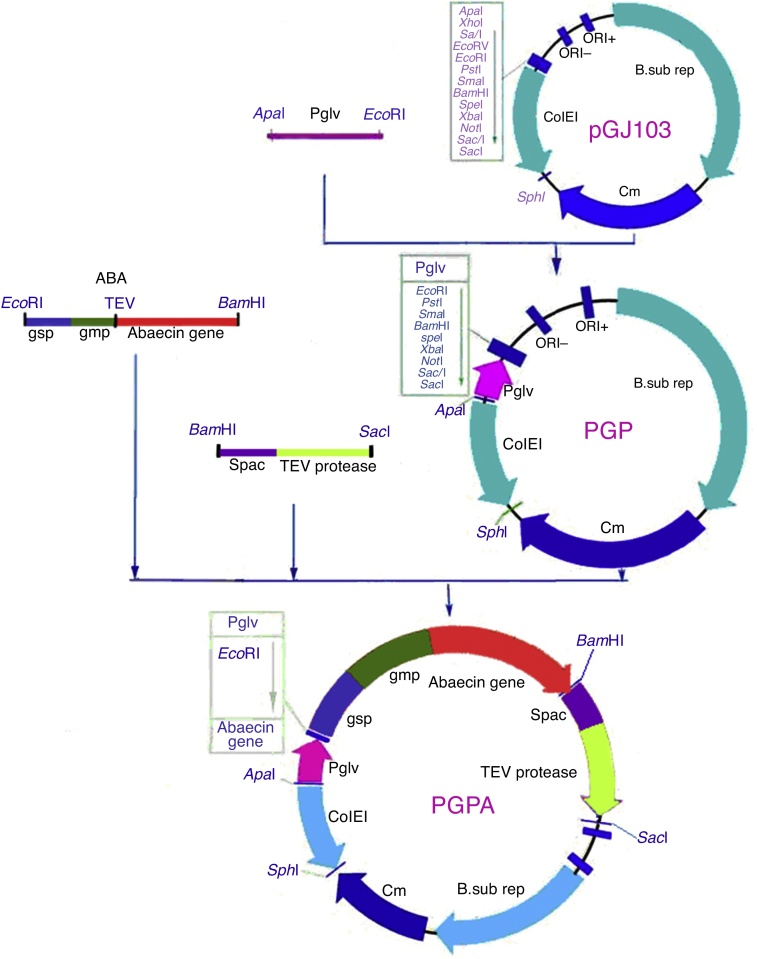
After cloning using T vector, an intact plasmid including the promoter Pglv was inserted into plasmid pGJ103 (3.2 kb), resulting in the construction of pGP (3.45 kb). The plasmid containing the genes of abaecin and TEV protein, with gmp and gsp, was digested with EcoRI and BamHI, while pGP was digested with EcoRI and BamHI. The two digested products were linked by T4 DNA ligase, yielding recombinant plasmid pGPA (Fig. 1). The operon, including the inducible promoter spac, and the TEV protease gene were inserted between the site BamHI and SacI of plasmid pGPA. The plasmid pGPA was transformed into E. coli DH5α, and the positive transformant was screened at a final concentration of 5 μg/mL chloromycin. The positive clones were further identified with the restriction endonuclease digestion of BamHI and SacI. The sequence was identified by the SinoGenoMax Company (Beijing, China).
Fig. 1.
Construction of recombinant plasmid pGPA. After cloning using T vector, an intact plasmid including the promoter Pglv was inserted into plasmid pGJ103 (3.2 kb), resulting in the construction of pGP (3.45 kb). The plasmid containing the genes of abaecin and TEV protein, with gmp and gsp, was digested with EcoRI and BamHI, while pGP was digested with EcoRI and BamHI. The two digested products were linked by T4 DNA ligase, yielding recombinant plasmid pGPA.
The plasmid pGPA extracted from the positive transformant was transformed into the expression vector B. subtilis 1A747 with chloramphenicol resistance. The clones were picked with shaking at 37 °C overnight in Luria-Bertani (LB) broth containing 50 μg/mL of chloramphenicol. After this, the culture was inoculated into new LB broth at a ratio of 1:100 with shaking at 37 °C until the optical density at 600 nm reached 0.8 − 1.0. The culture was induced with 1% glucose and incubated at 37 °C for an additional 24 h with a rotation speed of 250 rpm. Then, the supernatant was collected by centrifugation. Various pH values of the culture medium (pH 3.0 − 7.0) were tested for the optimal expression of recombinant proteins. It was found that pH 6.0 of the medium was the optimal condition for the expression of abaecin (data not shown).
Purification of the recombinant abaecin
The supernatant was first filtered twice using an Amicon ultrafiltration device (Millipore, MA, USA). The supernatant, containing proteins ranging from 3 to 10 kDa, was dialyzed overnight in 0.1 M sodium acetate and then applied to a CM-Sepharose CL-6B column (Pharmacia Biosciences, NJ, USA) pre-equilibrated with 0.1 M sodium acetate (pH 5.0). The column was washed with 0.1 M acetate buffer, and the proteins were eluted with a linear gradient of 0.1–1.0 M sodium acetate (pH 5.0). A sample (100 μL) of the filtrate was collected and applied to semi-preparative reversed-phase high-performance liquid chromatography (HPLC) on a C18 column (250 × 4.6 mm2, 5 μm, and 300 × Å), equilibrated in 0.1% trifluoroacetic acid and 18% acetonitrile. The bound protein was eluted with a linear gradient of acetonitrile (18% – 45%, v/v) in 0.1% trifluoroacetic acid. The flow rate was 1.0 mL/min, and the absorbance of eluted protein was monitored at 280 nm. The peak of the chimeric abaecin eluted at 31% acetonitrile from reversed-phase HPLC was determined by analyzing fractions on Tricine-sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).20 The fractions containing abaecin were collected for the next step. The sequence of recombinant abaecin was determined by the SinoGenoMax Company (Beijing, China)
Antimicrobial activities
Growth inhibition assays: E. coli K88ac was purchased from the China Veterinary Culture Collection Center (Beijing, China). The bacteria were cultivated and incubated in LB medium (0.5% NaCl, 0.5% yeast extract, 1% tryptone, pH 7). All strains were then stored at −80 °C with 20% sterile glycerol until needed.21
Growth inhibition was determined in sterilized 96-well plates in a final volume of 200 μL using the microdilution assay as described previously.22 A stock solution of the peptides was diluted tenfold in the culture medium and 100 μL of LB medium. The bacteria (2 × 107 to 4 × 107 cells/mL) were added to 100 μL of LB medium and peptide solution (serial twofold dilutions). The OD600 was measured every 20 min for 16 h in an Eon Microplate Spectrophotometer (BioTek Instruments, VT, USA). Control cultures with no AMPs were included in the assays.
Results
Plasmid constructions and expression
The recombinant plasmid ABA with a restriction enzyme EcoRI cleavage site at the 3′-terminus and a BamHI cleavage site at the 5′-terminus was linked with pGP (digested with EcoRI and BamHI) by T4 DNA ligase. As shown in Fig. 1, the constitutive expression vector pGPA, containing the operon of inducible promoter spac and TEV protease gene, was constructed. B. subtilis supplied a favorable base for expression. After a three-step construction process, the recombinant plasmid containing the fusion gene of abaecin and TEV was transformed into B. subtilis 1A747 for expression.
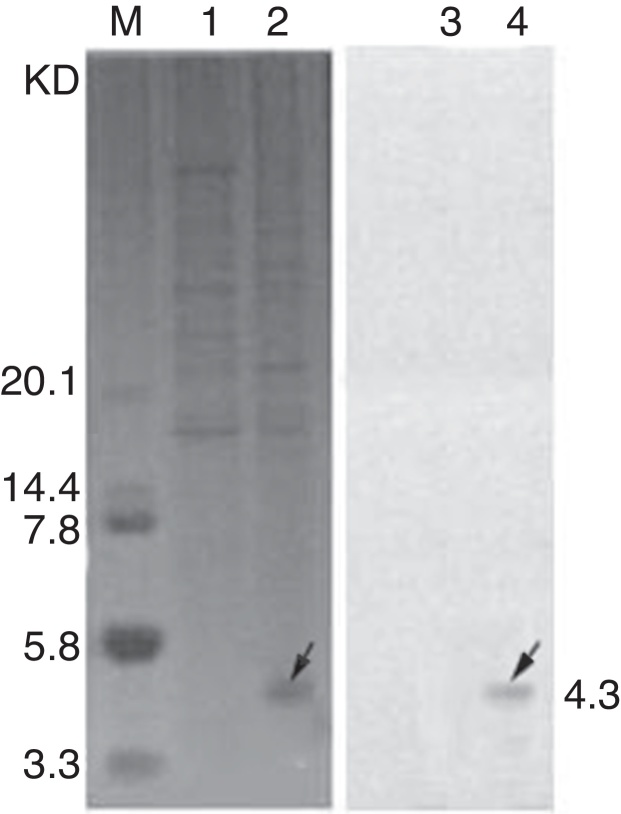
During the initial stage of expression, the expressed fusion protein including abaecin and TEV had no antimicrobial activity. This ensured that the expressed product would not exert a deleterious effect on B. subtilis host strain, and the host strain would continuously express the fusion protein within a certain period. Also, the fusion protein was expressed at a higher level when isopropyl β-d-1-thiogalactopyranoside (IPTG) was added. The cultivation was continued for a certain period after induction with IPTG, and then the culture conditions were adjusted (data not shown) to release TEV protease into the culture medium. The targeted expressed protein abaecin was activated on the hydrolysis of TEV protease to TEV. As shown in Fig. 2, the recombinant abaecin was expressed in B. subtilis at a quite high level. The expressed protein was released directly into the culture medium and obtained from the supernatant after centrifugation.
Fig. 2.
Expression of abaecin in Bacillus subtilis 1A747. The analysis of purified abaecin on Tricine-SDS-PAGE and Western Blot revealed its molecular mass to be about 4 kDa. (a) SDS-PAGE analysis of abaecin expressed. Lane 1, total cell protein (uninduced); lane 2, total cell protein (induced); lane M, low-range molecular mass marker (TaKaRa, Shiga, Japan). (b) Western blot analysis of abaecin expressed. Lane 3, total cell protein (uninduced); lane 4, total cell protein (induced). The arrow indicates the position of recombinant abaecin.
Purification of the modified abaecin
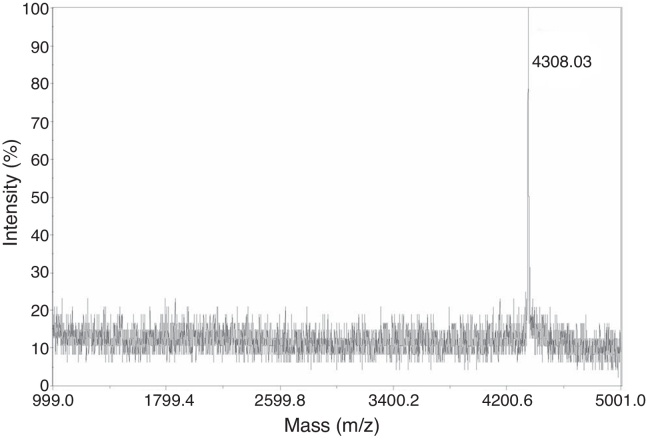
Following highly efficient purification including ultrafiltration and reversed-phase HPLC, pure chimeric antibacterial peptide abaecin was obtained from the culture medium. The analysis of purified abaecin on Tricine-SDS-PAGE revealed its molecular mass to be about 4 kDa (Fig. 2). Electrospray ionization mass spectrometry of the purified abaecin demonstrated a single, nondispersed signal with a molecular mass of 3.9 kDa (Fig. 3). The sequence of recombinant abaecin was identified as consistent with the theoretical sequence.
Fig. 3.
Analysis of purified recombinant abaecin by electrospray ionization mass spectrometry. Electrospray ionization mass spectrometry of the purified abaecin demonstrated a single, nondispersed signal with a molecular mass of 3.9 kDa.
Potency of the AMPs in bacterial growth inhibition assays
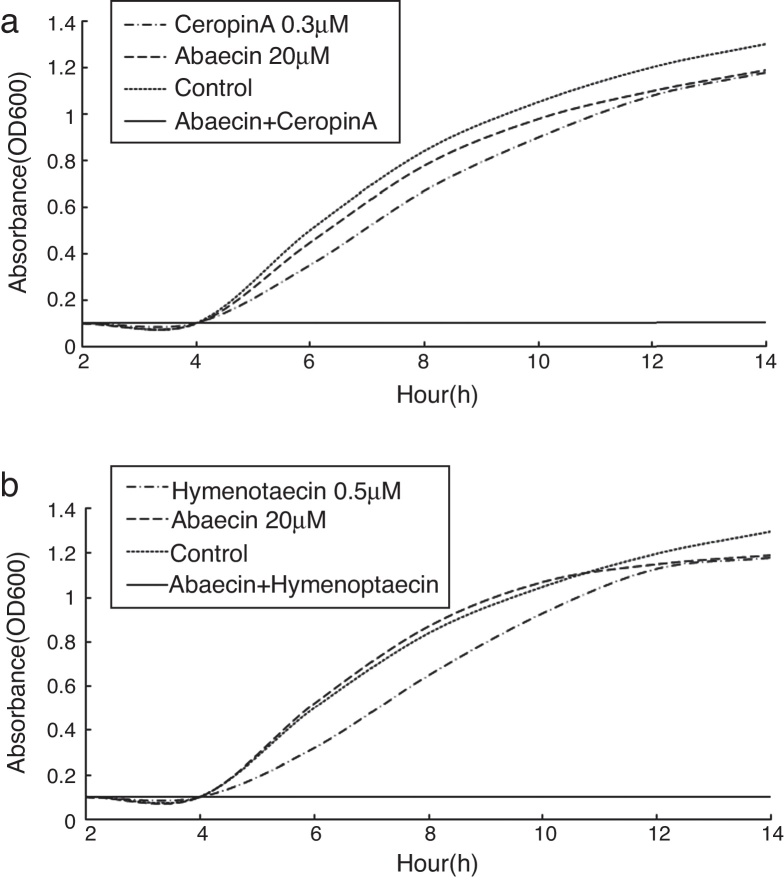
E. coli strain K88 showed no susceptibility in the presence of up to 200 μM abaecin and entered the exponential growth phase∼4 h after the initiation of cultivation. The conclusion was consistent with the findings of Rahnamaeian.23 In contrast, cecropin A and hymenoptaecin exhibited potent bactericidal activities at concentrations of 1 and 1.5 μM, respectively. The sublethal concentrations of the AMPs were determined by preparing serial dilutions and repeating the growth inhibition assays. This revealed that cecropin A and hymenoptaecin had sublethal concentrations of 0.3 and 0.5 μM, respectively (data not shown). Combinatorial assays were carried out by supplementing the sublethal doses of each pore-forming peptide with 20 μM abaecin (Fig. 4).
Fig. 4.
Escherichia coli growth inhibition assays. (a) E. coli strain K88 in the mid-logarithmic phase was incubated with the medium (control) or with abaecin and hymenoptaecin, alone or in combination. (b) E. coli strain K88 in the mid-logarithmic phase was incubated with the medium (control) or with abaecin and cecropin A, alone or in combination. The growth rate was determined by measuring the optical density of the culture at 600 nm.
Discussion
Honeybee AMPs are a family of small polypeptides with great potential for antibacterial application.24 However, to date, a few reports are available detailing the heterologous expression of recombinant honeybee AMPs. Therefore, in the present study, B. subtilis was used as a host for the high-level expression and secretion of chimeric peptide abaecin. The introduction of TEV into the fusion gene offered a favorable environment for the expression of abaecin. Abaecin was produced in B. subtilis with the help of a special operon without any damage to the host strain.
Producing AMPs (such as honeybee AMPs) with high efficiency is a significant challenge for developing commercial products.25 In the present study, the B. subtilis expression system was adopted for the constitutive expression of recombinant abaecin.25 To facilitate the expression of abaecin, this system included the TEV protease gene, which could release under appropriate conditions and ensure the cleavage of the target gene.26 Also, the modified expression system did not require an inducer during expression27 and did not produce inclusion bodies.23 Therefore, the system had the advantages of high efficiency of expression at low cost, easy operation, and application to industrial production. The recombinant abaecin was expressed at a quite high level (up to about 1 g/L) in bacterial cell culture.
The analysis of purified abaecin using Tricine-SDS-PAGE revealed its molecular mass to be about 4 kDa, which was consistent with the theoretical molecular mass of 3.966 kDa obtained from electrospray ionization mass spectrometry.
It has been shown previously that abaecin alone does not affect bacterial proliferation at concentrations of up to 200 μM, but can potentiate the activity of hymenoptaecin (a pore-forming peptide also from the bumblebee) and thus reduce the minimum inhibitory concentrations of hymenoptaecin required for membrane permeabilization.28 This interaction can be specific, given that the two AMPs are naturally co-expressed in response to a bacterial challenge, but can also represent a more general mechanism.
Antibiotics are currently widely used as therapeutic agents and growth stimulants for farm animals. However, the use of antibiotics in animal feed should be completely banned due to concerns regarding the presence of residues in animal products and the development of bacterial resistance to antibiotics.29 Consequently, the development of alternatives to antibiotics needs considerable attention.30 The fact that abaecin could enhance the antimicrobial activity against specific tested organisms t suggests that one potential application of abaecin may be in the livestock industry, since bacteria such as staphylococci, E. coli, and salmonella are some of the primary organisms causing infectious diseases in livestock.31, 32
In summary, a novel B. subtilis expression system for abaecin based on B. subtilis cells genetically modified with a TEV protease and the two-cistron expression vector gene of abaecin fused to TEV was developed in this study. Abaecin could enhance the effect of pore-forming peptides from other species on the inhibition of bacterial growth. These findings indicated that the B. subtilis expression system could be applied as a powerful tool for producing abaecin at a larger scale, which is expected to become a useful antimicrobial or even therapeutic agent in animals or even human beings. A further study would test this hypothesis through some practical feeding trials.
Conflict of interest
The authors declared that they have no conflict of interest.
Associate Editor: Gisele Monteiro de Souza
Contributor Information
Li Li, Email: 609532398@qq.com.
Ruiping Hu, Email: 783674348@qq.com.
References
- 1.Xu J., Zhong F., Zhang Y. Construction of Bacillus subtilis strain engineered for expression of porcine beta-defensin-2/cecropin P1 fusion antimicrobial peptides and its growth-promoting effect and antimicrobial activity. Asian-Australas J Anim Sci. 2016 doi: 10.5713/ajas.16.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casteels P., Ampe C., Riviere L. Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera) Eur J Biochem. 1990;187:381–386. doi: 10.1111/j.1432-1033.1990.tb15315.x. [DOI] [PubMed] [Google Scholar]
- 3.Gu M.J., Song S.K., Park S.M., Lee I.K., Yun C.H. Bacillus subtilis protects porcine intestinal barrier from deoxynivalenol via improved zonula occludens-1 expression. Asian-Australas J Anim Sci. 2014;27:580–586. doi: 10.5713/ajas.2013.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan C., Cui W., Cheng J. Construction of a highly active secretory expression system via an engineered dual promoter and a highly efficient signal peptide in Bacillus subtilis. N Biotechnol. 2016;33:372–379. doi: 10.1016/j.nbt.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Gupta M., Rao K.K. Phosphorylation of DegU is essential for activation of amyE expression in Bacillus subtilis. J Biosci. 2014;39:747–752. doi: 10.1007/s12038-014-9481-5. [DOI] [PubMed] [Google Scholar]
- 6.He Q., Fu A.Y., Li T.J. Expression and one-step purification of the antimicrobial peptide cathelicidin-BF using the intein system in Bacillus subtilis. J Ind Microbiol Biotechnol. 2015;42:647–653. doi: 10.1007/s10295-014-1582-5. [DOI] [PubMed] [Google Scholar]
- 7.Hemila H., Pakkanen R., Heikinheimo R., Palva E.T., Palva I. Expression of the Erwinia carotovora polygalacturonase-encoding gene in Bacillus subtilis: role of signal peptide fusions on production of a heterologous protein. Gene. 1992;116:27–33. doi: 10.1016/0378-1119(92)90625-y. [DOI] [PubMed] [Google Scholar]
- 8.Heng C., Chen Z., Du L., Lu F. Expression and secretion of an acid-stable alpha-amylase gene in Bacillus Subtilis by SacB promoter and signal peptide. Biotechnol Lett. 2005;27:1731–1737. doi: 10.1007/s10529-005-2743-4. [DOI] [PubMed] [Google Scholar]
- 9.Huang X., Li Z., Du C., Wang J., Li S. Improved expression and characterization of a multidomain xylanase from Thermoanaerobacterium aotearoense SCUT27 in Bacillus subtilis. J Agric Food Chem. 2015;63:6430–6439. doi: 10.1021/acs.jafc.5b01259. [DOI] [PubMed] [Google Scholar]
- 10.Ji S.H., Gururani M.A., Chun S.C. Expression analysis of rice pathogenesis-related proteins involved in stress response and endophytic colonization properties of gfp-tagged Bacillus subtilis CB-R05. Appl Biochem Biotechnol. 2014;174:231–241. doi: 10.1007/s12010-014-1047-3. [DOI] [PubMed] [Google Scholar]
- 11.Jia M., Xu M., He B., Rao Z. Cloning, expression, and characterization of l-asparaginase from a newly isolated Bacillus subtilis B11-06. J Agric Food Chem. 2013;61:9428–9434. doi: 10.1021/jf402636w. [DOI] [PubMed] [Google Scholar]
- 12.Joyet P., Derkaoui M., Poncet S., Deutscher J. Control of Bacillus subtilis mtl operon expression by complex phosphorylation-dependent regulation of the transcriptional activator MtlR. Mol Microbiol. 2010;76:1279–1294. doi: 10.1111/j.1365-2958.2010.07175.x. [DOI] [PubMed] [Google Scholar]
- 13.Khezri M., Jouzani G.S., Ahmadzadeh M. Fusarium culmorum affects expression of biofilm formation key genes in Bacillus subtilis. Braz J Microbiol. 2016;47:47–54. doi: 10.1016/j.bjm.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S.J., Pan J.G., Park S.H., Choi S.K. Development of a stationary phase-specific autoinducible expression system in Bacillus subtilis. J Biotechnol. 2010;149:16–20. doi: 10.1016/j.jbiotec.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 15.Li R.K., Chen P., Ng T.B. Highly efficient expression and characterization of a beta-mannanase from Bacillus subtilis in Pichia pastoris. Biotechnol Appl Biochem. 2015;62:64–70. doi: 10.1002/bab.1250. [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Li Z., Yamanaka K. Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Sci Rep. 2015;5:9383. doi: 10.1038/srep09383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Berg S., Lofdahl P.A., Hard T., Berglund H. Improved solubility of TEV protease by directed evolution. J Biotechnol. 2006;121:291–298. doi: 10.1016/j.jbiotec.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Lu F., Chen G. High-level expression, purification and characterization of a recombinant medium-temperature alpha-amylase from Bacillus subtilis. Biotechnol Lett. 2010;32:119–124. doi: 10.1007/s10529-009-0112-4. [DOI] [PubMed] [Google Scholar]
- 19.Lu Q., Ma J., Rong H. Cloning, expression, purification, crystallization and preliminary crystallographic analysis of 5-aminolaevulinic acid dehydratase from Bacillus subtilis. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1053–1055. doi: 10.1107/S1744309110027582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma P., Patching S.G., Ivanova E. Allantoin transport protein, PucI, from Bacillus subtilis: evolutionary relationships, amplified expression, activity and specificity. Microbiology. 2016;162:823–836. doi: 10.1099/mic.0.000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michna R.H., Commichau F.M., Todter D., Zschiedrich C.P., Stulke J. SubtiWiki-a database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res. 2014;42:D692–D698. doi: 10.1093/nar/gkt1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa T., Iwata T., Kaneko S., Itaya M., Hirota J. An inducible recA expression Bacillus subtilis genome vector for stable manipulation of large DNA fragments. BMC Genomics. 2015;16:209. doi: 10.1186/s12864-015-1425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahnamaeian M., Cytrynska M., Zdybicka-Barabas A., Vilcinskas A. The functional interaction between abaecin and pore-forming peptides indicates a general mechanism of antibacterial potentiation. Peptides. 2016;78:17–23. doi: 10.1016/j.peptides.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Saltykova E.S., L’Vov A.V., Ben’kovskaia G.V., Poskriakov A.V., Nikolenko A.G. [Difference in the gene expression of antibacterial peptides abaecin, hymenoptaecin, defensin in bees Apis mellifera and Apis mellifera caucasica] Zh Evol Biokhim Fiziol. 2005;41:404–407. [PubMed] [Google Scholar]
- 25.Phan T.T., Nguyen H.D., Schumann W. Development of a strong intracellular expression system for Bacillus subtilis by optimizing promoter elements. J Biotechnol. 2012;157:167–172. doi: 10.1016/j.jbiotec.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Philibert T., Rao Z., Yang T. Heterologous expression and characterization of a new heme-catalase in Bacillus subtilis 168. J Ind Microbiol Biotechnol. 2016;43:729–740. doi: 10.1007/s10295-016-1758-2. [DOI] [PubMed] [Google Scholar]
- 27.Pozsgai E.R., Blair K.M., Kearns D.B. Modified mariner transposons for random inducible-expression insertions and transcriptional reporter fusion insertions in Bacillus subtilis. Appl Environ Microbiol. 2012;78:778–785. doi: 10.1128/AEM.07098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinstein S.M., Kolodkin-Gal I., McLoon A. Osmotic pressure can regulate matrix gene expression in Bacillus subtilis. Mol Microbiol. 2012;86:426–436. doi: 10.1111/j.1365-2958.2012.08201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L., Li X., Wei D., Wang J., Shan A., Li Z. Expression of plectasin in Bacillus subtilis using SUMO technology by a maltose-inducible vector. J Ind Microbiol Biotechnol. 2015;42:1369–1376. doi: 10.1007/s10295-015-1673-y. [DOI] [PubMed] [Google Scholar]
- 30.Serrano M., Gao J., Bota J. Dual-specificity anti-sigma factor reinforces control of cell-type specific gene expression in Bacillus subtilis. PLoS Genet. 2015;11:e1005104. doi: 10.1371/journal.pgen.1005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song W., Nie Y., Mu X.Q., Xu Y. Enhancement of extracellular expression of Bacillus naganoensis pullulanase from recombinant Bacillus subtilis: effects of promoter and host. Protein Expr Purif. 2016;124:23–31. doi: 10.1016/j.pep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Summpunn P., Chaijan S., Isarangkul D., Wiyakrutta S., Meevootisom V. Characterization, gene cloning, and heterologous expression of beta-mannanase from a thermophilic Bacillus subtilis. J Microbiol. 2011;49:86–93. doi: 10.1007/s12275-011-0357-1. [DOI] [PubMed] [Google Scholar]