Abstract
Strain RT1 was isolated from root nodules of Lens culinaris (a lentil) and characterized as Rhizobium etli (a Gram-negative soil-borne bacterium) by 16S rDNA sequencing and phylogenetic analysis. The signaling molecules produced by R. etli (RT1) were detected and identified by high-performance liquid chromatography coupled with mass spectrometry. The most abundant and biologically active N-acyl homoserine lactone molecules (3-oxo-C8-HSL and 3-OH-C14-HSL) were detected in the ethyl acetate extract of RT1. The biological role of 3-oxo-C8-HSL was evaluated in RT1. Bacterial motility and biofilm formation were affected or modified on increasing concentrations of 3-oxo-C8-HSL. Results confirmed the existence of cell communication in RT1 mediated by 3-oxo-C8-HSL, and positive correlations were found among quorum sensing, motility and biofilm formation in RT1.
Keywords: Rhizobium etli, Quorum sensing, Motility, Biofilm, Lens culinaris
Introduction
Several endophytic bacteria reside within the living tissues of host plants and carry out a number of beneficial functions.1 Rhizobium etli is a Gram-negative α-proteobacterium found in soil that forms root nodules in lentils (Lens culinaris Medik.) and in other agriculturally important legumes, such as Phaseolus vulgaris. L. culinaris is an economically important legume crop cultivated in tropical, sub-tropical, and temperate areas worldwide.2 Many endophytic bacterial isolates (RT1–RT27) have previously been screened and characterized from lentil root nodules.3
Swarming motility enables groups of bacteria to move in a coordinated manner on solid surfaces, and the swarming phenomenon has been demonstrated for a number of bacteria, including the members of Azospiriillum, Aeromonas, Burkholderia, Bacillus, Chromobacterium, Clostridium, Escherichia, Proteus, Pseudomonas, Rhizobium, Salmonella, Serratia, Sinorhizobium, Vibrio, and Yersinia.4, 5, 6, 7 Bacterial motility probably occurs as a dispersal strategy in nutrient-limited environments or for locating a new host.8, 9 Normally swarming requires the differentiation of vegetative cells into a specialized cell type called swarmer cells. Surface contact (change in viscosity), physiological parameters, and chemical signals (nutritional status) provide the stimuli that trigger the differentiation of swarmer cells. Although, pathways responsible for signal integration are poorly understood, it is believed that signals are sensed and transmitted by two-component regulatory systems, cytosolic regulators or flagella leading to a complex network. Differentiated swarmer cells are often hyper flagellated and elongated, and these cells move in groups or rafts organized in parallel to the long axis of cells to maximize cell–cell contact and maximally colonize available surfaces. The migration front is preceded by a visible layer of slime-like extracellular material, and as a result of embedding in this extracellular slime matrix, population densities are normally extremely high.10
Bacteria may develop within plant roots as isolated cells, micro-colonies, aggregates, or biofilms.11 Biofilms are bacterial communities surrounded by a self-produced polymeric matrix attached to inert or biotic surfaces.12 Bacterial surface components, particularly exopolysaccharides (EPS), flagella, and lipopolysaccharides (LPSs), in combination with bacterial functional signals, are crucial for the formation of rhizobial biofilms in all species studied to date,13 and polysaccharides at rhizobial surfaces play important roles in symbiosis and in formation of active root nodules.
Bacteria can monitor and respond to changes in environmental conditions via a cell density-related process known as quorum sensing.14 Quorum sensing (QS) is a form of cell–cell communication regulated at the gene expression level and employed by several bacterial species to coordinate behavior at a community level. QS depends on the production of various signaling molecules, though N-acyl homoserine lactones (AHLs) are most commonly utilized by Gram-negative bacteria. Furthermore, the presence of AHL-like signal molecules in Gram-negative bacteria may be due to the active participation of the Luxl family of AHL synthases, which differ in side chain length, degree of substitution, and saturation.15 The phenotypes regulated by AHLs in Rhizobium species are remarkably diverse and of profound biological and ecological significance. These phenotypes include motility and biofilm formation, the productions of antibiotics and exo-enzymes, synthesis of the plant growth promoting auxin indole-3-acetic acid (IAA), the promotion of plant colonization, and biocontrol against several plant diseases. Some previous reports have demonstrated the production of QS signaling molecules by various rhizobia,14, 16 but little is known regarding the mechanisms of communication among lentil-nodulating strains.
Hence, the objective of the present study was to identify and characterize QS signaling molecules produced by R. etli RT1 and to evaluate the biological effects of one AHL molecule, namely, 3-oxo-C8-HSL on processes related to cell communication.
Materials and methods
Microorganisms
The isolates RT1–RT27 were screened from the root nodules of L. culinaris collected from a farmer's field in Uttar Pradesh (India). Isolate RT1 was identified as Rhizobium species based on its phenotypic and biochemical characteristics.3 Since isolate RT1 had the best plant growth promoting (PGP) attributes, it was further studied in detail. RT1 was grown at 30 °C for 24 h in tryptone soy and yeast extract mannitol media for further use.
Genomic DNA extraction
Genomic DNA of RT1 was extracted as described by Sambrook and Russel (2001).17 Briefly, a bacterial culture was grown overnight in 5 mL tryptone soy broth, stirred at 150 rpm for 24 h at 30 °C. The culture was then centrifuged at 10,000 × g for 2 min and the pellets so obtained were re-suspended in 50 μL 1× TE buffer (10 mM Tris–HCl, 1 mM EDTA), centrifuged at 10,000 × g for 2 min, re-suspended in 350 μL 1× TE buffer containing 5 μL lysozyme (50 mg/mL) and 2 μL RNAse A (10 mg/mL), and incubated at 37 °C for 10 min. Finally, 3 μL proteinase K (20 mg/mL) and 30 μL SDS (10%) were added and incubated at 37 °C for 35 min. The crude DNA template was PCI (phenol–chloroform–isoamyl alcohol) (25:24:1) and CI (chloroform–isoamyl alcohol) (24:1) extracted, precipitated in 1 mL absolute ethanol, washed with 500 μL 70% ethanol, re-suspended in 50 μL of sterilized ultrapure water, and stored at −20 °C.
16S rRNA gene amplification
The amplification of a 1492 bp region of the 16S rRNA gene was performed using 5′-GGCTCAGAACGAACGCTGGCGGC-3′ as the forward primer and 5′-CCCACTGCTGCCTCCCGTAGGAGT-3′ as the reverse primer.18 The PCR procedure consisted of preparing a master mix containing polymerase buffer (2.5 μL), 10 mM MgCl2 (1.5 μL), 10 mM dNTP (1.2 μL) and sterile distilled water. The gene fragment was amplified using 5–25 ng of bacterial DNA, 100 ng of each primer mentioned above and 0.3 μL of Taq polymerase (1 U/μL). The reaction volume was adjusted to 25 μL using sterile distilled water. The chemicals used for PCR were acquired from Bangalore Genei (India). The reaction conditions used were: initial denaturation for 7 min at 94 °C, then 29 amplification cycles {denaturation for 1 min at 94 °C, extension for 1 min at 72 °C, annealing at 54 °C for seven cycles, at 53 °C and 52 °C for one cycle each, and at 51 °C for 20 cycles} and a final extension for 10 min at 72 °C. The amplified gene was visualized in 0.8% agarose after electrophoresis, and the PCR product was sequenced at the Institute of Microbial Technology (IMTECH), Chandigarh, India.
Phylogenetic analysis
Phylogeny and evolutionary analysis were carried out using the MEGA 6.0 package.19 The 16S rDNA sequence of RT1 was aligned using CLUSTAL W20 against corresponding nucleotide sequences of Rhizobium (type strain) retrieved from GenBank (NCBI). The evolutionary distance matrix was generated as described by Jukes and Cantor (1969),21 and the phylogenetic tree was inferred by the neighbor-joining method.22 Tree topologies were evaluated by bootstrap analysis based on 1000 re-samplings of the neighbor-joining data set.
Nucleotide accession number
The sequence of 16S RNA gene of RT1 has been deposited in the GenBank database under accession number R. etli RT1 (KC762618).
Extraction and detection of QS signal molecules
AHL molecules were extracted as described by Shaw et al. (1997).23 R. etli RT1 was grown in 500 mL of AB minimal mannitol liquid medium for 48 h to the stationary phase. After centrifugation at 7400 × g, the supernatant was extracted twice with an equal volume of ethyl acetate containing 1.5 mL of acetic acid per liter. The extract was then evaporated to dryness using a vacuum rotator at 42 °C and re-dissolved in a small volume of ethyl acetate. This culture extract of R. etli RT1 was then analyzed by liquid chromatography/positive electrospray ionization (ESI+) multiple MS. Liquid chromatography and electrospray mass spectrometry (LC-ESI–MS) were conducted using a Thermo Finnigan LCQ advantage max ion trap mass spectrometer and a Finnigan Surveyor HPLC system. The column was thermo ODS-2, 250 × 4.6, 5 μm and eluted with acetonitrile and water at 800 μL/min. A 20 μL sample was introduced into the ESI source using a Finnigan Surveyor Autosampler. Mass spectra were scanned in the range of 150–2000 Th. Maximum ion injection time was set at 150 ns, ion spray voltage at 5.3 kV, and capillary voltage at 40 VA. A general elution program was used to elute all putative AHLs.
Bacterial motility assay
For the swarming experiment, yeast extract mannitol (containing 0.4 g/L MgSO4·7H2O) soft agar plates (0.75%; 60/15 mm) supplemented with 3-oxo-C8-HSL at different concentrations (0.05, 0.5, or 5.0 μM), were dried at 16 °C for 5 h and spot-inoculated centrally on their surfaces with the appropriate RT1 culture (1.2 μL, OD of overnight culture 595 0.7). Swarming behavior was analyzed on swarmer plates containing 3-oxo-C8-HSL. Inoculated plates were incubated at 30 °C for 24 h,24 and swarming halo zones were measured. Experiments were performed 4 times.
Biofilm formation assay
Biofilm formation was assayed quantitatively as previously described by O’Toole and Kolter (1992)25 by staining biofilms with crystal violet. RT1 was grown in 96-well polystyrene plates at 30 °C for 24 h without shaking in tryptic soy broth (TSB) medium containing 0.05, 0.5, or 5.0 μM of 3-oxo-C8-HSL. Planktonic cells were then gently removed, and 180 μL of an aqueous solution of crystal violet (0.1%, w/v) was added for 15 min. Each crystal violet-stained well was then rinsed three times with sterile distilled water and scored for biofilm formation by adding 150 μL of 95% ethanol. Optical densities of solubilized crystal violet were measured at 560 nm using a Micro ELISA Auto Reader (Biotek). Non-inoculated YEMB medium was used as a control. Control tubes were vortexed for 30 s, and the initial optical density was determined at 600 nm. Experiments were performed 4 times.
Effect of 3-oxo-C8-HSL on the growth of RT1
A bacterial broth co-inoculation assay was performed to visualize the effect of 5 μM 3-oxo-C8-HSL in tryptic soy broth (TSB) medium in order to confirm its effect on the growth rate of RT1. Control sets were prepared without inoculating 3-oxo-C8-HSL and optical densities were measured at 600 nm.
Statistical analysis
One-way analysis of variance (ANOVA) followed by post hoc Fisher's least-significant difference (LSD) test was used to determine the significances of the different effects treatments. All statistical analyses were performed using Prism version 6.03.
Results and discussion
Phylogenetic analysis
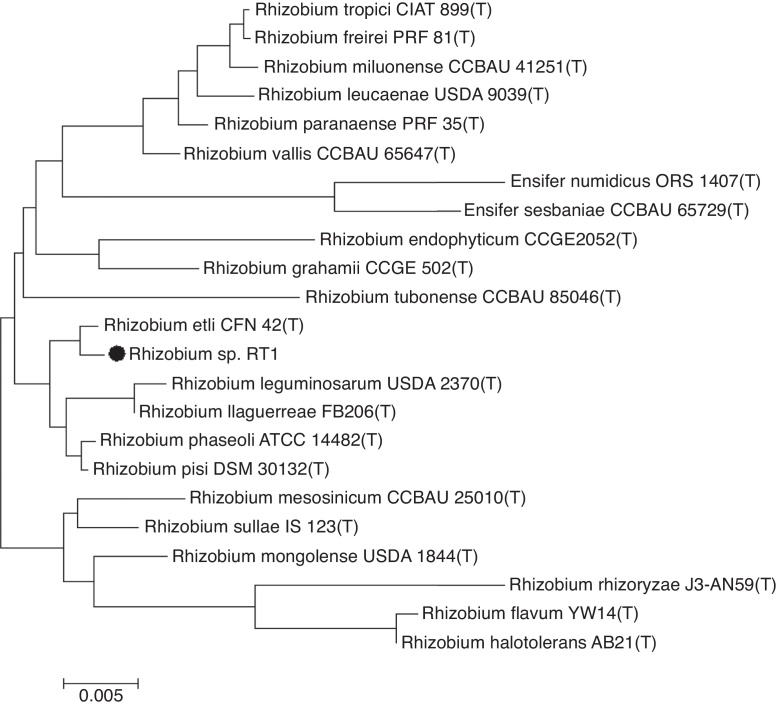
Sequencing of the 16S rRNA gene provided a 1492 bp long sequence of RT1. A phylogenetic tree constructed based on this sequence showed clustering of the RT1 isolate with R. etli CFN 42 (confidence = 99%) (Fig. 1), and thus, RT1 was tentatively classified as R. etli. Based on morphological and biochemical characteristics,3 and comparative analysis of the 16S rRNA gene sequence, the strain was unequivocally identified as R. etli RT1. It is generally accepted that organisms displaying 16S rRNA gene sequence similarity values of ≥97% belong to the same species.26 Therefore, Rhizobium RT1 was considered to be R. etli, since it exhibited 99.3% similarity with R. etli CFN 42.
Fig. 1.
Neighbor-joining phylogenetic tree based on 16S rDNA gene sequences, showing the relationship between strain RT1 and the most closely related type strains of Rhizobium etli CFN42. The bar represents 0.005 substitutions per nucleotide position.
Liquid chromatography electrospray mass spectrometry (LC–MS)
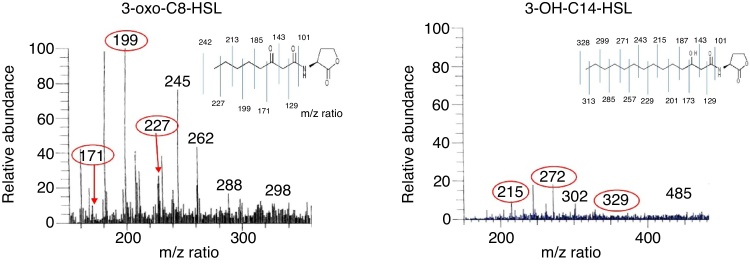
The AHL molecules 3-oxo-C8-HSL and 3-OH-C14-HSL were detected in the extract of RT1. The relative abundance (RA) of 3-oxo-C8-HSL was 100% at a retention time of 9.82–10.07 min (Fig. 2), and that of 3-OH-C14-HSL was 20% at an RT of 14.36–14.62 min. Comparative analysis of the AHL profile of RT1 by using LC–MS showed the presence of a broad range of AHL molecules with different acyl chains. AHL molecules are widely known to be intercellular communication signal molecules in bacterial populations.27 Several researchers have reported the presence of AHL-like signal molecules in an Acinetobacter strains that the strains are able to produce biofilms and that AHL-like molecules play a substantial role in quorum sensing in the context of metal tolerance.28, 29
Fig. 2.
LC–MS spectra showing the relative abundances of the molecular ions of the most abundant AHL molecules, 3-oxo-C8-HSL (a) and 3-OH-C14-HSL (b) in R. etli RT1 supernatant after 48 h of growth.
Motility assay
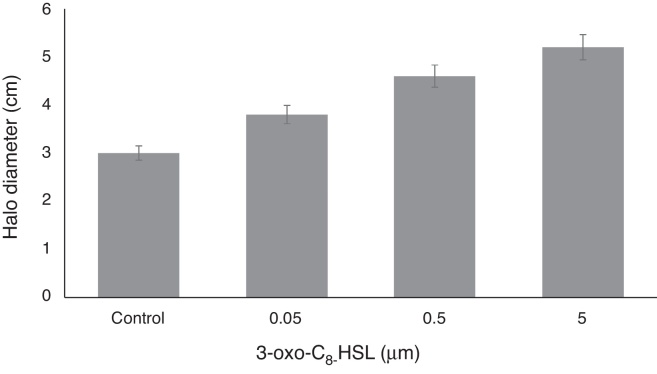
The motility of RT1 increased on increasing the concentration of 3-oxo-C8-HSL molecule. At a concentration of only 0.05 μM, 3-oxo-C8-HSL significantly increased motility, and at higher auto-inducer concentrations (0.5 or 5.0 μM) motility increased dose-dependently (Fig. 3). Microorganisms with the ability to move are more efficient at finding more favorable or less-hazardous niches for colonization in their environments,27 and although motility in rhizobia is not essential, it is needed to enable bacteria to find a specific host legume.27 Evidence has been previously presented regarding a connection between quorum sensing and motility control in R. etli,6 and in Pseudomonas fluorescens.30 In addition, Ahmer (2004) reported that a quorum sensing system (the LuxS/AI-2 system) present in Escherichia coli,31 synthesized AI-2 to increase motility and stimulate biofilm formation by wild-type E. coli K-12 strain.32
Fig. 3.
Effect of 3-oxo-C8-HSLon swimming motility by R. etli RT1. Values are means of experiments performed in quadruplicate. Bars represent SEMs (p < 0.05).
Biofilm formation assay
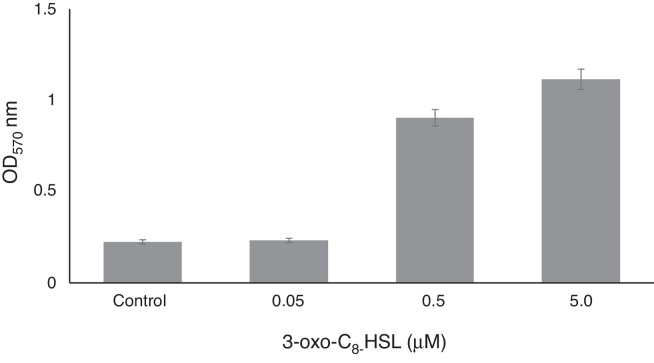
On polystyrene RT1 developed a sessile biomass with values ranging from 0.4 to ∼2.0 OD at 570 nm in the presence of exogenous 3-oxo-C8-HSL (Fig. 4), and at high concentration (5 μM), 3-OH-C12-HSL caused a significant 3- to 5-fold increase in biofilm formation. Lower concentrations (0.5 or 0.05 μM) had slight effects on biofilm formation (Fig. 4). When exogenous 3-oxo-C8-HSL was added 0.05 μM a non-significant 3-fold increase in biofilm formation was observed versus the non-treated control (non-inoculated YEMB medium). It is conceivable that one or more of the molecular signals may impact biofilm physiology.33, 34 Exposure of RT1 to exogenous additions of 3-oxo-C8-HSL or 3-OH-C14-HSL had positive effects on physiological processes related to survival and colonization ability, particularly the motility and biofilm formation. Rinaudi and Gonzalez (2009)35 explored the role played by AHL molecules in biofilm formation and their importance with respect to symbiotic relationships with the host.
Fig. 4.
Effect of different concentrations of 3-oxo-C8-HSL on biofilm formation by R. etli RT1 after 24 h of growth. Values are means of experiments performed in quadruplicate. Bars represent SEMs (p < 0.05).
The ability of rhizobia to produce AHLs is well known to be species and strain-dependent.6, 36 There are several different types of QS signaling mechanisms that use a range of different signal types and detection mechanisms.37 One of the most common of these mechanisms involves acyl-homoserine lactone (AHL) signal molecules. Furthermore, AHL-based quorum sensing appears to be highly conserved among bacteria, and more than 50 species have been identified to produce AHLs. AHL molecules are described as amphipathic because they contain both polar and non-polar regions, since the homoserine lactone ring is hydrophilic and the fatty acid side chain is hydrophobic, and this amphipathicity appears to facilitate the free diffusion of AHLs within the aqueous intra- and extracellular environments, and across the phospholipid bilayer.37
In addition to cell signaling, AHLs carrying long acyl-side chains possess significant surface activities at biologically relevant concentrations.24 Generally swarming motility is linked with biofilm formation, although the relationship between these phenomena appears to be rather dual.14 Attachment is pre-requisite of biofilm formation as it governs interactions between bacterial and plant tissues, and has been shown to rely on quorum sensing. Venturi (2006) reported that the extent of swarming depends on the structure of biofilms during the early stage of formation.38
Bacterial growth analysis
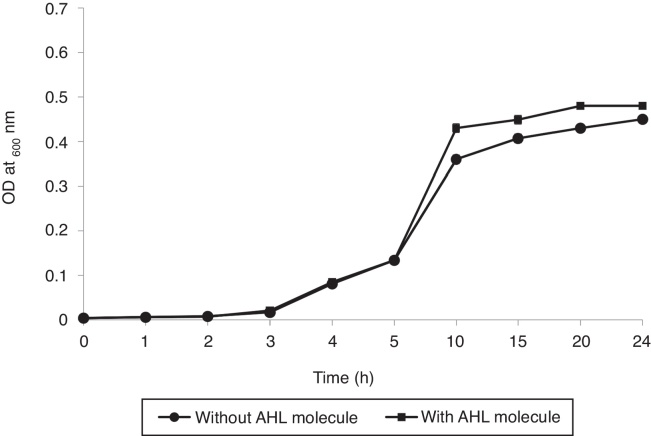
As shown in Fig. 5, the growth rate of RT1 was enhanced slightly by the presence of 3-oxo-C8-HSL at 5 μM. Furthermore, co-inoculation of RT1 with 3-oxo-C8-HSL caused no significant growth difference. Overall, our findings suggest that 3-oxo-C8-HSL neither promotes nor inhibits bacterial growth. Increased cell density (exponential phase) favors the release of chemical signals for social interactions in biofilms, which adds another level of complexity.39 Ideal approach use to confirm that a molecule is a quorum sensing molecule is to confirm that the molecule has no impact on final density after a suitable incubation period and that it has no effect on growth kinetics at defined concentrations.40
Fig. 5.
Growth curve of R. etli RT1 strain in TSB broth with or without the addition of 3-oxo-C8-HSL (5 μM). Values are means ± SEMs of experiments performed in quadruplicate (p < 0.05).
Conclusions
This study shows endophytic RT1 from the root nodules of lentils produces high levels of AHLs and that these regulate QS control of swarming motility and biofilm formation. Based on our findings, we conclude that quorum sensing, motility and biofilm formation observed in vitro are affected by AHL molecule type and concentration, and that the overall behavior of lentil-nodulating soil rhizobial populations is subjected to the fine and coordinated regulation of the synthesis of AHLs in response to specific environmental factors. Further studies are in progress to determine the putative quorum sensing mechanism of RT1.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
One of us (Swarnita Dixit) is thankful to the Chief Executive Officer, Biotech Park, Lucknow (India) for providing laboratory facilities.
Associate Editor: Cristiano Gallina Moreira
References
- 1.Ryan R.P., Germaine K., Franks A., Ryan D.J., Dowling D.N. Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett. 2008;278:1–9. doi: 10.1111/j.1574-6968.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- 2.Ribeiro R.A., Ormeno-Orrillo E., Dall Agnol R.F., Graham P.H., Martinez-Romero E., Hungria M. Novel Rhizobium lineages isolated from root nodules of the common bean (Phaseolus vulgaris L.) in Anden and Mesoamerican areas. Res Microbiol. 2013;164:740–748. doi: 10.1016/j.resmic.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Dixit S., Dubey R.C., Seth P.K. Characterization of plant growth promoting endophytic bacteria isolated from Lens culinaris Medik. with antagonistic potential against Fusarium oxysporum. Int J Bioinform Biol Sci. 2014;2:95–109. [Google Scholar]
- 4.Belas R., Schneider R., Melch M. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J Bacteriol. 1998;180:6126–6139. doi: 10.1128/jb.180.23.6126-6139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearns D.B., Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 6.Braeken K., Daniels R., Vos K. Genetic determinants of swarming in Rhizobium etli. Microb Ecol. 2008;55:54–64. doi: 10.1007/s00248-007-9250-1. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Heredia A., Garcia S., Merino-Mascorro J.A., Feng P., Heredia N. Natural plant products inhibits growth and alters the swarming motility, biofilm formation, and expression of virulence genes in enteroaggregative and enterohemorrhagic Escherichia coli. Food Microbiol. 2016;59:124–132. doi: 10.1016/j.fm.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Hussain M.B.B.M., Zhang H., Xu J., Liu Q., Jiang Z., Zhang L.H. The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J Bacteriol. 2008;190:1045–1053. doi: 10.1128/JB.01472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrout J.D., Chopp D.L., Just C.L., Hentzer M., Givskov M., Parsek M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 10.Huber B., Riedel K., Hentzer M. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology. 2001;147:2517–2528. doi: 10.1099/00221287-147-9-2517. [DOI] [PubMed] [Google Scholar]
- 11.Farrand S.K., Qin Y., Oger P. Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Methods Enzymol. 2002;358:452–484. doi: 10.1016/s0076-6879(02)58108-8. [DOI] [PubMed] [Google Scholar]
- 12.Waters C.M., Bassler B.L. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 13.Marketon M.M., Gronquist M.R., Eberhard A., González J.E. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J Bacteriol. 2002;184:5686–5695. doi: 10.1128/JB.184.20.5686-5695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels R., Vanderleyden J., Michiels J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol Rev. 2004;28:261–289. doi: 10.1016/j.femsre.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Sarkar S., Chakraborty R. Quorum sensing in metal tolerance of Acinetobacter junii BB1A is associated with Biofilm production. FEMS Microbiol Lett. 2008;282:160–165. doi: 10.1111/j.1574-6968.2008.01080.x. [DOI] [PubMed] [Google Scholar]
- 16.Wisniewski-Dyé F., Downie J.A., Wisniewski-dy F., Downie J.A. Quorum-sensing in Rhizobium. Ant Van Leeuwen. 2002;81:397–407. doi: 10.1023/a:1020501104051. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J., Russel D. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 18.Young J., Downer H., Eardly B. Phylogeny of the phototrophic Rhizobium strain BTAil by polymerase chain reaction-based sequencing of a16S rRNA gene segment. J Bacteriol. 1991;173:2271–2277. doi: 10.1128/jb.173.7.2271-2277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;11:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jukes T.H., Cantor C.R. Evolution of protein molecules. In: Munro H.N., editor. Mammalian Protein Metabolism. Academic Press; New York: 1969. pp. 21–123. [Google Scholar]
- 22.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Shaw P.D., Ping G., Daly S.L. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniels R., Reynaert S., Hoekstra H. Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc Natl Acad Sci USA. 2006;103:14965–14970. doi: 10.1073/pnas.0511037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 26.Stackebrandt E., Goebel B.M. Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequences analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 1994;44:846–849. [Google Scholar]
- 27.Nievas F., Bogino P., Sorroche F., Giordano W. Detection, characterization, and biological effect of quorum-sensing signaling molecules in peanut-nodulating Bradyrhizobia. Sensors. 2012;12:2851–2873. doi: 10.3390/s120302851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez R.H., Nusblat A., Nudel B.C. Detection and characterization of quorum sensing signal molecules in Acinetobacter strains. Microbiol Res. 2001;155:271–277. doi: 10.1016/S0944-5013(01)80004-5. [DOI] [PubMed] [Google Scholar]
- 29.Shakeri S., Kermanshahi R.K., Moghaddam M.M., Emtiazi G. Assessment of biofilm cell removal and killing and biocide efficacy using the microtiter plate test. Biofouling. 2007;23:79–86. doi: 10.1080/08927010701190011. [DOI] [PubMed] [Google Scholar]
- 30.Korber D.R., Lawrence J.R., Caldwell D.E. Effect of motility on surface colonization and reproductive success of Pseudomonas fluorescens in dual-dilution continuous culture and batch culture systems. Appl Environ Microbiol. 1994;60:1421–1429. doi: 10.1128/aem.60.5.1421-1429.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmer B.M.M. Cell-to-cell signaling in Escherichia coli and Salmonella enterica. Mol Microbiol. 2004;52:933–945. doi: 10.1111/j.1365-2958.2004.04054.x. [DOI] [PubMed] [Google Scholar]
- 32.González Barrios A.F., Zuo R., Hashimoto Y., Yang L., Bentleyh W.E., Wood T.K. Autoinducer 2 controls biofilm formation in E. coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labbate M., Queck S.Y., Koh K.S., Rice S.A., Givskov M., Kjelleberg S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J Bacteriol. 2004;186:692–698. doi: 10.1128/JB.186.3.692-698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alagely A., Krediet C.J., Ritchie K.B., Teplitski M. Signaling-mediated cross-talk modulates swarming and biofilm formation in a coral pathogen Serratia marcescens. ISME J. 2011;5:1609–1620. doi: 10.1038/ismej.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinaudi L.V., Gonzalez J.E. The low molecular weight fraction of exopolysaccharide II from Sinorhizobium meliloti is a crucial determinant of biofilm formation. J Bacteriol. 2009;191:7216–7224. doi: 10.1128/JB.01063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Houdt R., Givskov M., Michiels C.W. Quorum sensing in Serratia. FEMS Microbiol Rev. 2007;31:407–424. doi: 10.1111/j.1574-6976.2007.00071.x. [DOI] [PubMed] [Google Scholar]
- 37.Shrout J.D., Tolker-Nielsen T., Givskov M., Parsek M.R. The contribution of cell–cell signaling and motility to bacterial biofilm formation. MRS Bull. 2011;36:367–373. doi: 10.1557/mrs.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev. 2006;30:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 39.Li Y.H., Tian X. Quorum sensing and bacterial social interactions in biofilms. Sensors. 2012;12:2519–2538. doi: 10.3390/s120302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Defoirdt T., Brackman G., Coenye T. Quorum sensing inhibitors: how strong is the evidence. Trends Microbiol. 2013;21:619–624. doi: 10.1016/j.tim.2013.09.006. [DOI] [PubMed] [Google Scholar]