Abstract
The ability to adsorb zearalenone by five strain of lactic acid bacteria was evaluated: four strains of Lactobacillus spp. isolated from pig rectal swabs and one commercial strain (Lactobacillus rhamnosus). Several factors affecting the adsorption capacity were evaluated in order to improve the adsorption of the mycotoxin by bacteria. The stability of the zearalenone–bacteria complex was analyzed. In every case, bacterial adsorption capacity was higher than 40.0%. The strain showing the highest adsorption (68.2%) was selected for the following steps of this research. The adsorption percentages obtained after processing 6.5 and 7.5 mL MRS broth were 57.40% + 3.53 and 64.46% + 0.76, respectively. The stability of zearalenone–bacteria complex was evaluated by successively rinsing. In the first rinsing step 42.26% + 0.414 was still bound. In the second rinsing step 25.12% + 0.664 was still bound, whereas 15.82% + 0.675 remained in the pellet after the third rinse. Results obtained demonstrated that Lactic Acid Bacteria has capacity to adsorb zearalenone. Finally adsorption was increased using a higher volume of initial broth. These results could be used to design a new lyophilized powder for detoxification, using lactic acid bacteria as potential zearalenone adsorbents.
Keywords: Zearalenone, Adsorption, Lactic acid bacteria, Lactobacillus, Pigs
Introduction
Zearalenone (ZEA) is a worldwide distributed mycotoxin as indicated by the International Agency for Research on Cancer (IARC).1, 2, 3 Its toxicity and incidence was confirmed by recent reports.4, 5 This resorcylic lactone is produced mainly by Fusarium graminearum and is one of the most important toxins causing serious reproductive failures in pig production, due to its ability to couple 17-β-estradiol receptors. This interference with cytosolic estrogen receptors of target cells makes it an endocrine disruptor. When the dangerousness of this mycotoxin is considered, two factors are important: its stability to heat, milling, storage and processing of feedingstuffs,6 and the severity of the intoxication, even if the ingestion takes place only for a few days as indicated by Anadón et al.7
Several physicochemical methods for detoxification have been widely used, but the tendency is to use biological methods that do not cause nutritional and palatability changes in feed.8 There are several food additives for decontamination and/or detoxification of animal feedstuffs, such as the commercial adsorbent Mycosorb® (Alltech Inc.), in which the structure of yeast glucan layer is modified to increase its affinity and binding rate of mycotoxins above that of the native cell wall product.9 In spite of its greater efficacy, the use of Mycosorb is limited because of its high price.
Interactions between Fusarium mycotoxins, ZEA, its derivative α-zearalenol (α-ZOL) and food-grade strains of Lactobacillus have been reported by several researchers.10, 11, 12, 13, 14, 15, 16, 17 El-Nezami et al.13 demonstrated that, after co-incubation of ZEA and Lactobacillus, a considerable proportion (38.0%–46.0%) of these mycotoxins were recovered from bacterial pellets. Bacteria showed capacity to adsorb toxins and, as expected, results demonstrated that the binding depended on bacterial concentration. Co-incubation of ZEA and α-ZOL with bacteria significantly affected the percentage of toxin bound, suggesting that these toxins may share the same binding site. Recently Tinyiro et al.18 using some Bacillus strains, showed that the adsorption percentage was high (78.0% and 95.0%) and variation depended on the strain used. ZEA adsorption ability by Planococcus spp has also been demonstrated by Lu et al.19
Two fundamental rules are considered when bacterial strains are isolated in order to be administered to animals. The first is the host specificity, it is essential to achieve a good adaptation.20, 21 The second is the proximity of the ecosystem, it is vital that the microorganisms are isolated in the same place where it acts on the host.22, 23
For all these reasons, the purpose of the present work was to study the ZEA adsorption capacity of lactic acid bacteria isolated from pig rectal swabs under in vitro conditions, in order to broaden scientific knowledge on feed additives based on the use of lactic acid bacteria as ZEA binders.
Materials and methods
Bacterial strains and culture conditions
Lactobacillus rhamnosus (R1) (Lyofast LR B Sacco) was purchased from Sacco SRL (Cadorago, Italy). This strain was used as reference because of its demonstrated ability to adsorb ZEA.12, 13, 24
The tested Lactobacillus strains (L1, L2, L3, and L4) were isolated from 7 piglets kept on a single farm. The piglets, aged 21–60 days, were clinically healthy and no antibiotics had been administered during their lives. All samples were obtained within the same day. The strains were collected with a kit containing a swab and a transport medium (LAPTg semisolid agar without sugars). After being delivered to the laboratory, the swabs were broken off into peptone water, the dilutions were made and inoculated onto solid medium: LAPTg Agar,25, 26, 27 then the plates were incubated in microaerophilia (candle jar system) at 37 °C for 48 h.
Strains of lactic acid bacteria (LAB): L1, L2, L3 and L4 were isolated at the “Laboratorio de Toxicología” at the “Universidad Nacional del Centro de la Provincia de Buenos Aires” Tandil, Buenos Aires, Argentina. LAB strains were kept at −20 °C in LAPTg broth (1.5% peptone, 1% tryptone, 1% glucose, 1% yeast extract, and 0.1% Tween 80), pH 6.826 containing 15% glycerol (v/v) until use.
Reactivation process
The strains were defrosted at 37 °C for 15 min, then they were inoculated and incubated in LAPTg broth for 24 h at 37 °C in microaerophilia before genotypic an species identification. Before adsorption process two reactivation times were tested: 24 h (t1) and 72 h (t2).
Genotypic identification
The strains isolated for this work L1, L2, L3 and L4, were characterized as lactobacilli by PCR, according to the methodology described by Dubernet et al.28 Industrial strains provided by Sacco S.A.: L. rhamnosus (R1), Lactobacillus delbrueckii (R2), Lactobacillus casei (R3) and Lactobacillus helveticus (R4) were used only for genotypic identification as positive control. Identification was made by amplifying specific region of DNA. The amplification products were visualized by electrophoresis in agarose gels stained with ethidium bromide.29 General primers for lactobacilli LbLMA1-rev (5′-CTC AAA ACT TTC AAA CAA AGT-3′ and R16-1 5′-CTT GTA CAC ACC GCC CGT CA-3′) were used.28
Species identification
Genotypic identification was carried out by partial 16S rRNA gene sequencing. Genomic DNA was extracted according to Pospiech and Neumann.30 Oligonucleotide primers (PLB16, 5-AGAGTTTGATCCTGGCTCAG-3, and MLB16, 5-GGCTGCTGGCACGTAGTTAG-3) were used to amplify the variable (V1) region of the 16S ribosomal RNA gene according to the protocol described by several authors.31, 32, 33 PCR products were electrophoresed in 1% (wt/vol) agarose gels, stained and visualized as described above. Amplicons were excised from the gel and purified using a GFX PCR DNA gel band purification kit (GE Healthcare, UK). Purified PCR products were sequenced at CERELA-CONICET, Tucumán, Argentina, by using an ABI 3130 DNA sequencer (Applied Biosystems, Foster, CA). rRNA gene sequence alignments were performed using the multiple sequence alignment method34, 35 and identification queries were fulfilled by a BLAST search36 in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) and in the Ribosomal Database Project.37
Determination of Growth by Optical Density (OD560 nm)
The quantifying of the total number of cells for each strain (R1, L1, L2, L3, and L4) was made using a turbidimetric method. The OD560 nm of the bacterial suspension includes living cells, dead cells, and noncellular material that absorbs/scatters the light and indicates de total mass achieved. The wavelength of the spectrophotometer (UV-Vis Spectrofotometer Pharmacia LKB Biochrom Limited, Ultrospect III, Cambridge, England) was set at 560 nm. One milliliter of the sterile broth which was used to grow bacteria (LAPTg or MRS) was used to calibrate the equipment to zero absorbance and 1 mL of each sample was used to measure the OD560 nm.
Pellets adsorption process
In first place a stock ZEA solution was prepared by dissolving 25 mg ZEA (99.5% purity, Sigma, St. Louis, USA) in 125 mL chloroform (HPLC grade, Biopack, Argentina) to obtain a final concentration of 200 μg/mL. This solution was kept at −18 °C until its use. 95 μL of the stock ZEA solution were evaporated to dryness under an N2 current at 60 °C. The dry residue was resuspended in 50 μL of methanol (HPLC grade, Sintorgan, Argentina) and 1500 μL of a 0.9% NaCl (Sigma, St. Louis, USA) (SP1) or 0.9% CaCl2 (Sigma, St. Louis, USA) (SP2) to give a ZEA concentration of 12.3 μg/mL in both solutions.
In second place the adsorption pellets were prepared. LAB were activated and grown in LAPTg medium. Cultures were incubated in microaerophilia (candle jar system) for 72 h at 37 °C. OD560 nm was measured, then bacteria were harvested by centrifugation (Sigma Laborzentrifugen 1K15, St. Luis, United States) at 5900 × g, 4 °C, 15 min.
Finally bacterial pellets were rinsed twice with NaCl 0.9% solution and then resuspended in 1.5 mL of SP1 solution or SP2 solution accordingly and incubated for 1 hour at 37 °C, with gentle stirring every 30 min as indicated by Bueno.11
After incubation, the solutions were centrifugated (5900 × g, 4 °C, 15 min), then the supernatant was filtered through a 0.22 μm nylon membrane and analyzed by HPLC–UV.11, 38
As a control to demonstrate that the adsorption process recovers all the mycotoxin reference strain (R1) was used. ZEA in the supernatant and in the pellet (extracted with methanol) were quantified, these two concentrations were added and compared with the initial mycotoxin.
ZEA determination by HPLC
The HPLC system was equipped with a Gilson 151 UV-Vis detector, and Gilson 712 software (Gilson, Inc., Middleton, USA). Separation was achieved by HPLC, using a C18 stationary phase, 250 mm × 4.6 mm i.d., 5 μm column. The mobile phase consisted of acetonitrile (HPLC grade, Sintorgan, Villa Marteli, Argentina):water (ultra purified, Elga) mixture (55:45) working in isocratic mode at 1.2 mL/min. The column was maintained at 30 °C. The injection volume was 20 μL and the chromatographic run time was 20 min. Detection was carried out at 236 nm.39 To quantify ZEA using external standard calibration seven concentrations 0.63, 1.25, 2.5, 5, 10, 15 and 20 μg/mL were prepared with standards (Sigma St. Louis, USA), the responses were found to be linear (coefficient of correlation >0.998). Recorded retention times for β-ZOL, α-ZOL and ZEA were 6.70 min; 9.24 min; and 17.19 min respectively.
Adsorption percentage (ADS%) quantification
ADS% was quantified by comparing mycotoxins peak areas in the supernatant obtained after the adsorption process to peak areas for SP1 or SP2 solutions without bacterial addition, representing positive controls. The ADS% of ZEA was calculated according to Eq. (1).38, 40
| (1) |
Being:
ADS%: adsorption percentage.
AUC2: peak area of the mycotoxin in the supernatant after the adsorption process.
AUC1: peak area of the mycotoxin in the positive control (SP1 or SP2).
Strain selection
Two parameters were used for LAB strain selection: The OD560 nm achieved after the reactivation process, used as indicator of mass production, and ADS%. We used both variables following Eq. (2) because we consider that they have the same importance for strain selection.
| (2) |
Being:
X = mean of OD560 nm and ADS%.
OD560 nm = optical density.
ADS% = adsorption percentage.
Optimization of adsorption process conditions
With the aim of increasing adsorption percentage, using the selected strain, four variables of the technique developed by Bueno11 were evaluated.
-
(i)
Culture media used in the reactivation process: bacterial growth was compared in two culture media: MRS and LAPTg. First bacterial growth was determined by OD560 nm and cells were observed by scanning electron microscopy (SEM) (JEOL 35 CF from JEOL Ltd., Tokyo, Japan). Finally bacteria were cultured until OD560 nm = 1 was reached. This value was standardized for every strain. The adsorbent pellets were obtained for both culture media and the ADS% were calculated.
-
(ii)
Contact solution: the effects of solutions SP1 and SP2 on adsorption percentage were compared.
-
(iii)
Initial volume of culture media: the effects of an initial volume of 7.5 mL and 6.5 mL were compared.
-
(iv)
Viable and non-viable strain: the effects of sterilizing (120 °C, 15 min) and non-sterilizing strain were compared.
Evaluation of the selected method
Using data from 3 samples on 4 separate days of assay the selected method was also evaluated. The precision was expressed by the relative standard deviation (RSD). Note that the RSD is called as same as the coefficient of variation (CV) in statistics. Within day precision (repeatability) was calculated by the mean coefficient of variation (CV) which was required to be less than 15% in accordance with the Guidance for Industry – Bioanalytical Method Validation prepared by the Biopharmaceutics Coordinating Committee in the Center for Drug Evaluation and Research (CDER) in cooperation with the Center for Veterinary Medicine (CVM) at the Food and Drug Administration.41 Between days precision (intermediate precision) was expressed as between days coefficient of variation (CVbd), which was calculated using Eq. (3).42
| (3) |
Being:
μ = average media.
SDbd = between days standard deviation (calculated as the square root of between days variance).
Between days variance is obtained after subtracting the contribution of within day variability, using the following Eq. (4)
| (4) |
Being:
SD2(μ) = variance of every day mean.
n = number of observations per day.
SD2wd = average within day variance.
| (5) |
where:
D = total number of days.
n = total number of replicates per day.
xdr = result for replicate r on day d.
.
| (6) |
where:
D = total number of days.
Scanning electron microscopy (SEM)
The pellets obtained after the reactivation process and after the adsorption process were fixed with a modifier Karnofsky's solution (1.6% glutaraldehyde + 2.6% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2)). After 24 h at 4 °C, they were dehydrated by successive passages in solutions (ethyl alcohol–acetone) with greater graduation of ethyl alcohol each time (30%, 50%, 70%, 90% y 100%), the lapse for each passage was 10 min. After drying, the samples were glued on stubs using carbon tape and coated with gold (ion sputter JFC-1100). Bacteria were analyzed using a JEOL 35CF scanning electron microscope.
Stability of the ZEA-selected Lactoctobacillus complex
Using the strain with the highest ADS%, ZEA dissociation was study by successive rinses. In first place, the selected adsorption process was carried out, and then the pellet obtained was suspended in 1.0 mL of purified water and maintained at room temperature for 10 min. Then, the suspension obtained was centrifuged at 5900 × g, 4 °C, 15 min, and finally ZEA concentration was analyzed in the supernatant. This step was repeated up to three times. At the end, ZEA remaining bound was recovered with 1.0 mL of methanol. ZEA quantification in all supernatants was performed by HPLC as indicated in ZEA determination. ZEA bound to the bacterial pellet was calculated according to Eq. (7).
| (7) |
Being:
%ADS = adsorption percentage.
Initial ZEA = μM of Initial ZEA in the positive control SP1 = 38.64 μM.
Biotransformation of ZEA
The metabolization of ZEA into α-ZOL and β-ZOL was also investigated because ZEA concentration could be reduce as a consequence of biotransformation as indicated by Boswald et al.43 and not because of ZEA–bacteria complex formation. HPLC analysis of a mixed standard solution containing ZEA and both metabolites was used to identify the presence or absence of each compound.
Statistical analysis
All samples were analyzed in triplicate. Results were expressed as mean ± standard deviation (SD). Data were statistically evaluated using the InfoStat program, 2010. (InfoStat group FCA, Universidad Nacional de Córdoba, Argentina). A two-tailed Student's t-test for paired samples (p < 0.05) was used to determine significant differences between treatments.
Results and discussion
Genotypic identification
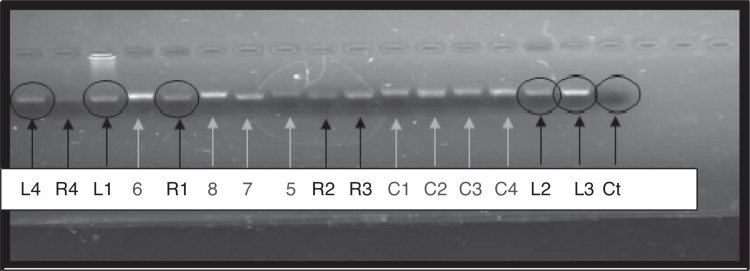
Results from the PCR molecular technique are shown in Fig. 1. All strains assayed and the reference strains L. rhamnosus (R1), L. delbrueckii (R2), L. casei (R3) and L. helveticus (R4) were positive (amplified) with general primers for Lactobacillus. In the negative control (Ct), as is expected, no amplification was observed.
Fig. 1.
PCR amplification products for Lactobacilli isolated from pig rectal swaps (L1–L2–L3–L4), and isolated from other sources (5–6–7–8–C1–C2–C3–C4) and reference strains (R1–R2–R3–R4) using general primers: LbLMA1-rev y R16-1. Ct, control (without any bacterial strain).
Species identification
The comparative analysis of the sequences obtained from L4 strain with those described in various databases, showed a high identity with strains belonging to Phylum “Firmicutes” Class “Bacilli” Order “Lactobacillales” Family “Lactobacillaceae” Gender “Lactobacillus” and within this genus to species “plantarum” and “pentosus”. Given the phylogenetic proximity of these species, additional studies were performed to differentiate between them. L4 strain did not ferment glycerol and D-xylose, which are typically fermented by L. pentosus but not by L. plantarum.44, 45 These results indicate that the selected strain (L4) was a L. plantarum.
Adsorption process
When time t2 (72 h) was used in the reactivation process the reference strain R1 gave an adsorption value of 79.8% ± 6.8. This result was similar to the results obtained by El-Nezami12 when nonviable bacteria were used (80%). With this reactivation time it was probable that waste and dead cells began to accumulate and it caused some changes in the adsorption process. In the next assay reactivation time t1 (24 h) was used. This change gave an adsorption value of 30.5% ± 5.4, it was close than values reported for viable cells.13
For R1 strain, the sum of the ZEA percentages, in the supernatant and in the methanol used to extract mycotoxin in the pellet was 97.73%. This value was close to 100% mycotoxin which indicated that the method achieved a high mycotoxin extraction.
Strain selection
The strains L1, L2, L3 and L4 showed adsorption percentages between 46% and 64% (Table 1), these were higher than those showed by Bueno11 and similar to those described by El-Nezami.13 The second parameter analyzed was OD560 nm, as Bueno11 indicated in order to achieve a good bacteria production, the value should be 1 or higher. Only in one case (L1 strain) the value was lower. According to our results, L4 strain achieved the highest average (X) between OD560 nm and ADS%, so it was decided to use this strain in subsequent research. Several genera of bacteria have been studied to evaluate their zearalenone adsorption ability. Bueno11 reported adsorption levels ranging from 0 to 36.44% for Bifidobacteria, 1.74 to 33.69% for Lactobacilli, and 0 to 59.09% for Actinomycetes. El-Nezami13 reported the ZEA adsorption percentages of two food grade strains (L. rhamnosus GG and L. rhamnosus LC705). Furthermore, Lu et al19 studied ZEA adsorption by Planococcus spp, achieving 21.82% adsorption for viable bacteria and 42.82% for inactivated bacteria.
Table 1.
Results of the parameters used for strain selection [OD560 nm, % ADS and average (X) of both values)]. Reactivation time used 24 h.
| Strain | Adsorption parameters |
||
|---|---|---|---|
| OD560 nm | % ADS | (X)average | |
| L4 | 1.062 + 0.085a | 64.2 + 1.5c | 32.618c |
| L3 | 1.361 + 0.118b | 47.6 + 1.5b | 24.501b |
| L2 | 1.426 + 0.143b | 49.1 + 0.0b | 25.278b |
| L1 | 0.905 + 0.014a | 46.5 + 5.8b | 23.698b |
| L. rhamnosus (R1) | 1.129 + 0.019a | 30.5 + 5.4a | 15.825a |
Results were represented as mean ± SD. Different superscript letters in the same column indicate statistical significant differences (p < 0.05).
Results were represented as mean ± SD. Different superscript letters in the same column indicate statistical significant differences (p < 0.05).
Evaluation of the adsorption process conditions
For the selected Lactobacillus strain L4 the change of various parameters in order to improve the adsorption process was evaluated.
Culture media comparison, L4 showed significantly higher OD560 nm in MRS (1.707 ± 0.16) than in LAPTg (1.101 ± 0.15) these differences were statistically significant (p < 0.05). With respect to adsorption percentages, using the same OD560 nm (1) higher results were also obtained using MRS media (57.38% ± 4.68) than LAPTg (49.33% ± 4.10), but these difference were not statistically significant (p > 0.05). Taking into account a future industrial use and observing that MRS broth achieved the higher production it was selected as culture media.
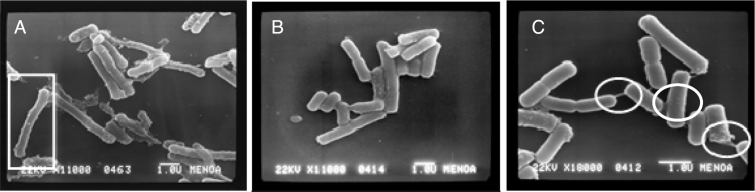
In addition, scanning electron microscopy revealed few differences comparing bacterial morphology during incubation in both broths (Fig. 2A and B), bacillus seemed longer and thinner when cultured in LAPTg than in MRS (marked in Fig. 2A with a square), that should be studied in future investigations. These changes would indicate different morphology of the cellular wall and could explain different adsorption percentage achieved when this different culture media were used in the reactivation process prior ZEA adsorption. Furthermore, after the adsorption process, L4 presents several intercellular bonds (marked in Fig. 2C with circles), this being an interesting strain property that should be studied in future investigations (Fig. 2C).
Fig. 2.
Scanning electron micrographs of L4 strain in (A) LAPTg media, (B) MRS media, (C) after the adsorption process.
A higher %ADS was obtained after incubation with SP1 than with SP2 solution, (57.28% ± 1.85 and 49.33% ± 6.11, respectively). These differences were not statistically significant (p > 0.05), which indicates that a change of contact solution does not produce a significant change in the adsorption. For this reason, the technique was not changed and SP1 was maintained as contact solution.
Adsorption percentages obtained after processing 6.5 and 7.5 mL MRS broth were 57.40% ± 3.53 and 64.46% ± 0.76, respectively. These differences were statistically significant (p < 0.05). Therefore the use of 7.5 mL of media was selected to carry out the adsorption process.
After sterilizing culture media, the ADS% was 82.4% ± 3.3 and 73.5% ± 1.0 for viable strain ADS%. These differences were statistically significant (p < 0.05). These findings could be considered in order to decide if non-viable bacteria may be used as mycotoxins adsorbent with higher ADS% or viable bacteria with lower ADS% but with possible probiotic effect at the same time. These findings confirm the results of other researches.13
Table 2 shows the ADS% obtained by processing three daily samples for four different days, under identical conditions. Relative standard deviations (%RSD) were calculated for each day, these values 1.81 to 7.23 were similar with that obtained by Kolfk-Clauw et al.,39 using the same technology for determination of ZEA (HPLC UV), and they never exceed the value of 15% that was our goal proposal. Repeatability (Sr), also known as within-run precision was calculated, under identical conditions successive measurements obtained were close in a value of 6.00. Using the overall mean obtained (70.10) and the relative standard deviation maximum allowed (15%), the maxim standard deviation claimed was 10.52. This value was less than SD achieved in each day and less than the repeatability obtained.
Table 2.
Adsorption percentage (%ADS) of L4 after the adsorption process for three samples in four different days. Estimation of repeatability (within day precision) and intermediate precision (between day precision).
| Day 1 | Day 2 | Day 3 | Day 4 | Mean | SD2(μ) | SD2wd | SD2bd | |
|---|---|---|---|---|---|---|---|---|
| % ADS | 64.46 | 65.30 | 75.61 | 75.01 | 70.095 (μ) | 31.409 | 36.042 | 55.437 |
| SD | 1.17a | 1.35a | 5.47a | 1.73a | ||||
| %RSD (CV) | 1.81 | 2.06 | 7.23 | 2.31 |
Results were represented as mean ± SD. Different superscript letters in the same row indicate significant differences (p < 0.05). Within day precision (repeatability) and between day precision were calculated and as required both values were less than 15%.
Within-day precision: 3.36%. Between-day precision: 10.62%.
Results were represented as mean ± SD. Different superscript letters in the same row indicate significant differences (p < 0.05).
Within day precision (repeatability) and between day precision were calculated and as required both values were less than 15%.
Stability of the ZEA–bacteria complex
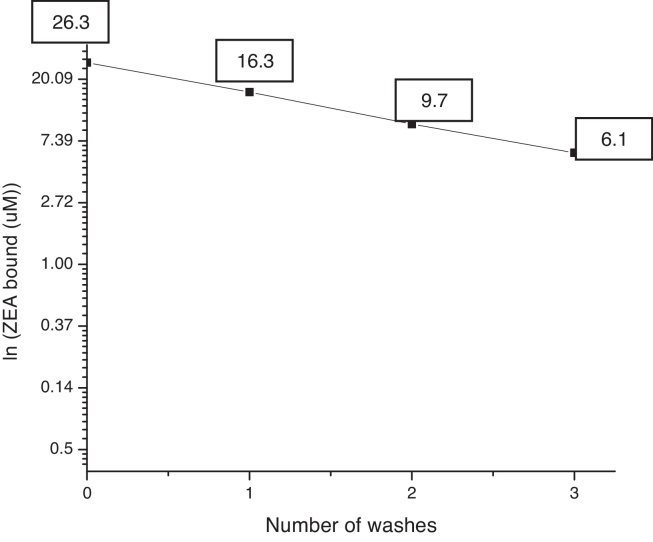
Using the improved process with an initial ZEA concentration of 38.7 μM, the initial adsorption percentage was 68.20% ± 1.74 (ZEA bound: 26.3 μM). In the first rinsing step 42.26% ± 0.414 was still bound to the pellet (ZEA bound: 16.3 μM). In the second rinsing step, 25.12% ± 0.664 was still bound to the pellet (ZEA bound: 9.7 μM), whereas 15.82% ± 0.675 remained in the pellet (ZEA bound 6.1 μM) after the third rinse.
Fig. 3 shows ln(ZEA bound) vs number of washes. Dissociation of ZEA was relatively linear. The dissociation constant, which could be estimated from the slope, was higher than that obtained by El-Nezami et al.12 These results could probably be due to the use of different bacteria strains than those used by El-Nezami et al.,12 specifically in terms of their differences in the cell wall. Some compounds of the cell wall are really important in the adsorption process as indicated by Yiannikouris et al.46, 47
Fig. 3.
ZEA remaining from ZEA–L4 complex after successive rinsing.
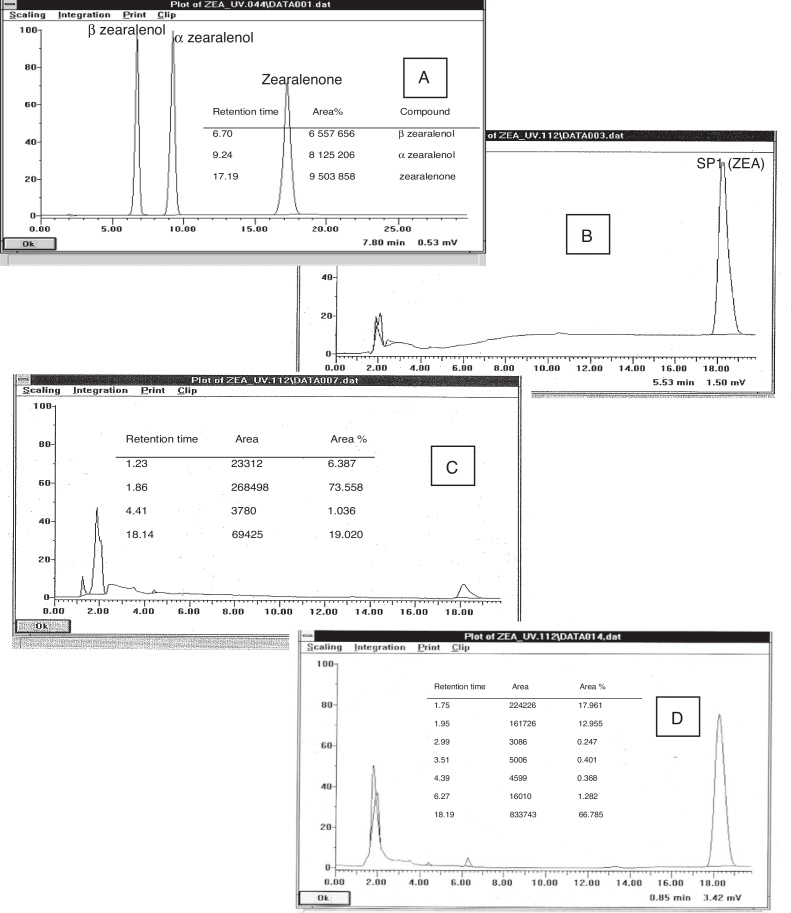
Zearalenone determination by HPLC chromatography
Results from a mixed standard solution of ZEA, α and β zearalenol are showed in Fig. 4 A. Results from SP1 solution without contact with adsorbent are showed in Fig. 4 B, peaks corresponding to α and β zearalenol were not observed. Results from the supernatant of L4 strain after incubation (37 °C, 1 h) with the mycotoxin are showed in Fig. 4C, peaks corresponding to α and β zearalenol were not observed. Results from L4 pellet after resuspension with methanol are showed in Fig. 4D. Peak corresponding to β zearalenol with a small Area % was observed, peak corresponding to α zearalenol was not observed, it was important to demonstrated that probably this Lactobacillus did not metabolize the mycotoxin as indicated by El-Nezami et al.13. The sum of ZEA in supernatant and in pellet was almost the initial ZEA added in SP1 solution, it showed that the initial mycotoxin in (SP1) solution was almost completely recovered.
Fig. 4.
Chromatographic profiles. (A) Chromatographic profile of standards: α zearalenol, β zearalenol and zearalenona. (B) Chromatographic profile of ZEA solution (SP1). (C) Chromatographic profile of L4 supernatant after co-incubating with ZEA (1 h, 37 °C). (D) Chromatographic profile of ZEA from L4 pellet after solubilization with methanol.
Conclusions
This work demonstrated that a Lactobacillus plantarum (L4 strain) isolated from pigs has great potential as adsorbent of the mycotoxin zearalenone. The results showed the best conditions for the adsorption process. The initial culture medium used for bacterial reactivation, the viability of the cells and the amount of bacterial mass measured by OD560 nm were strongly involved in the percentage of adsorption of L. plantarum to ZEA. The studies showed high stability of the complex bacteria–mycotoxin and the absence of metabolites of the toxin. This study is the first step in the development of a new ZEA adsorbent based on a lyophilized powder of a locally isolated strain and other components that help to improve reproductive health.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work is part of a project entitled “Detoxificación de la micotoxina zearalenona en alimentos para cerdos mediante métodos microbiológicos” which is financially supported by the “Comisión de Investigación Científica (CIC)” and developed in the following laboratories: Department of Public Health (FBQF – UNT – Tucuman, Tucuman, Argentina), Laboratory of Toxicology (SNITV) (FCV – UNCPBA, Tandil, Buenos Aires, Argentina), and Department of Technology and Quality of Food (FCV – UNCPBA, Tandil, Buenos Aires, Argentina).
We are grateful to Dr Peter Purslow from Canada, Departamento de Calidad y Tecnología de los Alimentos, Universidad Nacional del Centro de la Provincia de Buenos Aires, Tandil, Argentina, for his valuable revision of the manuscript.
Associate Editor: Mariza Landgraf
References
- 1.IARC Some naturally occurring substances: food items and constituents, heterocyclic aromatic amines and mycotoxins. IARC Monogr Eval Carcinog Risks Hum. 1993;56:397–444. [Google Scholar]
- 2.IARC Some food additives, feed additives and naturally occurring substances. IARC Monogr Eval Carcinog Risks Hum. 1983;31:279–291. [PubMed] [Google Scholar]
- 3.IARC Overall evaluation of carcinogenicity to humans. IARC Monogr. 1999;1:1–36. [Google Scholar]
- 4.EFSA Scientific opinion on the risks for public health related to the presence of zearalenone in food. EFSA J. 2011;9(6):2197. [Google Scholar]
- 5.Zinedine A., Soriano J.M., Molto J.C., Manes J. Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: an oestrogenic mycotoxin. Food Chem Toxicol. 2007;45(1):1–18. doi: 10.1016/j.fct.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 6.Ryu D., Hanna M.A., Eskridge K.M., Bullerman L.B. Heat stability of zearalenone in an aqueous buffered model system. J Agric Food Chem. 2003;51(6):1746–1748. doi: 10.1021/jf0210021. [DOI] [PubMed] [Google Scholar]
- 7.Anadón A., Céspedes A., Caballero V., Martínez-Larrañaga M.R., Martínez M.A. Mycotoxins of major impact in pig production and human health implications. Adv Tecnol Porcina. 2005;2(5):33–60. [Google Scholar]
- 8.Bata Á., Lásztity R. Detoxification of mycotoxin contaminated food and feed by microorganisms. Trends Food Sci Technol. 1999;10:223–228. [Google Scholar]
- 9.Fegan D.F., Spring P. Recognizing the reality of the aquaculture mycotoxin problem: searching for a common and effective solution; nutritional biotechnology in the feed and food industries. Proceedings of Alltech's 23rd Annual Symposium. The New Energy Crisis: Food, Feed or Fuel?; Lexington, KY, USA; 2007. [Google Scholar]
- 10.Abbes S., Ben Salah-Abbes J., Sharafi H., Noghabi K.A., Oueslati R. Interaction of Lactobacillus plantarum MON03 with Tunisian montmorillonite clay and ability of the composite to immobilize Zearalenone in vitro and counteract immunotoxicity in vivo. Immunopharm Immunotoxicol. 2012;34(6):944–950. doi: 10.3109/08923973.2012.674139. [DOI] [PubMed] [Google Scholar]
- 11.Bueno D.J. San Miguel de Tucumán (Tucumán); Centro de Referencia para Lactobacilos (CERELA), Universidad Nacional de Tucumán: 2003. Detoxificación de micotoxinas presentes en alimentos de aves de corral, in Centro de Referencia para Lactobacilos (CERELA) – Departamento de Biología Molecular, Facultad de Ciencias Exactas, Físico-Químicas y Naturales de la Universidad Nacional de Río Cuarto (UNRC) p. Capítulo V. [Google Scholar]
- 12.El-Nezami H., Polychronaki N., Lee Y.K. Chemical moieties and interactions involved in the binding of zearalenone to the surface of Lactobacillus rhamnosus strains GG. J Agric Food Chem. 2004;52(14):4577–4581. doi: 10.1021/jf049924m. [DOI] [PubMed] [Google Scholar]
- 13.El-Nezami H., Polychronaki N., Salminen S., Mykkanen H. Binding rather than metabolism may explain the interaction of two food-Grade Lactobacillus strains with zearalenone and its derivative (α)alpha-Zearalenol. Appl Environ Microbiol. 2002;68(7):3545–3549. doi: 10.1128/AEM.68.7.3545-3549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristensen N.B., Sloth K.H., Hojberg O., Spliid N.H., Jensen C., Thogersen R. Effects of microbial inoculants on corn silage fermentation, microbial contents, aerobic stability, and milk production under field conditions. J Dairy Sci. 2010;93(8):3764–3774. doi: 10.3168/jds.2010-3136. [DOI] [PubMed] [Google Scholar]
- 15.Mokoena M.P., Chelule P.K., Gqaleni N. Reduction of fumonisin B1 and zearalenone by lactic acid bacteria in fermented maize meal. J Food Prot. 2005;68(10):2095–2099. doi: 10.4315/0362-028x-68.10.2095. [DOI] [PubMed] [Google Scholar]
- 16.Niderkorn V., Boudra H., Morgavi D.P. Binding of Fusarium mycotoxins by fermentative bacteria in vitro. J Appl Microbiol. 2006;101(4):849–856. doi: 10.1111/j.1365-2672.2006.02958.x. [DOI] [PubMed] [Google Scholar]
- 17.Niderkorn V., Morgavi D.P., Pujos E., Tissandier A., Boudra H. Screening of fermentative bacteria for their ability to bind and biotransform deoxynivalenol, zearalenone and fumonisins in an in vitro simulated corn silage model. Food Addit Contam. 2007;24(4):406–415. doi: 10.1080/02652030601101110. [DOI] [PubMed] [Google Scholar]
- 18.Tinyiro S.E., Wokadala C., Xu D., Yao W. Adsorption and degradation of zearalenone by bacillus strains. Folia Microbiol (Praha) 2011;56(4):321–327. doi: 10.1007/s12223-011-0047-8. [DOI] [PubMed] [Google Scholar]
- 19.Lu Q., Liang X., Chen F. Detoxification of Zearalenone by viable and inactivated cells of Planococcus sp. Food Control. 2011;22(2):191–195. [Google Scholar]
- 20.Fuller R. Chapman & Hall; Londres: 1997. Probiotics 2: Applications and Practical Aspects. [Google Scholar]
- 21.Jurado G.H., Aguirre F.D., Ramírez T.C. Caracterización de bacterias probióticas aisladas del intestino grueso de cerdos como alternativa al uso de antibióticos. Rev MVZ Córdoba. 2009;14(2):1723–1735. [Google Scholar]
- 22.Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66(5):365–378. [PubMed] [Google Scholar]
- 23.Havenaar R., Ten Brink B., Huis in’t Veldt J.H.J. Chapman & Hall; London: 1992. Probiotics: The Scientific Basis. [Google Scholar]
- 24.Dogi C.A., Fochesato A., Armando R. Selection of lactic acid bacteria to promote an efficient silage fermentation capable of inhibiting the activity of Aspergillus parasiticus and Fusarium gramineraum and mycotoxin production. J Appl Microbiol. 2013;114(6):1650–1660. doi: 10.1111/jam.12173. [DOI] [PubMed] [Google Scholar]
- 25.Raibaud P., Caulet M. Lactobacilli predominant in the gastrointestinal tract of the adult swine. C R Hebd Seances Acad Sci. 1957;244(6):816–818. [PubMed] [Google Scholar]
- 26.Raibaud P., Caulet M., Galpin J.V., Moquot G. Studies on the bacterial flora of thealimentary tract of pigs. II. Streptococci: selective enumeration and differentiation of the dominant group. J Appl Bacteriol. 1961;24:285–306. [Google Scholar]
- 27.Raibaud P., Caulet M., Mocquot G. Quantitative study of lactobacilli and coliform bacteria in the digestive tract of the adult pig. C R Hebd Seances Acad Sci. 1957;244(5):683–685. [PubMed] [Google Scholar]
- 28.Dubernet S., Desmasures N., Gueguen M. A PCR-based method for identification of lactobacilli at the genus level. FEMS Microbiol Lett. 2002;214(2):271–275. doi: 10.1111/j.1574-6968.2002.tb11358.x. [DOI] [PubMed] [Google Scholar]
- 29.Kwon Y.M., Cox M.M. Improved efficacy of whole genome amplification from bacterial cells. Biotechniques. 2004;37(1) doi: 10.2144/04371BM03. 40, 42, 44. [DOI] [PubMed] [Google Scholar]
- 30.Pospiech A., Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995;11(6):217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 31.Hebert E.M., Raya R.R., Tailliez P., de Giori G.S. Characterization of natural isolates of Lactobacillus strains to be used as starter cultures in dairy fermentation. Int J Food Microbiol. 2000;59(1–2):19–27. doi: 10.1016/s0168-1605(00)00282-8. [DOI] [PubMed] [Google Scholar]
- 32.Klose V., Bayer K., Bruckbeck R., Schatzmayr G., Loibner A.P. In vitro antagonistic activities of animal intestinal strains against swine-associated pathogens. Vet Microbiol. 2010;144(3–4):515–521. doi: 10.1016/j.vetmic.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 33.Kullen M.J., Sanozky-Dawes R.B., Crowell D.C., Klaenhammer T.R. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J Appl Microbiol. 2000;89(3):511–516. doi: 10.1046/j.1365-2672.2000.01146.x. [DOI] [PubMed] [Google Scholar]
- 34.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Cole J.R., Wang Q., Cardenas E. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bueno D.J., Di Marco L., Oliver G., Bardon A. In vitro binding of zearalenone to different adsorbents. J Food Prot. 2005;68(3):613–615. doi: 10.4315/0362-028x-68.3.613. [DOI] [PubMed] [Google Scholar]
- 39.Kolfk-Clauw M., Ayouni F., Tardieu D. HPLC assay of zearalenone and reduced metabolites en S9 fractions of duck liver. Rev Med Veter. 2007;158(10):504–508. [Google Scholar]
- 40.Armando M.R., Pizzolitto R.P., Dogi C.A. Adsorption of ochratoxin A and zearalenone by potential probiotic Saccharomyces cerevisiae strains and its relation with cell wall thickness. J Appl Microbiol. 2012;113(2):256–264. doi: 10.1111/j.1365-2672.2012.05331.x. [DOI] [PubMed] [Google Scholar]
- 41.U. S. Department of Health and Human Services, Food and Drug Administration (FDA), Center for Drug Evaluation and Research (CDER), Center for Veterinary Medicine (CVM) 2001. Guidance for Industry – Bioanalytical Method Validation. Accessed 27.06.15. Available from: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm070107.pdf. [Google Scholar]
- 42.Chesher D. Evaluating assay precision. Clin Biochem Rev. 2008;29(suppl 1):S23–S26. [PMC free article] [PubMed] [Google Scholar]
- 43.Boswald C., Engelhardt G., Vogel H., Wallnofer P.R. Metabolism of the Fusarium mycotoxins zearalenone and deoxynivalenol by yeast strains of technological relevance. Nat Toxins. 1995;3(3):138–144. doi: 10.1002/nt.2620030304. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida S., Okano K., Tanaka T., Ogino C., Kondo A. Homo-d-lactic acid production from mixed sugars using xylose-assimilating operon-integrated Lactobacillus plantarum. Appl Microbiol Biotechnol. 2011;92(1):67–76. doi: 10.1007/s00253-011-3356-6. [DOI] [PubMed] [Google Scholar]
- 45.Zanoni P., Farrow J.A.E., Phillips B.A., Collins M.D. Lactobacillus pentosus (Fred Peterson, and Anderson) sp. nov., norn, rev. Int J Sistem Bacteriol. 1987;37(4):339–341. [Google Scholar]
- 46.Yiannikouris A., Andre G., Buleon A. Comprehensive conformational study of key interactions involved in zearalenone complexation with beta-d-glucans. Biomacromolecules. 2004;5(6):2176–2185. doi: 10.1021/bm049775g. [DOI] [PubMed] [Google Scholar]
- 47.Yiannikouris A., Francois J., Poughon L. Adsorption of Zearalenone by beta-d-glucans in the Saccharomyces cerevisiae cell wall. J Food Prot. 2004;67(6):1195–1200. doi: 10.4315/0362-028x-67.6.1195. [DOI] [PubMed] [Google Scholar]