Abstract
Food-borne diseases, caused by the pathogenic bacteria, are highly prevalent in the world. Salmonella is one of the most important bacterial genera responsible for this. Salmonella Enteritidis (SE) is one of the non-typhoid Salmonellae that can be transmitted to human from poultry products, water, and contaminated food. In recent years, new and rapid detection methods such as enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR) have been developed. In this study, recombinant FliC (rFliC) was produced to be used as an antigen. The immunization was conducted in mice with the purified recombinant FliC (rFliC). The mice were subcutaneously immunized with rFliC and elicited significant rFliC specific serum IgG antibodies. An indirect ELISA system was established for the detection of Salmonella Enteritidis. Our results confirmed that the recombinant flagellin can be one of the excellent indicators for the detection of Salmonella Enteritidis.
Keywords: Salmonella Enteritidis, Flagellin C, Indirect ELISA, Bacterial detection, Food contaminated
Introduction
Salmonella represents a group of important gram-negative bacterial pathogens that cause intestinal and systemic diseases in human and animal hosts after the ingestion of contaminated water and food such as poultry meat and eggs.1 Approximately one million cases of Salmonella infections are reported every year in the United States.2 In previous descriptive studies from different places and samples in Iran, the prevalence of Salmonella was found to be as 9.2% in 272 stool samples and 8% in 369 stool. In a study, 610 samples were obtained from children under 12 years of age with 37.5% prevalence of gastroenteritis, which is also caused by an important enteric pathogen bacterium.3, 4, 5, 6 More than 2500 serovars have been identified for Salmonella Enteritidis, based on antigenic differences in O, H1, and H2 antigens.7 Among the 30 Salmonella serovars that are responsible for 73% cases of salmonellosis in the United States, Salmonella enterica subsp. enterica serovar Enteritidis or Salmonella Enteritidis is an important and dominant bacterial pathogen. It was a prevalent cause of human salmonellosis and causative agents of foodborne illnesses worldwide during the early 1980s to the late 1990s.8, 9, 10 Different methods (e.g., conventional, immunological, and molecular-based methods) have been developed for the detection of Salmonella. Although the current culture-based methods for the detection of Salmonella are sensitive and inexpensive but at the same time they are very time and material-consuming and need initial enrichment. For example, the conventional method for the detection of Salmonellae, including Salmonella Enteritidis, from eggs takes 5–7 days, is labor-intensive and involves the isolation of the organism using pre-enrichment as well as selective enrichment procedures and serological confirmation tests. This method is useful for the detection of small numbers of Salmonella Enteritidis.
Molecular methods (PCR) are good but they also have few limitations. For PCR-based methods, the pathogen needs to be grown and a high concentration of nucleic acid is required to be extracted.11, 12 Bacterial flagellin is one of the outer membrane proteins that serve many functions like mobility, pathogenicity, and adjuvanticity and shows toll-like receptor (TLR)-ligand activity. It is effective at very low doses13, 14 and binds to toll-like-receptor 5 (TLR 5), which is present on the immune-system cells (epithelial cells, dendritic cells, and macrophage). One of the outmost flagellin proteins is FliC which has a molecular weight of 50–60 kDa.15, 16 The binding of FliC with TLR5 leads to a cascade of reactions that results in the production of pro-inflammatory cytokines like TNF-α, IL-6, and IL-12.17
In this study, we produced recombinant flagellin (r-FliC) for the detection of Salmonella Enteritidis (SE) using enzyme-linked immunosorbent assay (ELISA) and proposing its usefulness in ELISA for the detection of Salmonella.
Materials and methods
Kits, enzymes, and reagents
The plasmid extraction and gel purification kits were procured from iNtRON (Seongnam, Korea). Nickel-nitrilotriacetic acid (Ni-NTA) agarose was obtained from Qiagen (Maryland, State, USA). Primers were synthesized by Sinaclon (Tehran, Iran). Restriction endonucleases were obtained from Sinaclon (Tehran, Iran). T4 DNA ligase was supplied by Fermentas (Vilnius, Lithuania). All other reagents were of at least analytical grade and obtained from Sigma–Aldrich or Merck, Germany, including kanamycin (40 μg/mL, Sigma), nitrocellulose membrane (PROTRAN), anti-polyhistidine antibodies and anti-mouse IgG conjugated with horseradish peroxidase (HRP) (RAY Biotech), and an ELISA reader (Bio-Rad, Berkeley, CA, USA).
Bacterial strains and culture conditions
The standard strain of Salmonella enterica serovar Enteritidis (SE) (ATCC – 13076, Institute Pasteur of Iran) was used as the source of flic gene. It was grown in Luria-Bertani (LB) broth or LB agar at 37 °C. Bacterial genome was extracted by the CTAB-NaCl method, and the DNA concentration was measured by a spectrophotometer (Cecil, UK, OD 260 and 280 nm). The quality of the isolated DNA was assessed by electrophoresis on 1% agarose gel.
Amplification of flic gene
A colony of Salmonella Enteritidis was grown in Luria-Bertani broth (LB broth) overnight at 37 °C under constant agitation at 150 rpm. The genomic DNA was extracted from the Salmonella strain and flic gene was amplified by polymerase chain reaction (PCR) using the following two specific primers (flic F: 5-tatagaattcatggcacaagtcattaatac-3 containing an EcoRI-engineered restriction site and flic R: 5-tatataagcttttaacgcagtaaagagagg-3 containing a HindIII-engineered restriction site). The primers were designed to over the complete sequence of flic gene (1518 bp) located on the chromosomal DNA of Salmonella enteritidis as mentioned in the database available at the National Center for Biotechnology Information (NCBI). For the amplification of the flic gene, the polymerase chain reaction (PCR) was standardized using 10 pM of each gene specific primers, 2 μL of 25 mM MgCl2, 10 mM of each dNTPs, 2.5 μL of 10× enzyme buffer and 0.5 U of Taq DNA polymerase (Fermentas) in a-25 μL final reaction volume. The amplification was carried out with the initial denaturation of DNA at 95 °C for 5 min followed by 30 cycles at 95 °C for 1 min, annealing at 55 °C for 45 s, extension at 72 °C for 1 min, and then a final extension at 72 °C for 5 min. The amplified product was analyzed by the electrophoresis using 1% agarose gel and ethidium bromide as a tracking dye. For the cloning of flic gene, the amplification was carried out with pfu DNA polymerase (Fermentas, Lithuania) in a reaction mixture (25 μL) containing DNA in the presence of 4 mM magnesium sulfate and 10 pM of each primer. The cycling conditions were initial denaturation at 95 °C for 5 min, followed by 30 cycles at 95 °C for 1 min, 55 °C for 45 s, 72 °C for 60 s and the final extension at 72 °C for 5 min. The PCR products were analyzed by electrophoresis on 1% agarose gels and visualized by ethidium bromide staining.
Cloning and expression of flagellin
The amplified PCR product was double-digested with EcoRI and HindIII, and cloned into the prokaryotic expression vector pET-28a (+) at EcoRI/HindIII site to form the expression plasmid pET-fliC with kanamycin resistance as a selectable marker. Escherichia coli DH5α transformants grown overnight on LB plates containing kanamycin (20 μg/mL) were screened. For ligation, the flic gene PCR product and vector plasmid were used in the ratio of 2:1. The ligated product was initially propagated in E. coli DH5α competent cells.15 The transformed colonies were screened by colony PCR, restriction enzyme analysis, and sequencing. The recombinant pET-fliC plasmid extracted from E. coli DH5α cells was purified and transformed into E. coli BL21 (DE3) strain for the expression of flagellin. The expression was induced by adding 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) to growing culture of the transformed E. coli BL21 (DE3) when OD600 reached 0.6, at 37 °C, under constant shaking at 150 rpm. A zero-time aliquot (uninduced culture) was used as the control. The samples were collected after 24 h and analyzed for the protein expression in 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The recombinant protein was further confirmed by Western blot analysis.
Western blot analysis
Western blotting was performed to examine the FliC secretion from E. coli BL21 (DE3). In order to prepare samples, E. coli BL21 (DE3) strain was cultured overnight in LB broth at 37 °C. The cultured bacteria were harvested by centrifugation at 6000 rpm for 15 min. The secreted protein sample was separated on 12% SDS-PAGE. The separated proteins were transferred electrophoretically using transfer buffer (39 mM glycine, 48 mM Tris-base, 0.037% SDS, and 20% methanol, Bio-Rad) onto a polyvinylidene fluoride membrane (PVDF, Immobilon, Millipore) for immunoblotting. The membrane was blocked under constant shaking for 24 h with 5% (w/v) skimmed milk in phosphate-buffered saline (PBS), 0.05% Tween-20 (PBS/T), pH 7.4 at 4 °C. The membrane was incubated with a 1:3000 dilution of mice anti-His-tag specific antibody in the PBS containing 0.05% Tween-20 (PBS/T), with gentle shaking at room temperature for 1 h. After washing the membrane thrice with PBS/T, it was further incubated with goat anti-mouse IgG horseradish peroxidase conjugate, 1:1000 diluted in PBS/T, at room temperature for 1 h and then washed thrice with PBS/T. 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate was used to visualize the membrane.
Solubility and purification of the recombinant flagellin protein with His-tag
The solubility of the expressed protein was determined by resuspending the bacterial pellet (24 h post IPTG induction) in PBS. After sonication, the lysate was centrifuged at 12,000 × g (15 min) and the supernatant representing the soluble fraction was collected. The pellet containing the insoluble matter was again resuspended in 8 M urea. Both extracts were analyzed by resolving on 12% SDS-PAGE. The poly-His-tag containing recombinant flagellin was purified using nickel divalent ions (Ni-NTA chromatography) (Maryland, USA) under denaturation conditions as per the manufacturer's instructions. The protein concentration was estimated by Bradford's method18 and the positive elutes confirmed by SDS-PAGE were pooled and stored at −20 °C until future use.
Animal immunization
Female BALB/c mice (18–22 g) used in this study were obtained from Pasteur Institute of Iran, Tehran. The animals were housed and treated in accordance with the Research Ethics Committee according to the Declaration of Helsinki. The mice were divided into test (n = 5) and control groups (n = 3). In the test group, each mouse was injected subcutaneously, at the back of the neck, 10 μg of recombinant protein (rFliC) with complete Freund's adjuvant. Subsequently, 10 μg of rFliC protein was injected as the first, second, and third booster after 0, 14 and 28 days, using incomplete Freund's adjuvant. The control group was injected with sterile PBS following the same protocol. Blood samples were collected from the mice one week after the second and third booster dose (Table 1).
Table 1.
Summary of immunization protocols.
| Groups | Number | Initial Immunization components Quantity (μl) | Booster 1 components Quantity (μl) | Booster 2 components Quantity (μl) | |||
|---|---|---|---|---|---|---|---|
| Control | 3 | PBS | 200 μ1 | PBS | 200 μ1 | PBS | 200 μ1 |
| rFliC | 5 | rFliC | 10 μg | rFliC | 10 μg | rFliC | 10 μg |
| C.F.A | 100 μ1 | I.F.A | 100 μ1 | I.F.A | 100 μ1 | ||
C.F.A = Complete Freund > s Adjuvant, I.F.A = Incomplete Freund > s Adjuvant.
Enzyme-linked immunosorbent assay (ELISA) for plasma IgG
Antigen-specific antibody responses were determined by the enzyme-linked immunosorbent assay (ELISA). Polystyrene 8-well plates (MaxiSorp microtiter plates) were coated with 3 μg of rFliC diluted in 100 μL coating buffer (64 mM Na2CO3, 136 mM NaHCO3, pH 9.8) and then incubated overnight at 4 °C. The plates were washed three times with PBS/T (PBS containing 0.05%, w/v Tween-20). Nonspecific binding sites were blocked with 100 μL of 3% (w/v) skimmed milk in PBS/T (1 h, 37 °C). Mouse serum samples were serially diluted to 1:500 in PBS containing 0.03% Tween-20 and were added to the ELISA plates. After three washing steps, 100 μL of the goat anti-mouse IgG peroxidase conjugate (Sigma) diluted 1:5000 in PBS/T was added to the plates. The plates were incubated for 1 h at 37 °C and then washed thrice in PBS/T. To each well, 100 μL of TMB substrate (Sigma) was added and incubated at room temperature for 15 min. The reaction was stopped with 100 μL of 2 M H2SO4 and the result was read by an ELISA reader at the wavelength of 450 nm (Bio-Rad, Berkeley, CA, United States).
Purification of Anti-FliC IgG with Protein G Sepharose 4B column
IgG antibodies were purified using G Sepharose 4B column (Sigma). At first, the column was washed with 100 mM Tris–HCl buffer (pH = 8) followed by 10 mM Tris–HCl buffer (pH = 8) until the column was reached to equilibrium. The serum samples were loaded onto the column and it was washed with 100 mM Tris–HCl buffer (pH = 8) followed by 10 mM Tris–HCl buffer (pH = 8) until all other proteins were eluted. The antibodies were eluted from the column by passing 100 mM glycine buffer (pH = 3). At the end, the purity of IgG was confirmed by SDS-PAGE (12%).
The determination of optimum concentrations of the antibody and bacteria
ELISA was performed as described above using the different concentrations of the purified anti-FliC IgG antibody (2, 3, 4, 5, 10, 20, 30, 40, 50 and 60 μg) and a constant concentration of the antigen (3 μg). In another set of experiment, ELISA was performed with the different concentrations of the antigen (0.625, 1.25, 2.5, 5, 10, 15 and 20 μg) and a constant concentration of the antibody (10 μg). Subsequent to this, whole-bacterial cell-ELISA was carried out with the standard strain of Salmonella enteritidis according to McFarland standard (OD: 0.257 nm, cell density 3 × 108 CFU/mL). In this assay, the plates were coated with the different concentrations of bacterial cells (108, 107, 106, 105, 104, 103, 102, and 10 CFU/mL) and were exposed to a constant concentration (10 μg) of the antibody.
Specificity test
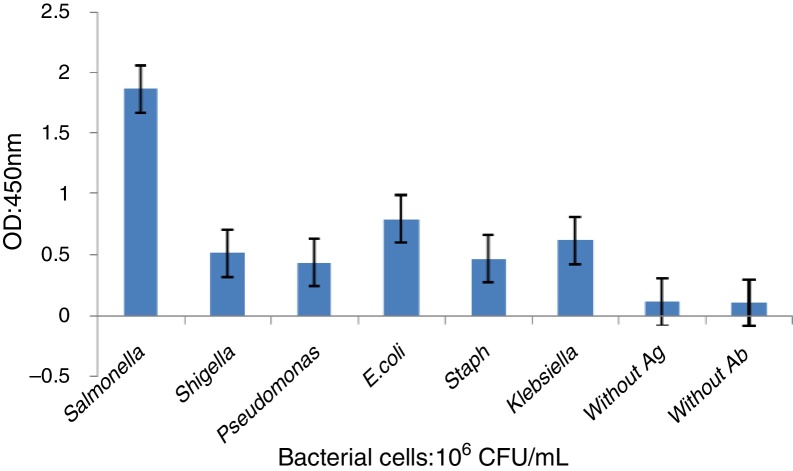
The specificity of the indirect-ELISA was checked using a variety of other bacteria, including E. coli, Shigella sp., Pseudomonas aeruginosa, Klebsiella sp., Staphylococcus aureus and Salmonella Enteritidis. The results revealed that ELISA was sensitive only against Salmonella Enteritidis with a detection sensitivity of 106 CFU/mL and showed no cross-reactivity with other tested bacterial species. The cut-off was OD 0.7 as determined on the basis of the average control samples plus two-fold standard deviations.
Clinical samples
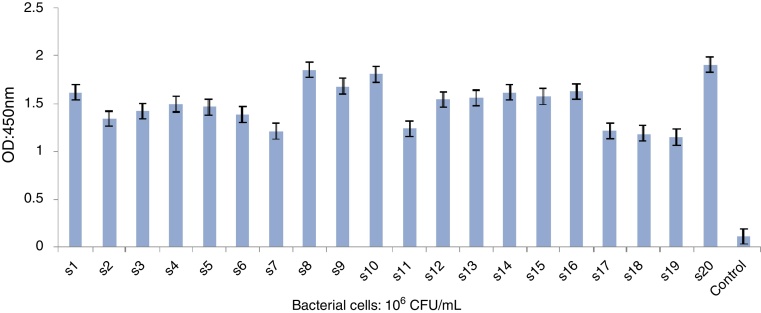
The infectious dose of Salmonella is 106–107 cells. Therefore, the ELISA was performed using clinical specimens collected from the patients with Salmonellosis (20 samples). The assay protocol was based on the previous ELISA, optimized by a standard strain of Salmonella Enteritidis with 106 CFU/mL cells.
Statistical analysis
All the data points were the mean (±SD) of three independent sets of experiments. SPSS and Excel software were used to determine means and standard deviations. The results were subjected to Student's t-test (Excel; Microsoft) for independent variables and the values were considered significant at p-values <0.05.
Results
Amplification and cloning of filc gene
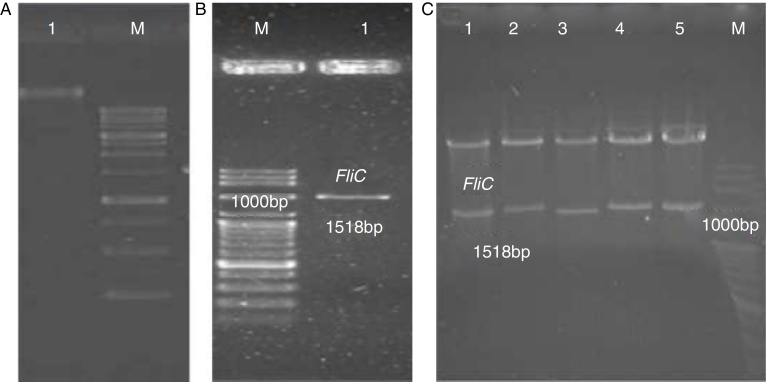
Salmonella Enteritidis genome was extracted (Fig. 1A) and the flic gene was amplified by PCR using the specific primers. The PCR product (1500 bp) is shown in Fig. 1B. The fragment was cloned in pET-28a (+) vector and then transformed into E. coli DH5α. pET28a-flic plasmids were extracted from E. coli and digested by EcoRI/HindIII and analyzed by agarose gel electrophoresis (Fig. 1C). The recombinant pET28a-fliC plasmid extracted from E. coli DH5α cells was purified and transformed to E. coli BL21 (DE3) cells for the expression of FliC protein.
Fig. 1.
Genomic DNA extraction, PCR products, and the digestion analysis on an agarose gel. (A) Lane M, DNA size marker; Lane 1, Salmonella genome. (B) Lane M, DNA size marker; Lane 1, fliC gene. (C) Lane M, DNA size marker; Lanes 1–5, digested constructs by EcoRI and HindIII restriction enzymes.
Expression and purification of recombinant protein
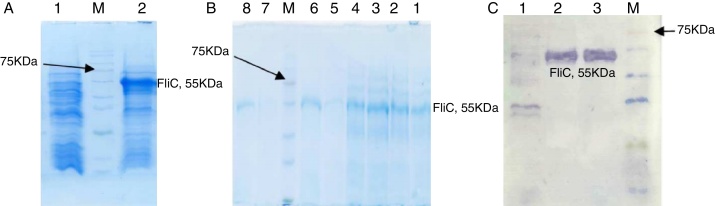
The recombinant FliC protein with N-terminal His-tag was expressed in E. coli BL21-DE3 cells (Fig. 2A) and purified by Ni-NTA affinity chromatography. The SDS-PAGE of the purified product is depicted in Fig. 2B. The Western blotting with the anti-His-tag antibody confirmed the presence of a 55-kDa protein (Fig. 2C).
Fig. 2.
Expression, purification, and identification of rFliC, stained with Coomassie blue R-250. (A) Expression of rFliC. M, protein size marker. Lane 1 non-induced transformed (pET28a without insert) BL21DE3 as control; Lane 2 induced transformed (pET28a with insert). (B) Purification steps of rFliC. M, protein size marker. Lane 1, cleared lysate before loading on the column; Lane 2, 3, flow through; Lane 4, 5, washing; Lane 6, 7, 8, elution with 250 mM imidazole. (C) Western blotting of rFliC. M, protein size marker; Lane 1, non-induced transformed (pET28a without insert) BL21 DE3 as control; Lane 2, the supernatant of induced transformed BL21DE3; Lane 3, purified recombinant protein.
Immune response elicited with rFliC
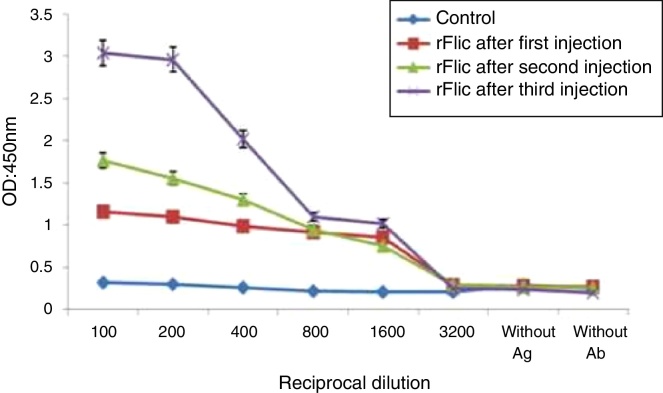
The patterns of antibody production followed by the immunization at three settings of the first, second and third injections were the same except for the increased antibody titer after each phase of the immunization (Fig. 3). This suggests that the adjuvanted recombinant flagellin injected into the BALB/c mice has good immunogenic properties. The sera obtained from the control mice that were injected with the same preparation without flagellin did not reveal any significant level of the anti-flagellin antibody.
Fig. 3.
Serum antibody response of BALB/c mice immunized with recombinant FliC proteins.
Purification of anti-FliC IgG with Protein G Sepharose 4B column
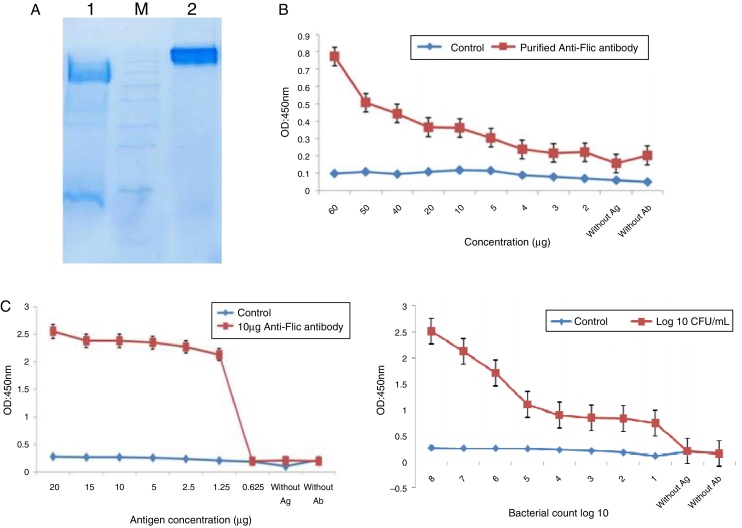
Anti-FliC IgG was purified with Protein G Sepharose 4B column chromatography and achieved a concentration of 2.339 mg/mL. In the 12% SDS-PAGE, the protein appeared as single band confirming its purity (Fig. 4A).
Fig. 4.
The purification of IgG antibody and establishing ELISA using the recombinant FliC antigen. (A) Purification of IgG antibody by G Sepharose 4B column. Lane 1, SDS-PAGE of purified IgG with 2ME; M, protein size marker. Lane 2, SDS-PAGE of purified IgG without 2ME. (B) Development of ELISA after IgG purification. (C) Development of antigen with the determination of purified antibody. (D) Bacterial ELISA.
The evaluation of the optimum concentrations of antibody and Salmonella Enteritidis for specific ELISA
A specific ELISA was designed with a constant concentration of antigen (3 μg) and varying antibody concentrations (2, 3, 4, 5, 10, 20, 30, 40, 50 and 60 μg) for the determination of the optimum concentration of antibody. A concentration of 10 μg antibody was found to be the optimum (Fig. 4B). Another ELISA was performed for the determination of antigen with a serially diluted concentration of recombinant flagellin (0.625, 1.25, 2.5, 5, 10, 15 and 20 μg) and a constant concentration of antibody (10 μg). On the basis of this result, 3 μg was considered for antigen coating (Fig. 4C). For the detection of Salmonella Enteritidis infection, bacterial-ELISA was designed with the standard strain of Salmonella (106 CFU/mL cells) (Fig. 4D).
Specificity test
The specificity of the indirect-ELISA was established using a variety of other microbial strains and showed no cross-reactivity with other tested bacteria. Based on the control samples the cut-off was determined to be 0.7 and the ELISA test assayed on 106 CFU/mL bacteria. The results of the ELISA are shown in Fig. 5.
Fig. 5.
Cross-reactivity of mouse anti-S. enteritidis IgG with other microbial strains. Assays were conducted in triplicate.
Samples results
The clinical samples were tested with ELISA for the detection of Salmonella antigen. The results of ELISA are shown in Fig. 6.
Fig. 6.
ELISA of clinical samples for the detection of Salmonella Enteritidis. Negative controls are also indicated. Assay cut-off was calculated by replicate analysis of negative samples.
Discussion
Non-typhoidal Salmonella enterica serovars cause most of the cases of bacterial food-borne diseases worldwide. Salmonella Enteritidis, which is one of the most common causes of food poisoning in humans, can cause diseases such as gastroenteritis, bacteremia, fever, intestinal, and fecal infection.19 Over the past decade, a number of investigations have reported an increasing incidence of non-typhoid Salmonella bacteremia.20 Therefore, a rapid and proper detection of this bacterium is important for food safety. The bacterial culture based methods for the identification of Salmonella are time-consuming (5–7 days) and require enrichment of the culture.21 As such, several screening methods such as nucleic acid-based assays and immunology-based assays have been examined for a rapid detection of Salmonella in the recent decades.20, 22, 23, 24 In earlier studies, lipopolysaccharides (LPS) present on the bacterial surface were used in the ELISA-assay for the detection of Salmonella, but the problem with lipopolysaccharides is the cross-reactivity with other gram-negative bacteria.25, 26, 27 Therefore, flagellin (FliC) was sought as an alternative for rapid diagnosis of Salmonella and, during the last decade, several studies have used it for the detection of Salmonella. For the production of recombinant FliC (rFlic), in this study, a standard strain of Salmonella enterica serovar Enteritidis was used that produces an antigen with high immunogenicity.28 A DNA fragment of the expected size (near 1518 bp) was amplified by PCR and expressed using chemically inducible T7 promoter (pET28a). The best condition for the recombinant rFliC expression was achieved at 1 mM IPTG concentration, the culture temperature of 37 °C and with overnight induction. However, the protein expression showed an increase with the incubation time up to 18 h. rFliC was separated on SDS-PAGE and its biochemical nature was confirmed by immunoblotting using mice anti-His-tag specific antibody. rFliC was purified based on 6XHis-tag using the Ni-NTA column. The purified rFliC was used for the immunization of mice. The ELISA test results showed that all immunized mice produced a high titer of anti-FliC antibody upon injecting rFliC three times. Then an ELISA system was designed for the detection of Salmonella Enteritidis, which is an important pathogen of human and poultry. Accurate and quick tests are essential to ensure food safety by the governments.29 The availability of sufficient quantities of recombinant antigens for the rapid detection of Salmonella can be achieved in serum samples in a short time.30 Thus, new methods for quick detection of Salmonella have been developed in the past decade. Among the molecular techniques, PCR is a common and accurate method and can also be used for the detection of low levels of pathogenic bacteria.31 ELISA can be used to screen a variety of samples and components in the presence of Salmonella spp.
The ELISA test, designed in this study, did not show any cross-reaction with other bacteria. Sensitivity analysis of this test showed that it could detect 103 bacteria in clinical samples, given that the infectious dose of Salmonella is about 106 approximately.
Besides ours, the studies of Minicozzi (2013),5 Jindal (2012)13 and Bang (2012)21 have also reported the production of recombinant flagellar antigen (r-FliC) and development of ELISA for the detection of Salmonella. In previous works, the investigations were carried out mainly with sera obtained from infected flocks and birds but, in this study, we used mice for the production of antibody and Protein G Sepharose 4B column for the purification. The flic gene of Salmonella Enteritidis was amplified, cloned successfully using pET-28a(+) expression vector and was introduced into E. coli BL21 (DE3) as a suitable host. The recombinant flagellin was purified successfully and demonstrated good immunogenic properties based on the immunization of BALB/c mice and its ability to induce a significant serum flagellin-specific IgG immune response, which was measured by ELISA. We provide further evidence that the recombinant flagellin can be one of the excellent indicators for the detection of Salmonella Enteritidis.
Conflicts of interest
The authors declare that they have no competing interest.
Acknowledgments
This article is a part a Ph.D. thesis being prepared by Ali Mirhosseini. The study was supported by the grants from Applied Microbiology Research Center Baqiyatallah University of Medical Sciences.
Associate Editor: Roxane Maria Fontes Piazza
References
- 1.Hyeon J.-Y., Chon J.-W., Choi I.-S., Park C., Kim D.-E., Seo K.-H. Development of RNA aptamers for detection of Salmonella Enteritidis. J Microbiol Methods. 2012;89(1):79–82. doi: 10.1016/j.mimet.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Kilic A., Bedir O., Kocak N. Analysis of an outbreak of Salmonella enteritidis by repetitive-sequence-based PCR and pulsed-field gel electrophoresis. Intern Med. 2009;49(1):31–36. doi: 10.2169/internalmedicine.49.2743. [DOI] [PubMed] [Google Scholar]
- 3.Ranjbar R., Giammanco G.M., Farshad S., Owlia P., Aleo A., Mammina C. Serotypes, antibiotic resistance, and class 1 integrons in Salmonella isolates from pediatric cases of enteritis in Tehran, Iran. Foodborne Pathog Dis. 2011;8(4):547–553. doi: 10.1089/fpd.2010.0736. [DOI] [PubMed] [Google Scholar]
- 4.Chashni S., Hassanzadeh M., Fard M., Mirzaie S. Characterization of the Salmonella isolates from backyard chickens in north of Iran, by serotyping, multiplex PCR and antibiotic resistance analysis. Arch Razi Inst. 2009;64(2):77–83. [Google Scholar]
- 5.Nosraty S., Fallah F., Sabokbar A., Dezfoolyan M., Adabian S., Esmaeilnejad N. Detection of Salmonella enteritidis, typhi and typhimurium in foods by multiplex PCR in children hospital. BMC Infect Dis. 2012;12(suppl 1):P95. [Google Scholar]
- 6.Dallal M.M.S. Prevalence of Salmonella spp. in packed and unpacked red meat and chicken in south of Tehran. Jundishapur J Microbiol. 2014;7(4) doi: 10.5812/jjm.9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fierer J., Guiney D.G. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J Clin Invest. 2001;107(7):775–780. doi: 10.1172/JCI12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minicozzi J., Sanchez S., Lee M.D., Holt P.S., Hofacre C.L., Maurer J.J. Development of recombinant flagellar antigens for serological detection of Salmonella enterica serotypes Enteritidis, Hadar, Heidelberg, and Typhimurium in poultry. Agriculture. 2013;3(3):381–397. [Google Scholar]
- 9.Desin T.S., Köster W., Potter A.A. Salmonella vaccines in poultry: past, present and future. Expert Rev Vaccines. 2013;12(1):87–96. doi: 10.1586/erv.12.138. [DOI] [PubMed] [Google Scholar]
- 10.Oracz G., Feleszko W., Golicka D., Maksymiuk J., Klonowska A., Szajewska H. Rapid diagnosis of acute Salmonella gastrointestinal infection. Clin Infect Dis. 2003;36(1):112–115. doi: 10.1086/344953. [DOI] [PubMed] [Google Scholar]
- 11.Siregar T.H., Elliman J., Owens L. Development of real time polymerase chain reaction for detection of Salmonella typhimurium and Salmonella enteritidis in fish. Squalen. 2013;7(2):51–58. [Google Scholar]
- 12.Amani J., Ahmadpour A., Fooladi A.A.I., Nazarian S. Detection of E. coli O157: H7 and Shigella dysenteriae toxins in clinical samples by PCR-ELISA. Braz J Infect Dis. 2015;19(3):278–284. doi: 10.1016/j.bjid.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jindal G., Tewari R., Gautam A., Pandey S.K., Rishi P. Immunological characterization of recombinant Salmonella enterica serovar Typhi FliC protein expressed in Escherichia coli. AMB Express. 2012;2(1):1–9. doi: 10.1186/2191-0855-2-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergman M.A., Cummings L.A., Barrett S.L.R. CD4+ T cells and toll-like receptors recognize Salmonella antigens expressed in bacterial surface organelles. Infect Immun. 2005;73(3):1350–1356. doi: 10.1128/IAI.73.3.1350-1356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajam I.A., Dar P.A., Sekar S.C. Expression, purification, and functional characterisation of flagellin, a TLR5-ligand. Vet Ital. 2013;49(2):181–186. [PubMed] [Google Scholar]
- 16.Cummings L.A., Wilkerson W.D., Bergsbaken T., Cookson B.T. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol. 2006;61(3):795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 17.Haiko J., Westerlund-Wikström B. The role of the bacterial flagellum in adhesion and virulence. Biology. 2013;2(4):1242–1267. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bollag D., Michael D., Stuart J. 2nd ed. John Wiley and Sons, Inc Pub; New York: 1996. Protein Methods. [Google Scholar]
- 19.Dehghani B., Rasooli I., Gargari S.L.M., Nadooshan M.R.J., Owlia P., Nazarian S. Immunogenicity of Salmonella enterica serovar Enteritidis virulence protein, InvH, and cross-reactivity of its antisera with Salmonella strains. Microbiol Res. 2013;168(2):84–90. doi: 10.1016/j.micres.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn K.G., Falkenhorst G., Ceper T.H. Detecting non-typhoid Salmonella in humans by ELISAs: a literature review. J Med Microbiol. 2012;61(1):1–7. doi: 10.1099/jmm.0.034447-0. [DOI] [PubMed] [Google Scholar]
- 21.Bang J., Shukla S., Kim Y., Kim M. Development of indirect competitive ELISA for the detection of Salmonella Typhimurium. Rom Biotechol Lett. 2012;17:7194–7204. [Google Scholar]
- 22.Rodpaia E., Moongkarndia P., Tungrugsasutb W., Phisannoradeja R., Kanaratc S. Comparison of multiplex polymerase chain reaction and immunoassay to detect Salmonella spp., S. Typhimurium, and S. Enteritidis in Thai chicken meat. ScienceAsia. 2013;39(2):150–159. [Google Scholar]
- 23.Majeed L.J., Znad K.H., Thamir M.K., Baqir H.I. Study of ELISA and antibiotic sensitivity test for Salmonella enteritidis as experimental infection in mice. Um-Salama Sci J. 2009;6(1):114–122. [Google Scholar]
- 24.Mandal J., Akhter M.Z., Kabir M.S., Ahsan S. Development of a multiplex polymerase chain reaction protocol for the simultaneous detection of Salmonella enterica serovar Typhi and Class 1 integron. Asian Pac J Trop Dis. 2014;4:S808–S812. [Google Scholar]
- 25.Isomaki O., Vuento R., Granfors K. Serological diagnosis of Salmonella infections by enzyme immunoassay. Lancet. 1989;333(8652):1411–1414. doi: 10.1016/s0140-6736(89)90124-4. [DOI] [PubMed] [Google Scholar]
- 26.Dalby T., Strid M.A., Beyer N.H., Blom J., Mølbak K., Krogfelt K.A. Rapid decay of Salmonella flagella antibodies during human gastroenteritis: a follow up study. J Microbiol Methods. 2005;62(2):233–243. doi: 10.1016/j.mimet.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Strid M.A., Dalby T., Mølbak K., Krogfelt K.A. Kinetics of the human antibody response against Salmonella enterica serovars Enteritidis and Typhimurium determined by lipopolysaccharide enzyme-linked immunosorbent assay. Clin Vaccine Immunol. 2007;14(6):741–747. doi: 10.1128/CVI.00192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crayford G., Coombes J.L., Humphrey T.J., Wigley P. Monophasic expression of FliC by Salmonella 4,[5], 12: i:-DT193 does not alter its pathogenicity during infection of porcine intestinal epithelial cells. Microbiology. 2014;160(Pt 11):2507–2516. doi: 10.1099/mic.0.081349-0. [DOI] [PubMed] [Google Scholar]
- 29.Lee K.-M., Runyon M., Herrman T.J., Phillips R., Hsieh J. Review of Salmonella detection and identification methods: aspects of rapid emergency response and food safety. Food Control. 2015;47:264–276. [Google Scholar]
- 30.Kremer C., O’Meara K., Layton S., Hargis B., Cole K. Evaluation of recombinant Salmonella expressing the flagellar protein fliC for persistence and enhanced antibody response in commercial turkeys. Poult Sci. 2011;90(4):752–758. doi: 10.3382/ps.2010-01076. [DOI] [PubMed] [Google Scholar]
- 31.Ricke S., Pillai S., Norton R., Maciorowski K., Jones F. Applicability of rapid methods for detection of Salmonella spp. in poultry feeds: a review. J Rapid Methods Autom Microbiol. 1998;6(4):239–258. [Google Scholar]