Abstract
Background
Panax ginseng is a physiologically active plant widely used in traditional medicine that is characterized by the presence of ginsenosides. Rb1, a major ginsenoside, is used as the starting material for producing ginsenoside derivatives with enhanced pharmaceutical potentials through chemical, enzymatic, or microbial transformation.
Methods
To investigate the bioconversion of ginsenoside Rb1, we prepared kimchi originated bacterial strains Leuconostoc mensenteroides WiKim19, Pediococcus pentosaceus WiKim20, Lactobacillus brevis WiKim47, Leuconostoc lactis WiKim48, and Lactobacillus sakei WiKim49 and analyzed bioconversion products using LC-MS/MS mass spectrometer.
Results
L. mesenteroides WiKim19 and Pediococcus pentosaceus WiKim20 converted ginsenoside Rb1 into the ginsenoside Rg3 approximately five times more than Lactobacillus brevis WiKim47, Leuconostoc lactis WiKim48, and Lactobacillus sakei WiKim49. L mesenteroides WIKim19 showed positive correlation with β-glucosidase activity and higher transformation ability of ginsenoside Rb1 into Rg3 than the other strains whereas, P. pentosaceus WiKim20 showed an elevated production of Rb3 even with lack of β-glucosidase activity but have the highest acidity among the five lactic acid bacteria (LAB).
Conclusion
Ginsenoside Rg5 concentration of five LABs have ranged from ∼2.6 μg/mL to 6.5 μg/mL and increased in accordance with the incubation periods. Our results indicate that the enzymatic activity along with acidic condition contribute to the production of minor ginsenoside from lactic acid bacteria.
Keywords: ginsenoside bioconversion, ginsenoside Rg3, ginsenoside Rg5, LC-MS/MS
1. Introduction
Lactic acid bacteria (LAB) have been used as probiotics and are present in many fermented foods (cheese, yogurt, butter, and kimchi), where they influence the taste and preservation by producing lactic acid and/or alcohol. Some enzymes produced by LABs can efficiently utilize ingested nutrients to benefit the host, i.e., linoleic acid isomerase from Lactobacillus acidophilus produces conjugated linoleic acid, which has biological properties, from linoleic acid [1], and β-glucosidase from Lactobacillus paraplantarum converts isoflavone glucosides, which are not absorbed by enterocytes [2], to absorbable aglycones [3]. Recent studies reported the bioconversion of ginsenosides using Lactobacillus pentosus and Leuconostoc citreum isolated from fermented foods due to β-glucosidase activity [4], [5].
The major commercial ginsengs such as Panax ginseng Meyer (Korean Red Ginseng), Panax quinquifolium (American ginseng), and Panax notoginseng (Burk.) F.H. Chen (Notoginseng) have been widely used as traditional herbal medicines [6]. Ginsenosides (ginseng saponins) are the major pharmacological constituents of ginseng, and over 100 ginsenosides have been identified [5], [7]. Major ginsenosides (80% of the ginsenosides) are composed of Rb1, Rb2, Rc, Rd, Re, and Rg1; minor ginsenosides are their deglycosylated forms and composed of Rg3, Rh2, Rh1, F2, C-K, Rg2, Rh1, Rg5, and F1 [8].
Minor ginsenosides are known to have a greater pharmaceutical potential than major ginsenosides [9], [10], [11], [12], [13], [14]. However, naturally occurring minor ginsenosides are present at very low concentrations. Therefore, hydrolysis of sugar moieties from abundant major ginsenosides are needed to produce minor ginsenosides. Gut microbiota metabolize orally administered ginseng and help transport across the epithelial membrane [15] and human intestinal microbiota convert major ginsenosides to minor ginsenosides [10], [16], [17]. However, ginsenoside metabolism varies between individuals depending on the population of gut microbiota, such as Ruminococcus spp., Bacteroides spp., and Bifidobacterium spp. [17]. Acidic environments as well as intestinal microbiota have important influences on the bioconversion of ginsenoside and the low pH of gastrointestinal environment could activate the deglycosylation of ginsenoside by acidic hydrolysis response [18], [19], [20].
Kimchi is a traditional Korean food that is fermented vegetables including cabbage and various seasonings. Kimchi has antioxidative and antidiabetic properties and bacteria isolated from kimchi produce beneficial enzymes [21], [22], [23]. Various LABs play important roles during kimchi fermentation: Lactobacillus and Leuconostoc are the predominant genera of the kimchi microbiome in the kimchi fermentation [24]. Lactobacillus species have neuroprotective, antifungal, and anticolitic properties [25], [26], [27] and Leuconostoc species play key roles in decreasing foodborne pathogen growth, viral activity, and the effects of lipid profiles [28], [29], [30]. Recent studies have suggested that LABs from kimchi produce hydrolytic enzymes that catalyze ginsenoside bioconversion by removing the glycosyl group of major ginsenosides [4], [5].
In this study, we isolated LABs associated with kimchi fermentation from homemade kimchi, and compared availability for ginsenoside bioconversion of five LABs such as Leuconostoc mensenteroides WiKim19, Pediococcus pentosaceus WiKim20, Lactobacillus brevis WiKim47, Leuconostoc lactis WiKim48, and Lactobacillus sakei WiKim49 by quantitating transformed ginsenoside using a sensitive and reliable LC-MS/MS method.
2. Materials and methods
2.1. Materials
Leuconostoc mensenteroides WiKim19, Pediococcus pentosaceus WiKim20, Lactobacillus brevis WiKim47, Leuconostoc lactis WiKim48, and Lactobacillus sakei WiKim49 were isolated from homemade kimchi using de Man, Rogosa, and Sharpe (MRS) media. MRS broth was purchased from Difco (Miller, Becton Dickinson, and Co., Sparks, MD, USA). Ginsenosides Rb1, Rg3, digoxin (internal standard), and β-glucosidase activity assay kit were purchased from Sigma Aldrich (St. Louis, MO, USA). Ginsenoside -F2, -Rg1, -Rf, -Ro, -Rg2, -R1, -Ra1, -Rb2, -Rb3, -F1, -Rd, -Rg5, -compound K; -Rh2, -Rh4, and gypenoside XVII were purchased from Ambo Institute (Daejeon, Korea). API 50 CH and inoculating fluid was purchased from bioMérieux (Lyon, France) [31].
2.2. Determination of 16S rRNA gene sequences and phylogenetic analysis
To identify the isolates using 16S rRNA sequencing, the isolates were sent to Macrogen Inc., Korea sequencing service (www.macrogen.com, Seoul, Korea). The obtained sequences were compared with available 16S rRNA sequences in the EzTaxon Server [32] to evaluate sequence similarity. Multiple sequence alignment of the 16S rRNA sequences from five lactic acid bacteria and these related species were performed with CLUSTAL W [33]. The phylogenetic trees were constructed using MEGA6 [34] with neighbor-joining [35] based on 1,000 random bootstrap replicates for each.
2.3. Assay of ginsenoside Rb1 bioconversion by lactic acid bacteria
L. mensenteroides WiKim19, P. pentosaceus WiKim20, L. brevis WiKim47, L. lactis WiKim48, and L. sakei WiKim49 were inoculated in MRS broth, until absorbance reached 600 nm of 1.0. The strains were cultured at 30°C for 1 d, 3 d, and 7 d with ginsenoside Rb1 (a final concentration of 200μM) dissolved in MeOH. After centrifugation at 5,000g for 10 min, 2 mg/mL digoxin as an internal standard was added and purified using Sep-Pak Light C18 cartridges (Waters, Milford, MA, USA) and then dissolved in MeOH.
2.4. Assay of ginsenosides by LC-MS/MS
Ginsenosides Rb1 and minor ginsenosides in the reactions were analyzed by using UPLC (Waters), coupled to a TripleTOF 5600 plus system with electrospray ionization (ESI; AB SCIEX, Framingham, MA, USA). To investigate and separate the precursor and fragmentation ions of ginsenosides Rb1, minor ginsenosides, an Acquity UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 μm particle size) from Waters was used at a flow rate of 0.5 mL/min. UPLC conditions were as follows: solvent A, water containing 10mM ammonium acetate; solvent B, acetonitrile containing 10mM ammonium acetate; gradient, 0–0.5 min (5% B), 0.5–14.5 min (5–30% B), 14.5–15.5 min (30–32% B), 15.5–16.5 min (32–40% B), 16.5–17 min (40–55% B), 17–19 min (55% B), 19–25 min (90% B), and 25–30 min (5% B). Two microliters of each sample were injected for the UPLC analysis, and peaks were identified by comparing their retention times and fragment ion with that of reference compound.
The mass spectrometry conditions were optimized under the negative ion mode as follows: curtain gas, 30; collision energy, −30; declustering potential, −80; nebulizer gas (Gas 1), 40 at MRM mode; heater gas (Gas 2), 50. The ion spray voltage was −4,500 V. Ginsenosides in all reaction mixtures were quantified with multiple reaction monitoring (MRM) using selected transitions as follows: Rb1, m/z 1,107→945; Rg3, F2, m/z 783→621; Rg5, m/z 765→603; digoxin, m/z 779→649, Rg1, Rf m/z 799→637; Ro, m/z 955 → 793; Rg2, m/z 783→637; R1, m/z 931→769; Ra1, m/z 1,209→1,077; Rb2, Rb3, m/z 1,077→ 945; F1, m/z 637→475; Rd, m/z 945→783; XVII, m/z 945→323; compound K, m/z 621→459; Rh2, m/z 621→459; and Rh4 m/z 619→161.
Data acquisition and processing were carried out using Analyst TF 1.6 and PeakVeiw 1.2 software (AB SCIEX), respectively. The data obtained from multiple reaction monitoring (MRM) mode were quantitated using MultiQuant software (AB SCIEX). The standard solutions containing 10–200μM were injected into the UPLC with 2 mg/mL digoxin. The linear calibration curve for peak area ratio (ginsenoside/digoxin) was obtained for the quantification of ginsenoside. The amounts of the ginsenosides in each sample were determined from corresponding calibration curves.
2.5. Assay of β-glucosidase activities using cell lysates
L. mensenteroides WiKim19, P. pentosaceus WiKim20, L. brevis WiKim47, L. lactis WiKim48, and L. sakei WiKim49 were cultured for 1 d in MRS broth at 30°C. The supernatant was removed after centrifugation at 12,000g for 10 min, and cell lysates including intracellular β-glucosidase were prepared by bead beating in 50mM sodium phosphate buffer (pH 7.0). Protein concentrations of cell lysates were determined using a Pierce BCA Protein Assay Kit (Thermo, Rockford, IL, USA). The proteins were diluted to the concentration of ∼0.5 mg/mL to assay enzyme activity. The enzyme activity was determined using β-glucosidase activity assay kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's protocol. The release of p-nitrophenol was measured at 405 nm (SPECTROstar Nano, BMG Labtech, Ortenberg, Germany). Analysis was performed in duplicate for each strain. One unit of β-glucosidase is the amount of enzyme that catalyzes the hydrolysis of 1.0 μmole substrate per min at pH 7.0. The results were reported as mean ± standard deviation. Statistical analyses were performed using GraphPad Prism 6 (GraphPad software, La Jolla, CA, USA). One-tailed Student t test for unpaired samples were used to assess significance of the differences between the three LABs. Differences with p values < 0.05 were considered statistically significant.
2.6. Measurement of pH and acidity
The LABs were cultivated in the MRS broth at a temperature of 30°C for 0 d and 3 d. The pH about 10 mL of each culture broth was measured using electrode pH meter (Mettler Toledo, Columbus, USA). Acidity was measured by titration against 0.1N NaOH to a phenolphthalein endpoint, pH 8.3.
3. Results and discussion
Although minor ginsenosides have various beneficial effects, only small percentages are found in ginseng. Therefore, many studies have investigated methods such as heating, mild acid hydrolysis, and enzymatic transformation for conversion of major ginsenosides [36], [37], [38].
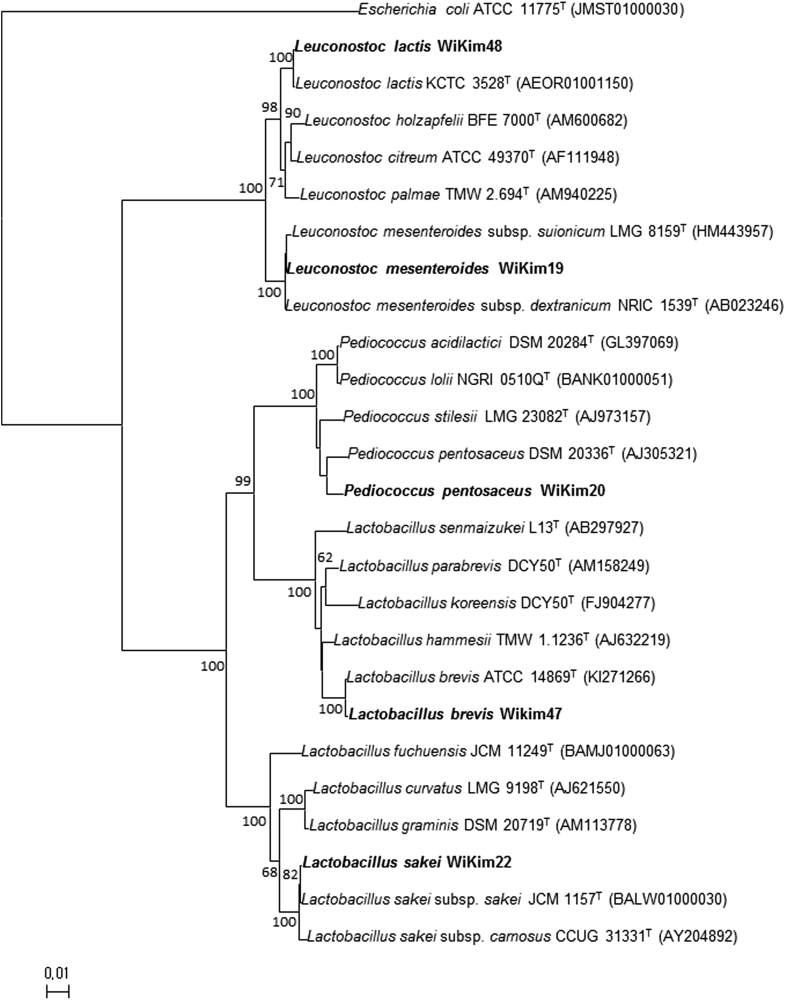
Lactic acid bacteria strains were isolated from kimchi previously and the strains were identified by 16S rRNA sequencing. The identified 16S rRNA sequences were deposited on NCBI GenBank under accession No. KT759681 (L. mesenteroides WiKim19), KX890131 (P. pentosaceus WiKim20), KX886794 (L. brevis WiKim47), KX886799 (L. lactis WiKim48), and KX886806 (L. sakei WiKim49). The constructed phylogenetic trees clustered WiKim 19, 20, 47, 48, and WiKim 49 with the Lactobacillus, Leuconostoc, and Pediococcus genera and well matched with reference strains (Fig. 1).
Fig. 1.
Phylogenetic trees constructed from 16S rRNA gene sequences. The phylogenetic relationships of Leuconostoc mensenteroides WiKim19, Pediococcus pentosaceus WiKim20, Lactobacillus brevis WiKim47, Leuconostoc lactis WiKim48 and Lactobacillus sakei WiKim49 with other species are shown. Trees were constructed using the neighbor-joining method. The numbers at the nodes represent bootstrap values (> 60%) are expressed as percentages of 1,000 replicates. Escherichia coli ATCC 11775 T was used as an outgroup. Bar, 0.01 accumulated changes per nucleotide.
The API 50 CH (bioMérieux) was used to determine the carbohydrate assimilation profile of five LABs. The test preparations were incubated at 30°, and readings were made after 48 h. The biochemical characteristics are listed in Table 1. L. mesenteroides WiKim19 showed the utilization of diverse α (1→4), β (1→4), α (1→6), α (1→2) linkage. L. brevis WiKim47 could not use maltose, lactose, inulin, and cellobiose which have α (1→4) and diverse β (1→4) linkage. P. pentosaceus WiKim20 showed the similar carbon utilizing profile with L. mesenteroides WiKim19. Both could use inulin and gentiobiose, which have β (1→2) and β (1→4) linkage, respectively. However, P. pentosaceus WiKim20 did not use lactose, which has galactopyranosyl β (1→4) linkage.
Table 1.
Distinctive features of the carbohydrate fermentation profiles of five LABs determined using API 50 CH (bioMérieux, Lyon, France)
| Characteristic |
Lactobacillus brevis WiKim47 |
Pediococcus pentosaceus WiKim20 |
Leuconostoc Lactis WiKim48 |
Lactobacillus sakei WiKim49 |
Leuconostoc mesenteroides WiKim19 |
|---|---|---|---|---|---|
| Glycerol | − | w | − | − | − |
| Erythritol | − | w | − | − | − |
| −D-arabinose | − | w | − | − | − |
| L-Arabinose | + | + | + | + | + |
| D-Ribose | + | + | + | + | + |
| D-Xylose | + | + | + | − | + |
| D-Galactose | + | + | + | + | + |
| D-Glucose | + | + | + | + | + |
| D-Fructose | + | + | + | + | + |
| D-Mannose | + | + | + | + | + |
| D-Mannitol | w | − | − | − | + |
| D-Sorbitol | − | − | − | − | + |
| Methyl-α D-glucopyranoside | + | − | + | − | + |
| N-Acetylglucosamine | + | + | + | + | + |
| Amygdalin | − | + | − | − | + |
| Arbutin | − | + | + | − | + |
| Esculin ferric citrate | − | + | + | + | + |
| Salicin | − | + | + | + | + |
| D-Cellobiose | − | + | + | + | + |
| D-Maltose | + | + | + | − | + |
| D-Lactose (bovine origin) | + | − | − | + | + |
| D-Melibiose | + | + | + | + | + |
| D-Saccharose (sucrose) | + | + | + | + | + |
| D-Trehalose | − | + | + | + | + |
| Inulin | − | + | − | − | + |
| D-Melezitose | − | − | − | − | + |
| D-Raffinose | + | + | + | − | + |
| Amidon (starch) | − | − | w | − | − |
| Gentiobiose | − | + | − | − | + |
| D-Turanose | − | − | + | − | + |
| D-Tagatose | − | + | − | − | + |
| D-Arabitol | − | − | − | − | − |
| Potassium gluconate | − | − | + | w | + |
| Potassium 2-ketogluconate | − | W | − | − | + |
−, negative; +, positive; 1, Leuconostoc mesenteroides WiKim19; Strain 2, Pediococcus pentosaceus WiKim20; Strain 3, Lactobacillus brevis WiKim47; Strain 4. Leuconostoc lactis WiKim48; Strain 5, Lactobacillus Sakei WiKim49; w, weak reaction
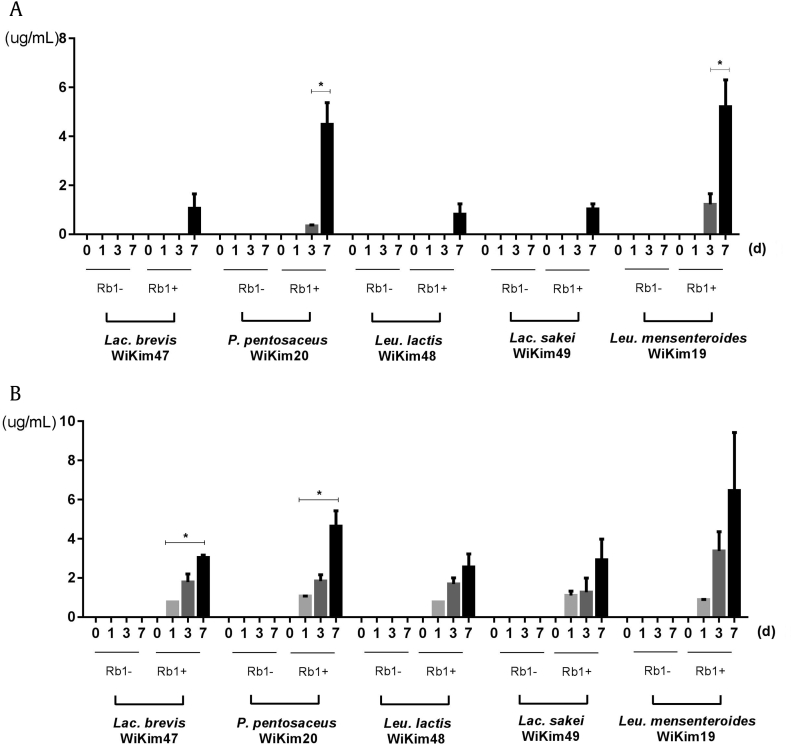
To investigate the bacterial bioconversion of ginsenoside Rb1 into minor ginsenosides, ginsenoside bioconversion was performed by incubation of five LABs with 200μM of Rb1 and measured the 15 minor ginsenoside components (ginsenoside -Rg3, -F2, -Rg5, -Rg1, -Rf, -Ro, -Rg2, -R1, -Ra1, -F1, -Rd, -compound K, -Rh2, -Rh4, gypenoside XVII).
In the results, five LABs did not produce the other minor ginsenosides except ginsenoside Rg3 and Rg5 (Figs. 1A and 1B). The content of ginsenoside Rg3 increased in L. mesenteroides WiKim19 (5.2 ± 1.1 μg/mL), P. pentosaceus WiKim20 (4.5 ± 0.9 μg/mL) and other lactic acid bacteria produced from ∼0.8 ± 0.4 μg/mL to 1.1 ± 0.6 μg/mL. Interestingly, we detected the ginsenoside Rg5 from five LABs ranged from 2.6 ± 0.7 μg/mL to 6.5 ± 3.0 μg/mL concentration at 7 d of incubation. L. mesenteroides WiKim19 (6.5 ± 3.0 μg/mL) and P. pentosaceus WiKim20 (4.6 ± 0.8 μg/mL) showed the higher ginsenoside Rg5 content among the five LABs (Fig. 2B).
Fig. 2.
Comparison of ginsenoside Rb1 bioconversion into Rg3 in cell culture supernatant of Leu. mesenteroides WiKim19, P. pentosaceus WiKim20, Lac. brevis WiKim47, Leu. lactis WiKim48 and Lac. sakei WiKim49. Concentrations of ginsenoside Rg3 (A) and Rg5 (B) after 1 d, 3 d, and 7 d incubation of each LAB strain with Rb1 at a concentration of 200μM. Data are presented as mean ± standard deviation. *p < 0.05.
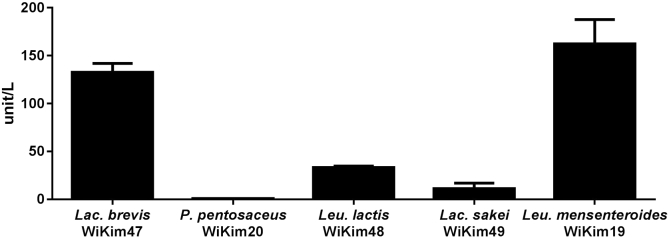
It is reported that bacterial β-glucosidase activity has been particularly involved among many glycosyl hydrolases to transform ginsenoside Rb1, which removes two glucose molecules at 20-C of protopanaxadiol into ginsenoside Rg3 [39]. To determine whether β-glucosidase influences the bioconversion of ginsenoside, we tested the β-glucosidase enzyme activity. L. mesenteroides WiKim19, which used diverse glycosidic linkage substrates (Table 1), showed the highest β-glucosidase activity supporting the maximum bioconversion capacity. L. lactis WiKim48 and L. sakei WiKim49 exhibited the reduced productions of Rg3 along with the low β-glucosidase activities. Interestingly P. pentosaceus WiKim20 showed an elevated production of Rb3 in spite of lack of β-glucosidase activity (Fig. 3).
Fig. 3.
Comparison of β-glucosidase activity in different LABs. The proteins were diluted to the concentration of ∼0.5 mg/mL to assay enzyme activity and the release of p-nitrophenol was measured at 405 nm. One-tailed Student t test for unpaired samples was used to assess significance of the differences between the three LABs. One unit of β-glucosidase is the amount of enzyme that catalyzes the hydrolysis of 1.0 μmole substrate per min at pH 7.0.
A recent study reported that the conversion of ginsenoside Rb1 into Rg3 and Rg5 with organic acid such as D-, L-tartaric acid, citric acid, and acetic acid [40]. Under acidic conditions, the low pH environment enhances the deglycosylation of ginsenoside by acidic hydrolysis response. Lactic acid bacteria are well known for their organic acid production, which lowered the pH during the fermentation and help the long-term storage by preventing contamination. Even with varying degrees of acidity among the LABs, all culture showed the acidic property and increased accordance to the growth of bacteria.
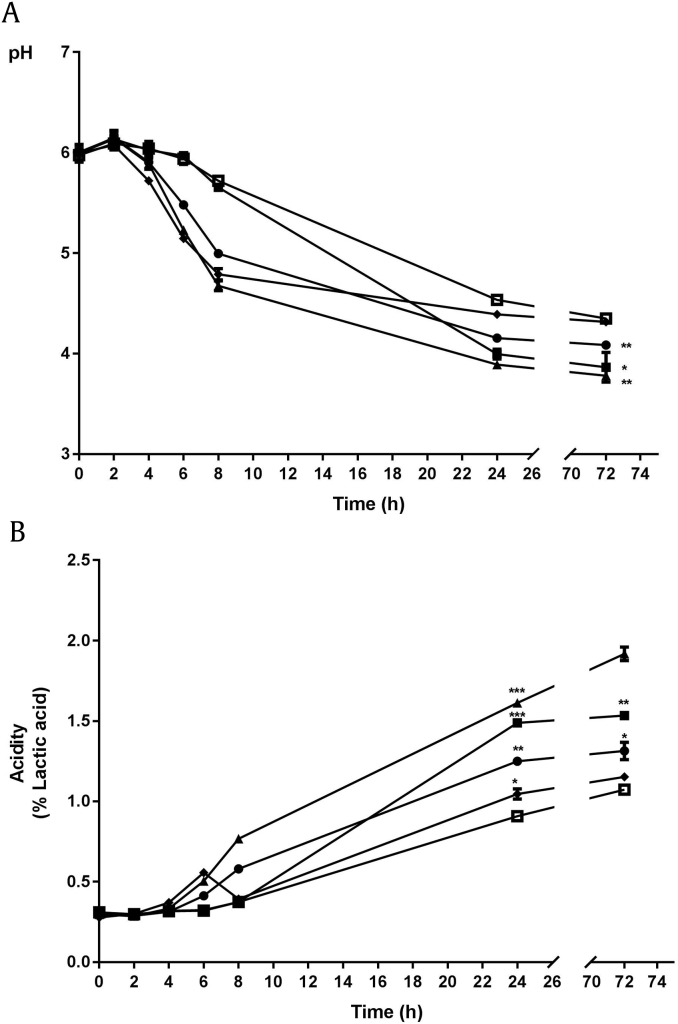
To determine whether media acidity influences the bioconversion of ginsenoside, we measured pH and acidity of bacterial cultures (Figs. 4A and 4B). The culture supernatant of P. pentosaceus WiKim20 had a pH ∼3.8, and acidity is 1.9, while L. mensenteroides WiKim19 had pH 4.4, and acidity is 1.1. The β-glucosidase activity and ginsenoside Rg3 production was correlated with L. mensenteroides WiKim19 which have higher enzyme activity and relatively low acidity among the five LABs. The elevated Rg3 production of P. pentosaceus WiKim20 could be explained by acidic hydrolysis instead of β-glucosidase activity. The five LABs could produce the ginsenoside Rg5 and this increase did not show the correlation with β-glucosidase enzymatic activity, supporting the theory that the organic acid could contribute to the production of Rg5.
Fig. 4.
pH (A) and acidity (B) during bioconversion of ginsenoside in different LABs. The pH of 10 mL of each culture broth was measured using electrode pH meter and acidity was measured by titration against 0.1N NaOH to a phenolphthalein endpoint, pH 8.3. * p < 0.05. **p < 0.01. ***p < 0.001. ■ Lactobacillus brevis WiKim47; ▲ Pediococcus pentosaceus WiKim20; ◆Leuconostoc lactis WiKim48; ● Lactobacillus sakei WiKim49; □ Leuconostoc mesenteroides WiKim19.
In the present study, we monitored the ginsenoside Rb1 bioconversion in LABs isolated from Kimchi with sensitive and accurate LC-MS/MS techniques. Among them, L. mesenteroides WIKim19 transformed ginsenoside Rb1 into ginsenoside Rg3 and showed a correlation with enzymatic activity, whereas, P. pentosaceus WiKim20 showed an elevated production of Rb3 in spite of lack of β-glucosidase activity but have the highest acidity among the five LABs. This result suggests that ginsenoside bioconversion by microorganisms might review with the enzyme activities and the environmental conditions such as production of organic acid. Also, lactic acid bacteria are useful to convert minor ginsenoside by enzymatic conversion together with mild acidic condition.
Conflicts of interest
The authors declare that there is no conflicts of interest.
Acknowledgments
This research was supported by grant KE1601-1 from the World Institute of Kimchi, Korea.
References
- 1.Lin T.Y., Lin C.W., Wang Y.J. Linoleic acid isomerase activity in enzyme extracts from Lactobacillus acidophilus and Propionibacterium freudenreichii ssp. Shermanii. J Food Sci. 2002;67:1502–1505. [Google Scholar]
- 2.Setchell K.D., Brown N.M., Zimmer-Nechemias L., Brashear W.T., Wolfe B.E., Kirschner A.S., Heubi J.E. Evidence for lack of absorption of soy isoflavone glycosides in humans, supporting the crucial role of intestinal metabolism for bioavailability. Am J Clin Nutr. 2002;76:447–453. doi: 10.1093/ajcn/76.2.447. [DOI] [PubMed] [Google Scholar]
- 3.Chun J., Kim G.M., Lee K.W., Choi I.D., Kwon G.H., Park J.Y., Jeong S.J., Kim J.S., Kim J.H. Conversion of isoflavone glucosides to aglycones in soymilk by fermentation with lactic acid bacteria. J Food Sci. 2007;72:M39–M44. doi: 10.1111/j.1750-3841.2007.00276.x. [DOI] [PubMed] [Google Scholar]
- 4.Kim S.H., Min J.W., Quan L.H., Lee S., Yang D.U., Yang D.C. Enzymatic transformation of ginsenoside Rb1 by Lactobacillus pentosus Strain 6105 from Kimchi. J Ginseng Res. 2012;36:291–297. doi: 10.5142/jgr.2012.36.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quan L.H., Piao J.Y., Min J.W., Yang D.U., Lee H.N., Yang D.C. Bioconversion of ginsenoside rb1 into compound k by Leuconostoc citreum LH1 isolated from kimchi. Braz J Microbiol. 2011;42:1227–1237. doi: 10.1590/S1517-838220110003000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D.H. Chemical diversity of Panax ginseng, Panax quinquifolium, and Panax notoginseng. J Ginseng Res. 2012;36:1–15. doi: 10.5142/jgr.2012.36.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan C.S., Wang C.Z., Wicks S.M., Qi L.W. Chemical and pharmacological studies of saponins with a focus on American ginseng. J Ginseng Res. 2010;34:160–167. doi: 10.5142/jgr.2010.34.3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui C.H., Kim J.K., Kim S.C., Im W.T. Characterization of a ginsenoside-transforming beta-glucosidase from Paenibacillus mucilaginosus and its application for enhanced production of minor ginsenoside F(2) PLoS One. 2014;9:e85727. doi: 10.1371/journal.pone.0085727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi S., Kim T.W., Singh S.V. Ginsenoside Rh2-mediated G1 phase cell cycle arrest in human breast cancer cells is caused by p15 Ink4B and p27 Kip1-dependent inhibition of cyclin-dependent kinases. Pharm Res. 2009;26:2280–2288. doi: 10.1007/s11095-009-9944-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi J.R., Hong S.W., Kim Y., Jang S.E., Kim N.J., Han M.J., Kim D.H. Metabolic activities of ginseng and its constituents, ginsenoside rb1 and rg1, by human intestinal microflora. J Ginseng Res. 2011;35:301–307. doi: 10.5142/jgr.2011.35.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim M., Ahn B.Y., Lee J.S., Chung S.S., Lim S., Park S.G., Jung H.S., Lee H.K., Park K.S. The ginsenoside Rg3 has a stimulatory effect on insulin signaling in L6 myotubes. Biochem Biophys Res Commun. 2009;389:70–73. doi: 10.1016/j.bbrc.2009.08.088. [DOI] [PubMed] [Google Scholar]
- 12.Leung K.W., Wong A.S. Pharmacology of ginsenosides: a literature review. Chin Med. 2010;5:20. doi: 10.1186/1749-8546-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park M.W., Ha J., Chung S.H. 20(S)-ginsenoside Rg3 enhances glucose-stimulated insulin secretion and activates AMPK. Biol Pharm Bull. 2008;31:748–751. doi: 10.1248/bpb.31.748. [DOI] [PubMed] [Google Scholar]
- 14.Qi L.W., Wang C.Z., Yuan C.S. Ginsenosides from American ginseng: chemical and pharmacological diversity. Phytochemistry. 2011;72:689–699. doi: 10.1016/j.phytochem.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen H., Leung W.I., Ruan J.Q., Li S.L., Lei J.P., Wang Y.T., Yan R. Biotransformation of ginsenoside Rb1 via the gypenoside pathway by human gut bacteria. Chin Med. 2013;8:22. doi: 10.1186/1749-8546-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae E.A., Choo M.K., Park E.K., Park S.Y., Shin H.Y., Kim D.H. Metabolism of ginsenoside R(c) by human intestinal bacteria and its related antiallergic activity. Biol Pharm Bull. 2002;25:743–747. doi: 10.1248/bpb.25.743. [DOI] [PubMed] [Google Scholar]
- 17.Kim K.A., Jung I.H., Park S.H., Ahn Y.T., Huh C.S., Kim D.H. Comparative analysis of the gut microbiota in people with different levels of ginsenoside Rb1 degradation to compound K. PLoS One. 2013;8:e62409. doi: 10.1371/journal.pone.0062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L., Deng Y., Xu S., Zeng X. In vivo pharmacokinetic and metabolism studies of ginsenoside Rd. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;854:77–84. doi: 10.1016/j.jchromb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa H., Sung J.-H., Benno Y. Role of human intestinal Prevotella oris in hydrolyzing ginseng saponins. Planta Med. 1997;63:436–440. doi: 10.1055/s-2006-957729. [DOI] [PubMed] [Google Scholar]
- 20.Kong H., Wang M., Venema K., Maathuis A., van der Heijden R., van der Greef J., Xu G., Hankemeier T. Bioconversion of red ginseng saponins in the gastro-intestinal tract in vitro model studied by high-performance liquid chromatography-high resolution Fourier transform ion cyclotron resonance mass spectrometry. J Chromatogr A. 2009;1216:2195–2203. doi: 10.1016/j.chroma.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 21.Eom H.J., Seo D.M., Han N.S. Selection of psychrotrophic Leuconostoc spp. producing highly active dextransucrase from lactate fermented vegetables. Int J Food Microbiol. 2007;117:61–67. doi: 10.1016/j.ijfoodmicro.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 22.Islam M.S., Choi H. Antidiabetic effect of Korean traditional Baechu (Chinese cabbage) kimchi in a type 2 diabetes model of rats. J Med Food. 2009;12:292–297. doi: 10.1089/jmf.2008.0181. [DOI] [PubMed] [Google Scholar]
- 23.Yoo E.J., Lim H.S., Park K.O., Choi M.R. Cytotoxic, antioxidative, and ACE inhibiting activities of Dolsan Leaf Mustard Juice (DLMJ) treated with lactic acid bacteria. Biotechno Bioprocess Eng. 2005;10:60–66. [Google Scholar]
- 24.Jung J.Y., Lee S.H., Kim J.M., Park M.S., Bae J.W., Hahn Y., Madsen E.L., Jeon C.O. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl Environ Microbiol. 2011;77:2264–2274. doi: 10.1128/AEM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho Y.R., Chang J.Y., Chang H.C. Production of gamma-aminobutyric acid (GABA) by Lactobacillus buchneri isolated from kimchi and its neuroprotective effect on neuronal cells. J Microbiol Biotechnol. 2007;17:104–109. [PubMed] [Google Scholar]
- 26.Kim J.D. Antifungal activity of lactic acid bacteria isolated from Kimchi against Aspergillus fumigatus. Mycobiology. 2005;33:210–214. doi: 10.4489/MYCO.2005.33.4.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang S.E., Han M.J., Kim S.Y., Kim D.H. Lactobacillus plantarum CLP-0611 ameliorates colitis in mice by polarizing M1 to M2-like macrophages. Int Immunopharmacol. 2014;21:186–192. doi: 10.1016/j.intimp.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 28.Chang J.Y., Chang H.C. Growth inhibition of foodborne pathogens by kimchi prepared with bacteriocin-producing starter culture. J Food Sci. 2011;76:M72–M78. doi: 10.1111/j.1750-3841.2010.01965.x. [DOI] [PubMed] [Google Scholar]
- 29.Seo B.J., Rather I.A., Kumar V.J., Choi U.H., Moon M.R., Lim J.H., Park Y.H. Evaluation of Leuconostoc mesenteroides YML003 as a probiotic against low-pathogenic avian influenza (H9N2) virus in chickens. J Appl Microbiol. 2012;113:163–171. doi: 10.1111/j.1365-2672.2012.05326.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim N.H., Moon P.D., Kim S.J., Choi I.Y., An H.J., Myung N.Y., Jeong H.J., Um J.Y., Hong S.H., Kim H.M. Lipid profile lowering effect of Soypro fermented with lactic acid bacteria isolated from Kimchi in high-fat diet-induced obese rats. Biofactors. 2008;33:49–60. doi: 10.1002/biof.5520330105. [DOI] [PubMed] [Google Scholar]
- 31.Richard Y., Favier C., Oudar J. Differentiation of glucidolytic mycoplasmas isolated from goats by the API 50 CH system and electrophoresis. Ann Rech Vet. 1991;22:353–358. [PubMed] [Google Scholar]
- 32.Chun J., Lee J.H., Jung Y., Kim M., Kim S., Kim B.K., Lim Y.W. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Kwon S.W., Han S.B., Park I.H., Kim J.M., Park M.K., Park J.H. Liquid chromatographic determination of less polar ginsenosides in processed ginseng. J Chromatogr A. 2001;921:335–339. doi: 10.1016/s0021-9673(01)00869-x. [DOI] [PubMed] [Google Scholar]
- 37.Han B.H., Park M.H., Han Y.N., Woo L.K., Sankawa U., Yahara S., Tanaka O. Degradation of ginseng saponins under mild acidic conditions. Planta Med. 1982;44:146–149. doi: 10.1055/s-2007-971425. [DOI] [PubMed] [Google Scholar]
- 38.Ko S.R., Choi K.J., Suzuki K., Suzuki Y. Enzymatic preparation of ginsenosides Rg2, Rh1, and F1. Chem Pharm Bull (Tokyo) 2003;51:404–408. doi: 10.1248/cpb.51.404. [DOI] [PubMed] [Google Scholar]
- 39.Chang K.H., Jo M.N., Kim K.T., Paik H.D. Evaluation of glucosidases of Aspergillus niger strain comparing with other glucosidases in transformation of ginsenoside Rb1 to ginsenosides Rg3. J Ginseng Res. 2014;38:47–51. doi: 10.1016/j.jgr.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang D., Li Y., Zhang M., Ruan S., Zhang H., Wang Y., Hu P. Tartaric acid induced conversion of protopanaxadiol to ginsenosides Rg3 and Rg5 and their in situ recoveries by integrated expanded bed adsorption chromatography. J Sep Sci. 2016;39:2995–3001. doi: 10.1002/jssc.201600269. [DOI] [PubMed] [Google Scholar]