Abstract
Aim:
This study was carried out to determine the bacteriological profile of infected diabetic foot ulcers (DFUs) and the antibiotic resistance pattern from the isolates. An attempt was made to suggest an empiric antibiotic regimen to treat such patients.
Materials and Methods:
Tissue samples were collected from 150 patients between February 2015 and January 2016 with DFUs under aseptic precautions and they were processed as per the Clinical and Laboratory Standards Institute guidelines.
Results:
A total of 185 bacterial isolates were obtained from 150 persons with diabetic and with foot ulcers. The age group of these persons ranged from 35 to 80 years and the maximum number of persons with DFUs was in the age group of 60–65 years. Among the isolates, Gram-negative bacilli were isolated in 112/185 (61%) and Gram-positive cocci in 73/185 (39%) cases. The most common isolate was Staphylococcus spp. 46 (25%), followed by Escherichia coli (20%) and Enterococcus spp. (15%). The antibiotic sensitivity profile of the bacteria was also studied. Among the isolates, 59/112 (53%) of the Gram-negative bacilli were extended spectrum beta-lactamase producers, 19/46 (41%) were methicillin-resistant Staphylococcus aureus, and 5/27 (19%) were vancomycin-resistant Enterococcus.
Conclusions:
This study showed a preponderance of multidrug-resistant strains among the isolates from the DFUs. Knowledge on the antibiotic sensitivity pattern of the isolates will be helpful in determining the drugs for the empirical treatment of diabetic ulcers. Thus, indiscriminate use of antibiotics and chances of subsequent development of antibiotic resistance can also be reduced.
Keywords: Bacterial profile, diabetic mellitus, drug resistance, ulcer
INTRODUCTION
Diabetic foot ulcer (DFU) is a serious and common complication of diabetes mellitus (DM) that significantly increases the cost of treatment.[1] In the United States, DM currently affects approximately 8.3% of the population and more than 79 million people have prediabetes.[2] And among persons with diabetes (PWD), 12%–25% have a risk of developing a foot ulcer during their lifetime.[3,4,5] The most common cause of morbidity and mortality in DFU is infections, which are seen in 40%–80% of the cases.[6] Diabetic neuropathy and micro- or macro-ischemia are the two main risk factors that cause DFU.[7] Impaired microvascular circulation limits the access of phagocytic cells to infected area, and this results in poor concentration of antibiotics in infected tissue.[8] Hence, diabetic foot wounds are commonly infected, and hence infection leads to the formation of microthrombi causing further ischemia, necrosis, and progressive gangrene. These types of situations necessitate limb amputation. Thus, accurate diagnosis of the causative organism is essential for the management of these cases. The burden of PWD in India is expected to increase to 57 million by the year 2025.[4] PWD has a 10-fold higher chance of hospitalization due to soft tissue and bone infection when compared with the nondiabetic individuals.[9] The blood supply to the lower extremities is further compromised by local injuries and inadequate foot care.[10] Diabetic neuropathy leads to repeated nonrecognized trauma to the insensate feet and this causes callosities, cracks, fissures, and ulcer formation. Secondary infection of the ulcer with arterial abnormalities further complicates the condition leading to gangrene and limb loss. A compromised immune state in PWD favors rapid and relentless development of local sepsis and even life-threatening septicemia. Massive infection is the most common factor leading to limb amputation.
DFUs are chronic in nature and patients with DFUs usually require several episodes of hospitalization. PWD are often exposed to several antibiotics which increase their risk of developing multidrug-resistant infection. Mostly, the diabetic foot infections (DFIs) are mixed bacterial infections, and the proper management of these infections requires an appropriate antibiotic selection, based on the culture and the antimicrobial susceptibility testing results.[11]
The present study was an attempt to evaluate the different microorganisms infecting the DFU and to know the antibiotic susceptibility patterns to the isolates. There was an increase in the population of multidrug organisms among the DFU isolates. And at present, there is a paucity of data on extended spectrum beta-lactamase (ESBL)-producing organisms, vancomycin-resistant Enterococcus (VRE), and methicillin-resistant Staphylococcus aureus (MRSA) from DFU isolates from North-East India. The knowledge of bacterial isolates from DFU is crucial for planning treatment with appropriate empirical antibiotics, reducing resistance pattern, and minimizing the cost of health care.
MATERIALS AND METHODS
One hundred and fifty PWD with foot ulcers were included in this study. The study was conducted for 1 year in a tertiary diabetic care hospital. This was a prospective and observational hospital-based study. The Institutional Ethical Committee's clearance was obtained prior to conducting the study. The study was carried out on PWD with foot infections from February 2015 to January 2016. For the present study, patients who attended the outpatient department of the study center with foot ulcer or infection, referred from other hospitals of North-East India, and lastly, patients who attended as self-referral after failure to respond to ongoing treatment outside the institute were selected.
Inclusion criteria
PWD who had foot ulcers or foot infection were included.
Exclusion criteria
The exclusion criteria were other foot ulcers and foot infection in persons without diabetes.
A clinical history was elicited with regard to the demographic profile of PWD, duration of the diabetes and foot problem, the type of treatment for diabetes earlier received, and the presence of other systemic illnesses.
PWD were also assessed clinically, and the foot ulcers were graded according to Wagner's grade (Wagner and Meggitt, 1970) as follows:
0 - No ulceration in a high-risk foot
1 - Superficial ulcer of skin or subcutaneous tissue
2 - Ulcers extend into tendon, bone, or capsule
3 - Deep ulcer with osteomyelitis or abscess
4 - Gangrene of toes or forefoot (localized gangrene)
5 - Extensive gangrene requiring a major amputation.
PWD having DFI along with systemic features of toxicity (Infectious Disease Society of America [IDSA] grade – severe)[12] were considered to have systemic sepsis. Foot type was ascertained on the presence of neuropathy, ischemia, and sepsis. For this, in addition to clinical history and examination of PWD, investigations such as monofilament, biothesiometry, and Doppler-based ankle brachial index estimation were carried out.
The samples were collected after obtaining informed consent from the PWD. Tissue samples were obtained after the wound was debrided.[13] No antimicrobial agent or antiseptic was used in the wound before collection of the tissue specimen. In addition, a deep tissue specimen (fat/fascia/muscles/bone) was obtained from the wound. The specimens were placed into sterile transport containers and sent to the microbiology laboratory for aerobic microbial culture as soon as possible. Anaerobic and fungal cultures were not performed for this study. Cultures were processed following the standard procedures for the tissue sample processing. Most of the bacterial isolates were identified using VITEK 2 Compact system (BioMérieux, Marcy l’Etoile, France), and a few isolates were identified manually. A direct Gram-stained smear of the specimen was examined. The specimens were inoculated onto blood agar, chocolate agar, Mac Conkey's agar, and thioglycollate medium. The inoculated plates were incubated at a temperature of 37°C overnight, and the plates were examined for growth on the following day. Further processing was done according to the nature of the isolate and as determined by Gram-staining and colony morphology. The organisms were identified on the basis of their Gram-staining properties, and further analysis was done in VITEK 2 Compact system (BioMérieux).[14]
Antibiotic susceptibility testing
A bacterial suspension was adjusted to a McFarland standard of 0.5 in 2.5 ml of a 0.45% sodium chloride solution with a VITEK 2 DensiChek instrument (BioMérieux). The time between preparation of the inoculums and the filling of the card was always <30 min. The format of the nonfermenting gram-negative cards (NGNC), i.e., a 64-well plastic card, is the same as that of the gram-negative cards (GNC), but the NGNC contains 47 tests, while the GNC contains 41 tests. The NGNC is a fully closed system to which no reagents have to be added. The card was put on the cassette designed for VITEK 2, placed in the instrument, automatically filled in a vacuum chamber, sealed, incubated at 35.5°C, and automatically subjected to a colorimetric measurement by the use of a new optical reading head every 15 min for a maximum incubation period of 10 h. Data were analyzed using VITEK 2 database version 4.01, which allows for organism identification in the kinetic mode after 2 h of incubation. The interpretations provided by the software were considered for the analysis.[15]
Each organism suspension was prepared from the growth of pure cultures of bacteria cultivated for 18–24 h on Columbia agar with 5% sheep blood. The handling time was very short, and suspensions were prepared in sterile saline (0.45% NaCl) to a turbidity equivalent to that of a 0.5 McFarland standard. These suspensions were used for the inoculation of both cards (ID-Gram-positive cocci [GPC] GN and AST-P628, N235, N280, N281). The cards were manually situated, as were the suspensions in plastic racks that were inserted in the VITEK 2 system's reader-incubator module (incubation temperature, 35.5°C). The cards were automatically filled by a vacuum device and were automatically sealed and subjected to a kinetic fluorescence measurement every 15 min. The results were interpreted by the ID-GPC database after an incubation period of 4 h, and the final results were obtained automatically after a minimum of 4 h and a maximum of 15 h of incubation. All cards used were automatically discarded in a waste container.
Quality control strains
Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, E. coli ATCC 35218, Enterococcus faecalis ATCC 29212, S. aureus ATCC 29213, and Enterococcus faecium ATCC 29212 were used as quality control strains during the evaluation of the VITEK 2 system.
RESULTS
In the present study, out of 150 PWD, 56 patients were below 50 years and 94 patients were above 50 years. In our study, 122 patients were males and 28 were females. Wagner's grading of DFI with Grade I–V was included. The baseline demographic and clinical characteristics of patients are shown in Table 1. In the present study, Grade I DFU was seen in 9 (6%) PWD, Grade II in 32 (21.3%), Grade III in 73 (48.6%), Grade IV in 30 (20%), and Grade V in 6 (4%) PWD. In this study, 93 (62%) PWD were neuropathic cases, 27 (18%) PWD were associated neuropathic cases with sepsis, 17 (11.3%) PWD were neuro-ischemic cases, and 13 (8.6%) PWD presented with neuro-ischemia and sepsis. The average duration of diabetes in these patients at the time of reporting was 9.87 years and average control was poor (mean hemoglobin A1c - 8.95).
Table 1.
Baseline demographic and clinical characteristics of the study cohort

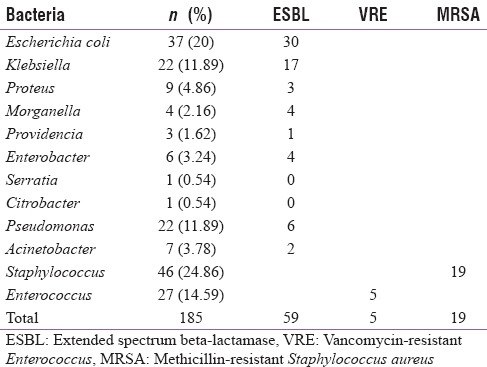
In this study, there were 43 (28.6%) polymicrobial cases, 96 (64%) monomicrobial cases, and in 11 (7.3%) cases, the culture was sterile. Gram-positive bacterial growths were present in 73 (41%) cases, whereas Gram-negative growth was seen in 106 (59%) cases. The most common single bacterial growth was that of S. aureus (27%), followed by E. coli (20%), and Enterococcus spp. (15.7%), as shown in Table 2. Among the isolates, 59/112 (53%) of the Gram-negative bacilli were ESBL producers, 19/46 (41%) were MRSA, and 5/27 (19%) were VRE as shown in Table 2.
Table 2.
Bacterial isolates of the study group of patients
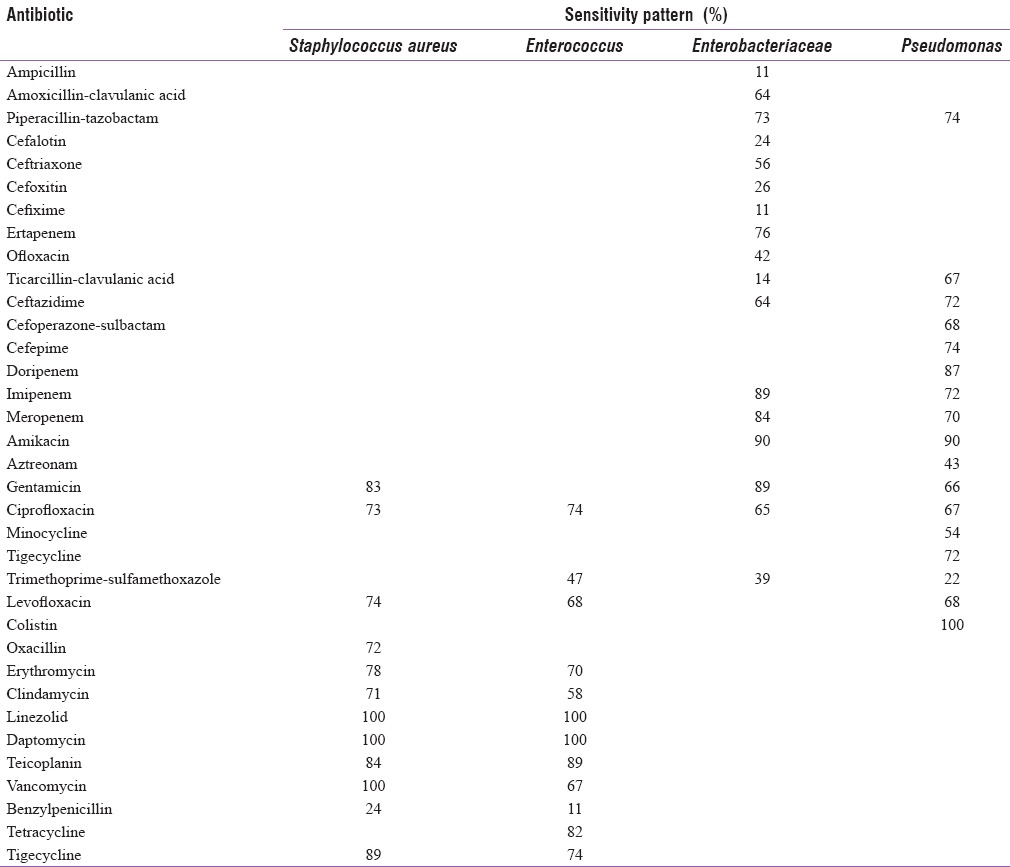
In the present study, most of the Enterobacteriaceae culture isolates were sensitive to amikacin (90%), imipenem (89%), meropenem (84%), ertapenem (76%), and piperacillin-tazobactam combination (73%), as shown in Table 3. In our study, most of the Pseudomonas culture isolates were sensitive to amikacin (90%), imipenem (72%), meropenem (70%), and piperacillin-tazobactam combination (74%) [Table 3]. Most of the Staphylococcus culture isolates were sensitive to linezolid (100%), daptomycin (100%), tigecycline (89%), teicoplanin (84%), and gentamicin (83%). In our study, most of the Enterococcus culture isolates were sensitive to linezolid (100%), daptomycin (100%), teicoplanin (89%), and tigecycline (74%), as shown in Table 3.
Table 3.
Bacterial isolates along with their sensitivity pattern
Bacterial patterns in amputation
Major amputations (below or above knee) were done in eight patients. In patients where a major amputation was carried out, E. coli (ESBL positive), Morganella morganii (ESBL positive), and Acinetobacter (multidrug resistance [MDR]) were isolated on two occasions. S. aureus (MRSA positive) and Enterococcus (VRE positive) were isolated on one occasion [Figure 1].
Figure 1.
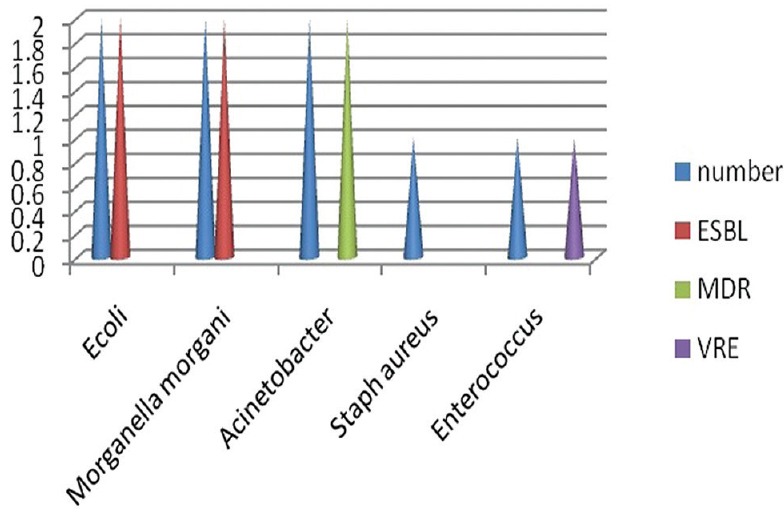
Bar diagram showing bacterial isolates in major amputation procedures
Forty-two minor amputations were done. In patients where a minor amputation (distal to ankle) was carried out, E. coli was isolated in seven (four ESBL positive), Klebsiella in 10 (all ESBL positive), Pseudomonas in six (all MDR positive), Proteus in four (two ESBL positive), Providencia in one, Enterobacter in two (all ESBL positive), Enterococcus in six, of which five were VRE positive, and Staphylococcus was isolated in four samples, of which, all were MRSA positive, as shown in Figure 2.
Figure 2.
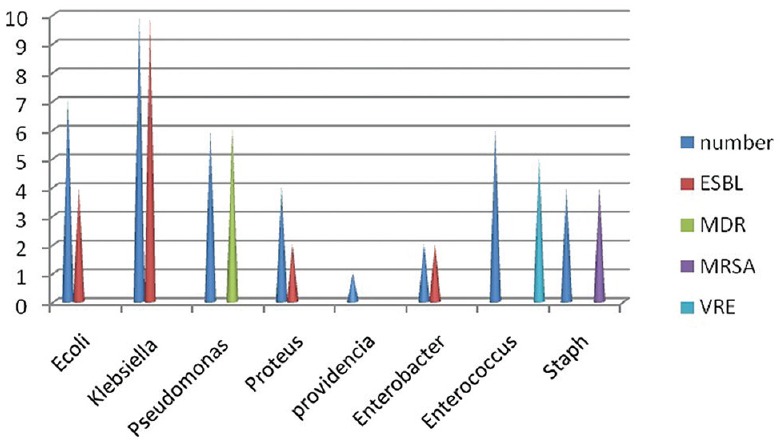
Bar diagram showing bacterial isolates in minor amputation procedures
DISCUSSION
In the present study, the majority of the PWD were above 50 years (63%). This may be an indication of higher level of physical activities undertaken by aging PWD to run their family and increased prevalence of comorbidities such as neuropathy, peripheral vascular disease, and kidney diseases in this age group. A study by King et al. in 1998 mentioned that the majority of people with diabetic foot were in 45–64 years’ age range in developing countries.[16] Among the 150 DFU patients in the present study, 81% (122/150) of patients were males and 18% (28/150) of patients were females. Higher male prevalence has been reported by Harrison and Lederberg.[17] This may be due to the higher level of outdoor physical activity in hot humid environment with inadequate and improper feet care among males in comparison to females.
Our study revealed that most of the patients with DFI reported late in advanced grade of infection, i.e., Wagner Grade III and above. This is usually attributed to the lack of awareness about feet care among the public and medical professionals.[18]
In the present study, majority of patients with diabetic foot ulcers were due to neuropathic cause (80%), with or without sepsis. Neuroischemic foot problem was seen in 20% of patients. The presence of sepsis was more commonly associated with neuro-ischemic condition (76%) than with neuropathic foot problem (29%). Diabetic foot wounds are commonly infected and infection leads to formation of microthrombi causing further ischemia, necrosis, and progressive gangrene.[19] In the present study, there was a preponderance of monomicrobial culture growth (64%). Polymicrobial growths were seen in 29% of patients whereas 7% of the specimens were sterile. Perim et al. revealed that infection in DFU was predominantly polymicrobial.[20] This could be due to the fact of stringent technique of obtaining tissue culture from the ulcer, taking care of skin commensals and contaminants.
In this study, microbiological evaluation of DFI showed a preponderance of Gram-negative organisms (59%, 106/185) over Gram-positive organisms (41%, 73/185), which is in accordance with earlier studies.[5,11] Another earlier study showed that Gram-negative organisms were more prevalent (63.8%, 65/102) than Gram-positive aerobes (36.1%, 37/102).[11] Gram-negative organisms were more prevalent, and the predominant organisms isolated were members of the Enterobacteriaceae.[21,22]
S. aureus was the single most frequent pathogen (26%, 46/179), followed by E. coli (20%, 37/179). Other studies have also found the same.[22,23,24] In contrast, another study carried out by Ako-Nai et al.[25] showed E. coli as the frequent bacterial pathogen, while P. aeruginosa was reported as the most common pathogen by Shankar et al.[26] Source of infection, use of antibiotic drug for treatment, sample collection method, and different types of infection can influence pathogen diversity in DFI.[22,23,25,27] In addition to S. aureus, our study showed other GPC such as Enterococcus in patients with DFU. This finding was similar with the results obtained by other researches.[23,25] Previous use of antimicrobial drug may increase the prevalence of Enterococcus spp. in DFI.[22] The increased prevalence of Enterococci has now emerged as a public health concern. Enterococci are frequently detected in immunocompromised patients, like PWD and in PWD with foot ulcers, but their role in infection at these sites has not been clearly defined.[28]
With the emergence of ESBL-producing bacteria, the wound condition deteriorates and treatment becomes difficult resulting in a poor outcome. In the present study, ESBL-producing Gram-negative bacteria were seen in 79.16% of patients. The highest prevalence of ESBL was observed in E. coli (79.16%) followed by P. aeruginosa (35%), which was consistent with the study carried out by Shobha et al.[29] In our study, 48.14% of S. aureus isolates were methicillin resistant that was determined by VITEK 2 Compact system. This finding was similar with studies reported earlier.[9,24] This could be due to the prolonged and indiscriminate use of antibiotic therapy and administration of broad-spectrum antibiotics that may increase the prevalence of antibiotic resistance organism such as MRSA or VRE in DFI.[28]
Gram-negative bacilli and mixed infection were more evident in Grade III and Grade IV, whereas GPC were most common in Grade I and Grade II, indicating that Gram-negative infections increase the severity and make the patients prone to undergo limb amputation. As revealed, fifty patients underwent amputation, of which thirty patients were infected with Gram-negative bacteria. GPC were the most common cause of mild-to-moderate infections whereas mixed Gram-negative bacilli and GPC tend to cause chronic infections.[30]
Nowadays, clinical microbiologists and clinicians are both equally concerned about this emerging menace of MDR organisms and their associated complications in developing countries.[31] In the present study, 91% of the bacteria were resistant to three or more antibiotics (VRE 33.33%, MRSA 48.14%, and ESBL 77.67%). These rates were in contrast to a study from Iran.[32] Infections with these isolates are more difficult to manage.
Factors responsible for MDR may be frequent hospitalization, recent use of broad-spectrum antibiotics, inadequate surgical source reduction, chronic wounds, irrational use of antibiotics, and the transfer of resistance genes by transport means. To alleviate this situation and also reduce the rate of amputation, clinicians should prescribe antibiotics rationally, timely, and sufficiently and there should be periodic supervisions on the drug consumption by the respective organizations.[33] Clinicians should switch to culture report-based use of narrower spectrum therapy. An adequate and timely surgical intervention is essential to achieve infection source reduction. These also help in reducing the indiscriminate and prolonged use of antimicrobials.
In this study, in most of the Enterobacteriaceae culture isolates, organisms were sensitive to amikacin (90%), imipenem (89%), meropenem (84%), ertapenem (76%), and piperacillin-tazobactam (73%). Among these isolates, the Enterobacteriaceae family was resistant to the majority of antibiotics tested, except colistin, imipenem, amikacin, and meropenem, partially consistent with the results of other studies.[34,35] However, the nonfermenting Gram-negative bacteria culture isolates showed the following sensitivity pattern - amikacin (90%), imipenem (72%), meropenem (70%), and piperacillin-tazobactam (74%). This was similar to the findings of Al Benwan et al.[21]
As for the Staphylococcus culture isolates, linezolid (100%), vancomycin (100%), daptomycin (100%), tigecycline (89%), teicoplanin (84%), and gentamicin (83%) were the most effective antibiotics. Other studies have shown different antibiotic susceptibility patterns, and in most, vancomycin and linezolid have shown good activity against the strains.[35] Furthermore, most of the Enterococcus culture isolates were sensitive to linezolid (100%), daptomycin (100%), teicoplanin (89%), and tigecycline (74%). A study by Al Benwan et al. showed vancomycin as the most effective antibiotic for Gram-positive bacteria.[21]
Based on the findings of the present study and IDSA 2012 guidelines[36] for the treatment of DFI, the following empiric antibiotic regimen can be suggested:
Clinically noninfected DFU - No antibiotic. Appropriate wound care
-
Clinically infected DFU - Antibiotic therapy + appropriate wound care
- For mild-to-moderate infections in patients who have not recently received antibiotic treatment - Antibiotic-targeting aerobic GPC, for example, amoxicillin/clindamycin
- For most severe infections, we recommend broad-spectrum antibiotic therapy, for example, linezolid/daptomycin with imipenem/meropenem/piperacillin tazobactam/cefepime along with metronidazole pending culture results and antibiotic susceptibility data
- Empiric therapy, for example, imipenem/meropenem/piperacillin tazobactam/cefepime directed at P. aeruginosa is for patients with risk factors for true infection with this organism. Provided the renal function is adequate, amikacin is also found to be a good option
- Consider providing empiric therapy, for example, linezolid/daptomycin directed against MRSA in a patient with a prior history of MRSA infection, when the local prevalence of MRSA colonization or infection is high or if the infection is clinically severe.
In the present study, due to late reporting or referral, nonspecific antibiotic protocol without doing culture and sensitivity, no or inadequate attempt for surgical source reduction, and self-medication, there remained some possibility of confounding. On most occasions, it was impossible to record the exact history of prior treatment and medications.
CONCLUSION
This study showed that most common organisms present in the PWD and with foot ulcer were Gram-negative aerobes. S. aureus was the most predominant single organism isolated from the lesions. Monomicrobial infection was more common than polymicrobial infection in the DFI cases. Presence of MDR organisms was alarmingly high in the PWD and with foot ulcer. These observations are important, especially for patient management and development of empirical antibiotic guidelines.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Lipsky BA. A report from the international consensus on diagnosing and treating the infected diabetic foot. Diabetes Metab Res Rev. 2004;20(Suppl 1):68–77. doi: 10.1002/dmrr.453. [DOI] [PubMed] [Google Scholar]
- 2.Center for Disease Control and Prevention. National Diabetes Fact Sheet: National Estimates and General Information on Diabetes and Prediabetes in the United States. Atlanta, GA, USA: Department of Health and Human Services, Center for Disease Control and Prevention; 2011. [Google Scholar]
- 3.Huang Y, Cao Y, Zou M, Luo X, Jiang Y, Xue Y, et al. Acomparison of tissue versus swab culturing of infected diabetic foot wounds. Int J Endocrinol 2016. 2016:8198714. doi: 10.1155/2016/8198714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh N, Armstrong DG, Lipsky BA. Preventing foot ulcers in patients with diabetes. JAMA. 2005;293:217–28. doi: 10.1001/jama.293.2.217. [DOI] [PubMed] [Google Scholar]
- 5.Andersen CA, Roukis TS. The diabetic foot. Surg Clin North Am. 2007;87:1149–77, x. doi: 10.1016/j.suc.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Richard JL, Sotto A, Lavigne JP. New insights in diabetic foot infection. World J Diabetes. 2011;2:24–32. doi: 10.4239/wjd.v2.i2.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ismail K, Winkley K, Stahl D, Chalder T, Edmonds M. A cohort study of people with diabetes and their first foot ulcer: The role of depression on mortality. Diabetes Care. 2007;30:1473–9. doi: 10.2337/dc06-2313. [DOI] [PubMed] [Google Scholar]
- 8.Bronze MS, Khardori R, editors. Diabetic foot infections treatment and management. Medscape. 2016. [Last accessed on 2016 Dec 15]. Available form: http://emedicine.medscape.com/article/237378-treatment .
- 9.Shakil S, Khan AU. Infected foot ulcers in male and female diabetic patients: A clinico-bioinformative study. Ann Clin Microbiol Antimicrob. 2010;9:2. doi: 10.1186/1476-0711-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pappu AK, Sinha A, Johnson A. Microbiological profile of diabetic foot ulcer. Calicut Med J. 2011;9:e1–4. [Google Scholar]
- 11.Zubair M, Malik A, Ahmad J. Clinico-bacteriology and risk factors for the diabetic foot infection with multidrug resistant microorganisms in North India. Biol Med. 2010;2:22–34. [Google Scholar]
- 12.Lipsky BA, Berendt AR, Deery HG, Embil JM, Joseph WS, Karchmer AW, et al. Diagnosis and treatment of diabetic foot infections. Clin Infect Dis. 2004;39:885–910. doi: 10.1086/424846. [DOI] [PubMed] [Google Scholar]
- 13.Nelson EA, Backhouse MR, Bhogal MS, Wright-Hughes A, Lipsky BA, Nixon J, et al. Concordance in diabetic foot ulcer infection. BMJ Open. 2013;3 doi: 10.1136/bmjopen-2012-002370. pii: E002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Garrote F, Cercenado E, Bouza E. Evaluation of a new system, VITEK 2, for identification and antimicrobial susceptibility testing of enterococci. J Clin Microbiol. 2000;38:2108–11. doi: 10.1128/jcm.38.6.2108-2111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pincus DH. Microbial Identification Using the Biomérieux VITEK® 2 System. Hazelwood, MO, USA: BioMérieux, Inc; [Last accessed on 2016 Dec 15]. Available form: https://store.pda.org/tableofcontents/ermm_v2_ch01.pdf . [Google Scholar]
- 16.King H, Aubert RE, Herman WH. Global burden of diabetes, 1995-2025: Prevalence, numerical estimates, and projections. Diabetes Care. 1998;21:1414–31. doi: 10.2337/diacare.21.9.1414. [DOI] [PubMed] [Google Scholar]
- 17.Harrison PF, Lederberg J. Antimicrobial Resistance: Issues and Options. Washington, DC: Forum on Emerging Infection; 1998. pp. 8–74. [Google Scholar]
- 18.Kishore S, Upadhyay AD, VP J. Awareness of foot care among patients with diabetes attending a tertiary care hospital. Natl Med J India. 2015;28:122–5. [PubMed] [Google Scholar]
- 19.James GA, Swogger E, Wolcott R, Pulcini Ed, Secor P, Sestrich J, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 20.Perim MC, Borges Jda C, Celeste SR, Orsolin Ede F, Mendes RR, Mendes GO, et al. Aerobic bacterial profile and antibiotic resistance in patients with diabetic foot infections. Rev Soc Bras Med Trop. 2015;48:546–54. doi: 10.1590/0037-8682-0146-2015. [DOI] [PubMed] [Google Scholar]
- 21.Al Benwan K, Al Mulla A, Rotimi VO. A study of the microbiology of diabetic foot infections in a teaching hospital in Kuwait. J Infect Public Health. 2012;5:1–8. doi: 10.1016/j.jiph.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Abdulrazak A, Bitar ZI, Al-Shamali AA, Mobasher LA. Bacteriological study of diabetic foot infections. J Diabetes Complications. 2005;19:138–41. doi: 10.1016/j.jdiacomp.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 23.El-Tahawy AT. Bacteriology of diabetic foot. Saudi Med J. 2000;21:344–7. [PubMed] [Google Scholar]
- 24.Amini M, Davati A, Piri M. Determination of the resistance pattern of prevalent aerobic bacterial infections of diabetic foot ulcer. Iran J Pathol. 2013;8:21–6. [Google Scholar]
- 25.Ako-Nai A, Ikem I, Akinloye O, Aboderin A, Ikem R, Kassim O. Characterization of bacterial isolates from diabetic foot infections in Ile-Ife, Southwestern Nigeria. Foot (Edinb) 2006;16:158–64. [Google Scholar]
- 26.Shankar EM, Mohan V, Premalatha G, Srinivasan RS, Usha AR. Bacterial etiology of diabetic foot infections in South India. Eur J Intern Med. 2005;16:567–70. doi: 10.1016/j.ejim.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 27.Osariemen IJ, Olowu SS, Adevbo E, Omon EE, Victoria O, Imuetinyan EJ, et al. Aerobic bacteria associated with diabetic wounds in patients attending clinic in a rural community in Nigeria. Glob Res J Microbiol. 2013;3:8–11. [Google Scholar]
- 28.Agudelo Higuita NI, Huycke MM. Enterococcal disease, epidemiology, and implications for treatment. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston, Massachusetts, USA: Massachusetts Eye and Ear Infirmary; 2014. [Google Scholar]
- 29.Shobha K, Ramachandra L, Rao G, Majumder S, Rao S. Extended Spectrum Beta-Lactamases (ESBL) in gram negative bacilli at a tertiary care hospital. J Clin Diagn Res. 2009;3:1307–12. [Google Scholar]
- 30.Chahine EB. Diabetic foot infections: An update on treatment. US Pharm. 2013;38:23–6. [Google Scholar]
- 31.Japoni A, Vazin A, Hamedi M, Davarpanah MA, Alborzi A, Rafaatpour N. Multidrug-resistant bacteria isolated from intensive-care-unit patient samples. Braz J Infect Dis. 2009;13:118–22. doi: 10.1590/s1413-86702009000200009. [DOI] [PubMed] [Google Scholar]
- 32.Akhi MT, Ghotaslou R, Asgharzadeh M, Varshochi M, Pirzadeh T, Memar MY, et al. Bacterial etiology and antibiotic susceptibility pattern of diabetic foot infections in Tabriz, Iran. GMS Hyg Infect Control. 2015;10:Doc02. doi: 10.3205/dgkh000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farshad S, Anvarinejad M, Tavana AM, Ranjbar R, Japoni A, I Zadegan RM, et al. Molecular epidemiology of Escherichia coli strains isolated from children with community acquired urinary tract infections. Afr J Microbiol Res. 2011;5:4476–83. [Google Scholar]
- 34.Banashankari G, Rudresh H, Harsha A. Prevalence of gram negative bacteria in diabetic foot-a clinico-microbiological study. Al Ameen J Med Sci. 2012;5:224–32. [Google Scholar]
- 35.Hefni AA, Ibrahim AM, Attia KM, Moawad MM, El-ramah AF, Shahin MM, et al. Bacteriological study of diabetic foot infection in Egypt. J Arab Soc Med Res. 2013;8:26–32. [Google Scholar]
- 36.Lipsky BA, Berendt AR, Cornia PB, Pile JC, Peters EJ, Armstrong DG, et al. 2012 Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin Infect Dis. 2012;54:e132–73. doi: 10.1093/cid/cis346. [DOI] [PubMed] [Google Scholar]


