Abstract
Background: Because delta-9-tetrahydrocannabinol (THC), the primary psychoactive ingredient in cannabis, binds to cannabinoid 1 (CB1) receptors, levels of CB1 protein could serve as a potential biomarker for response to THC. To date, available techniques to characterize CB1 expression and function in vivo are limited. In this study, we developed an assay to quantify CB1 in lymphocytes to determine how it relates to cannabis use in 58 daily cannabis users compared with 47 nonusers. Furthermore, we tested whether CB1 levels are associated with mutations in a single nucleotide polymorphism known to regulate CB1 functioning (i.e., rs2023239).
Methods: Total protein concentration was analyzed through the Pierce BCA Protein assay kit. CB1 protein was quantified through CNR1 enzyme-linked immunosorbent assay (ELISA) kit from MyBioSource. CB1 concentration and total protein concentration were quantified and used to calculate a ratio of CB1 to total protein.
Results: Inherent levels of peripheral lymphocyte CB1 were sufficient for quantification through ELISA without protein amplification. We found a group×genotype interaction such that users with the G allele had greater CB1 concentration than users with the A/A genotype, and a trend-level difference between genotypes in nonusers.
Conclusions: This study demonstrates a minimally invasive technique of CB1 quantification that holds promise for the use of CB1 protein concentration, along with rs2023239 genotype, as a potential biomarker for susceptibility to cannabis use. These results suggest a gene (rs2023239 G)×environment (cannabis use) effect on CB1 density.
Keywords: : biomarker, cannabis, cannabinoid receptor 1, orbitofrontal cortex, peripheral lymphocytes
Introduction
The human cannabinoid 1 (CB1) receptor is part of the endocannabinoid system (ECS), which is highly regulatory in various functions throughout the body, including the central nervous system (CNS) and the digestive, reproductive, and urinary tracts.1,2 The primary psychoactive component of cannabis, delta-9-tetrahydrocannabinol (THC), binds to CB1, and this binding in the CNS is responsible for the psychoactive effects of cannabis. Specifically, THC binds to CB1 receptors in the ventral tegmental area, which disinhibits dopaminergic signaling and results in a reward response.3 Evidence for CB1 variability exists in the single nucleotide polymorphism (SNP) rs2023239, which may code for an alternative of the CB1 protein.4 The G (risk) allele at this locus causes alternative splicing of CNR1.4,5 Variability at this location has been associated with craving and withdrawal in heavy cannabis users6 as well as proclivity toward cannabis use disorders.7 Thus, on a molecular level, the rewarding effects of cannabis may be associated with the G allele and CB1 concentration.
Results from animal studies examining CB1 expression after long-term exposure to THC are mixed. Burston et al. reported an overall downregulation of CB1 in adolescent female rats after THC administration for ∼10 days, mimicking long-term use in humans. This downregulation was more pronounced in females, particularly in the ventral midbrain, hippocampus, and prefrontal cortex, indicating variable effects of THC throughout the CNS.8 Conversely, Zhuang et al. found an increase in CB1 mRNA in the cerebellum and hippocampus of male rats after 21 days of exposure, whereas it decreased in the striatum.9 The incongruity of these results in animals may be because mRNA and protein do not always change in the same direction, depending on timing of measurements and other factors.
In humans, there is evidence for altered CB1 expression in the brain after long-term cannabis use. One postmortem in situ radioligand binding study found increased CB1 binding in the caudate and putamen of individuals who had consumed cannabis within 5 days of death compared with controls.10 Another group demonstrated that CB1 binding in postmortem pituitary adenomas correlated with levels of endogenous cannabinoids, indicating a positive relationship between cannabinoid availability and receptor expression.11 However, in known daily users, radioligand binding demonstrated decreased CB1 binding across all brain regions compared with controls.12 Similarly, studies using positron emission tomography (PET) have reported that CB1 expression in the brains of heavy cannabis users negatively correlates with years of smoking, returns to normal levels upon abstinence,13 and a negative correlation exists between general CB1 binding and age in all regions examined.14 In sum, these studies suggest that CB1 density is related to THC exposure, but characterization of CB1 expression and density is still in its infancy.15,16 Furthermore, the effect of rs2023239 on CB1 density, compounded with frequent cannabis use, remains unknown. Thus, a noninvasive measurement of CB1 changes in living humans is necessary and would facilitate the understanding of how cannabis affects cellular mechanisms over time.
Although CB1 is most dense in the CNS,17 it also exists in peripheral tissues18 where it is similarly regulatory. THC's binding to CB1 has been shown to alter immune function in rodents,19 indicating that exogenous cannabinoids may induce changes in these lymphocytes directly through cannabinoid receptor activation. In addition, THC has been shown to downregulate proinflammatory cytokines in human peripheral lymphocytes.20 Thus, a measure of peripheral CB1 could be a valuable tool with implications for CNS function, particularly when examined in cannabis users versus nonusers.
One avenue that holds promise for quantifying in vivo human CB1 expression is through lymphocytes, as they are easily accessible and the role of cannabinoids in immune function suggests high CB1 expression.21–23 Despite methodological difficulties,24 the presence and activity of CB1 in peripheral lymphocytes have been established through various means in previous studies.25–27 However, quantification methods are limited, as available antibodies are notably unreliable, and other methods (e.g., flow cytometry and radioactive ligand assays) are expensive. Furthermore, changes in peripheral CB1 levels in relation to cannabis use have not been directly investigated. This warrants investigation because CB1 promoter region methylation and mRNA expression in other human tissues have exhibited a difference between cannabis users and nonusers. For example, Rotter et al.28 used quantitative real-time polymerase chain reaction (PCR) to study potential changes in CB1 expression in peripheral blood cells of 77 subjects with varying degrees of cigarette and cannabis use and found a decrease in expression in THC-dependent participants, as well as a negative correlation between promoter methylation and CB1 mRNA. Methylation and mRNA analysis provide information about up- or downregulation in a protein's expression, but do not unequivocally indicate the result of that protein's expression. Thus, although these methods are informative, they require corroboration with actual CB1 concentration.
The aim of this study was to test a novel method of CB1 quantification through lymphocytes in humans and determine how peripheral CB1 levels may relate to known genetic and environmental modulators. Specifically, because CB1 agonists, such as THC, have been shown to downregulate CB1 receptors, we hypothesized that peripheral CB1 would be decreased in chronic cannabis users relative to nonusing controls and that CB1 would be negatively correlated with THC metabolite levels. Furthermore, because rs2023239 genotype has been associated with CB1 expression,4–6 we also expected that carriers of the rs2023239 G allele would show downregulation of CB1. Last, we expected a genotype (rs2023239)×environment (THC exposure) interaction similar to what has been previously reported,29 such that rs2023239 will moderate the effects of THC exposure on CB1 lymphocytes.
Materials and Methods
The Institutional Review Board of the University of Texas at Dallas and University of Texas Southwestern Medical Center approved these study procedures. All experiments were conducted according to the principles expressed in the Declaration of Helsinki.
Participants
One hundred twenty-one participants from a larger study of cannabis use30 took part in this study. The participants were recruited from the general community in the Dallas/Fort Worth metro area through media advertisements and were financially compensated for their participation. As the larger study included neuroimaging, all participants were right handed and had no MRI contraindications. All participants spoke English as their primary language, and reported no neurological conditions, axis I psychiatric disorders, or traumatic brain injury. The absence of all other drug use (besides cannabis in users) was verified through urinalysis and all participants reported smoking <20 cigarettes per month.
Peripheral blood was obtained from 66 daily cannabis-using adults and 55 nonusing adults (Table 1). Users met inclusion criteria of >5000 lifetime cannabis use occurrences, as well as daily use in the previous 60 days. Nonusers met the inclusion criteria of having no consecutive 2 days of cannabis use in their lifetime, as well as no cannabis use in the previous 60 days. Urine THC and creatinine metabolites were quantified from urine through gas chromatography/mass spectroscopy in the users. CB1 data were successfully obtained from 105 of these individuals, resulting in a sample of 47 nonusers and 58 users.
Table 1.
Demographics of Participants
| All (N=105) | Cannabis users (N=58) | Healthy controls (N=47) | |
|---|---|---|---|
| Males n (%) | 67 (64) | 41 (71) | 26 (55) |
| Age—mean (SD) | 29.74 (8.73) | 29.62 (7.52) | 29.89 (10.12) |
| Age of onset of regular cannabis use mean (SD) | — | 16.96 (11.43) | — |
| Education in years mean (SD) | 14.50 (2.40) | 13.45 (2.26) | 15.79 (1.89) |
| Race | |||
| Caucasian | 55 | 33 | 22 |
| African American | 21 | 13 | 8 |
| American Indian/Alaska Native | 3 | 0 | 2 |
| Asian | 10 | 4 | 9 |
| Native Hawaiian or Pacific Islander | 1 | 0 | 1 |
| Multiracial | 10 | 8 | 2 |
| Other | 6 | 3 | 3 |
| Lifetime cannabis problem symptom countb | — | 1.22 (2.32) | — |
| Current cannabis problem symptom countb | — | 1.19 (2.32) | — |
| Rs2023239 Genotype | GG: 3 | GG: 3 | GG: 0 |
| GA: 18 | GA: 14 | GA: 4 | |
| AA: 51 | AA: 27 | AA: 24 | |
| 33 NA | 14 NA | 19 NAs | |
From the Structured Clinical Interview for DSM-IV.
SD, standard deviation.
Blood draw and lymphocyte isolation
After informed consent, participants had their blood drawn from antecubital veins according to the standard phlebotomy technique. Approximately 20 mL of blood was drawn from each participant into two 10 mL acid–citrate–dextroxse tubes. Lymphocytes were isolated within 24 h through BD Vacutainer® CPT™ Mononuclear Cell Preparation Tubes with sodium heparin (BD Biosciences), which uses an inert gel of a molecular weight less than that of erythrocytes and granulocytes, but greater than that of mononuclear blood cells to separate these layers. After 25 min of centrifugation at 1.5 relative centrifugal force (RCF), the lymphocyte layer has a distinct cloudy appearance within the mononuclear cell layer and can be siphoned out through transfer pipette. Lymphocytes were then washed with Dulbecco's phosphate-buffered saline (DPBS) and centrifuged again for 25 min at 1.5 RCF, creating a pellet. Thirty-eight of the samples were resuspended in RIPA buffer (Catalog No. R0278-50ML) before a protocol change, in accordance with enzyme-linked immunosorbent assay (ELISA) guidelines. The remaining 42 of the samples were resuspended in 3 mL DPBS to avoid interactions with sodium dodecyl sulfide, a reagent in RIPA, and horseradish peroxidase used in the ELISA.31 All samples were preserved with 30 μL Phosphatase Inhibitor Cocktail 3 (Sigma Aldrich Catalog No. P0044), 30 μL Phosphatase Inhibitor Cocktail 2 (Sigma Aldrich Catalog No. P5726), and 30 μL Protease Inhibitor Cocktail (Sigma Aldrich Catalog No. P8340). The resulting suspension was then divided into three equal aliquots and frozen at −80°C until CB1 protein quantification.
Quantification preparation
Samples were thawed on ice the day of the CB1 and total protein quantification assays. To facilitate sonication, 400 μL samples were combined with 100 μL sonication buffer (2% Protease Inhibitor Cocktail, 0.5% Phosphatase Inhibitor Cocktail 2, 0.5% Phosphatase Inhibitor Cocktail 3, 10% glycerol, and 87% sonication solution), and sonicated in two short bursts. To minimize freeze/thaw cycles and prevent protein shearing, all samples' CB1 and total protein assays were assayed on the same day.
Protein concentration
Total protein concentration was quantified through Pierce BCA Protein assay kit (Thermo Fisher Scientific Catalog No. 23225).32 Optical density was measured at 562 nm with a BioTek Absorbance Reader (model ELX800).
CB1 receptors were quantified through the Human CNR1 ELISA Kit (My BioSource Catalog No: MBS2503052), which has demonstrated consistent high antibody specificity for CB1 during tests for protein cross-reactivity, particularly with analogues of CB1 as reported by the data sheet from MyBioSource found here.31 All samples were diluted 1:60 in DPBS, so that the final CB1 concentration would fall within the detectable range determined by the standard curve. CB1 quantification then occurred according to the kit protocol. Samples were run in duplicate and all reagents were prepared immediately before use. Optical density was determined by absorbance at 450 nm.
Sample concentrations were determined in accordance with each plate's standard curve, and results were calculated according to a four-parameter logistic regression curve. A ratio of CB1 concentration to total protein concentration was calculated to account for variability between samples that may have been incurred through sample preparation.
Calculation
Total protein concentration and CB1 concentration were calculated according to each assay's standard curves. These concentrations were then back calculated to determine CB1/total protein ratio in the initial sample concentration, for a normalized CB1 concentration across all subjects.
DNA sample and analysis
Saliva samples were collected through Oragene Discover kit.33 DNA was isolated and CNR1 rs2023239 genotype was determined on a 7900HT Fast Real-Time PCR System.34
Group analyses
An independent samples t-test was run between users and controls to determine any preliminary differences between groups. In users, correlations were performed between normalized CB1 and THC metabolites (Supplementary Fig. S1) in urine, as well as total years of cannabis use (Supplementary Fig. S2). A two-way ANOVA was run between cannabis use status (users vs. nonusing controls) and CNR1 rs2023239 genotype (G allele carrier vs. noncarriers homozygous for the A allele) to determine the effects of cannabis use and CNR1 genotype on CB1 density (Table 2). Finally, a t-test was run to compare CNR1 levels between users who have at least one risk (G) allele and users who do not. Because there have been reports of differences between male and female cannabis users,35 we performed a post hoc t-test to ensure no sex effects were occurring in our sample. Similarly, we performed a post hoc correlation between age and normalized CB1 concentration.14
Table 2.
Demographics of Participants by Genotype
| All participants | Cannabis users | Controls | ||||
|---|---|---|---|---|---|---|
| AA (N=51) | G carrier (N=21) | AA (N=27) | G carrier (N=17) | AA (N=24) | G carrier (N=4) | |
| Males, n (%) | 33 (65) | 15 (71) | 20 (74) | 13 (76) | 13 (54) | 2 (50) |
| Age | 29.67 (9.48) | 31.10 (6.88) | 29.41 (8.53) | 31.24 (7.05) | 29.96 (10.64) | 30.5 (7.05) |
| Age of onset | — | — | 15.69 (3.40) | 15.30 (3.05) | — | — |
| Education | 14.14 (2.78) | 14.61 (2.48) | 13.33 (2.34) | 13.53 (2.58) | 16.04 (1.78) | 16.75 (2.22) |
| Race | ||||||
| Caucasian | 32 | 10 | 17 | 9 | 15 | 1 |
| African American | 4 | 11 | 3 | 8 | 1 | 3 |
| American Indian or Alaska Native | 0 | 0 | 0 | 0 | 0 | 0 |
| Asian | 6 | 0 | 1 | 0 | 5 | 0 |
| Native Hawaiian or Pacific Islander | 0 | 0 | 0 | 0 | 0 | 0 |
| Multiracial | 6 | 0 | 4 | 0 | 2 | 0 |
| Other | 3 | 0 | 2 | 0 | 1 | 0 |
Results
Main effects
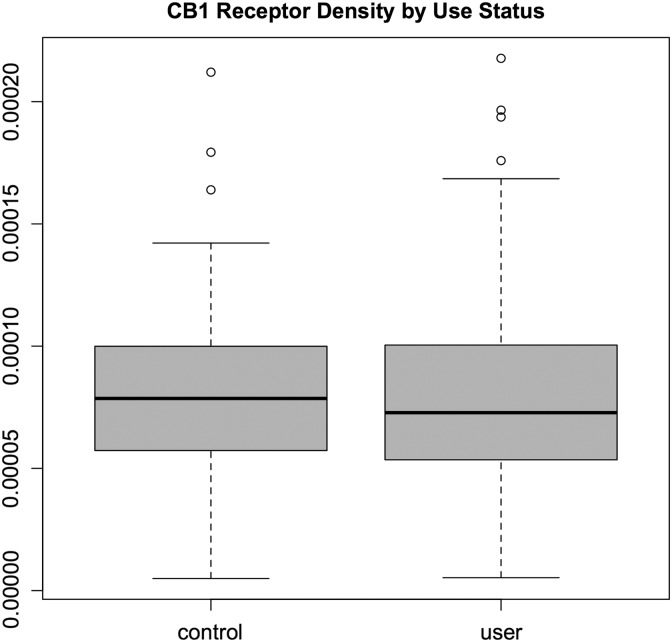
The protocol resulted in quantification of CB1 protein from human lymphocytes to establish a ratio of CB1 to total protein. Samples were highly concentrated and required dilutions between 40× and 80×. Cannabis users had an average concentration of 8.10E-5 (standard deviation [SD]=4.90E-5) and nonusing controls had an average concentration of 8.23E-5 (SD=4.02E-5) (Fig. 1). There was no difference in CB1 concentration between cannabis users and nonusing controls, t(103)=−0.145, p=0.89 (Fig. 1). In cannabis users, there was no significant correlation between normalized CB1 and THC metabolites, R(55)=−0.11, p=0.43. In addition, there was no correlation between normalized CB1 and years of cannabis use, R(55)=0.09, p=0.48 (Fig. 1). An independent samples t-test indicated that there was no difference in CB1 between males and females, t(78)=1.81, p=0.07. There was no relationship between age and CB1, R(103)=0.045, p=0.64.
FIG. 1.
CB1 protein levels do not differ between daily cannabis users (mean=8.10E-5, SD=4.90E-5) and healthy controls (mean=8.23E-5, SD=4.02E-5). CB1 protein was divided by overall protein concentration to normalize across participants. CB1, cannabinoid 1; SD, standard deviation.
A two-way ANOVA between users and nonusers carrying the risk allele (GG or GA) indicated a trend-level interaction between cannabis use status and genotype, such that users who were carriers for the G allele trended toward greater CB1 protein than both nonusers and users without a G allele F(1,67)=3.56, p=0.06 (Fig. 2).
FIG. 2.
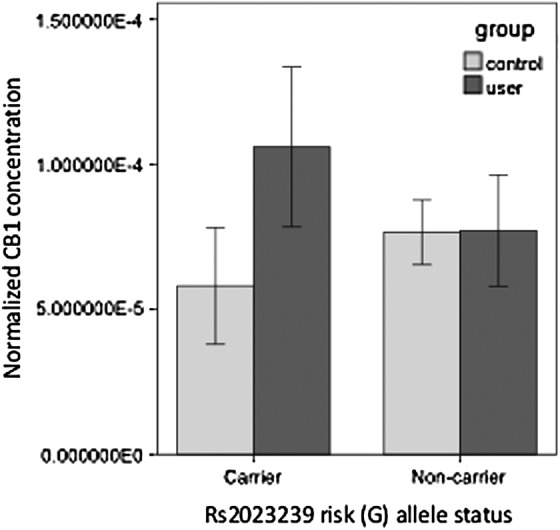
There is a trend toward a gene by group interaction between cannabis use and rs2023239 risk (G) allele carrier status F(1,67)=3.56, p=0.06. Light gray bars indicate control participants; dark gray bars indicate daily cannabis users. Error bars indicate 95% confidence intervals.
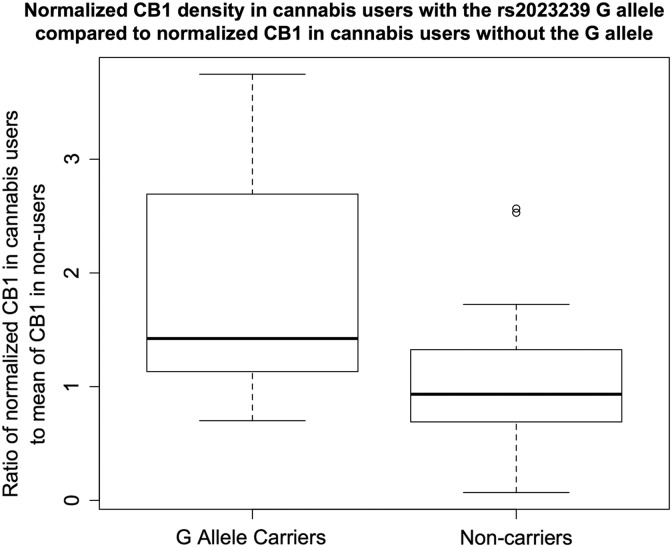
A manipulation check using a one-tailed t-test was used to examine the differences in individuals with the rs2023239 G allele compared with those without. We calculated a ratio of normalized CB1 protein in users with the G allele to the average normalized CB1 protein in nonusers with the G allele, and a ratio of CB1 protein in nonusers with the G allele to the average of normalized CB1 protein in nonusers without the G allele. A t-test between these two ratios, with a corrected standard error to account for the full sample size including nonusers, indicated that cannabis users with the rs2023239 G allele had greater CB1 density than users not carrying the G allele, t(68)=2.1, p=0.038 (Fig. 3).
FIG. 3.
A t-test between the ratio of protein in cannabis users to the average value in nonusers showed that rs2023239 risk (G) allele carriers had greater normalized CB1 than noncarriers t(68)=2.1, p=0.038.
Post hoc analysis
To ensure consistent results across the three plates used in this experiment and thereby further control for potential antibody cross-reactivity,36 we ran an ANOVA on normalized CB1 concentration across the plates. The results indicated that there was no effect of plate order on protein concentration, F(3,59)=1.09, p=0.28, contributing to our confidence in antibody specificity.
Discussion
This study demonstrates that peripheral lymphocytes express adequate CB1 for quantification without need for additional signal amplification and can be normalized into a ratio with total protein concentration for comparisons. These results suggest a gene×environment effect, such that cannabis users carrying the rs2023239 G allele had more CB1 than nonusers and non-G allele carriers.
Previous studies have reported brain CB1 receptor protein downregulation by repeated CB agonist treatment in the literature,8,12,13,37 but findings have not been consistent.38 Our findings in lymphocytes demonstrating increased CB1 density in daily cannabis users are aligned with findings in the brain reporting increased CB1 density postcannabis exposure.10,11 However, we acknowledge that lymphocyte CB1 may be under different post-translational regulatory mechanisms than CB1 in the CNS, such as internalization, degradation, or transport to the membrane.
Other studies have quantified human CB1 density through PET and postmortem methods, but to date, this study is the only documented method for quantifying how CB1 protein in human peripheral lymphocytes might be affected by rs2023239 genotype and cannabis use. Because measurement of receptor density in the human CNS is difficult, our results expand upon this body of literature by employing a minimally invasive technique to quantify CB1 in humans. Although reports of CB1 concentrations in the CNS after chronic cannabis exposure are variable, the majority of the literature indicates downregulation of CB1.37 Thus, our findings suggest that in participants with the rs2023239 G allele, lymphocyte CB1 may be regulated differently than reports showing CB1 downregulation in the CNS. This could be because of the differential roles of the ECS in these different tissues, or different post-translational mechanisms, for example, degradation. Furthermore, these results may reflect molecular effects of chronic and substantial THC exposure in peripheral tissue, which if statistically detectable as our results suggest may be informative for the effects of cannabis use overall.
Although data regarding the effects of chronic heavy cannabis use on CB1 density are sparse, they suggest that CB1 concentration is affected by modulation of downstream signaling as CB1 modulates GABAergic and glutamatergic signaling pathways. However, to date, it has been unclear how this affects all cannabinoid-mediated processes in the long term, and whether there could be a connection between peripheral data and CNS function.39–41 This assay provides the first evidence that measurement of peripheral CB1 lymphocytes is a cost-effective, minimally invasive method for measuring susceptibility to cannabis use.
Changes in CB1 as a result of cannabis use are widely reported in the literature. Alternative splicing of CNR1 in those with rs2023239 G allele is associated with greater binding of CB1 agonists.4,5 Thus, the rs2023239 G allele may indicate greater CB1 concentrations. CB1 density is increased postmortem in cannabis users relative to nonusers.38 Rs2023239 G allele carriers report greater sensitivity to the rewarding effects of cannabis and exhibit greater withdrawal after abstinence.7 Taken together, our finding of a gene by environment interaction, together with the absence of a difference between CB1 density in cannabis users versus nonusers, suggests the possibility that cannabis use leads to an upregulation of CB1 receptors in those predisposed for greater CB1 binding (i.e., rs2023239 G allele carriers). In addition, these results support previous findings by our group, which identified a cannabis use by rs2023239 genotype interaction on hippocampal volume.42 Together, these studies may indicate that the rs2023239 genotype moderates the effects of CB1 agonists, such as cannabis use.
The relationship between expression of CB1 in the periphery and CB1 in the CNS has not yet been elucidated, but extant literature supports the importance of genetic variation in neurological functions related to this gene.6 Similar work in the fatty acid amide hydrolase gene has shown effects of SNP genotype on peripheral protein density, with associated protein concentrations in the CNS and increased risk for cannabis use disorders.43,44 Although the relationship between peripheral and CNS receptor expression is complicated, it is valuable and necessary to characterize, because these proteins are highly regulated by internal and environmental factors.45 Thus, CB1 density, in conjunction with the rs2023239 genotype, holds promise for measurement of change in receptor density over time that can be corroborated with CNS CB1. Future work should examine CB1 density in conjunction with other measures of cannabis use, particularly to elucidate the relationship between CB1 expression and behavior. Such a relationship would facilitate the development of the Research Domain Criteria46 as well as treatment options for cannabis use disorders.
Limitations
Lymphocytes are a heterogeneous classification of several cell subtypes and CB1 may be altered in one cell type, but not another. Although this variability may be detectable on an individual basis, it is likely to be averaged out throughout our sample. However, to ensure that lymphocyte heterogeneity is not an issue, Fluorescence Activated Cell Sorting could differentiate subtypes for clear understanding of CB1 in each subtype. This study would benefit from additional specificity controls such as an antigen blockage of the signal or non-CB1R expressing cells and high-CB1R expressing cells, to serve as a negative and positive control, respectively. As our results reached trend-level significance, this study warrants further examination of these effects in a larger sample size. Finally, future work is necessary to divulge the relationship between CB1 expressed in peripheral lymphocytes and CB1 expressed in the brain, as well as how these concentrations are affected by daily chronic cannabis use. Importantly, this study serves as the first step toward understanding this relationship.
Conclusions
In sum, this is the first documented method for quantification of CB1 receptors from peripheral lymphocytes in long-term, daily cannabis users. Using this method, we found a gene by environment interaction indicating CB1 upregulation in cannabis users with the rs2023239 G allele. This protocol provides a minimally invasive method for examining potential up- or downregulation of a receptor with important psychoactive, immune, and digestive regulatory functions, and holds promise as a potential research and diagnostic tool.
Supplementary Material
Abbreviations Used
- CB1
cannabinoid 1
- DPBS
Dulbecco's phosphate-buffered saline
- ECS
endocannabinoid system
- ELISA
enzyme-linked immunosorbent assay
- PCR
polymerase chain reaction
- PET
positron emission tomography
- SD
standard deviation
- SNP
single nucleotide polymorphism
- THC
delta-9-tetrahydrocannabinol
Acknowledgments
The authors would like to thank the Filbey Lab research assistants for the data collection and processing of samples and Dr. Michael Burton for proofreading the article. This research was funded by NIH R01 1R01DA030344-01A1 (Filbey).
Authors Contribution
F.F. designed the study. A.K. carried out assays, analyses, and wrote the initial draft of the article. L.N. provided technical assistance for the assays and reviewed the article. F.F. and C.M. provided input and helped write the article.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Elphick MR, Egertová M. The neurobiology and evolution of cannabinoid signalling. Philos Trans R Soc Lond Ser B. 2001;356:381–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mackie K. Distribution of cannabinoid receptors in the central and peripheral nervous system. Handb Exp Pharmacol. 2005;168:299–325 [DOI] [PubMed] [Google Scholar]
- 3.Lupica CR, Riegel AC, Hoffman AF. Marijuana and cannabinoid regulation of brain reward circuits. Br J Pharmacol. 2004;143:227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang P-W, et al. . Human cannabinoid receptor 1: 5′ exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Mol Psychiatry. 2004;9:916–931 [DOI] [PubMed] [Google Scholar]
- 5.Hutchison KE, et al. . The incentive salience of alcohol: translating the effects of genetic variant in CNR1. Arch Gen Psychiatry. 2008;65:84–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filbey FM, Schacht JP, Myers US., et al. . Individual and additive effects of the CNR1 and FAAH genes on brain response to marijuana cues. Neuropsychopharmacology. 2010;35:967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haughey HM, Marshall E, Schacht JP, et al. . Marijuana withdrawal and craving: influence of the cannabinoid receptor 1 (CNR1) and fatty acid amide hydrolase (FAAH) genes. Addiction. 2008;103:1678–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burston JJ, Wiley JL, Craig AA, et al. . Regional enhancement of cannabinoid CB1 receptor desensitization in female adolescent rats following repeated Δ9-tetrahydrocannabinol exposure: THC and adolescent CB1 receptor desensitization. Br J Pharmacol. 2010;161:103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang S, et al. . Effects of long-term exposure to Δ9-THC on expression of cannabinoid receptor (CB1) mRNA in different rat brain regions. Mol Brain Res. 1998;62:141–149 [DOI] [PubMed] [Google Scholar]
- 10.Dean B, Bradbury R, Copolov DL. Cannabis-sensitive dopaminergic markers in postmortem central nervous system: changes in schizophrenia. Biol Psychiatry. 2003;53:585–592 [DOI] [PubMed] [Google Scholar]
- 11.Pagotto U., et al. . Normal human pituitary gland and pituitary adenomas express cannabinoid receptor type 1 and synthesize endogenous cannabinoids: first evidence for a direct role of cannabinoids on hormone modulation at the human pituitary level. J Clin Endocrinol Metab. 2001;86:2687–2696 [DOI] [PubMed] [Google Scholar]
- 12.Villares J. Chronic use of marijuana decreases cannabinoid receptor binding and mRNA expression in the human brain. Neuroscience. 2007;145:323–334 [DOI] [PubMed] [Google Scholar]
- 13.Hirvonen J., et al. . Reversible and regionally selective downregulation of brain cannabinoid CB1 receptors in chronic daily cannabis smokers. Mol. Psychiatry. 2012;17:642–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong DF, et al. . Quantification of cerebral cannabinoid receptors subtype 1 (CB1) in healthy subjects and schizophrenia by the novel PET radioligand [11C]OMAR. NeuroImage. 2010;52:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breivogel CS, Selley DE, Childers SR. Cannabinoid receptor agonist efficacy for stimulating [35S] GTPγS binding to rat cerebellar membranes correlates with agonist-induced decreases in GDP affinity. J Biol Chem. 1998;273:16865–16873 [DOI] [PubMed] [Google Scholar]
- 16.Izzo AA, Sharkey KA. Cannabinoids and the gut: new developments and emerging concepts. Pharmacol Ther. 2010;126:21–38 [DOI] [PubMed] [Google Scholar]
- 17.Löfgren P, Sjölin E, Wåhlen K, et al. . Human adipose tissue cannabinoid receptor 1 gene expression is not related to fat cell function or adiponectin level. J Clin Endocrinol Metab. 2007;92:1555–1559 [DOI] [PubMed] [Google Scholar]
- 18.Galiègue S., et al. . Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur J Biochem FEBS. 1995;232:54–61 [DOI] [PubMed] [Google Scholar]
- 19.Schatz AR, Koh W, Kaminski N. Delta-9-Tetrahydrocanabinol selectively inhibits T-cell dependent humoral immune responses through direct inhibition of accessory T-cell function. Immunopharmacology. 1993;26:129–137 [DOI] [PubMed] [Google Scholar]
- 20.Srivastava MD, Srivastava BIS, Brouhard B. Delta-9-tetrahydrocannabinol and cannabidiol alter cytokine production by human immune cells. Immunopharmacology. 1998;40:179–185 [DOI] [PubMed] [Google Scholar]
- 21.Börner C, Bedini A, Höllt V, et al. . Analysis of promoter regions regulating basal and interleukin-4-inducible expression of the human CB1 receptor gene in T lymphocytes. Mol Pharmacol. 2008;73:1013–1019 [DOI] [PubMed] [Google Scholar]
- 22.Bouaboula M., et al. . Cannabinoid-receptor expression in human leukocytes. Eur J Biochem. 1993;214:173–180 [DOI] [PubMed] [Google Scholar]
- 23.Gladkevich A, Kauffman HF, Korf J. Lymphocytes as a neural probe: potential for studying psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28:559–576 [DOI] [PubMed] [Google Scholar]
- 24.Grimsey NL, Goodfellow CE, Scotter EL, et al. . Specific detection of CB1 receptors; cannabinoid CB1 receptor antibodies are not all created equal! J Neurosci Methods. 2008;171:78–86 [DOI] [PubMed] [Google Scholar]
- 25.Maccarrone M, Valensise H, Bari M, et al. . Progesterone up-regulates anandamide hydrolase in human lymphocytes: role of cytokines and implications for fertility. J Immunol. 2001;166:7183–7189 [DOI] [PubMed] [Google Scholar]
- 26.Onaivi ES, et al. . Expression of cannabinoid receptors and their gene transcripts in human blood cells. Prog Neuropsychopharmacol Biol Psychiatry. 1999;23:1063–1077 [DOI] [PubMed] [Google Scholar]
- 27.Piszcz JA, Lemancewicz D, Kloczko J, et al. . Cannabinoid receptors expression in bone marrow trephine biopsy of chronic lymphocytic leukaemia patients treated with purine analogues. Exp Oncol. 2007;29:221–225 [PubMed] [Google Scholar]
- 28.Rotter A, Bayerlein K, Hansbauer M, Weiland J, Sperling W, Kornhuber J, Biermann T. CB1 and CB2 receptor expression and promoter methylation in patients with cannabis dependence. Eur Addiction Res. 2013;19:13–20 [DOI] [PubMed] [Google Scholar]
- 29.Hirvonen J., et al. . Reduced cannabinoid CB1 receptor binding in alcohol dependence measured with positron emission tomography. Mol Psychiatry. 2013;18:916–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filbey FM, et al. . fMRI study of neural sensitization to hedonic stimuli in long-term, daily cannabis users. Hum Brain Mapp. 2016;37:3431–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MyBiosource, Human CNR1 (Cannabinoid Receptor 1, Brain) ELISA Kit.pdf Available at: https://www.mybiosource.com/images/tds/protocol_manuals/800000-9999999/MBS2503052.pdf
- 32.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254 [DOI] [PubMed] [Google Scholar]
- 33.Birnboim HC. DNA yield with an Oragene® self-collection kit. DNA Genotek: Ottawa, Canada, 2011 [Google Scholar]
- 34.Rogers NL, Cole SA, Lan H-C, et al. . New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am J Hum Biol Off J Hum Biol Counc. 2007;19:319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ketcherside A, Baine J, Filbey F. Sex effects of marijuana on brain structure and function. Curr Addict Rep. 2016;3:323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cécyre B, Thomas S, Ptito M, et al. . Evaluation of the specificity of antibodies raised against cannabinoid receptor type 2 in the mouse retina. Naunyn Schmiedebergs Arch Pharmacol. 2014;387:175–184 [DOI] [PubMed] [Google Scholar]
- 37.Sim-Selley LJ. Regulation of cannabinoid CB1 receptors in the central nervous system by chronic cannabinoids. Crit Rev Neurobiol. 2003;15:91–119 [DOI] [PubMed] [Google Scholar]
- 38.Dean B, Sundram S, Bradbury R, et al. . Studies on [3H]CP-55940 binding in the human central nervous system: regional specific changes in density of cannabinoid-1 receptors associated with schizophrenia and cannabis use. Neuroscience. 2001;103:9–15 [DOI] [PubMed] [Google Scholar]
- 39.Bass CE, Welch SP, Martin BR. Reversal of Δ9-tetrahydrocannabinol-induced tolerance by specific kinase inhibitors. Eur J Pharmacol. 2004;496:99–108 [DOI] [PubMed] [Google Scholar]
- 40.Martin BR. Role of lipids and lipid signaling in the development of cannabinoid tolerance. Life Sci. 2005;77:1543–1558 [DOI] [PubMed] [Google Scholar]
- 41.Whitlow CT, Freedland CS, Porrino LJ. Functional consequences of the repeated administration of Δ9-tetrahydrocannabinol in the rat. Drug Alcohol Depend. 2003;71:169–177 [DOI] [PubMed] [Google Scholar]
- 42.Schacht JP, Hutchison KE, Filbey FM. Associations between cannabinoid receptor-1 (CNR1) variation and hippocampus and amygdala volumes in heavy cannabis users. Neuropsychopharmacology. 2012;37:2368–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boileau I, et al. . The fatty acid amide hydrolase C385A variant affects brain binding of the positron emission tomography tracer [11C]CURB. J Cereb Blood Flow Metab. 2015;35:1237–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boileau I, et al. . Fatty acid amide hydrolase binding in brain of cannabis users: imaging with the novel radiotracer [(11)C]CURB. Biol Psychiatry. 2016;80:691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matias I, Bisogno T, Di Marzo V. Endogenous cannabinoids in the brain and peripheral tissues: regulation of their levels and control of food intake. Int J Obes. 2006;30S1:S7–S12 [DOI] [PubMed] [Google Scholar]
- 46.Sanislow CA, Quinn KJ, Sypher I. NIMH Research Domain Criteria (RDoC). In The encyclopedia of clinical psychology. Cautin RL, Lilienfeld SO, (Eds.) John Wiley & Sons, Inc.: Hoboken, NJ, 2015, pp. 1–6 [Google Scholar]
References
Cite this article as: Ketcherside A, Noble LJ, McIntyre CK, Filbey FM (2017) Cannabinoid receptor 1 gene by cannabis use interaction on CB1 receptor density, Cannabis and Cannabinoid Research 2:1, 202–209, DOI: 10.1089/can.2017.0007.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.