Abstract
Gene therapy is a rapidly evolving field. So far, there have been >2,400 gene therapy products in clinical trials and four products on the market. A prerequisite for producing gene therapy products is ensuring their quality and safety. This requires appropriately controlled and standardized production and testing procedures that result in consistent safety and efficacy. Assuring the quality and safety of lentivirus-based gene therapy products in particular presents a great challenge because they are cell-based multigene products that include viral and therapeutic proteins as well as modified cells. In addition to the continuous refinement of a product, changes in production sites and manufacturing processes have become more and more common, posing challenges to developers regarding reproducibility and comparability of results. This paper discusses the concept of developing a first World Health Organization International Standard, suitable for the standardization of assays and enabling comparison of cross-trial and cross-manufacturing results for this important vector platform. The standard will be expected to optimize the development of gene therapy medicinal products, which is especially important, given the usually orphan nature of the diseases to be treated, naturally hampering reproducibility and comparability of results.
Keywords: : LV production, WHO standard, integration analysis, genomic DNA, qPCR quantitation
Introduction
Lentiviral vectors (LV) have emerged as the benchmark for gene therapy applications and have been successfully used in a cure for monogenic immunodeficiency disorders and chimeric antigen receptor (CAR) T-cell cancer immunotherapies. The number of clinical developments involving integrating vectors is expanding rapidly. There are currently a total of 72 LV clinical trials worldwide (six in Phase III/IV), 224 retroviral (RV) trials (19 in Phase II/IV), and 58 CAR T-cell trials (18 in Phase II/IV; https://clinicaltrials.gov/). It is generally anticipated that the first CAR T-cell product will come to market soon. In addition, products based on nuclease or zinc finger technologies and some induced pluripotent stem cell (iPSC)-derived products are also based on LV or RV. Therefore, the potential number of patients using LV-based products would be significant. Nevertheless, the number of patients is only one of the factors important for estimating the public-health impact. Other factors, including the severity of disease, impact of a given disease on both the patient and society and its unmet medical needs, are important determinants on the impact of LV-based products. This is confirmed by experiences across jurisdictions from orphan medicinal products where the number of patients is by definition very small but the impact of medicines can be considerable.
The safety level of LVs today is high, achieved by partial deletion of 3′ long terminal repeat (LTR).1 However, uncontrolled integration can still cause insertional mutagenesis and lead to overexpression or disruption of adjacent genes at the site of integration. For example, the LV was found to disrupt the HMGA2 transcript in a thalassemia trial.2 Intragenic insertion of LVs have been identified in preclinical studies that can lead to aberrant gene regulation.3–5 Therefore, before a fully controlled LV integration can be achieved, lowering the integration copy number is, at current status, an effective measure to reduce the genotoxic and tumorigenic potential. In fact, developers often aim at as low as possible integration copy numbers for a desirable clinical efficacy, and usually provide data on copy numbers as a minimum in clinical trial applications and marketing authorization application dossiers to support the efficacy and safety of their products.
Regulatory authorities such as the European Medicines Agency (EMA) and the U.S. Food and Drug Administration (FDA) require integration studies and long-term follow-up on products such as LVs that have a capacity to integrate permanently into the host cells and to persist for a long time in treated individuals. The information required by the EMA to support marketing authorization applications includes integrated vector copies per genome, integration profile, and integration sites (EMA/CAT/190186/2012, EMA/CAT/80183/2014). Although there are currently no regulatory requirements for the minimum or maximum integration copy number, an EMA reflection paper (EMA/CAT/190186/2012) on the management of clinical risks deriving from insertional mutagenesis highlighted the high vector copy number as a risk factor for oncogenesis and recommended risk assessment and management of the integration copy numbers, integration profile, and sites in products. The FDA recommends that the integration copy number shall be <5 copies per genome (presentation by Dr. Vatsan/FDA at ISBioTech conference on March 7, 2017).
Methods used in integration studies, for example linear amplification–mediated polymerase chain reaction (LAM-PCR) and high-throughput sequencing, still have important limitations in terms of sensitivity, accurate quantification, and data interpretation. A recent study has shown that using the currently available methods, the integration copy numbers (VCN) have often been underestimated.6 This is because the specificity and sensitivity of PCR-based methods are largely dependent on the choice and design of amplification target sequences and the conditions of reaction, which makes it difficult to compare data across clinical trials and assays.
At the current status, LV-based products are commonly used in ex vivo autologous therapies. Therefore, manufacture of LV-based products is often a decentralized or near-patient process. The complexity and intrinsic disparity of decentralized manufacture requires enhanced in-process control and standardization to ensure product consistency. In the field of CAR T-cell therapies using LVs, significant efforts have been made to move from autologous to allogenic therapies and from near-patient to centralized manufacturing processes. Well-controlled and standardized processes lead to well-understood and consistent products, and this is particularly important for LV-based ex vivo therapies. Therefore, standardization of manufacturing processes for ex vivo lentiviral products is one that is best put in place early in product development.
Historical effort toward standardization of gene therapy
The development of standards for gene therapy is at an early stage, with only a few accessible reference materials (RMs) being available. So far, there has been one RM for retroviral vectors,7 one for adenovirus vectors (ATCC_RSM_VR-1516),8,9 and two for adeno-associated virus (AAV) vectors (ATCC_RSM_VR1616 and VR1816).10–15 Demand for the RMs by the gene therapy community has been low, for example only ∼50 vials of adenovirus RMs were used during the 5 years following 2,000 vials being established (internal survey). In addition to the lack of community awareness of RMs, the fact that evolution in gene therapy products has been faster than the production of RMs may be another reason for the low uptake of the available RMs, especially for AAV-based systems where host immunity against adenoviruses necessitated finding and adapting the platform on a continuous basis. For example, AAV2 RMs (ATCC_RSM_VR1616) were first discussed in 2002. It took 8 years before they were ready for distribution in 2010, during which a significant amount of time was taken to decide on serotype choice, and a number of studies had indicated that AAV2 products showed lower than expected potency due to host immune responses, which makes AAV2 a less favorable choice for therapies and the RMs “redundant.” It is recognized that the production of early RMs under Good Manufacturing Practice (GMP)/Good Laboratory Practice (GLP) conditions might have unnecessarily led to increased time/cost of the RM development. RMs are used for assay development, optimization, and validation, and therefore may not necessarily have to be manufactured under GMP/GLP conditions (since they are not administered as medicines).
The utility of standards may be higher for other vector systems, and AAV vectors may actually have been much more prone to change than other vector systems. Such standards would not be mandatory for marketing authorization applications but would be useful tools to validate assays and enable interpretation of research results across studies—thus solving a commonly encountered comparability problem and enabling innovation rather than stifling it. Likewise, orphan medicines are often “entry portals” for technologies for more frequent diseases, which could well be true for this powerful LV platform. This is especially challenging in orphan disease scenarios for (long-term) correction of gene defects, since the potential to repeat a clinical trial and obtain missing data is difficult if not impossible. This holds for standards directly supporting such developments and has approached a level of regulation that justifies an International Standard.
Since the initiation of biological standardization by Paul Ehrlich in 1897, the World Health Organization (WHO) has set out criteria for selection as a WHO International Standard, including the requirements for (1) the stability of RMs, (2) adequate performance of the materials in as wide a range of assay methods as possible, and (3) linearity and parallelism of dose–response in order to compare and calibrate the primary standard and secondary standards/unknowns. In terms of the intended functions, WHO standards/RMs can be divided into three major groups: (1) the determination of dosage toxicity of vaccine products, (2) the determination of therapeutic potency of recombinant proteins, and (3) for the detection of adventitious agents. Accordingly, requirements for measurement and precision of read-out in each group of RMs are different, including threshold measurement of dosage toxicity, quantitative assessment of potency, or diagnostic detection of adventitious contaminants. Over the past 90 years, >1,000 International Standards/RMs have been produced for a wide range of biologicals by the National Institute for Biological Standards and Control (NIBSC) (www.nibsc.org), and >60,000 vials of RMs are distributed every year to laboratories in some 60 countries.
This article discusses the concept of the development of a first WHO International Standard, suitable for standardization of assays and enabling for cross-trial and cross-manufacturing results for this important vector platform. The standard will be expected to optimize development of gene therapy medicinal products, which is especially important, given the usually orphan nature of the diseases to be treated, naturally hampering reproducibility and comparability of results. The work performed to support an application at the WHO's Expert Committee on Biological Standardization is described, which was successful in October 2016. The data in this paper already at this point can assist developers of gene therapy medicinal products, since they will gain an understanding of how this standard, once available, will assist their product development.
Materials and Methods
LV production and transduction
Four plasmids—pMD2.G (env), pRSVRev (rev), pMDLg/pRRE (packaging), and pRRLSIN.cppt.PGK-GFP.WPRE (vector)—were kindly provided by Prof. Didier Trono (EPFL, Lausanne) and were used to produce LV particles by transient transfection of human embryonic kidney 293T cells (HEK293T) with a weight ratio of 3:2:1:1 vector:packaging:rev:envelope plasmids. Vector particles were then concentrated by ultracentrifugation at 12,500 g for 2 h and were titrated on HEK293T cells and analyzed by flow cytometry (BD FACSCanto II) to determine the percentage of LV-transduced cells.
For integration analysis, cells (e.g., 293T and MRC5 cells) were transduced with LV for 6 h in serum-free OptiMem™ medium with 8 μg/mL of polybrene (Sigma–Aldrich). Transduced cells were further cultured for 72 h in Dulbecco's modified Eagle's medium with 10% fetal calf serum (FCS) before being subjected to cell expansion and single cell sorting and being cryopreserved in FCS with 10% dimethyl sulfoxide (Sigma–Aldrich). Single-cell clones were established from LV-transduced cells at a multiplicity of infection (MOI) of 1 or 10 by sorting with a BD FACSCanto III cell sorter (BD Biosciences) based on the green fluorescent protein (GFP) fluorescence intensity of the cells. A LV unrelated spike plasmid DNA (pAAV2_hrGFP) was added to cells at 1 ng/5 × 106 cells (i.e., 1.77 × 106 copies of pAAV2_hrGFP/5 × 106 cells) before genomic DNA preparation to normalize DNA extraction-associated variations. Genomic DNA was then extracted from LV-transduced bulk cells and single-cell clones in the presence of 1 ng (1.77 × 106 copies) of a Spike plasmid DNA (pAAV2_hrGFP) using a Puregene Cell and Tissue kit and following the manufacturer's instructions (Qiagen).
PCR analysis
All PCR primers and dual-labeled probes with 6-FAM on the 5′ end and TAMRA on the 3′ end were purchased from Sigma–Aldrich. The sequences of primers and probes are given in Table 1. Primers were initially tested using endpoint PCR with an Eppendorf Mastercycler ProS. Briefly, 50 ng of plasmid DNA pRRLSIN.cppt.PGK-GFP.WPRE was incubated with 200 nM of each primer, 400 μM of dNTPs (Promega), 2 mM of MgCl2 (Promega), and 2.5 units of GoTaq® DNA polymerase (Promega). Cycling conditions were 94°C for 3 min followed by 30 cycles of 94°C for 30 s, annealing between 55°C and 65°C for 30 s, 72°C for 1 min. DNA amplicons were visualized by agarose gel electrophoresis.
Table 1.
Sequences of PCR primers and probes
| Name | Sequence | Start position | Length (bp) | Amplicon length | Primer Tm |
|---|---|---|---|---|---|
| 5′ LTR forward A | TGCCCGTCTGTTGTGTGACT | 108 | 20 | 140 | 59 |
| 5 LTR reverse A | CGAGTCCTGCGTCGAGAGAG | 228 | 20 | 140 | 59.18 |
| Psi forward B | CTCTCTCGACGCAGGACTCG | 228–247 | 20 | 126 (A to A) | 60 |
| Psi reverse B | GACGCTCTCGCACCCATCT | 335–353 | 19 | 126 (A to A) | 62 |
| Psi forward C | CAGGACTCGGCTTGCTGAAG | 239–258 | 20 | 107 (B to B) | 60 |
| Psi reverse C | CGCACCCATCTCTCTCCTTCT | 325–345 | 21 | 107 (B to B) | 61 |
| Psi forward D | GGCTTGCTGAAGCGCGC | 247–263 | 17 | 84 (C to C) | 61 |
| Psi reverse D | CCTTCTAGCCTCCGCTAGTCA | 310–330 | 21 | 84 (C to C) | 59 |
| Psi reverse E | CCGCTAGTCAAAATTTTTGGCGT | 297–319 | 23 | 73 (C to D) | 60 |
| RRE forward F | AGTAAGACCACCGCACAGCA | 669 | 20 | 134 | 58.78 |
| RRE reverse F | CCTTGGTGGGTGCTACTCCT | 783 | 20 | 134 | 58.18 |
| RRE forward G | GCACCCACCAAGGCAAAGAG | 790 | 20 | 198 | 58.87 |
| RRE reverse G | TTGCGCCTCAATAGCCCTCA | 968 | 20 | 198 | 58.92 |
| 5′ LTR A probe | AGTCCCTGTTCGGGCGCCACTGCT | 178–201 | 24 | 70 | |
| Psi B probe | CTCTTGCCGTGCGCGCTTCAGCAAGC | 248–273 | 26 | 70 | |
| RRE F probe | CCTCCAGGTCTGAAGATCAGCGGCCGC | 670–716 | 27 | 70 | |
| RRE G probe | ACGCTGCGCCCATAGTGCTTCCTGCT | 876–901 | 26 | 71 |
PCR, polymerase chain reaction.
Quantitative real-time PCR (qPCR) was performed using a Roche LightCycler®480 real-time PCR. All PCR reactions were performed using a LightCycler®480 probes master kit (Roche) in a 20 μL final volume containing 250 nM each of PCR primers, 75 nM of probe or a SensiMix SYBR No-Rox kit (Bioline), a 20 μL final volume containing 300 nM each of PCR primers, 100 ng of sample genomic DNA or 106 or 103 copies of sample plasmid DNA or genomic DNA from 1.39 × 105, cells and 1 × qPCR master mix. Seven serial dilutions of a plasmid standard (containing 108, 107, 106, 105, 104, 103, and 102 copies of pRRLSIN.cppt.PGK-GFP.WPRE plasmid DNA) were used to generate a standard curve for absolute quantification of vector samples. Plasmid DNA copy numbers were calculated based on the equation: copies/μL = {[concentration (ng/μL) × 6.022 × 1021]/[plasmid size (bp) × 109 × 650]}. All samples were tested in triplicate experiments, and a “no template DNA” negative control was included to monitor sample cross-contamination. qPCR was carried out with an initial denaturation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 30 s. Analysis of qPCR results was carried out using LightCycler® 480 Software 1.5.0 v1.5.0.39 (Roche). The fit points method for absolute quantification was used for analysis, and the noise band and threshold were set to Fluor.
Quantitation of cell karyotypes and integration copy numbers per cell
Cell karyotype (chromosome number/cell) was determined using the diploid MRC5 cells (48 chromosomes/cell) as calculation reference and based on an equal amount (e.g., 100 ng) of genomic DNA or an equal number of a testing cell line and the reference MRC5 cells. Human albumin, actin, and GapDH genes were used for the quantitation of the total copy numbers of a housekeeping gene. The chromosome numbers/cell was derived from the copy number ratio of a housekeeping gene between a testing cell line and the reference MRC5 cells. Integration copy numbers per cell was calculated based on the equation: LV copies/cell = (the copy numbers of integrated LV/the copy numbers of a housekeeping gene) × karyotype number (n). For example, the karyotype number of diploid MRC5 cells n = 48/24 = 2 and, when the chromosome numbers of 293T cells = 65, the karyotype number of the 293T cells n = 65/24 = 2.7.
Statistical analysis
Statistical analysis was carried out by performing a two-tailed paired Student's t-test. Significance was determined as p < 0.05 and p < 0.01. Error bars are expressed as the standard error of the mean.
Results and Discussion
First WHO International Standard
The proposal for developing a WHO International Standard for lentivirus-based gene therapy has been in consideration for several years, involving discussion with companies, clinicians, and the academic community in order to estimate where such a standard would be considered most useful, addressing unmet needs. A workshop was held at the NIBSC in June 2016, involving representatives of a number of LV manufacturers, clinical diagnostic laboratories from gene therapy centers, and regulators to assess the demand for a LV copy number standard and to consider what such a standard might comprise. The group was very positive about the proposal and articulated the need for standardization of LV products, and concluded that genomic DNA from a cell line with stable DNA content close to diploid would be ideal. As a result, the NIBSC proposed developing such standard to the WHO's Expert Committee on Biological Standardization (ECBS), which has to endorse any such proposal before work can commence. At their October 2016 meeting, the ECBS endorsed the development of the first WHO gene therapy genomic DNA standard for the quantitation of LV integration copy numbers—a historical event in the field of standardization of biological medicines. The development of the first WHO International Standard for LVs is built on existing NIBSC expertise and platform for making WHO genomic standards. So far, the NIBSC has established 11 genomic RMs for cancer diagnostics and genetic testing. One would also have to confirm that lessons learnt from AAV vector standards would be readily applicable to LV genomic standards.
The proposed LV genomic standard will be applicable to future product development because a highly homologous if not identical sequence has been identified to be biologically important and indispensable for lentiviral packaging and production, and is shared by three generations of LVs and most existing LV-based products. In particular, Fig. 1 shows that the homologous sequence highlighted in yellow is shared by all three generations of LV vectors and that in green is shared by second- and third-generation LV vectors. To test the presence and suitability of the predicted homologous sequences for genome copy quantitation, seven sets of PCR primers were designed within the identified homologous region, as detailed in Fig. 1, and were first tested by endpoint PCR using a third-generation of LV plasmid DNA pRRLSIN.cppt.PGK-GFP.WPRE. Figure 2A shows that all designed primers can specifically detect the homologous sequence, though four primer sets showed a greater efficiency than the others in PCR amplification.
Figure 1.
Schematic diagram showing the configuration of key gene elements in lentiviral vector pRRLSIN.cppt.PGK-GFP.WPRE and the locations of homologous sequences shared by three generations of lentiviral vectors (LVs) and the seven sets of primers and probes for polymerase chain reaction (PCR) detection and quantitative PCR quantitation.
Figure 2.
Image of agarose gel electrophoresis showing PCR detection of (A) homologous sequences in LV plasmid pRRLSIN.cppt.PGK-GFP.WPRE (pRRL) using seven sets of primers (A–G); (B) 18 different LV plasmids; and (C) confirmation of PCR detection of homologous sequences in the 18 LV vector plasmids. DNA molecular markers were given in the first lane indicating the size of PCR amplicons (bp). The generic annealing temperature (58°C) was used for all primer sets. −ve, negative control samples without input template DNA.
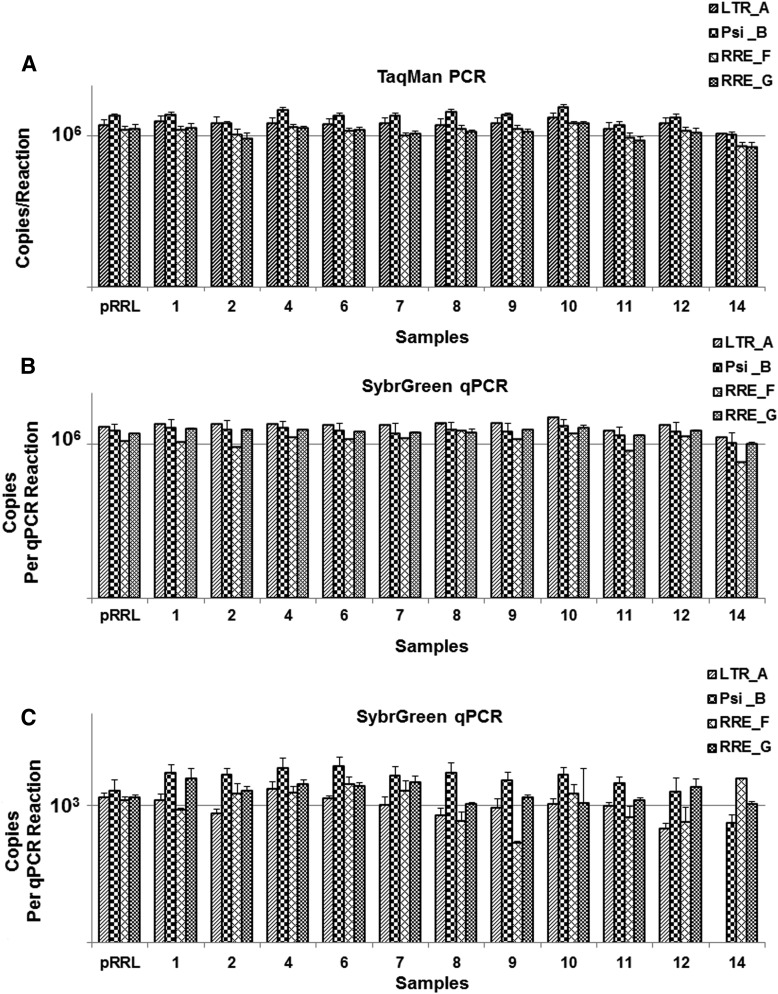
To evaluate whether the WHO standard will be suitable for integration study of existing LV vectors, 18 LV vector plasmids including early generations of LV vectors (e.g., pHV, pHR′-CMVLacZ, pHR′GFP, pSIN1.8, pCSFLW, and SINpHV; Fig. 2B) were obtained from various laboratories. Results from conventional endpoint PCR using four sets of PCR primers confirmed the presence and reliable detection of the predicted homologous sequence in the three generations of 18 LV vectors (Fig. 2C). qPCR using TaqMan (Fig. 3A) or Sybr Green primers/probes (Fig. 3B and C) was also performed on the 18 different LV vector plasmids, showing the quantitative detection of the predicted homologous sequence in all vector plasmids. The results also showed that the qPCR detection are independent of primer sets and assays used. As shown in Fig. 3, the detected copy numbers are comparable using four different sets of primers and using different detection methods of TaqMan or SybrGreen qPCR.
Figure 3.
qPCR analysis of 12 different LV plasmids, using (A) TaqMan and (B and C) SybrGreen qPCR and four sets of primers/probes (A, B, F, and G) targeting the homologous sequences among three generations of LV vectors. An equal amount of LV plasmid DNA, that is, (A) and (B) at 106 copies/PCR reaction and (C) at 103 copies/PCR reaction calculated based on individual plasmid molecular weight, was used for all LV plasmids tested. Vector copy numbers (copies/reaction) were calculated based on a circular pRRLSIN.cppt.PGK-GFP.WPRE plasmid. Data shown are the mean of three independent qPCR experiments of the same sample (n = 6).
To evaluate whether the WHO standard will be suitable for LV integration analysis, LVs were individually produced from seven selected LV vector plasmids (Fig. 4) and were then used to transduce 293T cells. LV-integrated genomic DNA was extracted from the transduced cells and was subjected to integration analysis. Figure 4 shows the detection of LV homologous sequences in the genomic DNA of LV transduced bulk cells (Fig. 4A) and single-cell clones that have been subjected to two rounds of single-cell sorting (Fig. 4B), demonstrating that the WHO genomic standard will be applicable for the integration study of three generations of LV vectors. A significant discrepancy was observed in bulk cell samples between vector copy numbers calculated from plasmid DNA and genomic DNA (Fig. 4A). Such a discrepancy becomes less apparent in single-cell clones (Fig. 4B), indicating the interference of bulk cell heterogeneity on absolute qPCR quantitation. The results highlight the significance of establishing a WHO genomic standard for the quantitation of LV integration. This is expected to increase regulatory certainty of products for an assessment of their oncogenic potential.
Figure 4.
qPCR analysis of LV integration (A) qPCR of bulk LV-transduced cells using four sets of primers/probes (A, B, F, and G), and (B) qPCR of LV-transduced single-cell clones after being subjected to two rounds of single-cell sorting and using the RRE-G primer set. An equal amount of LV-integrated genomic DNA (100 ng of gDNA/PCR reaction) was used for all samples tested. Integration copy numbers (copies/genome) were calculated based on a circular pRRLSIN.cppt.PGK-GFP.WPRE plasmid standard (A, left panel; B, dotted bars) or a LV_pRRL integrated genomic DNA standard (A, right panel; B, solid bars). Data shown are the mean of three independent qPCR experiments of the same sample (n = 6). *p < 0.05; **p < 0.01.
Current production of the standard
A number of cell lines, including diploid cell line (e.g., MRC5; NIBSC),16 293T cells, and T-cell lines (e.g., Jurkat and K652) have been evaluated as a candidate cell line for the standard production. The human diploid MRC5 cell line was initially selected because they are diploid cells with a competitive advantage over other cell lines in vector copy quantification. However, recent studies revealed that MRC5 cells require a certain cell density to grow, and it is impossible to establish single-cell clones from MRC5 cells. In addition, MRC5 cells are primary cells with limited genotype stability up to PDL32. Even if MRC5 cells can survive as a single-cell clone, it will immediately increase the PDL to 16, so there will never be enough cells before the MRC5 cells become unstable. Some T-cell lines have a similar limitation to the MRC5 cells plus undefined karyotypes of the T-cell lines.
Human HEK293T cells have been systemically evaluated and selected for the establishment of the WHO standard. This is because (1) 293T cells have been widely used in LV production and quantitation; (2) it has been recently reported that the commonly known heterogeneity and instability of HEK293 were largely due to a mix (bulk) cell population—in fact, the established single 293T cell clones show a high karyotype stability and homogeneity17,18; and (3) so far the karyotype of 293T bulk cells and two single 293T cell clones have been quantified using the diploid MRC5 cell karyotype of 48 chromosomes/cell as a reference. The results showed that the bulk 293T cells used have 65 chromosomes/cell, which is within the range of 62–72 in the 293 cells from European collection of cell cultures (ECACC).17 The two single 293T cell clones tested have 53 and 48 chromosomes/cell, respectively, one of which shows a diploid karyotype. The karyotype of the cell clones for the WHO standards will be karyotype quantified and monitored throughout the process, which will support the absolute quantitation of cell numbers and LV integration per cell and justify the use of 293T cells.
Four LV plasmids—pMD2.G, pRSVRev, pMDLg/pRRE, and pRRLSIN.cppt.PGK-GFP.WPRE—were kindly donated by Prof. Didier Trono (EPFL, Lausanne) for the establishment of the first WHO LV standard and will be used to produce HIV-1 LV particles. The production 293T cells were transduced with the LVs at a MOI of 1. A panel of single-cell clones was established and characterized for integration copy number using qPCR. Three single-cell clones with a LV integration copy number of 1 or 5 will be established as candidate cell lines for the production of the WHO standard and will be subjected to a full characterization study, including integration profile and integration site analysis. One of the fully characterized candidate cell clones will be subjected to further propagation to produce sufficient genomic DNA for 5,000 vials of WHO standard. Purified genomic DNA with defined LV integration will be vialled, lyophilized, and fully characterized using a range of methods, including qPCR, digital PCR, LAM-PCR, and deep sequencing to ensure no genomic alteration during single-cell clone propagation. The international unit of the WHO standard will be established and assigned from a collaborative study involving at least 15 laboratories worldwide. It is anticipated that it will take approximately 2 years to produce the WHO standard. An outline of the standard production procedures is given in Fig. 5.
Figure 5.
Outline of the process for establishing the first WHO LV genomic standard.
Intended use of the WHO standard
The first WHO lentiviral standard will comprise a panel of three standards: one vial of genomic DNA without LV integration, a second vial of genomic DNA with a single copy of LV integration, and a third vial with five copies per genome of LV integration. This standard will primarily be used for the validation of internal in-house standards used in different laboratories. Unit assignment of the standard, copy/genome, will be derived from different quantitative analysis, including TaqMan, SybrGreen, and digital PCR. Therefore, the standard will be applicable to a wide range of DNA detection methods. As the standard will be fully characterized for integration site analysis, it can also be used as a LAM-PCR and deep-sequencing control for integration profile and site analysis—a major step forward in obtaining reliable and reproducible data on insertional mutagenesis, the assessment of which is a major point of interest for regulatory authorities.
The development of the first WHO standard for LV integration will meet a high demand from the community. The WHO standard is primarily for standardizing three milestone stages of LV product development: establishment of the manufacturing process, clinical dose determination, and patient safety follow-up. During manufacture, the standard can be used to standardize the LV production process by measuring the number of vectors integrated in an in vitro testing cell line with methods that are independent of the therapeutic transgene or the promoter. Standardized LV potency assay is important for batch-to-batch comparability and consistency assessment, also allowing for establishing changes and optimization of the manufacturing process more reliably. Here, the standard could be used as a measurement of potency rather than safety (insertional mutagenesis is an expression of potency, albeit medically unwanted). Second, the genomic standard can be used to standardize the clinical dose of ex vivo LV products, where the medicinal product in these applications is the LV modified cells. The clinical protocols set an average copy number of vector integrants for such products, usually an average of approximately one copy, to ensure efficacy and an upper limit of four copies to minimize the risk of toxicity. In situations where the gene therapy medicinal product is intended to treat an orphan condition, this will be particularly useful, since access to patients for dose optimization is limited, and animal models may not be sufficiently relevant, depending on the situation. Finally, patients need to be monitored for long-term safety, often lifelong, for the level of vector copies in blood cells. This level may be used to determine further treatment options. Because cell modification occurs at multiple sites, assay standardization is vital to ensure delivery of a safe and efficacious dose, including allowing determining if a negative test result is “really” negative—a false-negative test result may have far reaching consequences for the patient and the benefit/risk assessment of a particular medicinal product. Global patient follow-up with rare disease needs to be consistent for the individual and for the patient population. The authors are convinced that the introduction of the first lentiviral gene therapy standard will be a decisive tool for developers of gene therapies.
Acknowledgments
We would like to thank Prof. Didier Trono (EPFL, Lausanne) for his donation of the LV plasmids to make the project possible. The project is funded by the Department of Health in the United Kingdom.
Author Disclosure
No competing financial interests exist for any of the authors.
References
- 1.Zufferey R, Dull T, Mandel RJ, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 1998;72:9873–9880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 2010;467:318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heckl D, Schwarzer A, Haemmerle R, et al. Lentiviral vector induced insertional haploinsufficiency of Ebf1 causes murine leukemia. Mol Ther 2012;20:1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arumugam PI, Higashimoto T, Urbinati F, et al. Genotoxic potential of lineage-specific lentivirus vectors carrying the β-globin locus control region. Mol Ther 2009;17:1929–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montini E, Cesana D, Schmidt M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest 2009;119:964–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charrier S, Ferrand M, Zerbato M, et al. Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction. Gene Ther 2011;18:479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson CA, Ng T-H, Miller AE. Evaluation of recommendations for replication-competent retrovirus testing associated with use of retroviral vectors. Hum Gene Ther 1997;8:869–874 [DOI] [PubMed] [Google Scholar]
- 8.Hutchins B, Sajjadi N, Seaver S, et al. Working toward an adenoviral vector testing standard. Mol Ther 2000;2:532. [DOI] [PubMed] [Google Scholar]
- 9.Hutchins B. Development of a reference material for characterizing adenovirus vectors. Bioprocessing J 2002;1:25–28 [Google Scholar]
- 10.Lock M, Alvira M, Vandenberghe LH, et al. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum Gene Ther 2010;21:1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayuso E, Blouin V, Lock M, et al. Manufacturing and characterization of a recombinant adeno-associated virus type 8 reference standard material. Hum Gene Ther 2014;25:977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moullier P, Snyder RO. International efforts for recombinant adeno-associated viral vector reference standards. Mol Ther 2008;16:1185. [DOI] [PubMed] [Google Scholar]
- 13.Werling NJ, Satkunanathan S, Thorpe R, et al. Systematic comparison and validation of quantitative real-time PCR methods for the quantitation of adeno-associated viral products. Hum Gene Therapy Methods 2015;26:82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Costa S, Blouin V, Broucque F, et al. Practical utilization of recombinant AAV vector reference standards: focus on vector genomes titration by free ITR qPCR. Mol Ther Methods Clin Dev 2016;3:16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WilBio. Conference on Reference Materials for Adeno-Associated Viruses, Working Group Meeting Minutes, June5, 2003 www.biotechnoblog.net/isbiotech-website/ReferenceMaterials/aav2.htm#Submitted 2003 [Google Scholar]
- 16.Jacobs JP, Jones CM, Baillie JP. Characteristics of a human diploid cell designated MRC-5. Nature 1970;227:168. [DOI] [PubMed] [Google Scholar]
- 17.Stepanenko AA, Dmitrenko VV. HEK293 in cell biology and cancer research: phenotype, karyotype, tumorigenicity, and stress-induced genome-phenotype evolution. Gene 2015;569:182–190 [DOI] [PubMed] [Google Scholar]
- 18.Lin Y-C, Boone M, Meuris L, et al. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. 2014;5:4767. [DOI] [PMC free article] [PubMed] [Google Scholar]