Abstract
Tirandamycin K (7), the first linear 7,13;9,13-diseco-tirandamycin derivative, was isolated from the tamI (encoding the TamI P450 monooxygenase) disruption mutant strain (ΔtamI) of marine Streptomyces sp. 307–9. Its chemical structure with relative and absolute configurations was elucidated by a combination of extensive spectroscopic analyses and biosynthetic inferences. Structural elucidation of this unusual compound provides new insights into tirandamycin biosynthesis. Moreover, examination of the biological activity of 7 confirms the essential function of the bicyclic ketal ring for antibiotic activities of tirandamycins.
Keywords: Tirandamycin, Natural product, Tetramic acid, Biosynthesis, Streptomyces
Tirandamycins belong to the tetramic acid family of natural products featured by a pyrrolidine-2,4-dione ring system.1 The antibiotic streptolydigin,2 the HIV-1 integrase inhibitor equisetin,3 the mycotoxin cyclopiazonic acid,4 the fungicidal dihydromaltophilin, 5 the quorum sensing related molecule 3-(1-hydroxydecylidene)- 5-(2-hydroxyethyl) pyrrolidine-2,4-dione,6 and the anti-tumor agents aurantosides,7 discodermides,8 and cylindramides, 9 are among this fast growing family of secondary metabolites. Collectively, they have been attracting continuous interest due to their broad spectrum of biological activities and structural diversity.
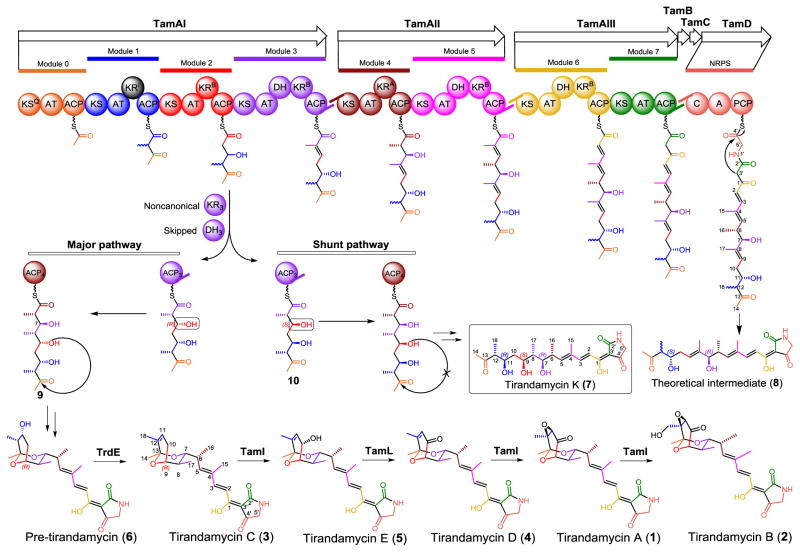
Tirandamycins usually contain an intriguing bicyclic ketal moiety in their chemical structures, and exhibit potent antimicrobial activity by targeting bacterial RNA polymerases and hence blocking the transcriptional elongation process.1,10 Tirandamycin A (1) and B (2) were first identified in the 1970s from a terrestrial Streptomyces species.11 From 2009 to 2011, we isolated and characterized three novel congeners, including tirandamycin C (3),12 D (4)12 and E (5)13 from marine Streptomyces sp. 307–9, together with the known compounds 1 and 2. More recently, the laboratories of Shen and Ju discovered more tirandamycin derivatives (tirandamycin F–J) from Streptomyces sp. 1794414,15 and Streptomyces sp. SCSIO 1666.16,17 All these tirandamycins differ from one another in the extent of oxidative tailoring of the bicyclic ketal moiety (Fig. 1), which has been found to be a key determinant of potency against vancomycin-resistant Enterococcus faecalis (VRE).12 This observation is consistent with the previous study18 revealing that the streptolol ring of streptolydigin (the structural analog to bicyclic ketal of tirandamycin) is involved in a number of key contacts with RNA polymerase (the molecular target of tetramic acid antibiotics including streptolydigin and tirandamycins) determined from co-crystal structure analysis.
Figure 1.
Tirandamycin biosynthetic pathway (Tam) and structures of tirandamycin derivatives. The color of the individual starter or extension unit installed by the hybrid PKS-NRPS assembly-line corresponds to the installing module.
To date, two biosynthetic gene clusters responsible for assembly of tirandamycins have been identified independently from two different species of Streptomyces.19,20 The hybrid polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene cluster (tam) is collinearly organized (Fig. 1). Based on detailed sequence analysis of individual domains using consensus sequence motifs as references, the predictions of the polyketide chain starter unit specified by the KSQ domain, the incorporation of a malonate or methylmalonate subunit by each acyltransferase domain (AT), the stereoselectivity of the ketoreductase domain (KR) catalyzed reduction at each β-keto position, and glycine incorporation by the NRPS module, seem to be consistent with tirandamycin structures. In vitro characterization of two tailoring enzymes encoded by tam, including the P450 monooxygenase TamI,13 and the FAD dependent dehydrogenase TamL,13,16 has ascertained the sequential post-PKS pathway from 3→5→4→1→2 (Fig. 1). TrdE, a glycoside hydrolase family enzyme, was recently identified to be responsible for elimination of the C-11 hydroxy group from the earliest post-PKS intermediate pre-tirandamycin (6) via an uncertain dehydration mechanism leading to the unusual β-γ cis double bond (C-11=C-12) in 3.17 This result indicates that the unusual olefin is not a prerequisite for bicyclic ketal formation.
Previous studies of tirandamycin biosynthesis,12,13,16,20 together with recent characterization of the streptolydigin biosynthetic system, 3 have significantly advanced understanding of the biogenesis of select tetramic acid natural products. However, a number of important biosynthetic questions remain unanswered. For example, formation of the bicyclic ketal ring represents the most intriguing issue, especially when the terpene cyclase analog TamF, and SlgX (TamF homolog in the streptolydigin gene cluster), were shown unlikely to be involved.3,19,20 It is also unclear whether the bicyclic ring is formed before or after release of the chain elongation intermediate from the terminal TamD NRPS module.
Herein, we report the identification of a novel tirandamycin derivative – tirandamycin K (7) that represents the first example of linear 7,13;9,13-diseco-tirandamycin analog (Figs. 1 and 2). Structure elucidation of this compound provides new insights into the remaining biosynthetic questions noted above. Moreover, examination of the biological activity of 7 confirms the essential function of the bicyclic ketal ring for the antibiotic activity of tirandamycins.
Figure 2.
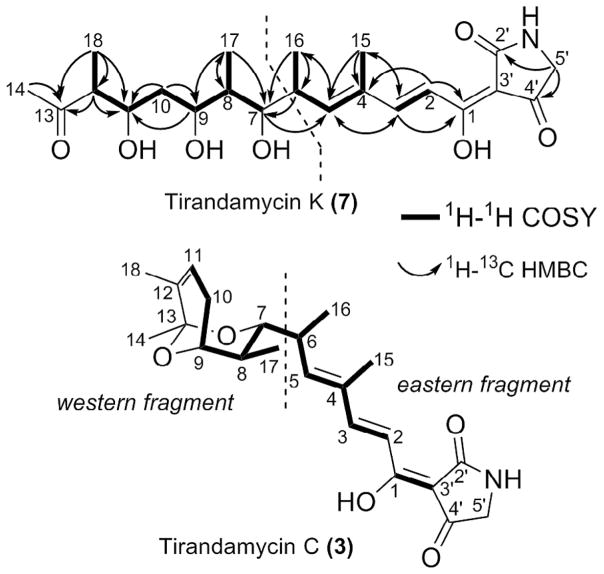
1H-1H COSY and 1H-13C HMBC correlations for tirandamycin K (7) and C (3).
During the high-performance liquid chromatography (HPLC) purification of 3 from the Md2 fermentation broth of the tamI (encoding the TamI P450 monooxygenase) disruption mutant strain (ΔtamI) of Streptomyces sp. 307–9,13 a minor metabolite 7 displaying a similar UV–Vis spectrum (Fig. S1) was eluted from the C18 reverse phase column. According to the comparative HPLC analysis, 7 is more polar than all tested tirandamycins except for 2 (Fig. S1).
A comparison of the 1D NMR data of 7 (Table 1) and 312 demonstrated that their eastern fragments (Fig. 2) including the tetramic acid ring (C-2′–C-5′) and the backbone C-1–C-6 are highly similar to each other. In 7, notably, the protons and carbon atoms at positions of C-2′–C-5′ and C-1–C-5 gave rise to two sets of peaks and the doubling signals disappeared from C-6, which is consistent with the well-known tautomeric behavior of tetramic acids.1 The split signals are generated by two pairs of slowly interconverting external tautomers (ab and cd) arising from the rotation of the acyl side chain (Fig. S2). The intensity ratio between tautomer ab and cd of 7 was found to be 1.5.
Table 1.
1H, 13C and 2D NMR data of tirandamycin K (7) in CDCl3.
| No. | δCa | Type | δH, mult (J in Hz)a | 1H-1H COSY | HMBC | NOESY |
|---|---|---|---|---|---|---|
| 1 | 175.4/177.0 | C | ||||
| 2 | 116.0/115.2 | CH | 7.15, d (15.6)/7.14, d (15.4) | 3 | 1, 4 | 15 |
| 3 | 150.7/152.1 | CH | 7.57, d (15.7)/7.64, d (15.3) | 2 | 1, 5, 15 | 5 |
| 4 | 134.1/134.3 | C | ||||
| 5 | 146.3/147.4 | CH | 6.16, d (10.3)/6.21, d (10.3) | 6, 15 | 3, 15 | 3, 8 |
| 6 | 34.8 | CH | 2.87, m | 5, 16 | 7 | 7, 8, 15, 16, 17 |
| 7 | 83.9 | CH | 2.94, dd (10.2, 2.5) | 8 | 5 | 6, 9, 11, 16, 17 |
| 8 | 42.1 | CH | 1.15, m | 7, 9, 17 | 17 | 5, 6, 10a |
| 9 | 73.5 | CH | 3.35, td (10.2, 4.6) | 8, 10a, 10b | 11 | 7, 8, 10a, 11, 17 |
| 10a | 38.3 | CH2 | 2.06, br dd (12.3, 3.7) | 9, 10b | 9 | 8, 9, 10b, 11, 12, 18 |
| 10b | 1.19, ddd (12.1, 11.7, 11.7) | 9, 10a, 11 | 9, 11 | 10a, 12, 17 | ||
| 11 | 77.9 | CH | 3.47, td (10.0, 1.9) | 10b, 12 | 7,b 9, 14, 10a, 18 | |
| 12 | 51.6 | CH | 2.71, m | 11, 18 | 11, 13 | 10a, 10b, 14 |
| 13 | 212.0 | C | ||||
| 14 | 30.3 | CH3 | 2.22, s | 13 | 12, 18 | |
| 15 | 12.2 | CH3 | 1.90, s | 5 | 3, 4, 5 | 2, 6, 16 |
| 16 | 12.8 | CH3 | 1.04, d (7.0) | 6 | 5, 6, 7 | 7, 15 |
| 17 | 12.4 | CH3 | 0.91, d (6.5) | 8 | 7, 8, 9 | 6, 7, 9, 10a |
| 18 | 17.9 | CH3 | 1.03, d (7.0) | 12 | 11, 12, 13 | 10a, 11, 14 |
| 2′ | 176.6/175.1 | C | ||||
| 3′ | 99.8/101.9c | C | ||||
| 4′ | 192.4/200.8 | C | ||||
| 5′ | 51.6/49.8 | CH2 | 3.97, s/3.84, s | 2′, 4′ |
Chemical shifts of tautomers of ab and cd, respectively; 400 MHz for 1H NMR and 150 MHz for 13C NMR spectra.
The NOESY correlation between H-11 and H-7 might be derived from conformational flexibility of the linear chain.
Signals were observed in 13C NMR spectrum but related HMBC correlations could not be observed.
When comparing the western fragment of 7 and 3, the proton chemical shift values of 7 (vs 3) for H-7 (δH 2.94 vs δH 3.49), H-8 (δH 1.15 vs δH 1.84), H-9 (δH 3.35 vs δH 3.90), H-10a (δH 2.06 vs δH 2.33), H-10b (δH 1.19 vs δH 1.96), H-11 (δH 3.47 vs δH 5.70), H3-14 (δH 2.22 vs δH 1.38), H3-17 (δH 0.91 vs δH 0.68), and H3-18 (δH 1.03 vs δH 1.61) were clearly distinct from the corresponding values of 312, demonstrating a different sub-structure.
Significantly, the absence of proton signals from 4 to 6 ppm indicated that 7 lacks the unique β-γ olefin (C-11=C-12) found in 3. Instead, an additional methine signal (H-12, δH 2.71, m) was observed, corresponding to the proton that would be removed upon a dehydration reaction leading to the C-11=C-12 double bond in 3.
Comparative analysis of the 1H-1H COSY spectra of 7 and 3 revealed similar correlation patterns (Fig. 2), suggesting that their backbone structures were closely related. Further examination of the 1H-13C HMBC spectrum revealed correlations from the protons H-12 (δH 2.71), H3-14 (δH 2.21), and H3-18 (δH 1.03) to a carbonyl carbon (C-13, δC 211.83), which strongly suggested that the bicyclic ring should not be present in 7. Thus, we proposed that 7 is a heretofore unknown tirandamycin pathway derived metabolite, and 7 would be analogous, yet distinct from the linear intermediate 8 (Fig. 1), which is predicted based on collinear annotation of the hybrid PKS-NRPS biosynthetic assembly-line.20
Using the structure of 8 as a template, extensive analyses of 1H and 13C NMR data, and 2D correlations including 1H-1H COSY, HSQC, and HMBC (Table 1 and Figs. S2–S6) enabled assignment of all protons and carbons for 7 in its planar structure (Fig. 2). The most remarkable difference is the appearance of the C-9 hydroxy functionality in 7, whereas the predicted structure 8 bears the C-8=C-9 olefin. The electrospray ionization-high resolution mass spectrometry (ESI-HRMS) data also supported the deduced chemical structure of 7. As observed, the m/z values of 428.2040, 388.2115 (positive ion mode), and 404.2077 (negative ion mode) should correspond to [M−H2O+Na]+ (calc. 428.2049), [M−2H2O +H]+ (calc. 388.2124) and [M−H2O−H]− (calc. 404.2073) of the molecular formula of 7 (C22H33NO7), respectively.
The relative configurations of the adjacent (1,2) or alternate (1,3) stereocenters in the acyclic carbon chain of 7 were determined using a combination of 3JH,H coupling constant and NOE correlation analyses for each fragment progressively (Table 1 and Figs. S7–S13).21 First, the small coupling constant between H-6 and H-7 (3JH-6,H-7 = 2.5 Hz) revealed a gauche arrangement of the two protons in Newman projection, and NOE correlations of H-5/H-8, H-6/H-8 and H-7/H3-16 agreed with the erythro configuration of C-6 and C-7 in Fisher projection, which defined the 6R*,7R* configurations (Fig. S8). Similarly, the large coupling constant between H-7 and H-8 (3JH-7,H-8 = 10.2 Hz) was consistent with an anti conformation in Newman projection, and NOE correlations of H-6/H-8, H-7/H-9, H-6/H-17 and H-7/H-17 revealed 7R*,8S* configurations (Fig. S9). The large coupling constant for H-8 and H-9 (3JH-8, H-9 = 10.4 Hz) and NOE correlations of H-7/H-9, H-8/H-10, H-9/H-17 and H-10/H-17 were consistent with 8S*,9S* configurations (Fig. S10). As to the 1,3 alternate stereocenter from C-9 to C-11, the two fragments of C-9—C-10 and C-10–C-11 were deduced separately with the same logic. The small coupling constant of 3JH-9,H- 10a (3.7 Hz) and the large value of 3JH-9,H-10b (10.2 Hz) revealed the gauche and anti arrangement for H-9/H-10a and H-9/H-10b in Newman projections (Fig. S11), respectively. The NOE correlation of H-8/H-10a, and H-9/H-11 revealed 9S*,10a-proR* configurations. Again, the small coupling constants between H-10a and H-11 (3JH-10a,H-11 = 1.9 Hz) and the large value for H-10b and H-11 (3JH-10b,H-11 = 10.0 Hz), together with the NOE correlations of H-9/H-11, H-10a/H-12, and H-10b/H12 revealed 10a-proR*,11R* configurations (Fig. S12). Finally, the large coupling constant value of 3JH-11,H-12 (10.1 Hz) and NOE correlations between H-10a and H3-18 confirmed 11R*,12R* configurations (Fig. S13). The sum of these analyses established relative configurations of 6R*, 7R*, 8S*, 9S*, 11R*, 12R* for 7. Based upon the absolute configuration of 2 (6R, 7R, 8S) determined from a crystal structure,22 we conclude that 7 takes the absolute configuration of 6R, 7R, 8S, 9S, 11R, 12R (Fig. 1).
The new tirandamycin derivative 7 was isolated from the Streptomyces sp. 307–9 ΔtamI strain with a yield of approximately 0.9 mg/L. To address whether its production is associated with the absence of tamI, we re-visited the metabolic profile of the wild type Streptomyces sp. 307–9 strain and found 7 was also produced, albeit in a significantly lower yield than the ΔtamI strain (data not shown).
Complete characterization of the structure of 7, the first identified linear tirandamycin derivative, enabled the determination of the stereochemistry at C-12, and allowed the unreduced keto group at C-13 (due to the putatively inactive KR domain in module 1) to be firstly observed, which is essential for formation of the bicyclic ring. The fact that 7 was released by the final NRPS TamD module strongly suggests that the chain release machinery does not require formation of the bicyclic ring. However, the possibility that the bicyclic ketal ring is formed prior to the chain release cannot be excluded. In addition, it is unclear if the formation of aberrant intermediate 7 is related to the type II thioesterase (TE) TamB presumably with proofreading activity20 or the conserved hypothetical protein TamC (Fig. 1).
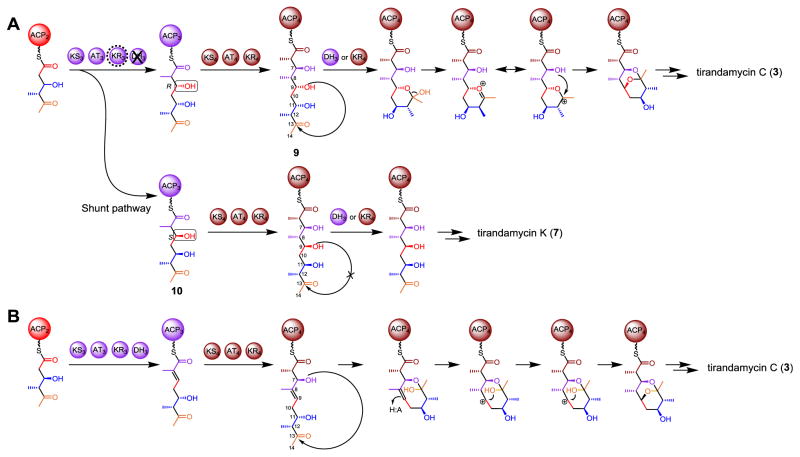
Compared to the structure of 8 predicted from the architecture of Tam PKS-NRPS assembly, 7 retains the C-9 hydroxyl group assumed to be removed by the dehydratase domain in module 3 (DH3). Therefore, it is likely that DH3 is skipped during biosynthesis of 7. More surprisingly, the stereochemistry of the C-9 (9S or 9“S”) and the C-11 (11R or 11“S”) alcohol in 7 contradicts the B-type KR3 and the B-type KR2 (Fig. 1) that are predicted to establish the 9“R” (9R) and 11“R” (11S) stereochemistry20 (the quotation marks denote a deviation from the classical RS system defined by Keatinge-Clay et al.23) based on the presence of LDD motif.24 Interestingly, this abnormal 11R configuration in 7 was also seen in 6 (Fig. 1).17 These inconsistencies point to modules 3 and 2, suggesting that atypical events might occur during these two rounds of polyketide elongation and processing. This also leads to re-consideration of a previously proposed mechanism for bicyclic ketal ring formation in tirandamycin19 (and streptolydigin3,25). Specifically, the 9R hydroxyl group was proposed to be required for nucleophilic attack of the C-13 carbonyl carbon. The resultant hemiketalization, dehydration and attack of the C-7 OH group on the oxonium ion would generate the bicyclic ketal (Fig. 3A).3,19 With respect to the catalyzing enzyme, it was hypothesized that DH3 might play a significant role,19 which if correct would require ACP4 to swing the linear pentaketide intermediate (9, Fig. 3) back to DH3 through contact between ACP4 and DH3. Although similar noncanonical contacts between ACP and DH domains from different modules have been previously reported in epothilone26 and curacin27 biosynthesis, the possibility of KR4 providing the active site for bicyclic ring formation cannot be excluded. This hypothesis (Fig. 3A) is plausible and overrides another mechanism20 (Fig. 3B) that we proposed before the publication of the streptolydigin biosynthetic gene cluster.3 The retention of C-9 oxygen is in good agreement with the previous [13C,18O]-labeled precursor feeding study for streptolydigin,25 showing that the C-9/C-13 bridging ether oxygen originates from the C-9/C-10 acetate unit. Due to the high similarity between the tirandamycin and streptolydigin biosynthetic gene clusters, it is likely these two pathways share a common spiroketalization mechanism. However, the presence of DH3 indeed raises the question whether and how the C-9 alcohol escapes the dehydratase activity. Identification of 7 bearing the C-9 hydroxy clearly indicates that DH3-catalyzed dehydration can be avoided, which provides important evidence to support the mechanism shown in Fig. 3A.
Figure 3.
Two proposed mechanisms for bicyclic ketal ring formation in tirandamycin biosynthesis. Tirandamycin K (7) is proposed to be a shunt pathway product in this study.
The abnormal stereochemistry of C-9 alcohol presumably installed by the B-type KR3 with noncanonical stereoselectivity may explain why 7 escapes from the cyclization step since the incorrect configuration (9S) of the C-9 OH group might disfavor cyclization, thus making the pentaketide 10 skip the ring formation steps (Fig. 1). To our knowledge, there have been no previous reports of an LDD motif containing a B-type ketoreductase domain aberrantly catalyzing keto group reduction to establish the opposite stereochemistry. Exceptions have only been observed when unnatural polyketide analogs were employed prior to this study.23,28 It could be speculated that an unusual event might occur when the triketide intermediate carried by ACP2 binds to the catalytic groove of KR3. Some specific active site residues, or an atypical dynamic conformation of KR3 may account for the “incorrect” stereochemistry.
Finally, the antibacterial activity of 7 against a select group of Gram-negative and Gram-positive strains including the Escherichia coli W3110 TolC disruption mutant strain, the Staphylococcus aureus 8325 NorA disruption mutant strain, the S. aureus ATCC 6538P, and the vancomycin-resistant Enterococci (VRE), was evaluated using 3 as a control. It was found that 7 was much less potent than 3 toward all four strains (Table 2). Together with the facts that all known bioactive tirandamycins contain a bicyclic ketal ring system, 11–17 and the analogous ketal ring in streptolydigin provides key contacts with its molecular target RNA polymerase,18 we suggest that the bicyclic ketal moiety should be necessary for the antibacterial activity of tirandamycins.
Table 2.
Biological activities of tirandamycin K (7) and C (3).
| Minimal inhibitory concentration (μM)
|
||
|---|---|---|
| Tirandamycin K (7) | Tirandamycin C (3) | |
| Escherichia coli TolC mutant W3110 | 100 | 0.78 |
| Staphylococcus aureus ATCC 6538P | >400 | 12.5 |
| Multidrug resistant S. aureus (MRSA) | >400 | 25 |
| Vancomycin-resistant enterococci (VRE) | 200 | 12.5 |
In summary, we have identified tirandamycin K (7) as the first tirandamycin derivative without the bicyclic ketal system. Its discovery has motivated us to re-interrogate the tirandamycin biosynthetic pathway, which suggests that the DH3 (or KR4) domain is likely to be a key element for bicyclic ketal formation. To test this hypothesis, in vitro and in vivo characterization of the enzymatic functions of DH3 and KR4 will be required, and are currently ongoing in our laboratories.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China under grant numbers of 21472204 to S.L. and 31300075 to W.Z., NIH of United States Grant R35 GM118101 to D.H.S., and the Youth Innovation Promotion Association of CAS (2015166) to W.Z. We also thank the financial support from the Applied Basic Research Programs of Science and Technology of Qingdao (14-2-4-10-jch).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tetlet.2016.11.080.
References
- 1.Royles BJL. Chem Rev. 1995;95:1981–2001. [Google Scholar]
- 2.Crum GF, Devries WH, Eble TE, Large CM, Shell JW. Antibiot Annu. 1955;3:893–896. [PubMed] [Google Scholar]
- 3.Olano C, Gómez C, Pérez M, et al. Chem Biol. 2009;16:1031–1044. doi: 10.1016/j.chembiol.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Holzapfel CW. Tetrahedron. 1968;24:2101–2119. doi: 10.1016/0040-4020(68)88113-x. [DOI] [PubMed] [Google Scholar]
- 5.Graupner PR, Thornburgh S, Mathieson JT, et al. J Antibiot. 1997;50:1014–1019. doi: 10.7164/antibiotics.50.1014. [DOI] [PubMed] [Google Scholar]
- 6.Lowery CA, Park J, Gloeckner C, et al. J Am Chem Soc. 2009;131:14473–14479. doi: 10.1021/ja9056079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsunaga S, Fusetani N, Kato Y, Hirota H. J AmChem Soc. 1991;113:9690–9692. [Google Scholar]
- 8.Gunasekera SP, Gunasekera MJ. J Org Chem. 1991;56:4830–4833. [Google Scholar]
- 9.Kanazawa S, Fusetani N, Matsunaga S. Tetrahedron Lett. 1993;34:1065–1068. [Google Scholar]
- 10.Reusser F. Antimicrob Agents Chem. 1976;10:618–622. doi: 10.1128/aac.10.4.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer CE. J Antibiot. 1971;24:2101–2119. [Google Scholar]
- 12.Carlson JC, Li S, Burr DA, Sherman DH. J Nat Prod. 2009;72:2076–2079. doi: 10.1021/np9005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson JC, Li S, Gunatilleke SS, Anzai Y, Burr DA, Podust LM. Nat Chem. 2011;3:628–633. doi: 10.1038/nchem.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Z, Vodanovic-Jankovic S, Ledeboer N, et al. Org Lett. 2011;13:2034–2037. doi: 10.1021/ol200420u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rateb ME, Yu ZG, Yan YJ, et al. J Antibiot. 2014;67:127–132. doi: 10.1038/ja.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mo X, Huang H, Ma J, et al. Org Lett. 2011;13:2212–2215. doi: 10.1021/ol200447h. [DOI] [PubMed] [Google Scholar]
- 17.Mo XH, Ma JY, Huang HB, et al. J Am Chem Soc. 2012;134:2844–2847. doi: 10.1021/ja206713a. [DOI] [PubMed] [Google Scholar]
- 18.Tuske S, Sarafianos SG, Wang X, et al. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo XH, Wang ZW, Wang B, et al. Biochem Biophys Res Commun. 2011;406:341–347. doi: 10.1016/j.bbrc.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 20.Carlson JC, Fortman JL, Anzai Y, Li S, Burr DA, Sherman DH. ChemBioChem. 2010;11:564–572. doi: 10.1002/cbic.200900658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bifulco G, Dambruoso P, Gomez-Paloma L, Riccio R. Chem Rev. 2007;107:3744–3779. doi: 10.1021/cr030733c. [DOI] [PubMed] [Google Scholar]
- 22.Duchamp DJ, Branfman AR, Button AC, Rinehart KL., Jr J Am Chem Soc. 1973;95:4077–4078. doi: 10.1021/ja00793a058. [DOI] [PubMed] [Google Scholar]
- 23.Keatinge-Clay AT, Stroud RM. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Caffrey P. ChemBioChem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Olesen SG, Harrison PHM. Org Lett. 2006;8:5329–5332. doi: 10.1021/ol0621304. [DOI] [PubMed] [Google Scholar]
- 26.Tang L, Ward S, Chung L, et al. J Am Chem Soc. 2004:126. doi: 10.1021/ja030503f. [DOI] [PubMed] [Google Scholar]
- 27.Akey DL, Razelun JR, Tehranisa J, Sherman DH, Gerwick WH, Smith JL. Structure. 2010;18:94–105. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calderone CT, Bumpus SB, Kelleher NL, Walsh CT, Magarvey NA. Proc Natl Acad Sci USA. 2008;105:12809–12814. doi: 10.1073/pnas.0806305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.