ABSTRACT
Mammalian body surfaces are inhabited by vast numbers of microbes, the commensal microbiota, which help the host to digest food, provide nutrients, and mature its immune system. For a long time, postnatal colonization was believed to be the main stimulus for microbial-induced immune development. Using a model of reversible colonization of germ-free mice during gestation, we recently showed that the microbial shaping of the neonatal immune system begins even before birth through molecular signals derived from the microbiota of the mother. Maternal microbiota was important to mature intestinal innate immune cells and to alter intestinal gene expression profiles in the offspring. These changes prepare the newborn for postnatal colonization. The majority of the gestational colonization-dependent effects required maternal antibodies. Here, we discuss and provide further evidence how maternal antibodies are important players in transferring a signal originating from the maternal intestinal microbiota to the offspring.
KEYWORDS: commensal microbiota, early life, gestation, host-microbial mutualism, innate immunity, maternal antibodies, maternal microbiota
Background
We all live in a microbial world, with microbes colonizing nearly all surfaces in our environment. Some of the highest microbial densities anywhere are found within the mammalian intestine. These microbes colonize the mammalian inner and outer body surfaces, such as the skin, the intestine, the urogenital tract and the airways.1 The microbial host relationship is mutual beneficial. The commensal microbes contribute to the digestion of nutrients and the synthesis of essential vitamins,2 and also protect the host from invasion by pathogens.3 By densely colonizing the mucosal surfaces, commensal bacteria occupy space and compete for nutrients thereby preventing pathogenic bacteria and viruses from taking up residence. In addition, the commensal microbiota has been shown to efficiently mature the host innate and adaptive immune systems, both at mucosal sites and generally in the body.4 Elegant models using germ-free mice have illustrated the important role of the microbiota in the development of immunity and other body systems.5 Indeed, in the absence of microbiota, intestinal IgA production and T helper cell differentiation are strongly diminished, secondary and tertiary lymphoid organs (lymph nodes, Peyer's patches and isolated lymphoid follicles) are underdeveloped and the innate immune system is hyporeactive.4
The birth of a newborn child is a tremendous life event. While the unborn child is well protected in the sterile environment of the uterus from external influences and infections by maternal immunity and by physical separation, birth exposes the newborn baby for the first time to a vast number of microbes – both commensal and potentially pathogenic in nature. The colonization of the body surfaces with an endogenous commensal microbiota starts immediately after birth and microbes living on the body soon reach numbers of astronomical proportions. Bacterial infections are the first cause of death of preterm babies and newborns,6,7 so understanding exactly when and how the immune system is prepared to deal with the enormous colonization that starts at birth is crucial. A long believed dogma stated that immune maturation is mainly shaped through postnatal endogenous colonization of the offspring.8,9 This is of course predicated on the knowledge that the developing fetus is sterile and not yet colonized by microbes. However, the unborn child is connected to the maternal blood stream for efficient nutrient supply and bacterial components or metabolites induced in the maternal organism by the microbiota can efficiently be transferred to the fetus. We hence challenged the old dogma and asked whether signals originating from the maternal microbiota during gestation contribute to the microbial shaping of the immunity of the offspring.10
Model of gestation-only colonization
An important aspect of our experimental approach was to uncouple maternal colonization with microbes during gestation from postnatal colonization of the offspring. The effects of commensal colonization of the mammalian body on the host immune system have been elegantly demonstrated by the specific colonization of germ-free mice. As colonization of a germ-free animal with a commensal bacteria is non-reversible, even if antibiotics are applied, offspring born to or nursed by a colonized mouse will lead to its immediate colonization, and the observed phenotype will no longer be attributable to either maternal or endogenous microbiota. To achieve gestation-only colonization, we made use of a system of reversible colonization of germ-free mice with the genetically modified strain Escherichia coli (E. coli) HA107, which had been developed in our laboratory several years ago.11 E. coli HA107 is an auxotrophic mutant of the non-pathogenic E. coli K-12 that is deficient in the biosynthesis of the D-isomer of alanine and meso-diaminopimelic acid, both of which are essential components of the bacterial cell wall and cannot be provided by the mammalian host. E. coli HA107 can thus only be grown in supplemented in vitro culture and colonizes a germ-free mouse reversibly for only 24–48 hours.
In our model of gestation-only colonization, we have delivered E. coli HA107 to pregnant dams every-other day starting from embryonic day (E) 8 until E16 by gavage. The dams regained germ-free status after treatment and therefore delivered their pups germ-free. The offspring of these gestationally colonized dams were then compared with offspring that were born to dams that were maintained germ-free throughout pregnancy.10
Effect of gestational colonization on offspring immunity
As a first approach to assess the effect of maternal microbiota on the immune system of the offspring, we screened various innate and adaptive immune cell populations in primary and secondary lymphoid organs and in the intestinal tissue of the newborn offspring using flow cytometry. While adaptive immune cells, such as B and T helper cell subsets were unaltered in the offspring of gestationally colonized mice compared with the offspring of germ-free control mice, we detected significant changes in the innate immune compartment.10 Specifically, small intestinal NKp46+ type 3 innate lymphoid cells (ILC3s) and intestinal F4/80+ CD11c+ mononuclear cells (iMNCs) in the small and large intestine were increased in relative and absolute numbers. These changes were apparent as early as 10 d after birth and were durable until adulthood. To appreciate the full effect of gestational colonization on the newborn intestinal immune system, we also performed RNA sequencing analysis from the intestine of 14 day-old pups born to either control or gestationally colonized dams. Astoundingly, more than 2000 genes were significantly altered in expression between the 2 groups. Among these, were not only immunologically relevant genes, but also genes involved in the establishment of host-microbial mutualism, for example those required for mucus production and secretion, and proliferation, differentiation and homeostasis of intestinal epithelial cells, including soluble factors like antimicrobial peptides and mucosal antibodies.10 Signals deriving from the maternal microbiota thus not only shape immunity of the offspring, but have pleiotropic effects, the function/influence of which on the physiology of the offspring throughout life remain to be elucidated in further research.
The microbiota composition in the first years of life differs from that in adults.12 Enterobacteriaceae and in particular E. coli are part of the initial composition of the microbiota of neonates.13,14 Knowing that antimicrobial peptides and intestinal ILC3s are important players in maintaining host-microbial mutualism and in restricting intestinal commensals to the intestinal lumen,15,16 we decided to address the impact of maternal microbiota in the ability of the newborn to restrain microbiota to the gut preserving the sterility of systemic organs. In our experimental set-up, the neonates of both the untreated germ-free control mothers and of the gestationally colonized mothers are born germ-free. To mimic colonization with commensal microbes that normally occurs at birth, we exposed the offspring on day 14 after birth to a benign E. coli strain. After 18 hours, bacteria had translocated to the mesenteric lymph nodes in the pups born to control dams but not in the offspring of gestationally colonized mothers.10 These results in combination with the RNA sequencing data show that signals of the maternal microbiota mainly serve to prepare the offspring for the enormous biomass of microbes that colonize the intestine and other body surfaces after birth.
How can signals originating from the maternal intestinal microbiota affect the offspring?
It is a well-established paradigm in host-microbial mutualism that intestinal bacteria hardly ever cross the intestinal barrier and if they do so they are efficiently stopped at the firewall of the mesenteric lymph nodes.17 Moreover, during pregnancy a second barrier, the placenta, prevents the colonization of the fetus in the uterus. There are studies suggesting that very low numbers of organisms from the maternal microbiota may be able to cross the placenta and reach fetal tissues.18,19 The technical challenge of such studies is to avoid contaminations during sample collection and processing in humans.20 In our model, we extensively checked the gestationally colonized dams as well as colonized pregnant mice by culturable and non-culturable methods and did not detect any bacteria either in the fetus or in the placenta. Therefore, in this model we found that the developing fetus was never exposed to live microbes. Several studies have illustrated the molecular transfer from the mother to the newborn organism during lactation via components present in the mother milk.21 However, in our system of only transient colonization during pregnancy, no live microbes remained in the maternal body at the time of delivery and the maternal milk in our experiments was always sterile. Although live bacteria from the maternal microbiota are not transferred to the offspring, bacterial compounds may well be. When we traced bacteria-derived metabolites by exposing pregnant dams to 14C-labeled E. coli HA107, we detected radioactivity both in the placenta and in the maternal milk,10 indicating that bacteria-derived metabolites can reach the offspring both during fetal development via the placenta and during lactation as components of the maternal milk (Fig. 1). This was also supported by cross-fostering experiments when the newborn litters of gestationally colonized dams were swapped to control dams for nursing and vice versa. Only if the offspring was born and raised by a transiently colonized mother, the effect on the newborn immune system was complete. However, 14C-labeling techniques do not allow the molecular identification of the bacteria-derived molecules and can also not distinguish between molecules of bacterial origin and 14C label incorporated into host molecules through secondary metabolism. To resolve the molecular identities of penetrant bacterial molecules, we labeled E. coli HA107 with the stable isotope 13C and transiently colonized pregnant dams. This enabled us to track bacterial products from the maternal intestine into the mother's milk and even into the newborn tissues. We identified several hundred partially or fully 13C-labeled potential metabolites based on accurate mass in the maternal milk of a gestationally colonized mouse.10 These tentatively annotated metabolites were members of a diverse range of different compound classes. However, of the many 13C-labeled bacteria-derived metabolites we focused our attention on potential ligands for the aryl hydrocarbon receptor (AhR). This cytoplasmic ligand-activated transcription factor is known to be involved in the metabolism of aromatic hydrocarbons and therefore in the detoxification of xenobiotics.22 AhR has a broad ligand range, including exogenous toxic compounds such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), food indoles present in cruciferous vegetables, and endogenous ligands generated during tryptophan metabolism.23 AhR is also required in the development and homeostasis of immune cells involved in microbiota-host mutualism, such as intraepithelial gamma delta T cells,24 regulatory T cells,25 T helper 17 cells26 and ILC3.27 We detected several partially or fully 13C-labeled AhR ligands to be significantly increased in the milk of gestationally colonized dams compared with the milk of control dams. We confirmed the functional AhR ligand effect by showing that the uptake of a purified AhR ligand, indole-3-carbinol, by germ-free dams during pregnancy was sufficient to recapitulate the immune phenotype observed in the offspring following gestational colonization.10 All together these results suggest that some of the multiparametric effects of maternal microbiota on intestinal immune education of the offspring could be driven by bacteria-derived AhR ligands.
Figure 1.
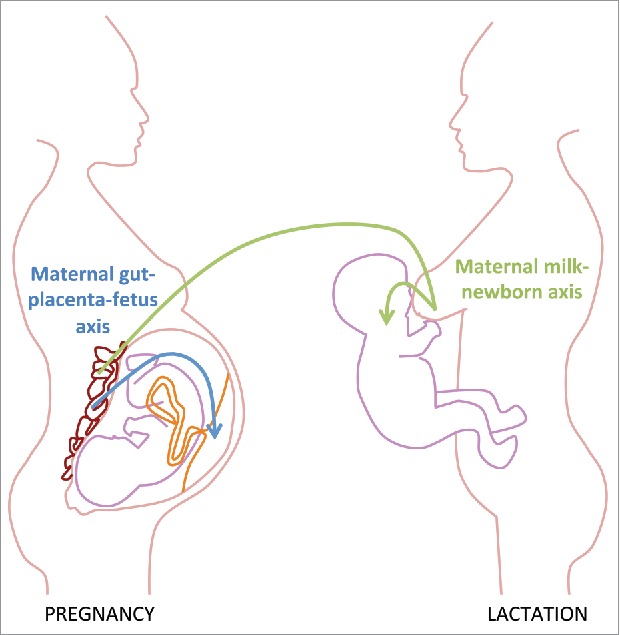
Maternal microbiota-derived signals are transferred to the offspring ante- and postnatally. During gestation signals derived from the maternal microbiota reach the offspring via the placenta (maternal gut-placenta-fetus axis). Signals originating from intestinal microbial colonization of the mother during pregnancy can also reach the offspring through the breast milk after birth (maternal milk-newborn axis).
The involvement of maternal antibodies in maternal microbiota-dependent imprinting of the neonatal immune system
It is well known that antibodies are transferred from the mother to her offspring to equip the child with passive immunity and protection.28,29 Immunoglobulin G is transferred via the placenta in mice and humans30 and taken up from the milk via the duodenum until about postnatal day 12 in mice.31 IgA is mainly secreted into the maternal milk.32 Given our observation that a transfer of signals from the maternal microbiota most probably occurs both in utero and during lactation, we hypothesized that maternal antibodies were involved in this process. We hence tested how the absence of maternal antibodies affected our observed phenotypes following transient gestational colonization through cross-breeding JH-deficient females, which are devoid of B cells, with wild type males. This showed that many of the changes observed in the immune system of the offspring born to a gestationally colonized dam compared with those born to a control dam were absent if the mother was deficient in B cells (e.g. the increase in intestinal NKp46+ ILC3s and most of the transcriptional changes observed in the RNA-Seq experiment10). Transfer of serum from an E. coli HA107-primed wild type mouse, but not from an E. coli HA107-primed antibody-deficient mouse, to a pregnant germ-free dam, induced similar changes in the offspring immune system as gestational colonization.
But how can maternal antibodies contribute to these immune changes in the offspring? There are 2 possibilities: i) mucosal priming of the mother during pregnancy significantly increases the concentration of immunoglobulins in the serum and the higher concentration of antibodies reaching the offspring may exert direct changes in the immune system of the offspring, or ii) specific antibodies may bind bacterial products, retain them in the maternal organism, and deliver them across the placenta or into the milk. We have tested both. By exposing female mice before pregnancy to the same dose of E. coli HA107 as we usually applied during gestation, we were able to increase serum and milk antibody levels to concentrations that equaled those after gestational priming. However, delivering the priming signal before gestation was insufficient to contribute to the maturation of the offspring immune system.10 Hence, solely increased antibody levels in the maternal serum are not the key to our observations. Our hypothesis is thus that the maternal antibodies in the serum, which are later secreted into the milk, are able to capture and retain microbial products in the maternal body. By labeling HA107 with radioactive 14C we tested for differences in uptake and retention of bacterial metabolites by the mother in antibody-sufficient and deficient mice. Consistent with our hypothesis, in the absence of antibodies, less radioactive products were retained in the maternal organism over time. Likewise, JH-deficient mothers that were gavaged with 13C-labeled HA107 during pregnancy, exhibited largely fewer fully 13C-labeled (microbial) metabolites in the maternal milk shortly after birth. Especially tryptophan metabolites, which are ligands of the AhR, were absent from the milk of antibody-deficient mice.10 To prove that metabolites of the maternal intestinal microbiota can bind to maternal serum antibodies, we made use of size exclusion chromatography to fractionate the serum 6 hours after gavaging 13C-labeled E. coli HA107. When we analyzed the fractions containing IgG and IgA, we were able to identify several partially or fully 13C-labeled metabolites.10 With high likelihood, these bacterial metabolites were bound to the antibodies and therefore co-eluted in the same fraction. It is known that intestinal microbiota-specific antibodies are present both in serum and mothers milk,33,34 and we have evidence that it is E. coli-specific antibodies in the maternal body that are responsible for the capture and transfer of bacterial products into the milk and subsequently into the offspring. Follow-up experiments will have to be performed to definitely prove the binding of bacterial metabolites to serum antibodies.
We believe that microbe-specific maternal antibodies, besides passively protecting the unborn and neonatal child from infections through immune exclusion, are an important player in the maternal-fetal/neonatal transfer of maternal microbiota-derived signals. Explicitly, microbiota-specific antibodies are able to bind bacterial fragments leading to a better retention of microbiota-derived metabolites in the maternal organism and hence a more efficient transport of these across the placenta and into the maternal milk, thereby preparing the offspring for immunological challenges after birth (Fig. 2).
Figure 2.
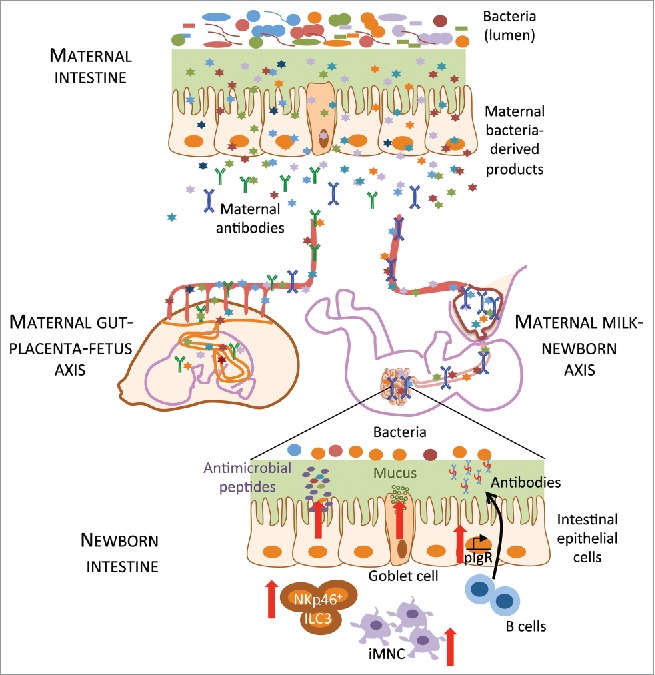
Maternal antibodies help in the transfer of microbial products to the offspring. Metabolites from the intestinal microbiota of the mother resulting from transient gestational colonization of the dam are transferred to the offspring antenatally via the placenta and postnatally through the maternal milk and induce immune maturation of the offspring intestinal mucosa, such as an increase in NKp46+ type 3 innate lymphoid cell (ILC3) and in intestinal mononuclear cell (iMNC) numbers. Expression of genes for antimicrobial peptides, mucus production and antibody secretion (pIgR: polymeric immunoglobuline receptor) were upregulated in intestinal epithelial cells in the offspring. Maternal antibodies (green: IgG, blue: dimeric IgA) mediated these effects, probably by more efficiently retaining microbial products in the maternal organism and transporting them to the placenta and/or the breast milk.
Retrospect
The work we published in 201610 and the additional considerations we describe here contribute to the understanding of the “critical window of opportunities,” which describes the importance of microbial influences on the host immune system happening early in life.35 Not only does postnatal colonization play an important role in setting the immune phenotype of the newborn child, but molecular signals originating from the microbiota of the mother, which can reach the offspring in utero via the placenta and postnatally through the breast milk, significantly contribute to the microbial shaping and setting up of the neonatal immune system. While we were able to demonstrate the importance of the maternal microbiota in preparing the offspring for the event of colonization in its first days of life, it now remains to show the consequences of this effect in adulthood if we colonize the offspring after birth.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
S.C.G-V. is holder of a long-term fellowship from the European Molecular Biology Organization. We thank Andrew J. Macpherson who directed the project described in this article and Kathy D. McCoy who played a key role in its execution. We also thank Uwe Sauer from the ETH Zurich.
Funding
A.J.M. is funded by the Swiss National Science Foundation (SNSF 310030B_160262, SNSF Sinergia CRSII3_154414) and the Swiss SystemsX program (GutX).
References
- [1].Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016; 164:337-40; PMID:26824647; http://dx.doi.org/ 10.1016/j.cell.2016.01.013 [DOI] [PubMed] [Google Scholar]
- [2].Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science 2001; 292:1115-8; PMID:11352068; http://dx.doi.org/ 10.1126/science.1058709 [DOI] [PubMed] [Google Scholar]
- [3].Stecher B, Macpherson AJ, Hapfelmeier S, Kremer M, Stallmach T, Hardt WD. Comparison of Salmonella enterica serovar Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect Immun 2005; 73:3228-41; PMID:15908347; http://dx.doi.org/ 10.1128/IAI.73.6.3228-3241.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012; 336:1268-73; PMID:22674334; http://dx.doi.org/ 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 2007; 19:59-69; PMID:17118672; http://dx.doi.org/ 10.1016/j.smim.2006.10.002 [DOI] [PubMed] [Google Scholar]
- [6].Ward DV, Scholz M, Zolfo M, Taft DH, Schibler KR, Tett A, et al.. Metagenomic Sequencing with Strain-Level Resolution Implicates Uropathogenic E. coli in Necrotizing Enterocolitis and Mortality in Preterm Infants. Cell Rep 2016; 14:2912-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bizzarro MJ, Shabanova V, Baltimore RS, Dembry LM, Ehrenkranz RA, Gallagher PG. Neonatal sepsis 2004–2013: the rise and fall of coagulase-negative staphylococci. J Pediatr 2015; 166:1193-9; PMID:25919728; http://dx.doi.org/ 10.1016/j.jpeds.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fulde M, Hornef MW. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol Rev 2014; 260:21-34; PMID:24942679; http://dx.doi.org/ 10.1111/imr.12190 [DOI] [PubMed] [Google Scholar]
- [9].Rakoff-Nahoum S, Kong Y, Kleinstein SH, Subramanian S, Ahern PP, Gordon JI, et al.. Analysis of gene-environment interactions in postnatal development of the mammalian intestine. Proc Natl Acad Sci U S A 2015; 112:1929-36; PMID:25691701; http://dx.doi.org/ 10.1073/pnas.1424886112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al.. The maternal microbiota drives early postnatal innate immune development. Science 2016; 351:1296-302; PMID:26989247; http://dx.doi.org/ 10.1126/science.aad2571 [DOI] [PubMed] [Google Scholar]
- [11].Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al.. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 2010; 328:1705-9; PMID:20576892; http://dx.doi.org/ 10.1126/science.1188454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ottman N, Smidt H, de Vos WM, Belzer C. The function of our microbiota: who is out there and what do they do? Front Cell Infect Microbiol 2012; 2:104; PMID:22919693; http://dx.doi.org/ 10.3389/fcimb.2012.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol 2014; 5:427; PMID:25250028; http://dx.doi.org/ 10.3389/fimmu.2014.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al.. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015; 17:690-703; PMID:25974306; http://dx.doi.org/ 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- [15].Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al.. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 2012; 336:1321-5; PMID:22674331; http://dx.doi.org/ 10.1126/science.1222551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al.. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011; 334:255-8; PMID:21998396; http://dx.doi.org/ 10.1126/science.1209791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Macpherson AJ, Uhr T. Compartmentalization of the mucosal immune responses to commensal intestinal bacteria. Ann N Y Acad Sci 2004; 1029:36-43; PMID:15681741; http://dx.doi.org/ 10.1196/annals.1309.005 [DOI] [PubMed] [Google Scholar]
- [18].Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med 2014; 6:237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Moles L, Gomez M, Heilig H, Bustos G, Fuentes S, de Vos W, et al.. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 2013; 8:e66986; PMID:23840569; http://dx.doi.org/ 10.1371/journal.pone.0066986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kliman HJ. Comment on “the placenta harbors a unique microbiome”. Sci Transl Med 2014; 6:254le4; http://dx.doi.org/ 10.1126/scitranslmed.3009864 [DOI] [PubMed] [Google Scholar]
- [21].Jenness R. The composition of human milk. Semin Perinatol 1979; 3:225-39. PMID:392766 [PubMed] [Google Scholar]
- [22].Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol 2014; 32:403-32; PMID:24655296; http://dx.doi.org/ 10.1146/annurev-immunol-032713-120245 [DOI] [PubMed] [Google Scholar]
- [23].Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev 2015; 67:259-79; PMID:25657351; http://dx.doi.org/ 10.1124/pr.114.009001 [DOI] [PubMed] [Google Scholar]
- [24].Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, et al.. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 2011; 147:629-40; PMID:21999944; http://dx.doi.org/ 10.1016/j.cell.2011.09.025 [DOI] [PubMed] [Google Scholar]
- [25].Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, et al.. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 2008; 453:65-71; PMID:18362915; http://dx.doi.org/ 10.1038/nature06880 [DOI] [PubMed] [Google Scholar]
- [26].Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U S A 2008; 105:9721-6; PMID:18607004; http://dx.doi.org/ 10.1073/pnas.0804231105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, et al.. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 2011; 334:1561-5; PMID:22033518; http://dx.doi.org/ 10.1126/science.1214914 [DOI] [PubMed] [Google Scholar]
- [28].Harris NL, Spoerri I, Schopfer JF, Nembrini C, Merky P, Massacand J, et al.. Mechanisms of neonatal mucosal antibody protection. J Immunol 2006; 177:6256-62; PMID:17056555; http://dx.doi.org/ 10.4049/jimmunol.177.9.6256 [DOI] [PubMed] [Google Scholar]
- [29].Zinkernagel RM. Maternal antibodies, childhood infections, and autoimmune diseases. N Engl J Med 2001; 345:1331-5; PMID:11794153; http://dx.doi.org/ 10.1056/NEJMra012493 [DOI] [PubMed] [Google Scholar]
- [30].Story CM, Mikulska JE, Simister NE. A major histocompatibility complex class I-like Fc receptor cloned from human placenta: possible role in transfer of immunoglobulin G from mother to fetus. J Exp Med 1994; 180:2377-81; PMID:7964511; http://dx.doi.org/ 10.1084/jem.180.6.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ghettie V, Ward ES. Multiple roles for the major histocompatibility complex Class I-related receptor FcRN. Ann Rev Immunol 2000; 18:739-66; http://dx.doi.org/ 10.1146/annurev.immunol.18.1.739 [DOI] [PubMed] [Google Scholar]
- [32].Wheeler TT, Hodgkinson AJ, Prosser CG, Davis SR. Immune components of colostrum and milk–a historical perspective. J Mammary Gland Biol Neoplasia 2007; 12:237-47; PMID:17992474; http://dx.doi.org/ 10.1007/s10911-007-9051-7 [DOI] [PubMed] [Google Scholar]
- [33].Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, et al.. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell 2016; 165:827-41; PMID:27153495; http://dx.doi.org/ 10.1016/j.cell.2016.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, et al.. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity 2016; 44:647-58; PMID:26944199; http://dx.doi.org/ 10.1016/j.immuni.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science 2016; 352:539-44; PMID:27126036; http://dx.doi.org/ 10.1126/science.aad9378 [DOI] [PMC free article] [PubMed] [Google Scholar]


