ABSTRACT
Mammalian Rap1, the most conserved telomere-interacting protein, beyond its role within nucleus for the maintenance of telomeric functions, is also well known for its pleiotropic functions in various physiological and pathological conditions associated with metabolism, inflammation and oxidative stress. For all these, nowadays Rap1 is the subject of critical investigations aimed to unveil its molecular signaling pathways and to scrutinize the applicability of its modulation as a promising therapeutic strategy with clinical relevance. However, the underlying intimate mechanisms of Rap1 are not extensively studied, but any modulation of this protein level has been associated with pathologies like inflammation, oxidative stress and deregulated metabolism. This is considerably important in light of the recent discovery of Rap1 modulation in diseases like cancer and cardiac metabolic disorders. In this review, we focus on both the telomeric and nontelomeric functions of Rap1 and its modulation in various health risks, especially on the heart.
KEYWORDS: diabetic cardiomyopathy, inflammation, metabolism, oxidative stress, Rap1
Introduction
Identification and structure of mammalian Rap1
Telomeres are specialized protein-DNA complex located at the terminal ends of chromosomes that ensure genomic stability and integrity.1,2 In mammalian cells, the telomeric DNA is bound by a specialized six protein subunits known as shelterin complex, which includes telomeric repeat-binding factor 1 (TRF1), telomeric repeat-binding factor 2 (TRF2), protection of telomeres 1 (POT1), TRF1-interacting nuclear protein 2 (TIN2), TIN2- and POT1-organizing protein (TPP1) and repressor-activator protein 1 (Rap1)2,3 (Fig. 1). The shelterin complex regulates the length of the telomeres and prevents chromosomal rearrangements or chromosomes fusing to each other which could lead to chromosomal breaks and recombination.2
Figure 1.
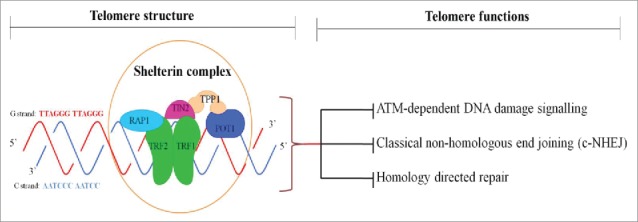
Telomere structure and functions. Mammalian telomere consists of TTAGGG repeats and end with 3′ G-strand overhang of up to a few nucleotides in length. Shelterin complex is formed by TRF1 (telomere repeat-binding factor 1), TRF2, Rap1 (repressor activator protein 1), TIN2 (TRF1-interaction factor 2), POT1 (protection of telomere 1) and TPP1 (POT1-organizing protein). Shelterin binds to double-stranded telomeric repeats and protects chromosome ends from ATM (ataxia telangiectasia mutated)- dependent DNA damage signaling, c-NHEJ (classical non-homologous end joining)-induced DNA repair, homology-directed repair.
The Rap1 subunit was first identified as a predominant telomere-binding protein in the budding yeast Saccharomyces cerevisiae (scRap1),4 where it directly recognizes telomeric DNA through the cooperation of its two internal tandem Myb-type domains.5 Unlike the budding yeast scRap1, mammalian Rap1 associates with telomeres exclusively through its interaction with TRF2.6 Human Rap1 was ascertained through a yeast two-hybrid screen of HeLa cDNAs (human cervical cancer cell) with human TRF2 as bait.6 This yields a 399 Amino acid protein (∼47kDa) that have significant sequence similarity (24%-25%) and three conserved sequence motifs in common with scRap1, featuring a domain of a breast cancer susceptibility protein that appears on its C-terminus (BRCT) in N-terminus, one central Myb DNA-binding motif and Rap1-specific protein-interaction domain (RCT) at the C terminus,6 indicating that it is a human ortholog of scRap1. Multiple protein- and DNA-interaction domains within Rap1 architecture suggest that Rap1 can work as an adaptor protein, introducing different protein/DNA factors into the shelterin complex with various functions.
Telomeric functions of Rap1
Rap1 and telomere length control
Because of the “end replication problem”, whereby the DNA replication leaves 50–200 base pairs DNA un-replicated at the 3' end leading to shorter telomere ends at each cycle of cell division, cumulative telomere attrition during ageing occurs in eukaryotes.7 Thus, telomere length reflects the life span of a cell and has been shown to be closely associated with several health outcomes, including, but not limited to myocardial infarction,8 atherosclerosis9 and cancer.10
In budding yeast, scRap1 binds to telomeric DNA through Myb domain and tethers Rif1 and Rif2 through protein interactions within the RCT domain, thereby inhibiting the telomere length elongation.11–13 The effect of Rap1 on telomere length control in mammalian cells was believed to be evolutionarily conserved. In human fibrosarcoma cell line HTC75, which has been widely used as telomere length control test system,14 inhibition of endogenous human Rap1 by RNA interference displayed a telomere elongation phenotype.15 It is likely that BRCT and Myb domains within the Rap1 structure recruit one or more protein factors, which together with endogenous Rap1 are required for the negative control of telomere length, as evidenced by higher telomere elongation rates in the four mutants of Rap1 (Rap1ΔM, Rap1ΔBr, Rap1ΔBrΔM, Rap1ΔMΔCT) than that in HTC75 cells overexpressing wild-type Rap1.16 However, recent studies challenged the role of mammalian Rap1 in telomere length control.17,18 Loss of Rap1 in various human cell lines [including Hela, HT1080, HTC116, ARPE-19] through transcription activator-like effector nuclease (TALEN) did not affect the telomere length dynamics, suggesting that Rap1 does not play a significant role in telomere length control.17 Furthermore, Rap1 knockout mice are alive and fertile.18 Quantitative telomere fluorescence in situ hybridization analysis of the liver and brown fat sections from both Rap1 wild-type and knockout mice showed that there was no change in telomere length between both genotypes.19 It is pointed out that the prior finding of altered telomere length upon over expression of Rap1 mutants could be due to the influence of nucleoplasmic titration of factors on the phenotypes.17 It is unclear why partial Rap1 inactivation by shRNA or TALEN-mediated Rap1 knockout showed contrary findings. The role of endogenous mammalian Rap1 in telomere length control still requires further investigation.
Rap1 and telomere end protection
The central function of eukaryotic telomeres is to protect chromosome ends from DNA degradation, unwanted DNA repair mechanisms and chromosome fusions20 (Fig. 1). The shelterin subunit TRF2 is life essential in telomere end protection since the deletion of TRF2 from telomeres results in the Ataxia Telangiectasia Mutated (ATM) kinase-dependent DNA damage signaling and the non-homologous end-joining (NHEJ) pathway-mediated massive end-to-end chromosome fusions.21,22 Furthermore, TRF2 also specifically inhibits homology-directed repair (HDR), which can create different telomeric sister chromatid exchanges, leading to the shortened telomere in one daughter cells.23
Mammalian Rap1, as an interacting partner of TRF2 in the telomere, is also removed upon the deletion of TRF2.6,24 Thus, Rap1 may contribute to the repression effect of TRF2 on ATM signaling, NHEJ and HDR pathway at the telomere. However, unlike TRF2−/− cells, deletion of Rap1 (Rap1Δex2/Δex2) in SV40LT-immortalized mouse embryonic fibroblasts (MEFs) did not affect the expression level and localization of other shelterin subunits and did not show ATM-dependent DNA damage response, phosphorylation of Chk1 and Chk2 and induction of Telomere Dysfunction-induced Foci (TIFs), which are hallmarks of telomere dysfunction.18,25 Furthermore, in Rap1Δex2/Δex2 and TRF2 knockout MEFs with TRF2 mutant (TRF2ΔRap1), that cannot bind to Rap1 in the telomere but retained its interaction with the TRF2-interacting protein Apollo,18 telomere fusions were also not induced like wild-type TRF2 MEFs because of the repressed NHEJ pathway mediated repair of telomeres.18,25 Collectively, these data indicated that mammalian Rap1 is dispensable in either the repression of NHEJ or ATM kinase signaling. In support of this notion, Rap1Δex2/Δex2 mice were born at the expected frequencies and were fertile, suggesting that deletion of Rap1 does not lead to lethal telomere dysfunction.18,25
In contrast with its role in ATM kinase signaling and NHEJ pathway, Rap1 is critical to repress aberrant HDR at telomeres.18,25 Indeed, in MEFs lacking TRF2 and Ku70 which displayed frequent telomere sister chromatid exchanges (T-SCEs), introduction of TRF2ΔRap1 did not down-regulate the level of T-SCEs and failed to suppress the telomeric HDR, whereas wild-type TRF2 significantly repressed the telomeric exchanges, suggesting that mammalian Rap1 may contribute to the repression of HDR pathway at telomere.18,25 This observation is further supported by Rap1 knockout studies, in which Cre-mediated Rap1 knockout in Ku70 deficient MEFs exhibit significantly increased T-SCEs.18 Taken together, these data indicate that mammalian Rap1 functions to repress HDR at the telomere. Despite the mechanism by which Rap1 supresses HDR pathway at telomeres remains largely unknown, the Rap1 Myb motif has been suggested to be essential to cooperate with primary domain of TRF2 to repress the localization of poly(ADP-ribose) polymerase 1 (PARP1) and SLX4 structure-specific endonuclease subunit (SLX4) to telomeres, both proteins have been demonstrated to be involved in HDR pathway.24,26 In support of this notion, a ∼35fold increase in PARP1 localization to telomeres was observed in MEFs lacking Rap1-TRF2 heterodimers.24 The murine SLX4 telomeric localization mainly depends on the presence of functional PARP1, since inhibition of PARP1 leads to ∼5fold reduction in SLX4 localization to telomeres. This suggests that increased telomeric localization of PARP1 and SLX4 in MEFs lacking Rap1-TRF2 heterodimers promote the rapid telomere resection, and formation of telomere-free chromosome ends.24 Thus, these findings collectively demonstrated the biological importance of Rap1 in telomere end protection by repressing the aberrant HDR pathway.
Nontelomeric functions of Rap1
Rap1 role in transcriptional control
In budding yeast, scRap1 can directly bind to specific sites on DNA through its two Myb motif and function as a transcriptional repressor or activator.27 This transcriptional effect of Rap1 is evolutionarily conserved in mammalian cells. Chromatin immune precipitation (ChIP)-sequencing reveals that Rap1-containing complex binds to both telomeric and extratelomeric TTAGGG repeats.28 In line with this, preferential derepression of subtelomeric genes which are enriched with Rap1-binding sites was observed in Rap1-deficient MEFs, as compared to wild-type cells.28 Furthermore, apart from the changes in subtelomeric gene expression, there are at least 16 genes involved in cell adhesion, cancer and metabolism, which were significantly upregulated or downregulated in Rap1-deficient MEFs.28 However, unlike scRap1, mammalian Rap1 cannot directly bind to telomeric DNA, since the surface of its Myb domain lacks positive charge.29 Supporting this notion, electrophoretic mobility shift assay studies have shown that Rap1 cannot bind to telomeric DNA until the molar ratio of Rap1:DNA reaches up to 40:1.30 It is, therefore, most likely that Rap1 exerts the transcriptional effect via its interacting partner, TRF2. Indeed, human Rap1 induces a partial release of TRF2 from telomeric DNA and with the higher concentration of Rap1, increasing amount of free DNA was observed.30 Furthermore, there are ∼2fold increased the selectivity of TRF2 binding to telomeric DNA in the presence of Rap130. Taken together, these findings indicate that Rap1 may play a role in the transcriptional control by modulating TRF2 attraction to telomeric and extratelomeric DNA.
Rap1 role in metabolism
Telomeric dysfunction and mitochondrial DNA damageare characteristic features of cellular senescence and organismal ageing, which are associated with age-related conditions like diabetes and atherosclerosis.31,32 Furthermore, it is remarkable that the function of the mitochondrial regulators (PGC-1α and PGC-1β) declines across ageing tissues. The significance of PGCs to insulin resistance and diabetes may stem from their control of salutary metabolic pathways such as fatty acid metabolism, gluconeogenesis and mitochondrial biogenesis.33,34 It is intriguing that, telomerase (TERT) depletion in mice repressed the expression of PGC-1α results in reduced mitochondrial mass and function in liver and heart tissues,35 indicating a relation between telomere dysfunction and mitochondrial compromise in mammalian cells.
Since mammalian Rap1is involved in telomeric stability and its regulation,16,36,37 it is anticipated that Rap1 may have regulatory control over metabolism through the telomere protection.Indeed, the role of mammalian Rap1 on metabolism has been characterized in both in vitroand in vivo system.19,28 Of particular interest, Rap1 knockout mice demonstrated reduced expression of PGC1α.19 However, it appears that mammalian Rap1 regulates metabolism independent of telomeric dysfunction since there was no evident of telomeric attrition upon Rap1 depletion in mice.
In budding yeast, scRap1 controls metabolism through regulation of enzymes and genes involved in glycolysis and ribosomal synthesis38 scRap1 participates in enhancer function of multiple genes implicated in glycolysis by mapping with upstream activation site elements (UAS) of glycolytic genes such as phosphoglycerate kinase (PGK1),39 pyruvate kinase (PYK1)40 and pyruvate decarboxylase (PDC1).41 Furthermore, scRap1 can tether with GCR1 (CTTCC sequence motif) for maximal Rap1 dependent glycolytic gene expression39,42 Thus, Rap1 plays a passive role in the gene regulation of glycolytic enzymes perhaps by allowing specific regulators to find their DNA-binding sites.
Although nontelomeric activities of scRap1 have been thoroughly studied in yeast, the role of mammalian Rap1 in metabolism is commencing to reveal through experiments in Rap1 knockout mice. Martinez et al. reported that mammalian Rap1 regulates metabolism through transcriptional regulation of the PPARα and PGC1α genes by binding to an intragenic F3 region in Pparα and Pgc1α loci.19 Furthermore, Rap1 deficiency leads to decreased activation of PPARα and PGC1α and eventually leading to deregulation of metabolic pathways involved in energy homeostasis.19 Livers of Rap1 deficient mice demonstrated reduced expressions of carnitine palmitoyl transferase (a rate-limiting step in fatty acid oxidation), phosphoenol pyruvate carboxykinase (a key gluconeogenic enzyme) and several fatty acid transporters (SLC27a2 and CD36).19 Deletion of Rap1 in mice causes dysfunction of the liver as well as adipose tissue, leading to excess fat accumulation, hepatic steatosis, glucose intolerance and insulin resistance.19,43 An interesting observation in this study is that Rap1 deficient mice exhibited sexual dimorphism in age-dependent obesity i.e. more accentuated in females, a similar feature observed with PPARα- and PGC1α-deficient mice.44,45 Furthermore, Rap1-null MEFs demonstrated significant down-regulation of metabolic genes, particularly genes related to insulin secretion (FABP4, PLA2G4A, ADCY8, MGLL, GDA, PYCR1, DPYD, PPAP2B) and peroxisome-proliferator-activated receptor signaling (LPL, UCP1, FABP4, ANGPTL4, PPARγ, ACSL4, PCK2). Besides, genes involved in type II diabetes (PIK3R5, CACNA1G, PIK3R3, SLC2A4) were up-regulated in Rap1-null MEF.28 These results underscore a direct physiologic role of Rap1 in regulation of metabolic signaling pathways and forging a link between mammalian Rap1 and metabolism (Fig. 2).
Figure 2.
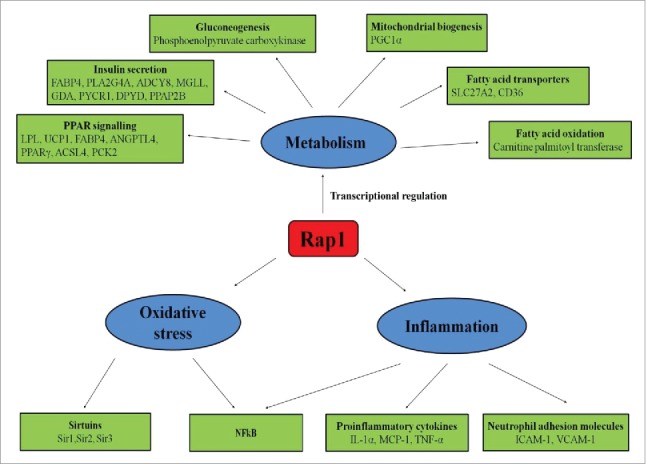
Nontelomeric functions of Rap1. Rap1 regulates various nontelomeric functions like metabolism, oxidative stress, and inflammation. Rap1controls energy metabolism by increasing fatty acid oxidation via increasing the expression of carnitine palmitoyltransferase, and fatty acid transporters such as SLC27A2 and CD36 and by increasing gluconeogenesis via increasing the expression of phosphoenolpyruvate carboxykinase. Rap1 also regulates mitochondrial biogenesis by increasing PGC-1α expression. Furthermore, Rap1 upregulates metabolic genes related to insulin secretion (FABP4, PLA2G4A, ADCY8, MGLL, GDA, PYCR1, DPYD, PPAP2B) and peroxisome-proliferator-activated receptor signaling (LPL, UCP1, FABP4, ANGPTL4, PPARγ, ACSL4, PCK2). In inflammation, Rap1 stimulates NFκB activations and upregulates the expression of proinflammatory cytokines such as IL-1α, MCP-1, TNF-α, and neutrophil adhesion molecules such as ICAM-1 and VCAM-1. Rap1 also modulates oxidative stress by increasing the expression of NFκB and sirtuins.
Rap1 role in inflammation
Inflammation is considered as a physiological homeostatic response of the organism to noxious stimuli. Inflammatory milieu comprises the synchronized action of various kinds of inflammatory mediators like cytokines, eicosanoids, chemokines and products of proteolytic cascades.46 The nuclear factor (NF)κB has long been recognized as “prototypical pro-inflammatory signaling pathway”, chiefly based on the role of NFκB in the expression of cytokines, chemokines and adhesion molecules.47 Genome-wide gain-of-function studies revealed that Rap1 as an inhibitor kinase (IKK) adaptor that controls NFκB activity in a dose-dependent manner.48–50 We and others demonstrated that cytoplasmic Rap1 is associated with NFκB mediated inflammation in macrophages and various cancer cell lines.48,51 Interestingly, cytoplasmic Rap1 in human cells does not associate with TRF2,52 perhaps it is this fraction that couples with IKK complex.51 Cytoplasmic Rap1 has been associated with several inflammatory-related diseases like cancer,48 atherosclerosis,51 corneal alkali injury53, myocardial infarction54, and liver ischemia-reperfusion injury.55
The non-canonical inflammatory role of cytoplasmic Rap1 has been comprehensibly deciphered in breast cancer BT476 and epithelial cancer HeLa S3 cell lines.48 In these human cancer cell lines, cytoplasmic Rap1 binds to IKK, thereby promotes subsequent degradation of inhibitor of kappa B (IκB) protein,56 which is the endogenous NFκB inhibitor.57 Thus, cytoplasmic Rap1 by recruiting IKKs ensures efficient NFκB activation via phosphorylation of p65 subunit.48 We further demonstrated that cytoplasmic Rap1 plays a role in inflammation in macrophages.51 Rap1 deficiency in pro-inflammatory phenotypic macrophages i.e. differentiated THP-1 cells and peritoneal macrophages downregulated the expression of pro-inflammatory cytokines (including interleukin (IL)-1α, monocyte chemoattractant protein 1 (MCP-1), tumor necrosis factor (TNF)-α) and enhanced IκBαexpression. In contrast, Rap1 deficiency in anti-inflammatory macrophages (bone marrow derived M2 macrophages)are less uniform, as it up-regulated the expression of IL-1α, IL-6,IκBα, while down-regulated MCP-1 expression. Moreover, the induction of NFκB-mediated pro-inflammatory cytokine production by Rap1 is restricted to macrophages as it does not appear in human endothelial and smooth muscle cell lines, which warrants further elucidation.51 Also, inhibition of Rap1 promotes better corneal recovery following alkali injury that was accompanied by inhibition of NFκB and reduced up-regulation of cytokine (IL-1α) and finely regulated levels of angiogenic factor (vascular endothelial growth factor) and antiangiogenic factor (pigment epithelium-derived factor).53 Therefore, the presence of Rap1 facilitates the pro-inflammatory environment by favoring the production of pro-inflammatory cytokines and by limiting the anti-inflammatory cytokines (Fig. 2).
We further observed persistent stimulation of NFκB with either lipopolysaccharide or TNFα enhanced Rap1 levels, indicating the presence of a positive feedback loop for Rap1 self-activation in macrophages.51 A similar observation was reported in murine embryonic fibroblast cells were abrogated NFκB stimulation associated with diminished Rap1 expression.48 However, more Rap1 levels were observed in the total cellular extracts, when the p65 subunit was over-expressed.48 Indeed, the Rap1 promoter region comprises binding sites for NFκB.48 Persistent stimulation of NFκB pathway in macrophages leading to augmented secretion of cytokines has been associated with the development and progression of atherosclerosis.58–60 The Rap1-NFkB positive feedback loop could be a possible mechanism which ensures chronic pro-inflammatory environment by constitutive activation of NFκB activation throughout the atherosclerosis progression and development.61,62
Interestingly, Rap1 has a significant role in mesenchymal stem cell (MSC) based therapy against myocardial infarction via regulation of NFκB activity.54 Knockdown of Rap1 with shRap1 led to decreased translocation of p65-NFκB from the cytoplasm to nuclei following TNF-activation. Furthermore, Rap1 knockout bone marrow-derived MSCs displayed a markedly reduced ratio of phosphorylated NFκB to NFκB-p65 and Bax to Bcl-2, and enhanced tolerance to hypoxia-induced apoptosis when compared to wild-type bone marrow-derived MSCs.54 Moreover, deletion of Rap1 in bone marrow-derived MSCs resulted in decreased activity of NFκB, escorted by reduced pro-inflammatory paracrine cytokine mediators TNF-α, IL-6 and MCP-1 compared with wild-type bone marrow-derived MSCs.54 In contrast, re-introduction of Rap1 in Rap1 knockout bone marrow-derived MSCs resulted in the loss of resistance to hypoxia-induced apoptotic death.54 These results suggest that abolition of Rap1 boosts MSC survival capacity under hypoxic conditions. Furthermore, in vivo transplantation of Rap1 knockout bone marrow-derived MSC contributed more to the recovery of heart function and had a better therapeutic efficacy than wild-type bone marrow-derived MSCs for cardiac repair post myocardial infarction.54 In summary, Rap1 has a crucial role in facilitating MSC paracrine component and MSC cell survival potential. Therefore, specific deletion of Rap1 in bone marrow-derived MSCs presents a novel approach to boost prospective MSC-based therapy by controlling paracrine profiling.
Similar to the above observation, Rap1 has been shown to be involved in accelerating liver ischemia/reperfusion injury (IRI) by elevating neutrophil inflammatory response.55 Overexpression of Rap1 was accompanied with severe neutrophil-mediated hepatic inflammatory response and has been shown a positive correlation with acute phase liver graft injury in both human and rat liver transplantation. Furthermore, in mice hepatic IRI model and primary neutrophils, knockout of Rap1 reduced liver damage and suppressed neutrophils migration activity and adhesion molecules (intracellular adhesion molecule-1 and vascular cell adhesion molecule-1) through decreasing neutrophils F-Actin expression and chemokine (C-X-C motif) ligand/receptor 2 (CXCL2/CXCR2) pathways.55 Besides, knockout Rap1 alleviated mouse liver IRI not only through suppressing migration of neutrophils but also through inhibiting the neutrophils function through NFκB and mitogen-activated protein kinase signaling pathways.55 Thus, Rap1 can be considered as a promising therapeutic target of attenuating liver IRI and myocardial infarction.
Rap1 role in oxidative stress
Oxidative stress plays a crucial role in the progression of diabetes and associated cardiomyopathy.63 Reactive oxygen species (ROS) like hydroxyl free radical, superoxide anion and hydrogen peroxide could cause damage to lipids, proteins, and DNA.64 It is well described that hyperglycemia provokes stimulation of ROS production due to impaired function of mitochondrial electron transport chain. ROS overproduction is the primary cause for hyperglycemia-induced cell damage.65,66 It is well established that oxidative stress and sirtuins are mutually linked together that oxidative stress can alter the sirtuins functions at different levels such as expression, posttranslational modifications, and protein-protein interactions67 whereas sirtuins can control oxidative stress by directly deacetylating several transcription factors that regulate antioxidant genes like FOXO and p65-subunit of NFκB.68–70 Furthermore, Rap1 association with sirtuins in yeast was characterized.71,72 A study conducted in wine yeasts demonstrated that ethanol-induced ROS and superoxide signals was accompanied by increased proteins levels of sirtuins (Sir1,Sir2, Sir3) and Rap1, that interacts with Sir complex,73 which suggests that Rap1 modulates oxidative stress in yeast and is speculated that can be conserved in mammals as well, though it warrants further investigation.
A recent study showed a link between telomere length, oxidative stress and the onset of diabetes.74 Telomere shortening due to oxidative stress eventually leads to beta cells senescence and the subsequent apoptosis which accelerates the inception of diabetes.74 Since Rap1 exert a direct effect on telomere homeostasis and regulation of telomere length, it is hypothesized that Rap1 may respond to oxidative stress. Swanson et al. observed that decrease in telomere length was associated with reduced levels of TRF2 and Rap1 in aged human dermal fibroblasts.75 Interestingly, only nuclear levels but not the cytoplasmic pool of Rap1 were decreased significantly in two different cell lines like human dermal fibroblasts (do not express telomerase) and U251 glioblastoma cells (express telomerase) when exposed to hydrogen peroxide or serum depleted medium.75 This study indicates that nuclear Rap1 responds to oxidative stress in every cell type irrespective of telomerase expression. The cytoplasmic pool of Rap1 remains constant and seems likely to facilitate the preservation of cell survival signaling NFκB pathway. One possible explanation is that under oxidative conditions, nuclear Rap1 translocates to the cytoplasm to maintain constant levels of Rap1 for continuous signaling of NFκB (Fig. 2).
Potential role of Rap1 in diabetic cardiomyopathy
From a clinical standpoint, an inverse relationship exists between leukocyte telomere length and chronological age. Genetic variation and sex have an additional influence on accelerated leukocyte telomere shortening.76 Furthermore, substantial evidence has demonstrated that telomere shortening has been linked with greater risk of obesity77 and cardiovascular disease.78 In particular, a cohort study conducted on 7,008 U.S. residents suggested that a unit increase in percent body fat, waist circumference (cm) and body mass index (kg/m2) contributed, respectively, to a 0.01%, 0.09% and 0.2% decrease in leukocyte telomere length (T/S ratio), indicating that life course adiposity dynamics are the determinant of telomere shortening.77 Also, the Strong Heart Family Study sheds light on the potential role of leukocyte telomere length in the prediction of adverse cardiovascular disease outcomes.78 It was observed that longer leukocyte telomere length was significantly linked with reduced arterial stiffness.78 In another observational cohort study on 257 kidney donors for transplantation provides evidence for the close association between shorter telomere length in peripheral blood leukocytes and kidney tissues and intrarenal arteriosclerosis, independent of cardiovascular risk factors.79 Thus, collectively, based on the available literature, it can be suggested that telomere attrition plays a central role in the development of renovascular and cardiovascular diseases in humans.
Diabetic cardiomyopathy is a heart muscle disorder associated with oxidative stress and inflammation in people with diabetes, which can lead to inability of the heart to circulate blood through the body effectively. Moreover, it is also previously reported that telomere shortening is observed in cardiomyopathy.80–83 As the review focuses on both the telomeric and non-telomeric functions of Rap1 and its role in the regulation of telomeric length, metabolism, oxidative stress and inflammation, collectively it can be anticipated that Rap1 plays an important role in the pathophysiology of diabetic cardiomyopathy either directly or indirectly which is yet to be explored.
Conclusion and future perspectives
During the past decade, substantial evidence has been accumulated for non-telomeric role for Rap1 in the regulation of inflammation, oxidative stress and metabolism. The greatest challenge at present is to comprehend what the downstream molecules are, that allow Rap1 to execute pleiotropic functions. Remarkably, some protein targets like NFκB and PPARα have emerged as immediate downstream effectors of Rap1 so far from genetic studies. NFκB activation is detrimental by triggering chronic inflammation through elevation of cytokines leading to cell death, including cardiac cells.84 Indeed, persistent activation of NFκB has been associated with myocardial IRI.85 Simultaneously, PPARα regulates cardiac energy metabolism and various cell signaling pathways concerning cardiovascular health and disease.86,87 Thus, a continuing need exists for getting definitive evidence for the Rap1 role in the cardiovascular physiology and its pathological diseases like myocardial infarction. It raises several immediate questions like whether Rap1 modulation exacerbates myocardial IRI? Does Rap1 contribute to inflammation through NFκB signaling during myocardial IRI? Whether Rap1 regulates cardiac energy metabolism through regulation of PPARα transcription? The enigma of Rap1 and its putative effects cannot be resolved by genetic studies only, but the development of suitable animal model systems will help to decipher the potentiality of Rap1 as a drug target in cardiac diseases. Thus, it is prime requisite to gain a comprehensive knowledge on the Rap1 regulatory role in the cardiovascular biology.
Abbreviations
- ADH
Alcohol dehydrogenase
- ATM
Ataxia telangiectasia mutated
- CXCL2/CXCR2
Chemokine (C-X-C motif) ligand/receptor 2
- ChIP
Chromatin immune precipitation
- GCR
Glycolytic genes transcriptional activator
- HDR
Homology-directed repair
- IKK
Inhibitor kinase
- IRI
Ischemia reperfusion injury
- IκB
Inhibitor of kappa B
- IL
Interleukin
- MEFs
Mouse embryonic fibroblasts
- MSC
Mesenchymal stem cell
- NHEJ
Non-homologous end-joining
- NF
Nuclear factor
- POT1
Protection of telomeres 1
- PARP1
Poly(ADP-ribose) polymerase 1
- PGC
Peroxisome proliferator-activated receptor gamma, coactivator
- PPARs
Peroxisome proliferator-activated receptors
- RCT
Rap1-specific protein-interaction domain
- ROS
Reactive oxygen species
- SLX4
SLX4 structure-specific endonuclease subunit
- TRF1
Telomeric repeat-binding factor 1
- TRF2
Telomeric repeat-binding factor 2
- TIN2
TRF1-interacting nuclear protein 2
- TPP1
TIN2- and POT1-organizing protein
- TALEN
Transcription activator-like effector nuclease
- TIFs
Telomere Dysfunction-induced foci
- T-SCEs
Telomere sister chromatid exchanges
- TNF
Tumor necrosis factor
- UAS
Upstream activation site elements
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have approved the submission for publication of the work.
Disclosure of interest
The authors report no conflict of interest.
Acknowledgments
The authors' work was supported by the Health and Medical Research Fund (04151816, Hong Kong) and General Research Fund (17121315M, 17123915M and 17117217M, Research Grants Council of Hong Kong).
Authors' contributions
All authors participated in drafting and revising the article. All authors read and approved the final manuscript.
References
- [1].de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532-40. doi: 10.1038/sj.onc.1205080. PMID:11850778 [DOI] [PubMed] [Google Scholar]
- [2].de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100-10. doi: 10.1101/gad.1346005. PMID:16166375 [DOI] [PubMed] [Google Scholar]
- [3].Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301-34. doi: 10.1146/annurev.genet.41.110306.130350. PMID:18680434 [DOI] [PubMed] [Google Scholar]
- [4].Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721-32. doi: 10.1016/0092-8674(87)90095-X. PMID:3315231 [DOI] [PubMed] [Google Scholar]
- [5].Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125-36. doi: 10.1016/S0092-8674(00)81088-0. PMID:8620531 [DOI] [PubMed] [Google Scholar]
- [6].Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471-83. doi: 10.1016/S0092-8674(00)80858-2. PMID:10850490 [DOI] [PubMed] [Google Scholar]
- [7].Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere end-replication problem and cell aging. J Mol Biol. 1992;225:951-60. doi: 10.1016/0022-2836(92)90096-3. PMID:1613801 [DOI] [PubMed] [Google Scholar]
- [8].Brouilette S, Singh RK, Thompson JR, Goodall AH, Samani NJ. White cell telomere length and risk of premature myocardial infarction. Arterioscler Thromb Vasc Biol. 2003;23:842-6. doi: 10.1161/01.ATV.0000067426.96344.32. PMID:12649083 [DOI] [PubMed] [Google Scholar]
- [9].Benetos A, Gardner JP, Zureik M, Labat C, Xiaobin L, Adamopoulos C, Temmar M, Bean KE, Thomas F, Aviv A. Short telomeres are associated with increased carotid atherosclerosis in hypertensive subjects. Hypertension. 2004;43:182-5. doi: 10.1161/01.HYP.0000113081.42868.f4. PMID:14732735 [DOI] [PubMed] [Google Scholar]
- [10].Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Semin Cancer Biol. 2011;21:349-53. doi: 10.1016/j.semcancer.2011.10.001. PMID:22015685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383-7. doi: 10.1016/S0955-0674(97)80011-7. PMID:9159071 [DOI] [PubMed] [Google Scholar]
- [12].Kaizer H, Connelly CJ, Bettridge K, Viggiani C, Greider CW. Regulation of Telomere Length Requires a Conserved N-Terminal Domain of Rif2 in Saccharomyces cerevisiae. Genetics. 2015;201:573-86. doi: 10.1534/genetics.115.177899. PMID:26294668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257-69. doi: 10.1101/gad.8.19.2257. PMID:7958893 [DOI] [PubMed] [Google Scholar]
- [14].van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740-3. doi: 10.1038/385740a0. PMID:9034193 [DOI] [PubMed] [Google Scholar]
- [15].O'Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585-91. doi: 10.1074/jbc.M312913200. PMID:15100233 [DOI] [PubMed] [Google Scholar]
- [16].Li B, de Lange T. Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell. 2003;14:5060-8. doi: 10.1091/mbc.E03-06-0403. PMID:14565979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kabir S, Hockemeyer D, de Lange T. TALEN gene knockouts reveal no requirement for the conserved human shelterin protein Rap1 in telomere protection and length regulation. Cell Reports. 2014;9:1273-80. doi: 10.1016/j.celrep.2014.10.014. PMID:25453752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657-61. doi: 10.1126/science.1185100. PMID:20339076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Martinez P, Gomez-Lopez G, Garcia F, Mercken E, Mitchell S, Flores JM, de Cabo R, Blasco MA. RAP1 protects from obesity through its extratelomeric role regulating gene expression. Cell Reports. 2013;3:2059-74. doi: 10.1016/j.celrep.2013.05.030. PMID:23791526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lu W, Zhang Y, Liu D, Songyang Z, Wan M. Telomeres-structure, function, and regulation. Exp Cell Res. 2013;319:133-41. doi: 10.1016/j.yexcr.2012.09.005. PMID:23006819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712-8. doi: 10.1038/ncb1275. PMID:15968270 [DOI] [PubMed] [Google Scholar]
- [22].van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401-13. doi: 10.1016/S0092-8674(00)80932-0. PMID:9476899 [DOI] [PubMed] [Google Scholar]
- [23].Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885-90. doi: 10.1038/ncb1444. PMID:16845382 [DOI] [PubMed] [Google Scholar]
- [24].Rai R, Chen Y, Lei M, Chang S. TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat Commun. 2016;7:10881. doi: 10.1038/ncomms10881. PMID:26941064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, et al.. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Structural Mol Biol. 2011;18:213-21. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hegan DC, Lu Y, Stachelek GC, Crosby ME, Bindra RS, Glazer PM. Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proc Natl Acad Sci U S A. 2010;107:2201-6. doi: 10.1073/pnas.0904783107. PMID:20133863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Azad GK, Tomar RS. The multifunctional transcription factor Rap1: a regulator of yeast physiology. Frontiers Biosci (Landmark edition). 2016;21:918-30. doi: 10.2741/4429. [DOI] [PubMed] [Google Scholar]
- [28].Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:768-80. doi: 10.1038/ncb2081. PMID:20622869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hanaoka S, Nagadoi A, Yoshimura S, Aimoto S, Li B, de Lange T, Nishimura Y. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J Mol Biol. 2001;312:167-75. doi: 10.1006/jmbi.2001.4924. PMID:11545594 [DOI] [PubMed] [Google Scholar]
- [30].Janoušková E, Nečasová I, Pavloušková J, Zimmermann M, Hluchý M, Marini V, Nováková M, Hofr C. Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res. 2015;43:2691-700. doi: 10.1093/nar/gkv097. PMID:25675958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Khan S, Chuturgoon AA, Naidoo DP. Telomeres and atherosclerosis. Cardiovascular J Africa. 2012;23:563-71. doi: 10.5830/CVJA-2012-056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Armanios M. Telomeres and age-related disease: how telomere biology informs clinical paradigms. J Clin Invest. 2013;123:996-1002. doi: 10.1172/JCI66370. PMID:23454763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Eisele PS, Salatino S, Sobek J, Hottiger MO, Handschin C. The peroxisome proliferator-activated receptor gamma coactivator 1alpha/beta (PGC-1) coactivators repress the transcriptional activity of NF-kappaB in skeletal muscle cells. J Biol Chem. 2013;288:2246-60. doi: 10.1074/jbc.M112.375253. PMID:23223635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Patten IS, Arany Z. PGC-1 coactivators in the cardiovascular system. Trends Endocrinol Metab TEM. 2012;23:90-7. doi: 10.1016/j.tem.2011.09.007. PMID:22047951 [DOI] [PubMed] [Google Scholar]
- [35].Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al.. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359-65. doi: 10.1038/nature09787. PMID:21307849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801-14. doi: 10.1101/gad.6.5.801. PMID:1577274 [DOI] [PubMed] [Google Scholar]
- [37].O'Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585-91. doi: 10.1074/jbc.M312913200. PMID:15100233 [DOI] [PubMed] [Google Scholar]
- [38].Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408-12. doi: 10.1016/0168-9525(94)90058-2. PMID:7809947 [DOI] [PubMed] [Google Scholar]
- [39].Chambers A, Stanway C, Tsang JS, Henry Y, Kingsman AJ, Kingsman SM. ARS binding factor 1 binds adjacent to RAP1 at the UASs of the yeast glycolytic genes PGK and PYK1. Nucleic Acids Res. 1990;18:5393-9. doi: 10.1093/nar/18.18.5393. PMID:2120676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nishizawa M, Araki R, Teranishi Y. Identification of an upstream activating sequence and an upstream repressible sequence of the pyruvate kinase gene of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:442-51. doi: 10.1128/MCB.9.2.442. PMID:2651900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Butler G, Dawes IW, McConnell DJ. TUF factor binds to the upstream region of the pyruvate decarboxylase structural gene (PDC1) of Saccharomyces cerevisiae. Mol Gen Genet. 1990;223:449-56. doi: 10.1007/BF00264453. PMID:2270085 [DOI] [PubMed] [Google Scholar]
- [42].Bitter GA, Chang KK, Egan KM. A multi-component upstream activation sequence of the Saccharomyces cerevisiae glyceraldehyde-3-phosphate dehydrogenase gene promoter. Mol Gen Genet. 1991;231:22-32. doi: 10.1007/BF00293817. PMID:1753943 [DOI] [PubMed] [Google Scholar]
- [43].Yeung F, Ramirez CM, Mateos-Gomez PA, Pinzaru A, Ceccarini G, Kabir S, Fernandez-Hernando C, Sfeir A. Nontelomeric role for Rap1 in regulating metabolism and protecting against obesity. Cell Reports. 2013;3:1847-56. doi: 10.1016/j.celrep.2013.05.032. PMID:23791522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor α (PPARα) in the cellular fasting response: The PPARα-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A. 1999;96:7473-8. doi: 10.1073/pnas.96.13.7473. PMID:10377439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Costet P, Legendre C, Moré J, Edgar A, Galtier P, Pineau T. Peroxisome proliferator-activated receptor α-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and steatosis. J Biol Chem. 1998;273:29577-85. doi: 10.1074/jbc.273.45.29577. PMID:9792666 [DOI] [PubMed] [Google Scholar]
- [46].Qatanani M, Lazar MA. Mechanisms of obesity-associated insulin resistance: many choices on the menu. Genes Dev. 2007;21:1443-55. doi: 10.1101/gad.1550907. PMID:17575046 [DOI] [PubMed] [Google Scholar]
- [47].Lawrence T. The Nuclear Factor NF-κB pathway in inflammation. Cold Spring Harbor Perspect Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, et al.. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat Cell Biol. 2010;12:758-67. doi: 10.1038/ncb2080. PMID:20622870 [DOI] [PubMed] [Google Scholar]
- [49].Ghosh AS, Tergaonkar V. Telomeres and inflammation: Rap1 joins the ends? Cell cycle (Georgetown, Tex). 2010;9:3834-5. doi: 10.4161/cc.9.19.13383. PMID:20890110 [DOI] [PubMed] [Google Scholar]
- [50].Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, Wong M, Zhang Z, Li G, Sung WK, et al.. Telomerase directly regulates NF-kappaB-dependent transcription. Nat Cell Biol. 2012;14:1270-81. doi: 10.1038/ncb2621. PMID:23159929 [DOI] [PubMed] [Google Scholar]
- [51].Cai Y, Sukhova GK, Wong HK, Xu A, Tergaonkar V, Vanhoutte PM, Tang EHC. Rap1 induces cytokine production in pro-inflammatory macrophages through NFκB signaling and is highly expressed in human atherosclerotic lesions. Cell Cycle (Georgetown, Tex). 2015;14:3580-92. doi: 10.1080/15384101.2015.1100771. PMID:26505215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457-67. doi: 10.1074/jbc.M109.038026. PMID:19864690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Poon MW, Yan L, Jiang D, Qin P, Tse HF, Wong IY, Wong DS, Tergaonkar V, Lian Q. Inhibition of RAP1 enhances corneal recovery following alkali injury. Invest Ophthalmol Visual Sci. 2015;56:711-21. doi: 10.1167/iovs.14-15268. [DOI] [PubMed] [Google Scholar]
- [54].Zhang Y, Chiu S, Liang X, Gao F, Zhang Z, Liao S, Liang Y, Chai YH, Low DJH, Tse HF, et al.. Rap1-mediated nuclear factor-kappaB (NF-κB) activity regulates the paracrine capacity of mesenchymal stem cells in heart repair following infarction. Cell Death Discovery. 2015;1:15007. doi: 10.1038/cddiscovery.2015.7. PMID:27551443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li CX, Lo CM, Lian Q, Ng KT, Liu XB, Ma YY, Qi X, Yeung OW, Tergaonkar V, Yang XX, et al.. Repressor and activator protein accelerates hepatic ischemia reperfusion injury by promoting neutrophil inflammatory response. Oncotarget. 2016;7:27711-23. doi: 10.18632/oncotarget.8509. PMID:27050284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225-60. doi: 10.1146/annurev.immunol.16.1.225. PMID:9597130 [DOI] [PubMed] [Google Scholar]
- [57].Tedgui A, Mallat Z. Cytokines in atherosclerosis: pathogenic and regulatory pathways. Physiol Rev. 2006;86:515-81. doi: 10.1152/physrev.00024.2005. PMID:16601268 [DOI] [PubMed] [Google Scholar]
- [58].Monaco C, Paleolog E. Nuclear factor kappaB: a potential therapeutic target in atherosclerosis and thrombosis. Cardiovasc Res. 2004;61:671-82. doi: 10.1016/j.cardiores.2003.11.038. PMID:14985064 [DOI] [PubMed] [Google Scholar]
- [59].Mallat Z, Tedgui A. NF-κB activation in atherosclerosis: a friend or a foe? Blood. 2004;103:754-5. doi: 10.1182/blood-2003-11-3818. [DOI] [Google Scholar]
- [60].Kanters E, Pasparakis M, Gijbels MJ, Vergouwe MN, Partouns-Hendriks I, Fijneman RJ, Clausen BE, Forster I, Kockx MM, Rajewsky K, et al.. Inhibition of NF-kappaB activation in macrophages increases atherosclerosis in LDL receptor-deficient mice. J Clin Invest. 2003;112:1176-85. doi: 10.1172/JCI200318580. PMID:14561702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Xanthoulea S, Curfs DM, Hofker MH, de Winther MP. Nuclear factor kappa B signaling in macrophage function and atherogenesis. Curr Opin Lipidol. 2005;16:536-42. doi: 10.1097/01.mol.0000180167.15820.ae. PMID:16148538 [DOI] [PubMed] [Google Scholar]
- [62].Libby P, Ridker PM, Hansson GK. Inflammation in Atherosclerosis: from pathophysiology to practice. J Am Coll Cardiol. 2009;54:2129-38. doi: 10.1016/j.jacc.2009.09.009. PMID:19942084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rosa CM, Xavier NP, Henrique Campos D, Fernandes AA, Cezar MD, Martinez PF, Cicogna AC, Gimenes C, Gimenes R, Okoshi MP, et al.. Diabetes mellitus activates fetal gene program and intensifies cardiac remodeling and oxidative stress in aged spontaneously hypertensive rats. Cardiovascular Diabetol. 2013;12:152. doi: 10.1186/1475-2840-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tabak O, Gelisgen R, Erman H, Erdenen F, Muderrisoglu C, Aral H, Uzun H. Oxidative lipid, protein, and DNA damage as oxidative stress markers in vascular complications of diabetes mellitus. Clin Invest Med. 2011;34:E163-71. doi: 10.25011/cim.v34i3.15189. PMID:21631993 [DOI] [PubMed] [Google Scholar]
- [65].Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813-20. doi: 10.1038/414813a. PMID:11742414 [DOI] [PubMed] [Google Scholar]
- [66].Nishikawa T, Edelstein D, Du XL, Yamagishi S, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes HP, et al.. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404:787-90. doi: 10.1038/35008121. PMID:10783895 [DOI] [PubMed] [Google Scholar]
- [67].Santos L, Escande C, Denicola A. Potential modulation of Sirtuins by oxidative stress. Oxidative Med Cell Longevity. 2016;2016:9831825. doi: 10.1155/2016/9831825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Merksamer PI, Liu Y, He W, Hirschey MD, Chen D, Verdin E. The sirtuins, oxidative stress and aging: an emerging link. Aging (Albany NY). 2013;5:144-50. doi: 10.18632/aging.100544. PMID:23474711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al.. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011-5. doi: 10.1126/science.1094637. PMID:14976264 [DOI] [PubMed] [Google Scholar]
- [70].Lee JH, Song MY, Song EK, Kim EK, Moon WS, Han MK, Park JW, Kwon KB, Park BH. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-kappaB signaling pathway. Diabetes. 2009;58:344-51. doi: 10.2337/db07-1795. PMID:19008341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Luo K, Vega-Palas MA, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528-39. doi: 10.1101/gad.988802. PMID:12080091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genetics. 2001;28:327-34. doi: 10.1038/ng569. PMID:11455386 [DOI] [PubMed] [Google Scholar]
- [73].Adamczyk J, Deregowska A, Skoneczny M, Skoneczna A, Kwiatkowska A, Potocki L, Rawska E, Pabian S, Kaplan J, Lewinska A, et al.. Adaptive response to chronic mild ethanol stress involves ROS, sirtuins and changes in chromosome dosage in wine yeasts. Oncotarget. 2016;7:29958-76. doi: 10.18632/oncotarget.8673. PMID:27074556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Salpea KD, Talmud PJ, Cooper JA, Maubaret CG, Stephens JW, Abelak K, Humphries SE. Association of telomere length with type 2 diabetes, oxidative stress and UCP2 gene variation. Atherosclerosis. 2010;209:42-50. doi: 10.1016/j.atherosclerosis.2009.09.070. PMID:19889414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Swanson MJ, Baribault ME, Israel JN, Bae NS. Telomere protein RAP1 levels are affected by cellular aging and oxidative stress. Biomedical Reports. 2016;5:181-7. doi: 10.3892/br.2016.707. PMID:27446538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Berglund K, Reynolds CA, Ploner A, Gerritsen L, Hovatta I, Pedersen NL, Hägg S. Longitudinal decline of leukocyte telomere length in old age and the association with sex and genetic risk. Aging (Albany NY). 2016;8:1398-407. doi: 10.18632/aging.100995. PMID:27391763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wulaningsih W, Watkins J, Matsuguchi T, Hardy R. Investigating the associations between adiposity, life course overweight trajectories, and telomere length. Aging (Albany NY). 2016;8:2689-701. doi: 10.18632/aging.101036. PMID:27650676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Peng H, Zhu Y, Yeh F, Cole SA, Best LG, Lin J, Blackburn E, Devereux RB, Roman MJ, Lee ET, et al.. Impact of biological aging on arterial aging in American Indians: findings from the Strong Heart Family Study. Aging (Albany NY). 2016;8:1583-92. doi: 10.18632/aging.101013. PMID:27540694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].De Vusser K Pieters N, Janssen B, Lerut E, Kuypers D, Jochmans I, Monbaliu D, Pirenne J, Nawrot T, Naesens M. Telomere length, cardiovascular risk and arteriosclerosis in human kidneys: an observational cohort study. Aging (Albany NY). 2015;7:766-75. doi: 10.18632/aging.100814. PMID:26539975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhao MX, Zhou B, Ling L, Xiong XQ, Zhang F, Chen Q, Li YH, Kang YM, Zhu GQ. Salusin-beta contributes to oxidative stress and inflammation in diabetic cardiomyopathy. Cell Death Dis. 2017;8(3), e2690 doi: 10.1038/cddis.2017.106. PMID:28333148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhang B, Shen Q, Chen Y, Pan R, Kuang S, Liu G, Sun G, Sun X. Myricitrin Alleviates Oxidative Stress-induced Inflammation and Apoptosis and Protects Mice against Diabetic Cardiomyopathy. Sci Rep. 2017;7, 44239 doi: 10.1038/srep44239. PMID:28287141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Prakoso D, DeBlasio MJ, Qin C, Rosli S, Kiriazis H, Qian H, Du XJ, Weeks KL, Gregorevic P, McMullen JR, Ritchie RH. Phosphoinositide 3-Kinase (p110alpha) gene delivery Limits Diabetes-induced Cardiac NADPH oxidase and cardiomyopathy in a mouse model with established Diastolic Dysfunction. Clin Sci (Lond). 2017;131:1345-60. doi: 10.1042/CS20170063. PMID:28487469 [DOI] [PubMed] [Google Scholar]
- [83].Chang AC, Ong SG, LaGory EL, Kraft PE, Giaccia AJ, Wu JC, Blau HM. Telomere shortening and metabolic compromise underlie dystrophic cardiomyopathy. Proc Natl Acad Sci U S A. 2016;113:13120-25. doi: 10.1073/pnas.1615340113. PMID:27799523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Gordon JW, Shaw JA, Kirshenbaum LA. Multiple facets of NF-kappaB in the heart: to be or not to NF-kappaB. Circulation Res. 2011;108:1122-32. doi: 10.1161/CIRCRESAHA.110.226928. PMID:21527742 [DOI] [PubMed] [Google Scholar]
- [85].Kim JW, Jin YC, Kim YM, Rhie S, Kim HJ, Seo HG, Lee JH, Ha YL, Chang KC. Daidzein administration in vivo reduces myocardial injury in a rat ischemia/reperfusion model by inhibiting NF-kappaB activation. Life Sci. 2009;84:227-34. doi: 10.1016/j.lfs.2008.12.005. PMID:19109981 [DOI] [PubMed] [Google Scholar]
- [86].Ravingerova T, Adameova A, Carnicka S, Nemcekova M, Kelly T, Matejikova J, Galatou E, Barlaka E, Lazou A. The role of PPAR in myocardial response to ischemia in normal and diseased heart. Gen Physiol Biophys. 2011;30:329-41. doi: 10.4149/gpb_2011_04_329. PMID:22131314 [DOI] [PubMed] [Google Scholar]
- [87].Kolwicz SC, Purohit S, Tian R. Cardiac metabolism and its interactions with contraction, growth, and survival of the cardiomyocte. Circulation Res. 2013;113:603-16. doi: 10.1161/CIRCRESAHA.113.302095. PMID:23948585 [DOI] [PMC free article] [PubMed] [Google Scholar]


