ABSTRACT
Many HIV-infected individuals on antiretroviral therapy (ART) exhibit persistent systemic inflammation, which predicts morbidity and mortality. ART-treated subjects concurrently exhibit marked compositional alterations in the gut bacterial microbiota and the degree of dysbiosis correlates with systemic inflammation. Whether interventions to modulate the microbiome can affect systemic inflammation is unknown. An open-label fecal microbial transplantation (FMT) was delivered by colonoscopy to asymptomatic HIV-infected ART-suppressed individuals without antibiotic pre-treatment. Stool was assessed before and after FMT for engraftment of donor microbes, and peripheral blood was assayed for immune activation biomarkers. Six participants received FMT and 2 participants served as controls. No serious adverse effects occurred during 24 weeks of follow-up. At baseline, HIV-infected individuals exhibited microbiota profiles distinct from uninfected donors. During the 8 weeks post-FMT, recipients demonstrated partial engraftment of the donor microbiome (P < 0.05). Recipient microbiota remained significantly distant from donors, unlike that observed following FMT for treatment of C. difficile infection. Systemic inflammatory markers showed no significant change post-FMT. FMT was well-tolerated in ART-treated, HIV-infected individuals. Engraftment was detectable but modest, and appeared to be limited to specific bacterial taxa. Whether antibiotic conditioning can enhance engraftment and the capacity of microbiota to modulate inflammation remains to be investigated.
KEYWORDS: Microbiota, fecal transplant, engraftment, HIV, inflammation, fecal microbiome transplant, microbiome engraftment
Introduction
HIV infection leads to the depletion of circulating and tissue-resident CD4+ T cells, with the earliest and most dramatic depletion observed within the gut-associated lymphoid tissue.1,2 In particular, HIV eliminates activated CD4+ T cells and preferentially depletes the subsets that produce IL-17 and IL-22 (Th17/22 cells).3 Since IL-17 and IL-22 play an important role in maintaining mucosal barrier integrity, containing microbial translocation, and regulating the intestinal microbiome, their loss in HIV infection may explain why microbial translocation, systemic immune activation, and microbial dysbiosis are all increased during HIV disease.4,5
We and others have previously shown that gut microbiota dysbiosis is characteristic of untreated HIV infection, and remains prevalent despite treatment.6-13 Furthermore, the relative degree of alteration in the gut microbiota positively correlates with peripheral inflammatory markers such as IL-6 and the kynurenine pathway of tryptophan catabolism,6 both of which have been linked to clinical outcomes.14-16 Given the relationship between gut microbiota and systemic inflammation,17 interventions aimed at reconstituting the distal gut microbiome have the potential to interrupt chronic immune activation.
Fecal microbiome transplantation (FMT) has proven durable and successful as a therapeutic strategy against gut dysbiosis, particularly in the treatment of recurrent Clostridium difficile infection (CDI),18,19 whereas other interventions such as probiotics appear to have more modest effects.20 It remains unknown whether donor microbial communities can successfully engraft in the recipients outside the setting of CDI. FMT has an established record of safety with limited adverse effects,18,21 even in the context of immunocompromised and HIV-infected subjects.22,23 Given that FMT can restructure the composition of the gut microbiome to resemble that of the healthy donor in CDI subjects,24 we hypothesized that this intervention might reverse gut microbial dysbiosis and reduce markers of immune activation and inflammation in ART-treated HIV-infected individuals. To test this possibility, we performed an open-label interventional study to evaluate the ability of FMT to reconstitute the gut microbiota and reduce systemic inflammation in the treated HIV population.
Methods
Study subjects
Inclusion criteria
FMT and control participants were included if they were older than 18 y of age, were on continuous ART with full viral load suppression, and provided written informed consent. While ART-suppressed HIV-infected individuals with high CD4 counts may have abnormally elevated biomarkers of inflammation,25,26 those with CD4+ T cell counts less than 500 cells/mm3 and CD4:CD8 ratios less than 1 tend to have greater immune activation and gut barrier defects,27 and were preferentially targeted for recruitment. Controls were recruited among HIV-infected individuals on ART who were scheduled for routine screening colonoscopy.
Exclusion criteria
Participants were excluded if the CD4+ T cell count was less than 200 cells/mm3 (as these individuals often receive trimethoprim-sulfamethoxazole prophylaxis), if they were pregnant, breastfeeding, or unwilling to practice birth control during participation, or if they had active gastrointestinal symptoms undergoing investigation (e.g., inflammatory bowel disease, abdominal pain, hematochezia, or other alarming symptoms), recent hospitalization or acute medical condition or antibiotics use within preceding 3 months, or severe co-morbidities (e.g., cirrhosis, heart failure, renal failure, or respiratory failure). A history of anaphylaxis or severe food allergies, major immunosuppressive medications (e.g., calcineurin inhibitors, exogenous glucocorticoids, biologic agents, etc.), or systemic antineoplastic agents were additional criteria for exclusion. At the screening visit, participant stool was screened and participants were excluded if tested positive for: Clostridium difficile toxin B gene, routine bacterial culture for enteric pathogens (E. coli, Salmonella, Shigella, Yersinia, Campylobacter), culture for Vibrio spp, Giardia antigen, Cryptosporidium, acid-fast stain for Cyclospora and Isospora, and microscopy for the detection of ova and parasites.
Stool donors
OpenBiome (Somerville, MA) performed the donor screen and supplied healthy donor stool samples. Usage of the donor samples was approved by the FDA via linking of our study to the OpenBiome Drug Master File. In brief, donors were derived from volunteers who were carefully screened. Each was thoroughly interviewed, had undergone a questionnaire using the Donor History Questionnaire (DHQ) used in screening blood donors, and had received laboratory testing of blood and stool according to FDA guidelines provided for donors of human cells,28 tissues, and cellular and tissue-based products (HCT/Ps) and recommendations by the Fecal Microbiota Transplant Workgroup.29 All tests are outsourced to third party Clinical Laboratory Improvement Amendments (CLIA) certified testing facilities. In addition, OpenBiome provided 16S rRNA sequencing data for donor stool samples, from which 2 donors were selected that harbored microbial signatures with low abundance of Proteobacteria and high abundance of Bacteroidetes, with these 2 taxa being specifically examined due to prior studies linking their abundance to HIV infection.
Study visits
Study visits were scheduled for each participant before (weeks -4, -2), at the time of (week 0), and after FMT (weeks 1, 2, 4, 8, 24). During each visit, stool and blood were collected, processed, and banked. Peripheral blood mononuclear cells (PBMCs) were separated from blood plasma and cryopreserved while stool was immedieately stored at −80°C. Control participants provided stool samples before colonoscopy and at weeks 1 and 8 following colonoscopy.
Study procedure
Donor fecal material (FMP250, OpenBiome, Somerville, MA) was stored at −20°C and thawed before use, according to protocol.30 Participants underwent standard bowel purge (Golytely) the day preceding FMT and the 250 mL stool suspension was introduced via colonoscopy and delivered into the ileum, cecum, and ascending colon. HIV-infected ART-treated control participants underwent standard bowel purge the preceding day and then colonoscopy, but did not receive the donor stool suspension.
Study measurements
Microbiota analysis
Microbiota profiling was performed on samples from donors, each FMT recipient, and control subjects. DNA was extracted using a protocol optimized for the isolation of bacterial DNA from feces.31 Universal 16S rRNA primers that target the V4 hypervariable region and bear unique dual-indexed barcode oligonucleotide sequences32 were used with the Illumina MiSeq platform to generate >92,000 high-quality paired-end 16S rRNA reads/sample. QIIME software33 was used to process 16S sequencing data and to collapse reads with 97% sequence similarity into discrete operational taxonomic units (OTUs) for microbial community analyses using the Greengenes 13_5 database.
The same process was used to analyze publically available stool microbiome profiling data from CDI subjects before and after FMT.34 The referenced study used the same primers targeting the V4 region of 16S rRNA, making it highly comparable to the present study. For analyses incorporating CDI data from the referenced study, sample data from these samples and the current study were concatenated before undergoing quality filtration and OTU picking as described above. Sample community profiles were rarefied to 10,000 reads per sample.
An unweighted UniFrac distance matrix,35 which considers phylogenetic similarity of microbial communities but not taxon relative abundances in the calculation of between-sample ecological distances, was constructed in QIIME to compare the microbiome of FMT recipients over time to those of the donor sample.36 This permitted assessment of whether engraftment of the donor microbiome is associated with a phylogenetic shift in microbiota composition. For each subject, the pairwise distance between 3 replicates of the donor microbiota profile (donor replicate samples obtained at each FMT procedure event) and those of the recipient patient at all time points was calculated. Mean distances were tested using a linear mixed effects model to assess whether significant differences in community composition existed before and after FMT. Numbers of shared OTUs between donors and recipients were also tested for significance using linear mixed effects models. Control subjects who provided stool samples pre- and post-colonoscopy were examined for changes in the microbiota that occur over time and changes that would be attributable to the laxative (e.g., Golytely).
The permutational multivariate analysis of variance (PERMANOVA) approach37 designed for ecological β-diversity distance matrices (generated using the weighted UniFrac distance metric) and implemented in the R package ‘adonis’ was used to test significance of differences in microbial communities based on subject groups.
Peripheral blood assays
Plasma kynurenine to tryptophan ratios were measured using high performance liquid chromatography/mass spectroscopy (LC/MS), and levels of the innate immune activation marker IL-6 or sCD14 using standard ELISA kits, as per established methods.3
The level of T cell activation and immunophenotyping were measured by flow cytometry. Frozen PBMCs were thawed and counted for batched analysis. Two million cells per sample were stained as follows: CD3-BV650 (SK7, BD Biosciences, San Jose, CA), CD4-BV711 (OKT4, BioLegend, San Diego, CA), CD8α-APC-R700 (RPA-T8, BD), CD45RA-APC-Cy7 (HI100, BioLegend), CCR7-BV785 (G043H7, BioLegend), CD27-BV570 (O323, BioLegend), HLA-DR-PE-Cy7 (G46–6, BD), CD38-FITC (HIT2), TCR Vα7.2-BV421 (3C10. A viability dye (eF506, Affymetrix, Santa Clara, CA) and dump gates (CD14-BV510 M5E2, CD19-BV510 HIB19, BioLegend) were used, and samples were fixed using 1% paraformaldehyde in phosphate buffered saline. Gating for positive populations were established based on FMO (fluorescence minus one) controls. Flow cytometry was done using an LSRII (BD) and data were analyzed using FlowJo 10.1 (Treestar, Ashland, OR). Statistical tests were completed using linear mixed effects modeling to account for the longitudinal study design and intra-individual co-variance, as implemented in the R package ‘lme4’.
Regulatory approval
The proposal was been approved by the FDA (IND #: 15926) and UCSF IRB (13–12675), and registered at ClinicalTrials.gov (NCT02256592). The safety monitoring board comprised of clinical trial investigators met regularly for this study.
Results
Participant characteristics
Nine individuals were screened and 6 were enrolled for FMT (Fig. 1). All enrolled FMT participants were men, with a median age of 61 (range 31–72), a median CD4+ T cell count of 431 (range 357–835), and a median CD4 to CD8 ratio of 0.44 (range 0.33–1.36, see Table 1 for detailed cohort characteristics). None of the 6 recipients had serious adverse effects post-FMT (follow-up 24 weeks). One of the control participants was female and the other was African-American.
Figure 1.
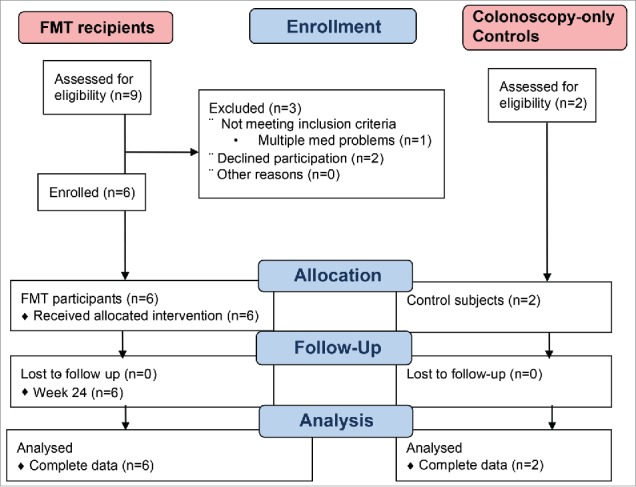
Flow diagram for recruitment of study participants. FMT, fecal microbiome transplant.
Table 1.
Characteristics of the study participants.
| FMT | ID | Age | Gender | Race | CD4 count (cells/μL) | CD8 count (cells/μL) | CD4/8 ratio |
|---|---|---|---|---|---|---|---|
| Yes | 1713 | 31 | Male | White | 463 | 1393 | 0.34 |
| Yes | 2112 | 61 | Male | White | 835 | 613 | 1.34 |
| Yes | 2150 | 53 | Male | White | 431 | 532 | 0.79 |
| Yes | 2294 | 70 | Male | White | 357 | 819 | 0.44 |
| Yes | 2356 | 72 | Male | White | 401 | 1027 | 0.39 |
| Yes | 3164 | 69 | Male | White | 622 | 1122 | 0.56 |
| No | 2447 | 57 | Female | White | 815 | 927 | 0.88 |
| No | 2558 | 71 | Male | Black | 257 | 301 | 0.85 |
Healthy donor gut microbiomes differ from HIV-infected recipients
At baseline before FMT, HIV-infected individuals exhibited microbiota profiles that were distinct from uninfected donors (PERMANOVA test P = 0.043, R2 = 0.253). Numerous taxa differed in abundance between uninfected donors and recipients before FMT. Notably, HIV-infected recipients exhibited an enrichment of Prevotella (P = 0.035) and a decreased abundance of Bacteroides (P = 0.020) as well as a trend toward decreased Faecalibacterium abundance (P = 0.07, Supplemental Figure 1a and 1b), which is consistent with prior reports comparing the HIV-infected gut microbiota to uninfected subjects.6-13
HIV-infected FMT recipient microbiota shift toward donor profiles
Bacterial community sequence analyses were performed using quantifications of β diversity, which assess compositional differences between pairs of ecological communities. The unweighted UniFrac β diversity metric, which measures similarity among communities by examining phylogenetic relationships of taxa within both samples, was selected for analyses, as it performed best in classifying donor replicates as belonging to their respective grouping via the PERMANOVA test (Supplemental Table 1). Unweighted UniFrac distances between the paired donor microbiota and each recipient microbiota profile generated from samples collected pre- and post-FMT were calculated. The calculated UniFrac distance between donor and recipient pairs decreased following FMT, indicating that that the fecal microbiota of FMT recipients became significantly more similar to that of the donor (Fig. 2A). In comparison, control subjects who underwent bowel lavage and colonoscopy alone showed no significant change with respect to their microbial compositional similarity to that of the donor microbial community. The compositional relatedness between donors and recipients were most significant at weeks 2 and 4 (P < 0.01) following FMT and less so at week 8 (P = 0.04). Canberra β diversity distances, which measure community similarity by examining numbers of shared taxa between samples, were also calculated and similarly exhibited significant shift toward donor profiles after FMT (Supplemental Figure 2). Furthermore, proportions of shared OTUs between donor and recipient microbiota profiles also increased following FMT (P = 0.0019, Fig. 2B). To understand which specific bacterial taxa contributed to this increase in similarity with the donors, we examined differences in all microbial genera before and after FMT. No changes were significant in this small cohort following adjustment for false discovery rates (Supplemental Table 3). However, nominal increases in Faecalibacterium and Rikenellaceae, and decreases in Erysipelotrichaceae were observed in recipients post-FMT (Fig. 3, Supplemental Table 3), taxa that have been found to exhibit consistent abundance shifts in HIV-infected subjects by prior reports.6-13
Figure 2.
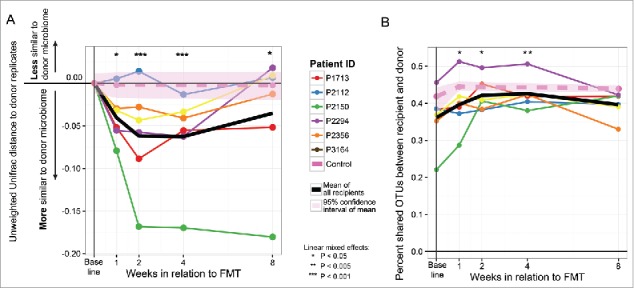
Change in UniFrac distance over time in recipients and controls relative to donors. A) Significant shifts in the microbiome occur and persist post-FMT, but are diminished by week 8. Statistical significance of change in ecological distance to donor profiles before FMT as compared with after FMT was assessed statistically using linear mixed effects modeling. Control subjects (pink dotted line, average of 2 subjects) exhibited no significant changes in their microbiome relative to donors before and after colonoscopy. B) Proportions of shared OTUs between each recipient profile and its respective donor were calculated and plotted across time. Linear mixed effects was used to assess significance of difference in shared OTU proportions pre and post-FMT (P = 0.0019).
Figure 3.
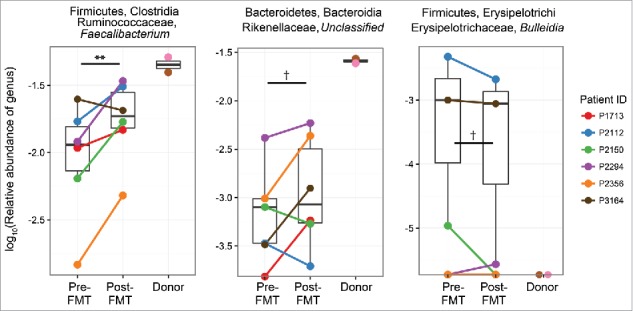
Changes in relative abundance of key gut-resident bacterial genera after FMT. Selected genera shown are Faecalibacterium, an unclassified Rikenellaceae family member genus, and the Bulleidia genus within the Erysipelotrichaceae family (**, P = 0.005; †, P < 0.10). FMT, fecal microbiome transplant.
Shifts toward donor microbiome profiles are modest in comparison to those observed in recurrent C. difficile infection
Ordination of microbiota profiles for HIV-infected recipients and uninfected donors was performed using principal coordinate analysis in conjunction with the unweighted UniFrac distance metric, which showed a retention of intra-individual clustering post-FMT in recipients with modest shifts toward respective donor microbiota compositions (Fig. 4A). Furthermore, microbiota profiles of recipients post-FMT remained significantly different from each of their donors by PERMANOVA (P < 0.05, Supplemental Table 2), suggesting an incomplete change in the recipient microbiome and a lack of full FMT engraftment.
Figure 4.
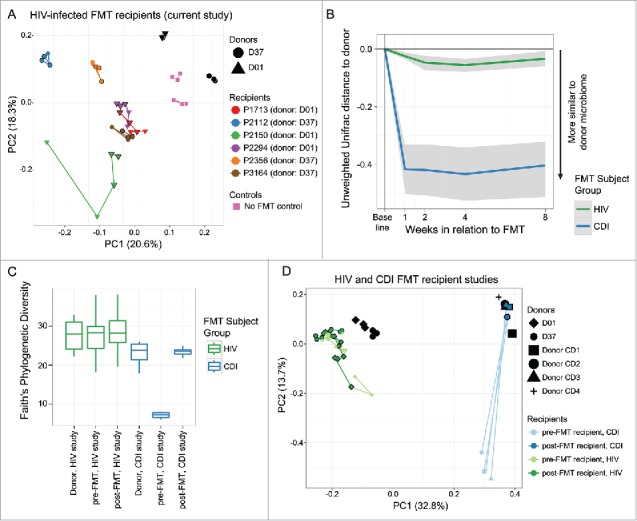
Microbiome shifts much more pronounced in recurrent C. difficile infection (CDI) subjects than in HIV-infected subjects post-FMT. A) Principal coordinate analysis (PCoA) representing triplicate donor microbiota profiles and HIV-infected recipient microbiota community dynamics pre- and post-FMT was generated using the Unweighted Unifrac distance metric. After FMT, recipient microbiota profiles remain distinct from the donors. Points outlined in black are post-FMT time points, and shapes of recipient points reflect which donor material was infused by FMT (Donor 01, circles; Donor 37, triangles), while control subjects that received only bowel lavage over the same time period are shown as squares. Lines connect subject sample time points in a temporally linear fashion. B) Unweighted UniFrac distances were calculated as in Figure. 2 between each recipient stool microbiota profile time point and its respective donor, using data in the current study for HIV-infected subjects and the study by Weingarden et al. for CDI subjects. C) α diversity was calculated using the Faith's Phylogenetic Diversity metric for each sample in each category shown. D) PCoA plot of data from recurrent CDI subjects given FMT by Khoruts et al.24 reveals that CDI subjects differ greatly from donor samples pre-FMT and cluster closely with donor samples post-FMT. E) PCoA plot of data from recurrent CDI subjects given FMT24 and samples from the current study in HIV-infected subjects shows that movement of the microbiome toward the donor samples is much more dramatic for CDI subjects than for HIV-infected subjects as quantified discretely in panel B.
As CDI is the main current indication for which FMT is performed clinically, we compared levels of engraftment in that state in a prior study34 to our current pilot trial in HIV-infected subjects. Significantly greater compositional shift in affected CDI recipients toward the donor community was observed as compared with HIV-infected subjects using unweighted UniFrac β diversity distances as used above (Fig. 4B). Furthermore, a significant increase in α diversity (defined as numbers of observed unique taxa and their evenness of abundance distribution) was seen in CDI subjects after FMT, which was not observed in our HIV-infected subjects post-FMT (Fig. 4C). Principal coordinates ordination also revealed a more dramatic shift toward donor profiles for CDI subjects than for HIV-infected subjects undergoing FMT (Fig. 4D).
Markers of HIV-associated inflammation remain stable after FMT
Markers of immune activation were assessed across time, and no trends were evident in changes in the expression of the activation markers CD38 and HLA-DR on CD8+ T cells, activity of the inflammation-associated indoleamine 2,3-dioxygenase pathway as measured by plasma ratios of kynurenine to tryptophan, or in plasma levels of the innate immune activation marker IL-6 or sCD14 (Supplemental Figure 3). A nominal decrease in expression of the immune exhaustion-associated marker PD-1 on CD8+ T cells was observed though this did not meet statistical significance (P = 0.07, Benjamini-Hochberg false discovery rate Q = 0.29, Supplemental Figure 3).
Discussion
Ongoing inflammation and immune activation persist in HIV-infected subjects despite optimal antiretroviral therapy, and markers of this inflammation remain among the strongest predictors of morbidity and mortality in treated HIV-infected subjects.14,38 Accordingly, identification of the etiology of this inflammation as well as development of novel strategies to mitigate inflammation are a high biomedical priority. Our prior work established a novel link between the altered gut microbiome in treated HIV infection and systemic markers of inflammation including the kynurenine pathway.6 Whether the gut microbiota causally contributes to chronic inflammation in treated HIV infection remains poorly understood. Methods to alter the gut microbiota in the form of fecal microbiome transplantation present an experimental tool that has been found to be safe and effective for the reversion of the dysbiotic inflammatory condition of recurrent Clostridium difficile infection. Here, we present evidence that a single introduction of fecal microbiome transplantation via colonoscopy can result in a modest degree of microbial engraftment into HIV-infected, ART-suppressed recipients. Within recipients, specific taxa that trended toward relative abundances found in the microbiota of the donors included Faecalibacterium, Rikenellaceae, and Erysipelotrichaceae. Though a statistically significant shift in overall community composition to donor profiles was observed in recipients following FMT, shifts in individual taxa were not significant after false discovery rate correction, likely due to the inter-individual heterogeneity as well as the very small number of subjects in this study. Furthermore, the immune profile was largely unchanged post-FMT and the nominal decrease in PD-1 expression on CD8 T cells in the context of multiple immune markers is likely attributable to false discovery.
While microbial engraftment was measurable in our study, the degree of engraftment did not resemble that observed in the treatment of recurrent Clostridium difficile infection (CDI), and was not seen in all subjects. In the setting of CDI, engraftment of donor stool was found to be significantly greater in magnitude than that in our HIV-infected subjects. A putative explanation for the modest engraftment in our study could be the vastly decreased α diversity seen in CDI as compared with healthy and HIV-infected subjects. A prominent macroecological phenomenon is “resilience in diversity,” in which a diverse community (defined by the richness and evenness of component member distribution) has greater capacity to restore its microbial composition after stress than one that is less ecologically diverse.39 Thus, the uniform and phylogenetically restricted community present during recurrent CDI may be susceptible to engraftment by the diverse donor microbial community. Another possible contributor for why CDI subjects experience more complete engraftment after FMT is that patients with recurrent CDI are invariably treated with antibiotics before FMT,18,40 and such antibiotic conditioning will destabilize the existing microbial community, promoting engraftment of another community. Indeed, the subject that experienced greatest engraftment by the 2 metrics used herein also had the greatest magnitude of microbiome change in the 2 time points before FMT, suggesting the subject's microbiome was less stable over time than the microbial communities of the other participants and perhaps more susceptible to engraftment of exogenously introduced microbes. Such ecological instability as introduced by the stressor of antibiotic-mediated microbiota disruption may augment engraftment of donor microbes during FMT procedures. However, studies in mice suggest that antibiotics may instead drive a microbiome shift according to the spectrum of the given antibiotic.41 Alternatively, coprophagic behavior in mice42 and repeated exposure to FMT in several ulcerative colitis trials43,44 can override the stability of one community by another, suggesting the importance of repeated inoculation in the absence of antibiotics. Efforts to understand the factors that stabilize or destabilize a microbial community will inform future interventions that attempt to introduce a consortium of microbes into the gastrointestinal tract.
We nevertheless observed changes in abundance of several taxa that have been previously reported to be altered in abundance in HIV-infected subjects as compared with healthy controls, including Faecalibacterium and Rikenellaceae, which are depleted in HIV, and Erysipelotrichaceae, which have been observed as being enriched in HIV.6-10,45,46 Notably, Erysipelotrichaceae exhibited an apparent modest depletion after FMT and Faecalibacterium and Rikenellaceae were increased after FMT in our pilot study. Intriguingly, the only subject not to exhibit an increase in Faecalibacterium post-FMT was the only subject not to exhibit a decrease in PD-1 expression on CD8+ T cells, consistent with the hypothesis that this clade of gut-resident bacteria may modulate systemic immune activation. Depletion of Faecalibacterium has been linked with inflammatory bowel disease in numerous studies and meta-analyses,47-50 and has been shown to exert anti-inflammatory effects in murine experimental colitis.47 Thus, an increased abundance of this bacterium in the gut may contribute to restoration of immune homeostasis in multiple gastrointestinal disease states, highlighting the potential for FMT as a therapeutic intervention in settings outside of HIV (and CDI).
Other taxa notably altered in HIV did not exhibit any changes in abundance as a result of FMT, including Bacteroides, which is decreased in abundance in HIV-infected individuals, Proteobacteria members of the Enterobacteriaceae family, which are more abundant in HIV infection. Prevotella was enriched in HIV-infected recipients as compared with donors, but did not change in abundance after FMT. This genus has been recently observed to associate with behavioral factors and not HIV status,51 suggesting that this clade of bacteria may not be causally related to HIV-associated inflammation. However, Bacteroides and Enterobacteriaceae may be important taxa that modulate gut mucosal immune homeostasis in HIV infection, as several gut-resident Enterobacteriaceae members have been shown causally to induce chronic inflammation,52-54 and Bacteroides members are associated with restricting immune activation in mouse models.55 Thus, strategies to increase engraftment of donor stool during FMT may alter abundance of the various aforementioned taxa in the gut microbial community, and may in turn produce improvements in markers of chronic inflammation.
FMT has been reported to be safe, with few reported adverse effects including patients who may be immunocompromised, such as HIV-infected and bone marrow transplant recipients.22,23 Although we excluded those who have CD4 counts less than 200 cells/mL and those who have untreated HIV infection, our study provides further information about the safety of this procedure. Several individuals who participated in our study experienced longstanding and idiopathic chronic loose stools. Anecdotally, several of these participants reported that they felt FMT improved the odor of their stool, improved stool form on the Bristol-Stool scale, and would repeat the procedure again because of the subjective health benefits.
In summary, FMT was well tolerated in ART-treated HIV-infected individuals. Engraftment was detectable though modest, and appeared to be limited to specific bacterial taxa. In light of successful engraftment during CDI, a protocol mimicking CDI treatment, where antibiotic conditioning occurs before FMT, would appear to be warranted. Given the association between specific microbial taxa and systemic inflammation, efforts to enhance engraftment and displace pro-inflammatory microbes may lead to reduction in systemic inflammation, thereby reducing excess morbidity and mortality observed during chronic HIV-infection.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Number R21DK104664. The work was also supported in part by the American College of Gastroenterology Pilot Research Award and the UCSF Academic Senate. I.V.C. received support from the California HIV/AIDS Research Grants Program Office of the University of California, D13-SF-388, and the Cancer Research Institute Irvington Postdoctoral Fellowship Award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- [1].Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol 2003; 77:11708-17; PMID:14557656; https://doi.org/ 10.1128/JVI.77.21.11708-11717.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 2005; 434:1148-52; PMID:15793562; https://doi.org/ 10.1038/nature03513 [DOI] [PubMed] [Google Scholar]
- [3].Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, Barbour JD, Lowe MM, Jayawardene A, Aweeka F, et al.. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med 2010; 2:32ra6; PMID:20484731; https://doi.org/ 10.1126/scitranslmed.3000632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, et al.. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 2008; 112:2826-35; PMID:18664624; https://doi.org/ 10.1182/blood-2008-05-159301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, et al.. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med 2008; 14:421-8; PMID:18376406; https://doi.org/ 10.1038/nm1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M, et al.. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 2013; 5:193ra91; PMID:23843452; https://doi.org/ 10.1126/scitranslmed.3006438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, French A, Demarais P, Sun Y, Koenig L, et al.. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014; 10:e1003829; PMID:24586144; https://doi.org/ 10.1371/journal.ppat.1003829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, et al.. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7(4):983-94; PMID:24399150; https://doi.org/ 10.1038/mi.2013.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, Anton P, Braun J. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 2013; 1:26; PMID:24451087; https://doi.org/ 10.1186/2049-2618-1-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, Palmer BE. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14:329-39; PMID:24034618; https://doi.org/ 10.1016/j.chom.2013.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, Wanke CA, Ward HD. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis 2015; 211:19-27; PMID:25057045; https://doi.org/ 10.1093/infdis/jiu409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, Lankowski A, Baldridge MT, Wilen CB, Flagg M, et al.. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 2016; 19:311-22; PMID:26962942; https://doi.org/ 10.1016/j.chom.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vazquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrús ML, Madrid N, Vallejo A, Sainz T, Martínez-Botas J, Ferrando-Martínez S, et al.. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol 2015; 8:760-72; PMID:25407519; https://doi.org/ 10.1038/mi.2014.107 [DOI] [PubMed] [Google Scholar]
- [14].Kalayjian RC, Machekano RN, Rizk N, Robbins GK, Gandhi RT, Rodriguez BA, Pollard RB, Lederman MM, Landay A. Pretreatment levels of soluble cellular receptors and interleukin-6 are associated with HIV disease progression in subjects treated with highly active antiretroviral therapy. J Infect Dis 2010; 201:1796-805; PMID:20446847; https://doi.org/ 10.1086/652750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, Barbour JD, Lowe MM, Jayawardene A, Aweeka F, et al.. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med 2010; 2:32ra6; PMID:20484731; https://doi.org/ 10.1126/scitranslmed.3000632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hunt PW. Th17, gut, and HIV: Therapeutic implications. Curr Opin HIV AIDS 2010; 5:189-93; PMID:20543599; https://doi.org/ 10.1097/COH.0b013e32833647d9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al.. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012; 482:179-85; PMID:22297845; https://doi.org/ 10.1038/nature10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al.. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407-15; PMID:23323867; https://doi.org/ 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- [19].Seekatz AM, Aas J, Gessert CE, Rubin TA, Saman DM, Bakken JS, Young VB. Recovery of the gut microbiome following fecal microbiota transplantation. MBio 2014; 5:e00893-14; PMID:24939885; https://doi.org/ 10.1128/mBio.00893-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hempel S, Newberry SJ, Maher AR, Wang Z, Miles JN, Shanman R, Johnsen B, Shekelle PG. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: A systematic review and meta-analysis. JAMA 2012; 307:1959-69; PMID:22570464; https://doi.org/ 10.1001/jama.2012.3507 [DOI] [PubMed] [Google Scholar]
- [21].Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 2011; 53:994-1002; PMID:22002980; https://doi.org/ 10.1093/cid/cir632 [DOI] [PubMed] [Google Scholar]
- [22].Kelly CR, Ihunnah C, Fischer M, et al.. Fecal Microbiota Transplantation (FMT) for Treatment of Clostridium difficile Infection (CDI) in Immunocompromised Patients. Am J Gastroenterol 2014. Jul; 109(7):1065-71; PMID:24890442; https://doi.org/ 10.1038/ajg.2014.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Elopre L, Rodriguez M. Fecal microbiota therapy for recurrent Clostridium difficile infection in HIV-infected persons. Ann Intern Med 2013; 158:779-80; PMID:23689775; https://doi.org/ 10.7326/0003-4819-158-10-201305210-00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010; 44:354-60; PMID:20048681; https://doi.org/ 10.1097/MCG.0b013e3181c87e02 [DOI] [PubMed] [Google Scholar]
- [25].Lederman MM, Calabrese L, Funderburg NT, Clagett B, Medvik K, Bonilla H, Gripshover B, Salata RA, Taege A, Lisgaris M, et al.. Immunologic failure despite suppressive antiretroviral therapy is related to activation and turnover of memory CD4 cells. J Infect Dis 2011; 204:1217-26; PMID:21917895; https://doi.org/ 10.1093/infdis/jir507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wada NI, Jacobson LP, Margolick JB, Breen EC, Macatangay B, Penugonda S, Martínez-Maza O, Bream JH. The effect of HAART-induced HIV suppression on circulating markers of inflammation and immune activation. AIDS 2015; 29:463-71; PMID:25630041; https://doi.org/ 10.1097/QAD.0000000000000545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Serrano-Villar S, Sainz T, Lee SA, Hunt PW, Sinclair E, Shacklett BL, Ferre AL, Hayes TL, Somsouk M, Hsue PY, et al.. HIV-infected individuals with low CD4/CD8 ratio despite effective antiretroviral therapy exhibit altered T cell subsets, heightened CD8+ T cell activation, and increased risk of non-AIDS morbidity and mortality. PLoS Pathog 2014; 10:e1004078; PMID:24831517; https://doi.org/ 10.1371/journal.ppat.1004078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen Y, Chen S, Kang J, Fang H, Dao H, Guo W, Lai C, Lai M, Fan J, Fu L, et al.. Evolving molecular epidemiological profile of human immunodeficiency virus 1 in the southwest border of China. PLoS One 2014; 9:e107578; PMID:25207977; https://doi.org/ 10.1371/journal.pone.0107578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, Kelly C, Khoruts A, Louie T, Martinelli LP, et al.. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 2011; 9:1044-9; PMID:21871249; https://doi.org/ 10.1016/j.cgh.2011.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].OpenBiome Stool donor criteria, treatment information, and safety documentation. Available at: 1. http://www.openbiome.org/stool-donation/ 2. http://www.openbiome.org/treatment-information 3. http://www.openbiome.org/safety/
- [31].Vujkovic-Cvijin I, Swainson LA, Chu SN, Ortiz AM, Santee CA, Petriello A, Dunham RM, Fadrosh DW, Lin DL, Faruqi AA, et al.. Gut-Resident Lactobacillus Abundance Associates with IDO1 Inhibition and Th17 Dynamics in SIV-Infected Macaques. Cell Rep 2015; 13:1589-97; PMID:26586432; https://doi.org/ 10.1016/j.celrep.2015.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011; 108:4516-22; PMID:20534432; https://doi.org/ 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al.. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335-6; PMID:20383131; https://doi.org/ 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Weingarden A, Gonzalez A, Vazquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D, Knights D, Unno T, Bobr A, Kang J, et al.. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 2015; 3:10; PMID:25825673; https://doi.org/ 10.1186/s40168-015-0070-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 2007; 73:1576-85; PMID:17220268; https://doi.org/ 10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. Forensic identification using skin bacterial communities. Proc Natl Acad Sci U S A 2010; 107:6477-81; PMID:20231444; https://doi.org/ 10.1073/pnas.1000162107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology; 2001; 26:32-46; https://doi.org/ 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- [38].Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, Pedersen C, Ruxrungtham K, Lewin SR, Emery S, et al.. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis 2011; 203:780-90; PMID:21252259; https://doi.org/ 10.1093/infdis/jiq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Folke C, Carpenter S, Walker B, et al.. Regime shifts, resilience, and biodiversity in ecosystem management. Annu Rev Ecol Evol S 2004; 35:557-81; ; https://doi.org/ 10.1146/annurev.ecolsys.35.021103.105711 [DOI] [Google Scholar]
- [40].McFarland LV, Elmer GW, Surawicz CM. Breaking the cycle: Treatment strategies for 163 cases of recurrent Clostridium difficile disease. Am J Gastroenterol 2002; 97:1769-75; PMID:12135033; https://doi.org/ 10.1111/j.1572-0241.2002.05839.x [DOI] [PubMed] [Google Scholar]
- [41].Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, Guigo R, Knight R, Guarner F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res 2010; 20:1411-9; PMID:20736229; https://doi.org/ 10.1101/gr.107987.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Laukens D, Brinkman BM, Raes J, De Vos M, Vandenabeele P. Heterogeneity of the gut microbiome in mice: Guidelines for optimizing experimental design. FEMS Microbiol Rev 2016; 40:117-32; PMID:26323480; https://doi.org/ 10.1093/femsre/fuv036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, et al.. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 2015; 149:102-9 e6; PMID:25857665; https://doi.org/ 10.1053/j.gastro.2015.04.001 [DOI] [PubMed] [Google Scholar]
- [44].Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RW, Connor S, Ng W, Paramsothy R, et al.. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: A randomised placebo-controlled trial. Lancet 2017; 389(10075):1218-28; PMID:28214091; https://doi.org/ 10.1016/S0140-6736(17)30182-4 [DOI] [PubMed] [Google Scholar]
- [45].Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, et al.. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 2012; 151:253-66; PMID:23063120; https://doi.org/ 10.1016/j.cell.2012.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ellis CL, Ma ZM, Mann SK, Li CS, Wu J, Knight TH, Yotter T, Hayes TL, Maniar AH, Troia-Cancio PV, et al.. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr 2011; 57:363-70; PMID:21436711; https://doi.org/ 10.1097/QAI.0b013e31821a603c [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al.. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008; 105:16731-6; PMID:18936492; https://doi.org/ 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al.. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014; 15:382-92; PMID:24629344; https://doi.org/ 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007; 104:13780-5; PMID:17699621; https://doi.org/ 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett 2014; 588:4223-33; PMID:25307765; https://doi.org/ 10.1016/j.febslet.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Noguera-Julian M, Rocafort M, Guillen Y, Rivera J, Casadellà M, Nowak P, Hildebrand F, Zeller G, Parera M, Bellido R, et al.. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine 2016; 5:135-46; PMID:27077120; https://doi.org/ 10.1016/j.ebiom.2016.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al.. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010; 8:292-300; PMID:20833380; https://doi.org/ 10.1016/j.chom.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007; 2:204; PMID:18030708; https://doi.org/ 10.1016/j.chom.2007.08.002 [DOI] [PubMed] [Google Scholar]
- [54].Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al.. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 2010; 467:426-9; PMID:20864996; https://doi.org/ 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008; 453:620-5; PMID:18509436; https://doi.org/ 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


