ABSTRACT
In humans, sleeping sickness (i.e. Human African Trypanosomiasis) is caused by the protozoan parasites Trypanosoma brucei gambiense (Tbg) in West and Central Africa, and T. b. rhodesiense in East Africa. We previously showed in vitro that Tbg is able to excrete/secrete a large number of proteins, including Translationally Controlled Tumor Protein (TCTP). Moreover, the tctp gene was described previously to be expressed in Tbg-infected flies. Aside from its involvement in diverse cellular processes, we have investigated a possible alternative role within the interactions occurring between the trypanosome parasite, its tsetse fly vector, and the associated midgut bacteria. In this context, the Tbg tctp gene was synthesized and cloned into the baculovirus vector pAcGHLT-A, and the corresponding protein was produced using the baculovirus Spodoptera frugicola (strain 9) / insect cell system. The purified recombinant protein rTbgTCTP was incubated together with bacteria isolated from the gut of tsetse flies, and was shown to bind to 24 out of the 39 tested bacteria strains belonging to several genera. Furthermore, it was shown to affect the growth of the majority of these bacteria, especially when cultivated under microaerobiosis and anaerobiosis. Finally, we discuss the potential for TCTP to modulate the fly microbiome composition toward favoring trypanosome survival.
KEYWORDS: bacteria midgut flora, Glossina, Trypanosoma brucei gambiense, Translationally Controlled Tumor Protein (TCTP)
Introduction
Trypanosoma brucei gambiense (Tbg) and T. b. rhodesiense are protozoan parasites that cause sleeping sickness (Human African Trypanosomiasis/HAT), respectively in West and Central Africa, and in East Africa. It is a neglected tropical disease endemic to many sub-Saharan African countries and responsible for severe social and economic issues. Several biologic control strategies have been developed over the years, including the control of tsetse fly populations by aerial insecticide applications1-3 and/or by the sterile insect technique.4 Moreover, various diagnostic tools have been developed and improved,5-12 as well as the treatments of infected humans.13 However, the disease still has a strong presence and continuing research is required to fight it.
Sleeping sickness is propagated by a hematophagous vector, the tsetse fly, which ingests the trypanosome parasite during a blood meal on an infected host. The trypanosome must then fulfill part of its complex biologic cycle in the fly before it is transmitted to yet another mammalian host when the fly consumes a subsequent blood meal. As a HAT control strategy, interrupting a step within the trypanosome life cycle inside the fly could become an effective approach to prevent its transmission. In this context, several studies have already investigated the molecular dialog occurring between the tsetse fly, its midgut bacteria and the ingested trypanosomes to identify factors involved in these interactions that could become potential targets to hinder vector competence.
Previous investigations have allowed identifying several proteins that are excreted/secreted by trypanosomes.14 A high number of midgut proteins were also identified as differentially expressed in Tbg-stimulated versus non-stimulated Glossina palpalis gambiensis flies.15 Similarly, numerous tsetse, midgut bacteria and trypanosome genes were found to be differentially expressed in Tbg-infected vs. non-infected tsetse flies.16 Among these proteins we identified the trypanosomal Translationally Controlled Tumor Protein (TCTP), for which the corresponding gene was found to be overexpressed in Tbg-infected flies.
TCTP is a ubiquitously distributed protein in eukaryotes that appears to be highly conserved across animals and plants.17 It has been reported to play multiple roles in cellular processes including cell proliferation,18,19 the cell cycle,20 stress response21 and apoptosis,22 and is involved in embryo development.23 As such, TCTP can be considered a good therapeutic target against several diseases or physiological disorders.24-26 Regarding the Tbg form of TCTP (TbgTCTP), the fact that the protein can be excreted/secreted by the trypanosome raises a question about its paracrine role, since the parasite develops in the fly gut where several bacteria are harbored, some of which have been shown to be involved in tsetse fly vector competence.27,28
Here we report on the production of the TbgTCTP recombinant protein (rTbgTCTP) and its biological effect on several bacterial strains that were previously isolated from the midgut of diverse Glossina species sampled in different HAT foci.29-31 These results will provide a basis for future research into controlling HAT from the perspective of the trypanosome parasite.
Results
Production, purification and molecular characterization of rTbgTCTP
The sequence of the synthesized Tbg 927.8.6750 gene exhibited a 100% match with the mRNA sequence from the NCBI database (AAX80036.1, EMBL-EBI). In addition, the amino acid sequence of the corresponding protein produced using the baculovirus Spodoptera frugiperda (Sf 9) / insect cell system displayed a 100% match with the TCTP amino acid sequence from the T. brucei gambiense reference strain TREU97 (Fig. 1A).
Figure 1.
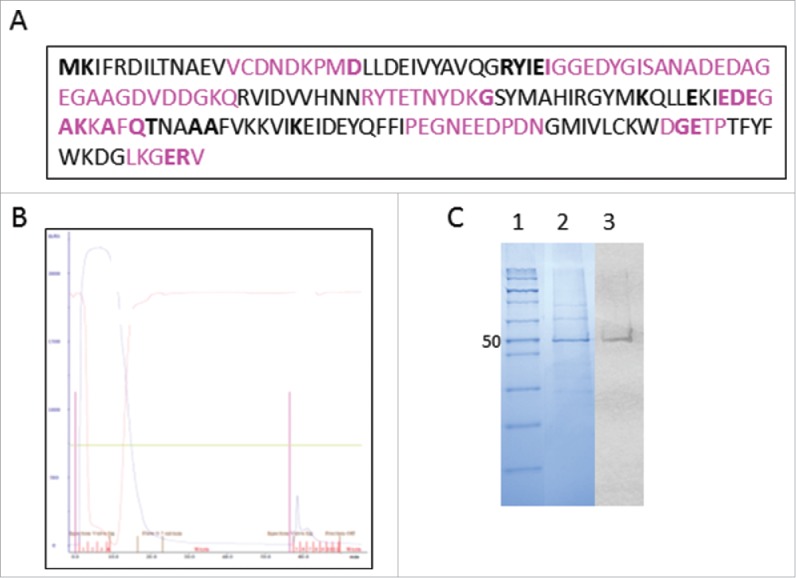
(A) Protein sequence of rTbgTCTP with linearized epitope (pink); amino acids involved in the epitope conformation are presented in bold type. (B) Purification profile for rTbgTCTP on a nickel-charged affinity resin (Ni-NTA) column. (C) sodium dodecyl sulfate - PAGE analysis of rTbgTCTP. Lane 1: protein molecular marker (Novex Sharp Pre-Stained Protein Standard, Thermo Fisher Scientific); lane 2: electrophoresis of rTbgTCTP (10 µg) purified on a Ni-NTA column stained with Coomassie Blue; lane 3: western blot of rTbgTCTP (10 µg) on nitrocellulose membrane using anti-His monoclonal antibody (mAb).
Figure 1B shows the result from purifying the produced rTbgTCTP on the Ni-NTA (nickel-charged affinity resin) agarose column. The purification process was continuously monitored (OD = 280 nm) and the first peak of the chromatogram (representing proteins unbound to the Ni-NTA column) corresponded to non-specific proteins produced by the insect cells; the second peak eluted from the column contained the rTbgTCTP. The purified fraction was then loaded onto 10% SDS-PAGE (sodium dodecyl sulfate - polyacrylamide gel electrophoresis) gels; after electrophoresis and staining, the presence of an intense protein band was shown at 50 KDa, corresponding to the molecular weight of rTbgTCTP (Fig. 1C, lane 2). The presence of rTbgTCTP in the 50 KDa protein band was further confirmed by immunodetection using HRP (horseradish peroxidase) - conjugated anti-histidine antibody (Fig. 1C, lane 3). The rTbgTCTP yield was approximately 174 mg per liter of insect cell culture medium, with a 95% purity rate (Bio 1D software).
Protein structure and sequence homologies with TCTP from other organisms
For our 3D structure analysis of TCTP, only a limited number of TCTP sequences were available in the EMBL-EBI database (Protein Data Bank in Europe) of 3D protein structures. Our analysis indicates that the T. b. gambiense rTbgTCTP amino acid sequence had an overall 35.5% homology with the TCTP model from Plasmodium falciparum (3P3K), although it displays homologous portions distributed homogeneously all along the protein sequence (Fig. 2A–C). The portions of sequences forming TCTP epitopes represent 46% of the total protein sequence; when the protein is linearized (i.e., the primary structure), these portions are distributed homogeneously all along the protein sequence (Fig. 1A and Fig. 2A). Our results also show that TCTP contains very few conformational epitope sites (Fig. 2B) or lysine sites (Fig. 2C).
Figure 2.

(A) The Tbg 927.8.6750 tertiary structure. The part corresponding to the epitope is in pink. (B) Linear epitope (pink) of Tbg 927.8.6750 from Trypanosoma brucei gambiense (Bepiprep 1.0 server) and the conformational epitope 3P3K (green; DiscoTope 2.0 server). (C) Lysine localization of 3P3K (red).
Figure 3 depicts a clustering of TCTPs from different species, based on differences in their respective amino acid sequences. The proteins from trypanosomatids cluster in a group that is clearly separate from that formed by the TCTP of different Glossina species, with 5 TCTPs from this latter group appearing almost identical. The genetic distance between this group and Musca domestica TCTP (used as a control) is shorter than the distance between TCTPs from flies and trypanosomatids. The TCTPs from trypanosomatids appear to be closely related (high homologies), even though those from Trypanosoma and Leishmania cluster into 2 distinct subgroups.
Figure 3.

Phylogenetic tree for TCTP among trypanosomatids and their corresponding host species (PhyML). The PhyML website was used to construct the phylogenetic tree based on the amino acid sequences of the corresponding TCTP and default parameters of the server. Numbers above branches indicate bootstrap values.
Evidence of rTbgTCTP binding to Glossina midgut bacteria
The occurrence of binding between rTbgTCTP and Glossina midgut bacteria was revealed by western blot analysis. Figure 4 presents a representative result of the binding capacity between rTbgTCTP and Providencia bacteria species from Glossina palpalis midguts. As shown in Figure 4, an rTbgTCTP band (lane 4) similar to the rTbgTCTP control (lane 2) was only revealed when bacteria were incubated in the presence of rTbgTCTP, even though the bacteria were washed 4 times with PBS before being boiled and treated with β-mercaptoethanol. The fact that the PBS washing did not eliminate the rTbgTCTP indicates that the protein was bound to the bacteria wall, possibly through the formation of disulfide bonds between –SH groups of rTbgTCTP and the bacteria. No band was detected when rTbgTCTP were absent during bacteria incubation (lane 3). Among the 39 bacteria species/strains that we examined (Table 1) for their capacity to bind rTbgTCTP, only 24 showed a positive result, whereas the others were unable to bind rTbgTCTP in our experimental conditions.
Figure 4.
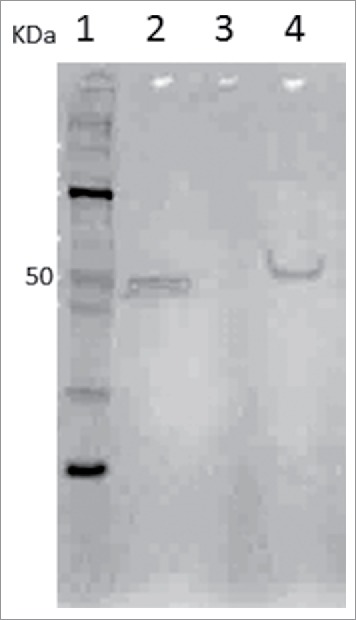
Binding between bacteria and rTbgTCTP as revealed by western blot using anti-His mAb. Lane 1: molecular weight markers (Novex Sharp Pre-Stained Protein Standard, Thermo Fisher Scientific); lane 2: purified rTbgTCTP; lane 3: bacteria (Providencia spp. from Glossina palpalis palpalis) incubated without rTbgTCTP as a control; lane 4: bacteria incubated with rTbgTCTP and subsequently treated with β-mercaptoethanol.
Table 1.
Designation and origin of the bacteria strains examined in this study. Descriptions include the bacterial identifying code and species, as well as the Glossina species from which they were isolated and the country where flies were collected.
| Strains | Results Western blot | Bacteria species | Glossina species | Localization |
|---|---|---|---|---|
| Anne B1 | – | Serratia sp. | Glossina palpalis gambiensis | Insectarium |
| Anne 17B | – | Enterobacter sp. | Glossina palpalis palpalis | Angola_HAT focus of Maria-Teresa (Bengo province) |
| Cam 29 B | + | Providencia sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Campo Beach/Ipono |
| Serratia glossinae | – | Serratia sp. | Glossina palpalis gambiensis | Insectarium |
| Anne L1 | – | Serratia sp. | Glossina palpalis gambiensis | Insectarium |
| Cam 24 C | – | Enterobacter sp. | Glossina nigrofusca | Cameroon_HAT focus of Campo_Campo Beach/Ipono |
| Cam 26 C | + | Enterococcus sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_ND |
| Angola 23 C | + | Serratia sp. | Glossina palpalis palpalis | Angola_HAT focus of Maria-Teresa (Bengo province) |
| Cam 29 A | + | Providencia sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Campo each/Ipono |
| Gmm 11 | + | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Cam 14 A | + | Lactococcus sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Akak |
| Angola 8 B | – | Acinetobacter sp. | Glossina palpalis palpalis | Angola_HAT focus of Maria-Teresa (Bengo province) |
| Angola 8 A | – | Enterobacter sp. | Glossina palpalis palpalis | Angola_HAT focus of Maria-Teresa (Bengo province) |
| Cam 33 B | – | Enterococcus sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_ND |
| Cam 20 A | – | Providencia sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo |
| Cam 32 A | + | Providencia sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo |
| Cam 24 B | – | Enterobacter sp. | Glossina nigrofusca | Cameroon_HAT focus of Campo |
| Cam 18 A | – | Enterobacter sp. | Glossina pallicera | Cameroon_HAT focus of Campo |
| Cam 32 C | + | Providencia sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo |
| Cam 32 A | + | Enterococcus sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Campo each/Ipono |
| Gmm 15 | – | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Cam 11 B | + | Enterobacter sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Akak |
| Cam 33 C | + | Enterococcus sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Campo each/Ipono |
| Angola 16 C | + | Enterococcus sp. | Glossina palpalis palpalis | Angola_HAT focus of Maria-Teresa (Bengo province) |
| Cam 15 A | + | Enterococcus sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Akak |
| Cam 15 B | + | Enterococcus sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Akak |
| Angola 24 C | + | Enterobacter sp. | Glossina nigrofusca | Cameroon_HAT focus of Campo_Campo Beach/Ipono |
| Gmm 09 | + | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Gmm10 | + | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Gmm 02 | + | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Cam 20 B | + | Providencia sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Akak |
| Gmm 04 | – | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Cam 20 C | + | Providencia sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Akak |
| Gmm 07 | + | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Gmm 08 | + | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Gmm 12 | – | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Cam 14 B | + | Lactococcus sp. | Glossina palpalis palpalis | Cameroon_HAT focus of Campo_Akak |
| Gmm 05 | + | Serratia sp. | Glossina morsitans morsitans | Insectarium |
| Gmm 06 | – | Serratia sp. | Glossina morsitans morsitans | Insectarium |
Effect of rTbgTCTP on bacterial growth
Among the 24 bacteria strains that were shown to bind rTbgTCTP, growth kinetics were only obtained for 14 strains (Fig. 5). Figure 5 shows the bacterial growth in the absence of rTbgTCTP, with the respective controls under the different culture conditions (Fig. 5A–C: culture at 25°C under aerobiosis, microaerobiosis, and anaerobiosis, respectively; Fig. 5D–F: similar culture conditions at 37°C). The effect of rTbgTCTP was then evaluated with respect to the growth kinetics of this control experiment.
Figure 5.
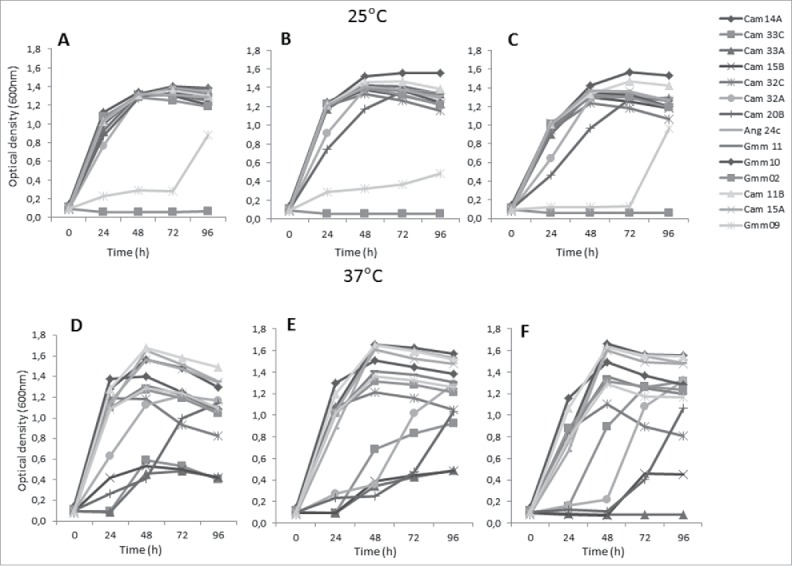
Growth kinetics of the bacteria strains grown in the absence of rTbgTCTP. The bacterial concentration was measured using the OD at 600 nm. Bacteria were cultivated in Mitsuhashi medium at 25°C (A-C) or 37°C (D-F), in either aerobiosis (A, D), microaerobiosis (B, E) or anaerobiosis (C, F).
When cultivated at 25°C, most of the tested bacteria strains were not affected by the presence of rTbgTCTP, regardless of the concentration (data not shown). Only the strains Gmm09 (Serratia sp.), Cam33C (Enterococcus sp) and Cam20B (Providencia sp) exhibited modified growth kinetics in the presence of rTbgTCTP (Fig. 6A).
Figure 6.
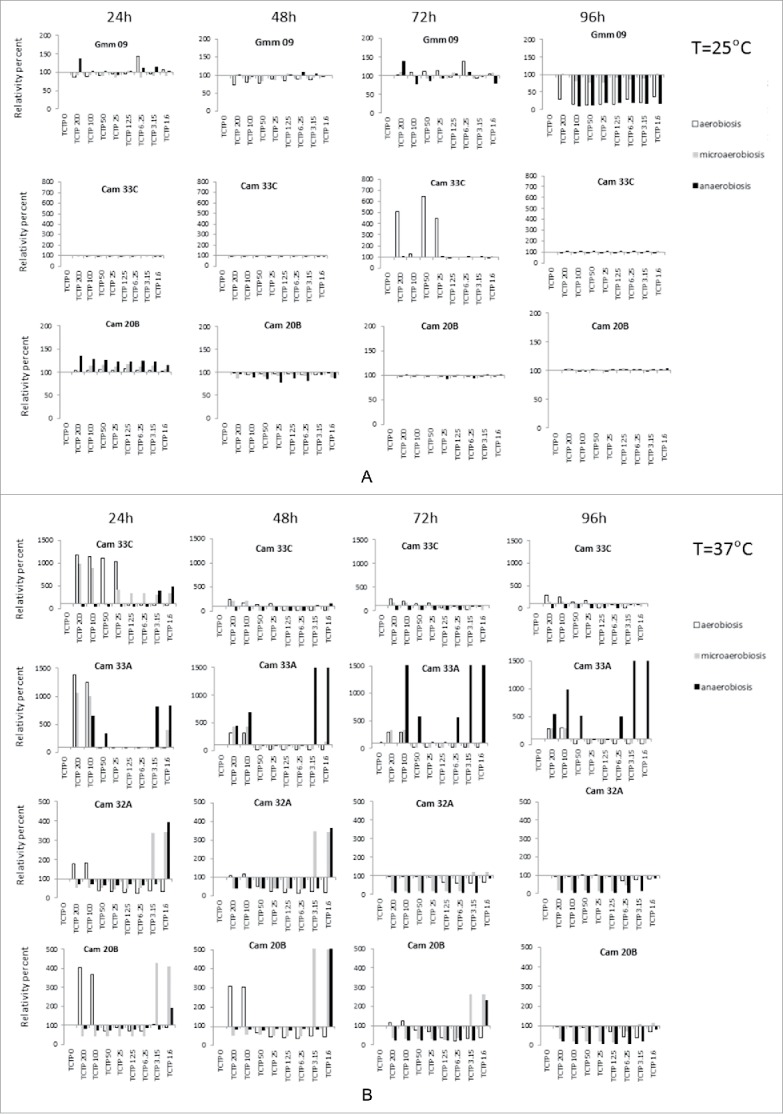
Effect of different concentrations of rTbgTCTP on bacteria growth. The effect was tested at 25°C (A) and 37°C (B-E), under 3 different atmosphere compositions: aerobiosis (white column), microaerobiosis (gray column), and anaerobiosis (black column). Histograms compare, for a given experimental condition (and for each time), the OD of the tested culture (with TCTP at a given concentration) with the OD of the corresponding control culture without TCTP (assigned to the value 100%). The results are the mean of triplicate experiments.
Figure 6.
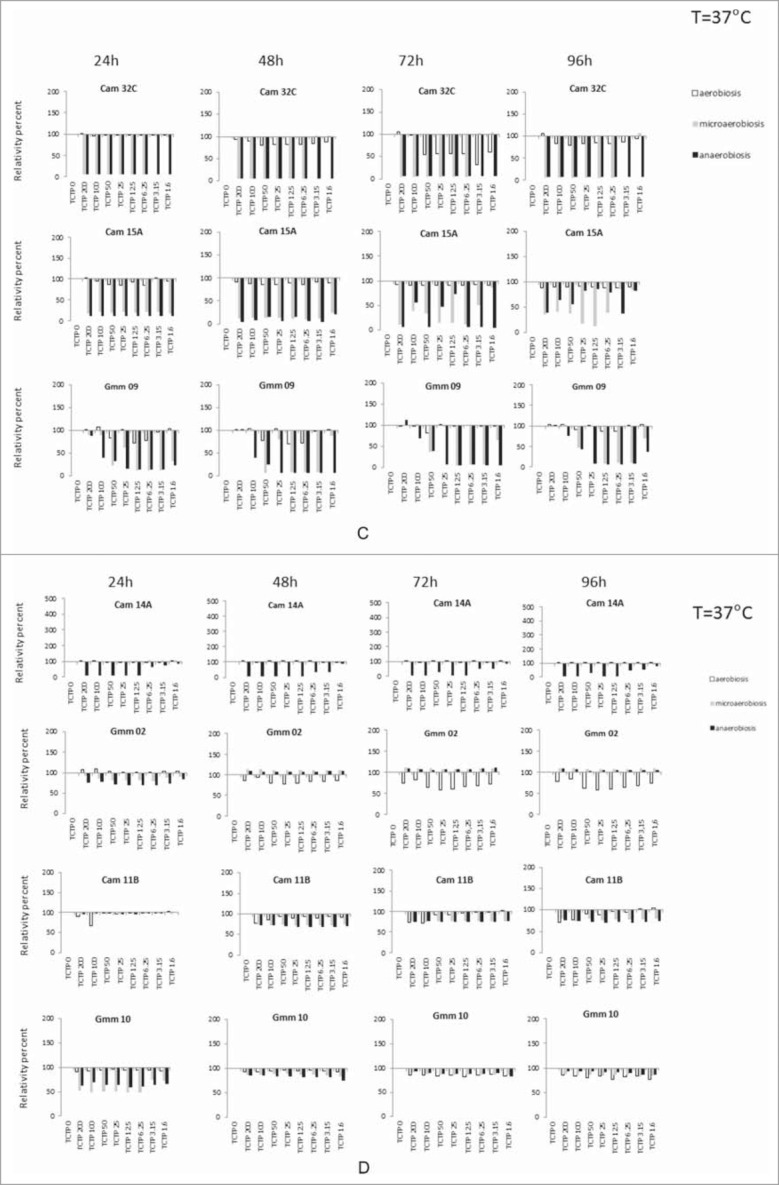
(Continued).
Figure 6.
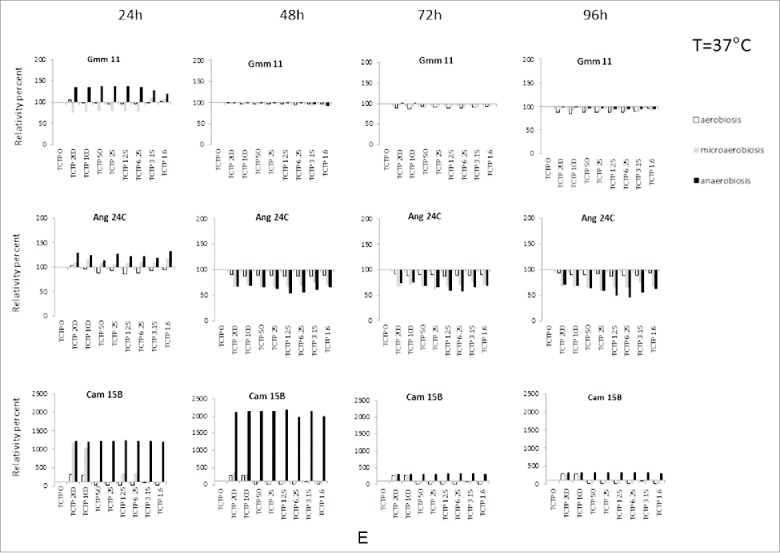
(Continued).
At 37°C, the effect of rTbgTCTP was very diverse depending on the bacterial strain, the culture conditions (aerobiosis, microaerobiosis or anaerobiosis) and the concentration of rTbgTCTP (Fig. 6B–E). With the exception of several strains (Cam33A, Cam33C and Cam20B) after 24 h of culture in aerobiosis, the effect of rTbgTCTP was stronger when the strains were cultured in anaerobiosis or microanaerobiosis. We observed that the presence of rTbgTCTP, no matter the concentration, frequently caused a dramatic decrease in bacterial growth (Fig. 6B–E), with a comparable effect irrespective of the bacterial strain or culture conditions. However, this was not the case for Cam 33C, Cam33A, Cam32A and Cam20B (Fig. 6B), or Gmm11, Ang24C and Cam15B (Fig. 6E). In these cases, growth was stimulated up to 10- or 20-fold the reference of the corresponding control, depending on the rTbgTCTP concentration and the culture conditions, as shown in Fig. 6C (Cam 32C, Cam 15A and Gmm09) and Fig. 6D (Cam14A, Gmm02, Cam11B and Gmm10). This positive or negative effect of rTbgTCTP was detectable with very low concentrations of the protein (i.e., 1.6 µM).
Discussion
TCTP is one of the proteins that we previously demonstrated are excreted/secreted by trypanosomes, whose gene is differentially expressed in trypanosome-infected vs. non-infected Glossina. This protein has been reported to play multiple roles, and we investigated its possible effect on bacteria from Glossina gut microflora using a recombinant TCTP from Tbg along with bacteria isolated from diverse Glossina species. This focus on gut microflora was based on the premises that (i) TCTP in vivo excretion by trypanosomes should be accompanied by its release in the gut, and that (ii) at least some portion of the midgut bacteria are involved in tsetse vector competence.
First, we fully succeeded in producing a recombinant TCTP that corresponds to the TCTP encoded by T. brucei gambiense (GST (glutathione S-transferase) - His-Tbg 927.8.6750 protein, referred to here as rTbgTCTP). As expected, given the data reported in the literature, the rTbgTCTP was very close to its homologs from other trypanosomatids. Of even greater interest was the demonstration that this protein can bind to bacteria isolated from the midgut of different tsetse fly species and interfere with bacteria growth, despite multiple variations in bacteria strain and culture conditions.
Binding of rTbgTCTP with bacteria
Recently, Hansenová Maňásková et al.32 reported on the incorporation of a protease substrate into the bacterial cell wall. This event is processed by a bacterial cell wall-associated protease and involves the valine-leucine-lysine (VLK) peptidyl sequence of the substrate that acts as an anchor for binding the substrate to the bacterium. Interestingly, the rTbgTCTP amino acid sequence comprises a nearly similar peptidyl motif valine-leucine-cysteine-lysine (VLCK), which differs by its inclusion of a cysteine. Further investigations must therefore address whether or not this cysteine could hinder the potential anchor effect attributed to the VLK motif. Nevertheless, the presence of a cysteine moiety in the rTbgTCTP primary structure is of great interest, since the –SH function of this amino acid frequently establishes a disulfide covalent bridge (-S-S-) with the –SH of another cysteine. Such a bridge is commonly involved in the formation and maintenance of protein tertiary (requiring the presence of at least 2 cysteines) or quaternary (requiring at least one cysteine in each subunit) conformations. This disulfide covalent bond is easily broken by β-mercaptoethanol (or dithiothreitol) treatment. This leads us to believe that the binding of rTbgTCTP with bacteria is most probably due to the formation of a disulfide bridge between –SH groups from cysteines in the rTbgTCTP and the bacterial cell wall. This covalent bond is strong enough to maintain the protein/bacterial link during a drastic washing procedure like we used here, although the protein was released when the rTbgTCTP/bacterium complex was treated with β-mercaptoethanol, as revealed in our electrophoresis and immunodetection experiments. Another type of binding could be available between the bacterial membrane and rTbgTCTP. In this scenario, lysin sites of the rTbgTCTP protein, localized on the 3D structural helix with an outside orientation, could play a role in binding to the bacteria membrane, thereby impacting bacteria growth as antimicrobial properties.33
The effect of rTbgTCTP
Our results indicate that the effect of rTbgTCTP can either inhibit or stimulate the growth of bacteria from the Glossina midgut, and that this effect depends mainly on the bacterium under consideration, the culture conditions and (to a lesser extent) the protein concentration. At this stage of research, it is difficult to determine how certain bacteria strains are sensitive to the presence of TCTP while others, sometimes belonging to the same species, are insensitive to this protein. Similarly, in the case of “susceptible” bacteria, it is difficult to speculate on the factors that direct this effect toward the stimulation or inhibition of bacterial growth. Nevertheless, the presence of rTbgTCTP is able to modify the microbial composition of the “ecological niche” constituted by the tsetse fly midgut. Modifying the microbial composition may in turn alter the ability of the fly to transmit the trypanosome responsible for sleeping sickness, given that several intestinal bacteria including Sodalis glossinidius were previously shown to be involved in tsetse fly vector competence.34,35
TCTP is known to play many physiological roles, raising the question of how its role could seemingly be limited to modifying bacterial growth as reported here. Wang et al.36 demonstrated the involvement of Bombyx mori (Bm) TCTP (BmTCTP) in the immune response of the silkworm Bm to viral infection. When added to the insect cell culture, BmTCTP induced the activation of the extracellular signal regulate kinase (ERK) signaling pathway along with the expression of several genes, including the anti-microbial peptide cecropin. It was previously shown that the tsetse cecropin gene (in addition to many others) is overexpressed in tsetse flies that are refractory to trypanosome infection, e.g. flies that are able to eliminate parasites ingested while taking a blood meal on an infected host.37 It could be argued that in the B. mori example the TCTP acting on the cell culture is the endogenous protein resulting from the expression of the gene encoded by the insect's own genome, whereas in the present study the TCTP that is expected to act on the fly cells is an exogenous protein produced and excreted by the trypanosome. However, this argument may be ruled out to the extent that the TCTP of one organism can supplement the TCTP deficiency of another organism, no matter how genetically distant. Indeed, this ability has been demonstrated by Brioudes et al.,17 who reported on the ability of Arabidopsis TCTP to supplement a defect in Drosophila TCTP (after disruption of the corresponding gene), and vice versa. Thus, it cannot be excluded that rTbgTCTP can “manipulate” genome expression in insect cells, targeted to specific genes including anti-microbial compounds and possibly also genes involved in the fly immune response. This is consistent with the number of proteins that are excreted/secreted by trypanosomes,38,39 moreover number of genes from the different partners involved in host (tsetse fly), midgut bacteria (e.g., Sodalis and Wigglesworthia), and the parasite (trypanosome) that have been shown to be differentially expressed.40 In this context, Aksoy et al.28 recently reported on the release of the trypanosome coat protein variable surface glycoprotein (VSG) into the tsetse fly gut. The internalization of VSG into the cardia cells causes an interference that ultimately leads to the reduction of the peritrophic matrix efficiency, consequently favoring the crossing of this barrier by the parasite and its establishment in the gut.
In conclusion, trypanosomes have developed a wide range of mechanisms allowing them to establish and survive within the midgut of their insect host, the tsetse fly. TbgTCTP may be part of this array, as it could, among other possible effects, eliminate intestinal bacteria, or reduce their growth, some of which being known to secrete anti-parasitic compounds such as prodigiosin produced by Serratia spp and Enterobacter spp, toxic to P. falciparum and T. cruzi,41,42,43 or active oxygen reported to be produced by Enterobacter spp, and to be toxic to P. falciparum44; this effect would favor trypanosome establishment into the tsetse fly gut. Thus, as such, TbgTCTP may constitute a target for controlling the survival and transmission of the trypanosome, with consequences for the mammalian hosts and the proliferation of sleeping sickness.
Future work should continue the investigations on TbgTCTP and its potential molecular partners, so as to decipher their role in fly vector competence. Furthermore a question will raise of how to counteract the possible favorable effect of TbgTCTP on trypanosome infection and how to manage it in the frame of a disease control strategy in the field. The use of compounds blocking the active site of the protein, or of antibodies anti-TbgTCTP could be explored. To deliver such products in situ, within the fly gut, a para-transgenic approach such as suggested by Rio et al.45 could be tested. This would need Sodalis glossinidius, a tsetse fly facultative symbiont, to be isolated, cultured in vitro, adequately genetically transformed, and transferred (micro-injection) into a female fly harboring Wolbacchia (W), another secondary tsetse symbiont which confers to the fly a reproductive advantage compared with wild-type (W-) flies. Since both symbionts are maternally transferred to the offspring, the dissemination in a focus of (W+) female tsetse flies hosting the modified Sodalis symbiont, will lead to the progressive replacement, over generations, of the natural population by the population of modified tsetse fly. According to Alam et al.,46 the dissemination of several flies corresponding to 10% of the natural population of a focus should lead to the replacement of 90% of this population just within 2 years, thus decreasing drastically the risk of the sleeping sickness spread. Thus, such approach could open new strategies to improve vector control and the fight against HAT.
Materials and methods
Glossina intestinal bacteria strains
The bacteria that were tested in our study were previously isolated from different Glossina species that were either amplified in the CIRAD's insectarium in Montpellier (France) or collected in HAT foci in different sub-Saharan African countries. The bacterial strains were preserved in the UMR IRD-CIRAD Intertryp cryobank in Montpellier (France) or the IRD-MIO in Marseille (France), and re-cultured as needed to perform the present study. The designation and origin of the bacteria strains are provided in Table 1.
Construction, in vitro production and purification of rTbgTCTP
The TbgTCTP gene was synthesized (Genecust Company; Ellange, Luxembourg) according to the Tbg 927.8.6750 sequence from the NCBI database. The gene was then cloned into the baculovirus vector pAcGHLT-A using the EcoRI and NotI restriction sites. Automated DNA sequencing was performed to confirm the integrity of the cloned sequence (GATC Biotech; Constance, Germany).
Two million Sf9 cells were co-transfected with 2 µg of Tbg927.8.6750-pAcGHLT-A and 0.5 µg of BD BaculoGold Baculovirus linearized DNA, according to the manufacturer's instructions (BD Biosciences PharMingen). The presence of the recombinant virus was detected by PCR after viral DNA extraction with QIAmp MiniElute Virus Spin (Qiagen). Infected Sf9 cells were cultured for 48 h, corresponding to the optimum time to reach the highest glutathione S-transferase-His tagged Tbg 927.8.6750 (rTbgTCTP) expression level (data not shown). The insect cells were collected, lysed with a buffer containing 50 mM Tris HCl, 150 mM NaCl, and nonyl phenoxypolyethoxylethanol.(NP40) (1%) at pH 8, incubated on ice for 30 min, and finally sonicated (3 10-s pulses of 12 V, each separated by 6-s intervals). The homogenate was then centrifuged at 14,000 g for 10 min at 4°C, and the supernatant was loaded at a flow rate of 0.5 ml/min onto a 1 ml Ni-NTA agarose column (ÄKTAprime system, GE Healthcare Life Sciences; Little Chalfont, UK) pre-equilibrated with buffer A (20 mM Tris HCl, pH 7.4; 500 mM NaCl; 20 mM imidazole) at room temperature. The column was subsequently washed (flow rate: 1 ml/min) with buffer A until the absorbance at 280 nm decreased to the basal level. The rTbgTCTP was then eluted (flow rate: 1 ml/min) with buffer B (20 mM Tris HCl buffer pH 7.4; 500 mM NaCl; 250 mM imidazole). The eluate was first dialyzed with spectra/Por 3 (Spectrumlabs; Rancho Dominguez, California, USA) against phosphate buffer saline (PBS) overnight at 4°C, and then concentrated by dialyzing against polyethylene glycol 3350 Da (Sigma-Aldrich; Darmstadt, Germany). The protein content of the concentrated eluate was quantified using the Bradford method (Pierce Coomassie Protein Assay Kit, Thermo Fisher; Waltham, Massachusetts, USA), and the purity of the eluted protein was verified after electrophoresis on 10% polyacrylamide SDS-PAGE gels47 stained with PAGE blue staining solution (Thermo Scientific; Waltham, Massachusetts, USA).
Western blot analysis
SDS-PAGE gels were blotted onto a 0.45-µm nitrocellulose™ membrane (Bio-Rad; Hercules, California, USA). Membranes were then incubated for 1 h at room temperature in 5% (w/v) nonfat skim milk agitated with shaking at 450 rpm. Next, the membranes were washed 3 times for 5 min with PBS containing 0.05% Tween 20. Finally, the membranes were incubated for 1 h at room temperature in PBS containing 5% nonfat skim milk and a 1:2000 diluted mouse anti-His tag antibody conjugated to horseradish peroxidase (Sigma-Aldrich; Darmstadt, Germany). After 3 washes of 5 min in PBS containing 0.05% Tween 20, membranes were developed using the Luminata crescendo system (Millipore; Molsheim, France).
Bioinformatics analysis
Several bioinformatics tools were used to investigate the structural characteristics and biochemical properties of the rTbgTCTP. 3D structure analysis was performed with the Chimera software. The CBS prediction servers were used to predict the post-translational modifications of rTbgTCTP, especially the N-linked glycosylation (NetNGlyc 1.0 Server)48 and O-glycosylation (NetOGlyc 4.0 Server)49 sites, but also immunological features such as Linear B-cell epitopes (Bepipred 1.0 Server)50 and discontinuous B-cell epitopes (DiscoTope 2.0 Server).51 The PhyML website was used to construct a phylogenetic tree based on differences in the amino acid sequences between TCTP and several trypanosomatids (including Tbg) as well as several mammalian and Glossina species.52-55
Interaction between TCTP and bacteria from Glossina midguts
In collaboration with the IRD-MIO team (Marseille, France), a total of 39 bacteria strains were obtained for use in this study. This includes species that we previously identified such as Acinetobacter sp, Serratia sp, Enterobacter sp, Enterococcus sp, Lactococcus sp, and Providencia sp, as well as species isolated from different Glossina species sampled in different countries such as G. p. palpalis (Angola and Cameroun), G. m. morsitans (CIRAD insectarium; Montpellier, France), G. pallicera (Cameroun), G. nigrofusca (Cameroun), and G. palpalis gambiensis (CIRAD insectarium; Montpellier, France).14,29,31 To test the interaction between rTbgTCTP and the bacteria, 5 µg of the protein were added to 100 µl of freshly cultured bacteria displaying an optical density (OD) of 0.3 at 600 nm, to obtain a 0.1 OD to start the growth kinetic for each bacterium tested; samples were then incubated 2 h at 25°C. The incubation medium was then centrifuged at 12,000 g for 10 min, after which the supernatant was discarded and the pellet (comprising bacteria coupled or not to rTbgTCTP) was washed 4 times in 500 µl PBS. The final pellet was re-suspended in 10 µl PBS. The bacteria-only control was processed similarly. β-mercaptoethanol (1 µl) and 10 µl buffer were added to each sample to reduce covalent disulfide bonds, and finally 10 µl of 2X loading buffer containing sodium dodecyl sulfate (SDS), glycerol and running buffer (Trizma base, SDS, Glycin) were added to the samples that had been boiled for 3 min to denature the rTbgTCTP. Samples (21 µl) were loaded onto a 10% SDS acrylamide gel, and after electrophoresis (30 mA for 1 h) the protein bands were transferred onto a 0.45 µm nitrocellulose™ membrane (Bio-Rad) and processed as described in section 1.3, to detect the presence of rTbgTCTP.
Effect of rTbgTCTP on the growth of bacteria isolated from Glossina midguts
To evaluate the effect of rTbgTCTP on bacteria growth, a solution containing 150 µl of ‘Mitsuhashi and Maramorosch’ insect culture medium, 50 µl of each bacterium pre-culture and 10 µl of rTbgTCTP at different concentrations (sterilized by filtration on a 0.22 µm membrane) were mixed in one well of a 96-well NUNC Elisa plate. The final rTbgTCTP concentration in the cultures ranged from 1.6 – 200 µM (0 µM for the control culture). This protocol was performed in triplicate at 25°C and 37°C, under 3 controlled atmosphere conditions: aerobiosis, microaerobiosis (atmosphere enriched with 5% CO2) and anaerobiosis. These different growth conditions have been chosen a) since in HAT foci the temperature may vary from around 22°C up to over 35°C thus influencing the corporeal temperature of the tsetse fly which is a poïkilotherm invertebrate (its corporeal temperature is not fixed and stable but depends on the temperature of its environment), and b) since the concentration in oxygen in the fly intestine (from which the different bacteria have been isolated) may vary from near anaerobic to microaerophilic conditions; the aerobiosis atmosphere has been tested to complete the spectrum. The evolution of bacterial growth was estimated by measuring the culture optical density (OD) at 600 nm at 0, 24, 48, 72 and 96 h after incubation, using an Enspire spectrophotometer (Perkin Elmer; Waltham, Massachusetts, USA). At t = 0, each bacterial culture was adjusted to OD = 0.1 at 600 nm. To present the effect of TCTP, histograms were constructed by comparing, for a given experimental condition (and for each time), the OD of the tested culture (with TCTP at a given concentration) with the OD of the corresponding control culture (without TCTP; arbitrarily assigned to 100%). This procedure facilitates comparisons across all culture conditions and times of growth.
Disclosure of potential conflicts of interest
No conflicts of interest were disclosed.
Funding
This work was supported by CIRAD and IRD (UMR INTERTRYP).
References
- [1].Hocking KS, Lamerton JF, Lewis EA. Tsetse-fly control and eradication. Bull. World Health Organ. 1963; 28:811-23; PMID:13963757 [PMC free article] [PubMed] [Google Scholar]
- [2].Lee CW. Aerial applications of insecticides for tsetse fly control in East Africa. Bull. World Health Organ. 1969; 41:261-8 [PMC free article] [PubMed] [Google Scholar]
- [3].Rogers DJ, Slingenbergh JH. Tsetse flies and their control. Rev Sci Tech 1994; 13:1075-124; PMID:7711306; https://doi.org/ 10.20506/rst.13.4.811 [DOI] [PubMed] [Google Scholar]
- [4].Vreysen MJ, Saleh KM, Ali MY, Abdulla AM, Zhu ZR, Juma KG, Dyck VA, Msangi AR, Mkonyi PA, Feldmann HU. Glossina a. J Econ Entomol 2000; 93:123-35; PMID:14658522; https://doi.org/ 10.1603/0022-0493-93.1.123 [DOI] [PubMed] [Google Scholar]
- [5].Bafort JM, Gathiram V. Specificity of the Testryp CATT card agglutination test in a non-sleeping-sickness area of Africa. S Afr Med J 1986; 69:541-2; PMID:3704869 [PubMed] [Google Scholar]
- [6].Chappuis F, Pittet A, Bovier PA, Adams K, Godineau V, Hwang SY, Magnus E, Büscher P. Field evaluation of the CATT/Trypanosoma brucei gambiense on blood-impregnated filter papers for diagnosis of human African trypanosomiasis in southern Sudan. Trop Med Int Heal 2002; 7:942-8; https://doi.org/ 10.1046/j.1365-3156.2002.00956.x [DOI] [PubMed] [Google Scholar]
- [7].Kanmogne GD, Asonganyi T, Gibson WC. Detection of Trypanosoma brucei gambiense, in serologically positive but aparasitaemic sleeping-sickness suspects in Cameroon, by PCR. Ann Trop Med Parasitol 1996; 90:475-83; PMID:8915123; https://doi.org/ 10.1080/00034983.1996.11813072 [DOI] [PubMed] [Google Scholar]
- [8].Moser DR, Cook GA, Ochs DE, Bailey CP, McKane MR, Donelson JE. Detection of Trypanosoma congolense and Trypanosoma brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitology 1989; 1:57-66; https://doi.org/ 10.1017/S0031182000061023 [DOI] [PubMed] [Google Scholar]
- [9].Penchenier L, Njokou F, Eboo Eyenga V, Büscher P. Evaluation of LATEX/T.b.gambiense for mass screening of Trypanosoma brucei gambiense sleeping sickness in Central Africa. Acta Trop 2003; 85:31-7; PMID:12505181; https://doi.org/ 10.1016/S0001-706X(02)00232-2 [DOI] [PubMed] [Google Scholar]
- [10].Van Meirvenne N, Magnus E, Büscher P. Evaluation of variant specific trypanolysis tests for serodiagnosis of human infections with Trypanosoma brucei gambiense. Acta Trop 1995; 60:189-99; PMID:8907397; https://doi.org/ 10.1016/0001-706X(95)00127-Z [DOI] [PubMed] [Google Scholar]
- [11].Büscher P, Mertens P, Leclipteux T, Gilleman Q, Jacquet D, Mumba-Ngoyi D, Pyana PP, Boelaert M, Lejon V. Sensitivity and specificity of HAT Sero-K-SeT, a rapid diagnostic test for serodiagnosis of sleeping sickness caused by Trypanosoma brucei gambiense: A case-control study. Lancet Glob Heal 2014; 2:359-63; https://doi.org/ 10.1016/S2214-109X(14)70203-7 [DOI] [PubMed] [Google Scholar]
- [12].Bisser S, Lumbala C, Nguertoum E, Kande V, Flevaud L, Vatunga G, Boelaert M, Büscher P, Josenando T, Bessell PR, et al.. Sensitivity and specificity of a prototype rapid diagnostic test for the detection of Trypanosoma brucei gambiense infection: A Multi-centric prospective study. PLoS Negl Trop Dis 2016; 10:1-16; https://doi.org/ 10.1371/journal.pntd.0004608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wenzler T, Yang S, Patrick DA, Braissant O, Ismail MA, Tidwell RR, Boykin DW, Wang MZ, Brun R. In vitro and in vivo evaluation of 28DAP010, a novel diamidine for treatment of second-stage African sleeping sickness. Antimicrob Agents Chemother 2014; 58:4452-63; PMID:24867978; https://doi.org/ 10.1128/AAC.02309-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Geiger A, Hirtz C, Bécue T, Bellard E, Centeno D, Gargani D, Rossignol M, Cuny G, Peltier JB. Exocytosis and protein secretion in Trypanosoma. BMC Microbiol. 2010a; 10:20; https://doi.org/ 10.1186/1471-2180-10-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Geiger A, Hamidou Soumana IH, Tchicaya B, Rofidal V, Decourcelle M, Santoni V, Hem S. Differential expression of midgut proteins in Trypanosoma brucei gambiense-stimulated vs. non-stimulated Glossina palpalis gambiensis flies. Front Microbiol 2015; 6:1-12; PMID:25653648; https://doi.org/ 10.3389/fmicb.2015.00444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hamidou Soumana IH, Klopp C, Ravel S, Nabihoudine I, Tchicaya B, Parrinello H, Abate L, Rialle S, Geiger A. RNA-seq de novo Assembly Reveals Differential Gene Expression in Glossina palpalis gambiensis infected with Trypanosoma brucei gambiense vs. non-infected and self-cured flies. Front Microbiol 2015; 6:1259; PMID:26617594; https://doi.org/ 10.3389/fmicb.2015.01259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M. Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc Natl Acad Sci U S A 2010; 107:16384-9; PMID:20736351; https://doi.org/ 10.1073/pnas.1007926107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stierum R, Gaspari M, Dommels Y, Ouatas T, Pluk H, Jespersen S, Vogels J, Verhoeckx K, Groten J, van Ommen B. Proteome analysis reveals novel proteins associated with proliferation and differentiation of the colorectal cancer cell line Caco-2. Biochim Biophys Acta 2003; 1650:73-91; PMID:12922171; https://doi.org/ 10.1016/S1570-9639(03)00204-8 [DOI] [PubMed] [Google Scholar]
- [19].Arcuri F, Papa S, Carducci A, Romagnoli R, Liberatori S, Riparbelli MG, Sanchez JC, Tosi P, del Vecchio MT. Translationally controlled tumor protein (TCTP) in the human prostate and prostate cancer cells: expression, distribution, and calcium binding activity. Prostate 2004; 60:130-40; PMID:15162379; https://doi.org/ 10.1002/pros.20054 [DOI] [PubMed] [Google Scholar]
- [20].Chen W, Wang H, Tao S, Zheng Y, Wu W, Lian F, Jaramillo M, Fang D, Zhang DD. Tumor protein translationally controlled 1 is a p53 target gene that promotes cell survival. Cell Cycle 2013; 12:2321-8; PMID:24067374; https://doi.org/ 10.4161/cc.25404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bommer U, Heng C, Perrin A, Dash P, Lobov S, Elia A, Clemens MJ. Roles of the translationally controlled tumour protein (TCTP) and the double-stranded RNA-dependent protein kinase, PKR, in cellular stress responses. Oncogene 2010; 29:763-73; PMID:19901967; https://doi.org/ 10.1038/onc.2009.380 [DOI] [PubMed] [Google Scholar]
- [22].Susini L, Besse S, Duflaut D, Lespagnol A, Beekman C, Fiucci G, Atkinson AR, Busso D, Poussin P, Marine JC, et al.. TCTP protects from apoptotic cell death by antagonizing bax function. Cell Death Differ 2008; 15:1211-20; PMID:18274553; https://doi.org/ 10.1038/cdd.2008.18 [DOI] [PubMed] [Google Scholar]
- [23].Roque CG, Wong HH, Lin JQ, Holt CE. Tumor protein Tctp regulates axon development in the embryonic visual system. Development 2016; 143:1134-48; PMID:26903505; https://doi.org/ 10.1242/dev.131060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Eichhorn T, Winter D, Büchele B, Dirdjaja N, Frank M, Lehmann WD, Mertens R, Krauth-Siegel RL, Simmet T, Granzin J, et al.. Molecular interaction of artemisinin with translationally controlled tumor protein (TCTP) of Plasmodium falciparum. Biochem Pharmacol 2013; 85:38-45; PMID:23085438; https://doi.org/ 10.1016/j.bcp.2012.10.006 [DOI] [PubMed] [Google Scholar]
- [25].Kadioglu O, Efferth T. Peptide aptamer identified by molecular docking targeting translationally controlled tumor protein in leukemia cells. Invest New Drugs 2016; 34:515-21; PMID:26972431; https://doi.org/ 10.1007/s10637-016-0339-6 [DOI] [PubMed] [Google Scholar]
- [26].Seo E-J, Efferth T. Interaction of antihistaminic drugs with human translationally controlled tumor protein (TCTP) as novel approach for differentiation therapy. Oncotarget 2016; 7:16818-39; PMID:26921194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Farikou O, Njiokou F, Mbida Mbida JA, Njitchouang GR, Djeunga HN, Asonganyi T, Simarro P, Cuny G, Geiger A. Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes—An epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect Genet Evol 2010; 10:115-21; PMID:19879380; https://doi.org/ 10.1016/j.meegid.2009.10.008 [DOI] [PubMed] [Google Scholar]
- [28].Aksoy E, Vigneron A, Bing X, Zhao X, O'Neill M, Wu YN, Bangs JD, Weiss BL, Aksoy S. Mammalian African trypanosome VSG coat enhances tsetse's vector competence. Proc Natl Acad Sci U S A 2016; 113:6961-6; PMID:27185908; https://doi.org/ 10.1073/pnas.1600304113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Geiger A, Fardeau ML, Grebaut P, Vatunga G, Josénando T, Herder S, Cuny G, Truc P, Ollivier B. First isolation of Enterobacter, Enterococcus, and Acinetobacter spp. as inhabitants of the tsetse fly (Glossina palpalis palpalis) midgut. Infect Genet Evol 2009; 9:1364-70; PMID:19800031; https://doi.org/ 10.1016/j.meegid.2009.09.013 [DOI] [PubMed] [Google Scholar]
- [30].Geiger A, Fardeau ML, Falsen E, Ollivier B, Cuny G. Serratia glossinae sp. nov., isolated from the midgut of the tsetse fly Glossina palpalis gambiensis. Int J Syst Evol Microbiol 2010b; 60:1261-5; https://doi.org/ 10.1099/ijs.0.013441-0 [DOI] [PubMed] [Google Scholar]
- [31].Geiger A, Fardeau ML, Njiokou F, Joseph M, Asonganyi T, Ollivier B, Cuny G. Bacterial diversity associated with populations of Glossina spp. from Cameroon and distribution within the campo sleeping sickness focus. Microb Ecol 2011; 62:632-43; PMID:21387098; https://doi.org/ 10.1007/s00248-011-9830-y [DOI] [PubMed] [Google Scholar]
- [32].Hansenová Maňásková S, Bikker FJ, Nazmi K, van Zuidam R, Slotman JA, van Cappellen WA, Houtsmuller AB, Veerman EC, Kaman WE. Incorporation of a Valine–Leucine–Lysine-containing substrate in the bacterial cell wall. Bioconjug Chem 2016; 27:2418-23; PMID:27611478; https://doi.org/ 10.1021/acs.bioconjchem.6b00381 [DOI] [PubMed] [Google Scholar]
- [33].Shah K, Weers PMM. Binding of lysine residues of apolipophorin III to phosphatidylglycerol membranes. FASEB journal 2016; 30(1):661.5 [Google Scholar]
- [34].Geiger A, Ravel S, Mateille T, Janelle J, Patrel D, Cuny G, Frutos R. Vector competence of Glossina palpalis gambiensis for Trypanosoma brucei s.l. and genetic diversity of the symbiont Sodalis glossinidius. Mol Biol Evol 2007; 24:102-9; PMID:17012373; https://doi.org/ 10.1093/molbev/msl135 [DOI] [PubMed] [Google Scholar]
- [35].Hamidou Soumana I, Simo G, Njiokou F, Tchicaya B, Abd-Alla AMM, Cuny G, Geiger A. The bacterial flora of tsetse fly midgut and its effect on trypanosome transmission. J Invertebr Pathol 2013a; 112:S89-93; https://doi.org/ 10.1016/j.jip.2012.03.029 [DOI] [PubMed] [Google Scholar]
- [36].Wang F, Hu C, Hua X, Song L, Xia Q. Translationally controlled tumor protein, a dual functional protein involved in the immune response of the silkworm, Bombyx mori. PLoS One 2013; 8:e69284; PMID:23894441; https://doi.org/ 10.1371/journal.pone.0069284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hu C, Aksoy S. Innate immune responses regulate trypanosome parasite infection of the tsetse fly Glossina morsitans morsitans. Mol Microbiol 2006; 60:1194-204; PMID:16689795; https://doi.org/ 10.1111/j.1365-2958.2006.05180.x [DOI] [PubMed] [Google Scholar]
- [38].Atyame Nten CM, Sommerer N, Rofidal V, Hirtz C, Rossignol M, Cuny G, Peltier JB, Geiger A. Excreted/secreted proteins from trypanosome procyclic strains. J Biomed Biotechnol 2010; 2010:212817; PMID:20011064; https://doi.org/ 10.1155/2010/212817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Grébaut P, Chuchana P, Brizard JP, Demettre E, Seveno M, Bossard G, Jouin P, Vincendeau P, Bengaly Z, Boulangé A, Cuny G, Holzmuller P. Identification of total and differentially expressed excreted-secreted proteins from Trypanosoma congolense strains exhibiting different virulence and pathogenicity. Int J Parasitol 2009; 39:1137-50; PMID:19285981; https://doi.org/ 10.1016/j.ijpara.2009.02.018 [DOI] [PubMed] [Google Scholar]
- [40].Hamidou Soumana I, Berthier D, Tchicaya B, Thevenon S, Njiokou F, Cuny G, Geiger A. Population dynamics of Glossina palpalis gambiensis symbionts, Sodalis glossinidius, and Wigglesworthia glossinidia, throughout host-fly development. Infect Genet Evol 2013b; 13:41-8; https://doi.org/ 10.1016/j.meegid.2012.10.003 [DOI] [PubMed] [Google Scholar]
- [41].Moss M. Bacterial pigments. Microbiologist 2002; 3:10-2 [Google Scholar]
- [42].Lazaro J.E, Nitcheu J, Predicala RZ, Mangalindan GC, Nesslany F, Marzin D, Concepcion GP, Diquet B. Heptyl prodigiosin, a bacterial metabolite is antimalarial in vivo and non-mutagenic in vitro. J Nat Toxins 2002; 11:367-77; PMID:12503881 [PubMed] [Google Scholar]
- [43].Azambuja P, Garcia ES, Ratcliffe NA. Gut microbiota and parasite transmission by insect vectors. Trends Parasitol 2005; 21:568-72; PMID:16226491; https://doi.org/ 10.1016/j.pt.2005.09.011 [DOI] [PubMed] [Google Scholar]
- [44].Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, Dimopoulos G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 2011; 332:855-8; PMID:21566196; https://doi.org/ 10.1126/science.1201618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rio RV, Hu Y, Aksoy S. Strategies of the home-team: symbioses exploited for vector-borne disease control. Trends Microbiol 2004; 12:325-36; PMID:15223060; https://doi.org/ 10.1016/j.tim.2004.05.001 [DOI] [PubMed] [Google Scholar]
- [46].Alam U, Medlock J, Brelsfoard C, Pais R, Lohs C, Balmand S, Carnogursky J, Heddi A, Takac P, Galvani A, et al.. Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog 2011; 7:e1002415; PMID:22174680; https://doi.org/ 10.1371/journal.ppat.1002415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schägger H. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 1987; 1:368-79; https://doi.org/ 10.1016/0003-2697(87)90587-2 [DOI] [PubMed] [Google Scholar]
- [48].Wenzler T, Yang S, Patrick DA Braissant O, Ismail MA, Tidwell RR, Boykin DW, Wang MZ, Brun R. In vitro and in vivo evaluation of 28DAP010, a novel diamidine for treatment of second-stage African sleeping sickness. Antimicrob Agents Chemother 2014; 58(8):4452–63. doi: 10.1128/AAC.02309-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al.. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 2013; 32:1478-88; PMID:23584533; https://doi.org/ 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res 2006; 2:2; PMID:16635264; https://doi.org/ 10.1186/1745-7580-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kringelum JV, Lundegaard C, Lund O, Nielsen M. Reliable B cell epitope predictions: Impacts of method development and improved benchmarking. PLoS Comput Biol 2012; 8:e1002829; PMID:23300419; https://doi.org/ 10.1371/journal.pcbi.1002829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al.. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008; 36(Suppl 2):W465-9; PMID:18424797; https://doi.org/ 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792-7; PMID:15034147; https://doi.org/ 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate Maximum-Likelihood Phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 2010; 59:307-21; PMID:20525638; https://doi.org/ 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- [55].Anisimova M, Gascuel O. Approximate likelihood ratio test for branch: A fast, accurate and powerful alternative. Syst Biol 2006; 55:539-52; PMID:16785212; https://doi.org/ 10.1080/10635150600755453 [DOI] [PubMed] [Google Scholar]


