ABSTRACT
The inter- and intra-tumoral metabolic phenotypes of tumors are heterogeneous, and this is related to microenvironments that select for increased glycolysis. Increased glycolysis leads to decreased pH, and these local microenvironment effects lead to further selection. Hence, heterogeneity of phenotypes is an indirect consequence of altering microenvironments during carcinogenesis. In early stages of growth, tumors are stratified, with the most aggressive cells developing within the acidic interior of the tumor. However, these cells eventually find themselves at the tumor edge, where they invade into the normal tissue via acid-mediated invasion. We believe acid adaptation during the evolution of cancer cells in their niche is a Rubicon that, once crossed, allows cells to invade into and outcompete normal stromal tissue. In this study, we illustrate some acid-induced phenotypic changes due to acidosis resulting in more aggressiveness and invasiveness of cancer cells.
KEYWORDS: acid adaptation, cancer microenvironment, evolution, phenotype
Introduction
In the last decades we, and others, have documented that the extracellular pH (pHe) of solid tumors is acidic1; that this acidity is encountered early in carcinogenesis, and that it has profound consequences on tumor progression. Most clinical cancer cells evolve and exist in a hostile microenvironment, characterized by acidosis, hypoxia, oxygen radicals (ROS) and nutrient deprivation.2,3 It has been known that this environment induces genomic instability and selects for emergence of aggressive clones (clades) of cells, leading to genomic diversity, evident in intratumoral genetic heterogeneity,4-7 which is a proximal cause of malignance and resistance.8 Using dorsal window chambers, we have demonstrated that acidic environments promote tumor invasion and growth by degrading adjacent extracellular matrix].9,10 Thus, we view the intratumoral environment as “niche engineering” wherein cancer cells, in response to Darwinian dynamics, generate an environment that promotes their own growth and invasion11 at the expense of the stromal cells into which they invade. Acidosis also inhibits anti-tumor immunity and promotes immune editing.12 From these observations, we have proposed the “acid-mediated invasion” model, wherein tumor-derived acid promotes remodeling of the extracellular matrix, while cancer cells contemporaneously become adapted and more resistant to acidity, compared to the stromal cells into which they invade.10 However, the mechanism by which tumor cells adapt to this chronic acidosis is not yet fully known. This is a critically important question, as it is virtually axiomatic that, in order to thrive, cells in tumors must develop mechanisms of defense that render them insensitive to this acid challenge. We have begun to address this question by chronically adapting pre-malignant cells in culture to growth in low pH conditions. These cells are chronically autophagic.13 Also, they have reproducibly altered protein expression patterns, including up-regulation of a large number of lysosomal proteins. One of these, the highly glycosylated lysosomal-associated membrane protein, LAMP2, is of particular interest as this heavily glycosylated protein serves to protect the lysosomal membranes from acid hydrolysis and plays a critical role in autophagy.14,15 We recently unraveled a mechanism by which cancer cells use a strategy to protect themselves form extracellular acidosis, that is “borrowed” from a strategy that lysosomes use to protect their membranes against high acidity of their lumen (pH∼4-5). This borrowed mechanism involves redistribution of LAMP2 to the cell surface, which we hypothesize serves to protect cells against extracellular acidosis inside the tumor. In clinical breast cancer specimens, we observed that LAMP2 expression increased significantly with grade and that it appeared to redistribute from lysosomes to the cell surface, especially in poorly perfused areas and those expressing the glucose transporter, GLUT1, which are expected to be acidic.15 The focus of this article is to illustrate some of the phenotypic changes that are associated with adaptation of cancer cells to chronic intratumoral acidosis. This is within the context of a broader investigation to define the role of the microenvironment on cancer evolution and therapy. We show acid adapted cancer cells acquire aggressive and invasive phenotypes; such as being more migratory and invasive during evolution in their acidic niche. We also show rearrangement of cancer cells in their tumor society as a defensive strategy against extracellular acidosis. Combination of these 2 phenotypes makes acid-adapted cancer cells key players in cancer invasion and metastasis. Therefor, developing a biomarker for detecting the acid adapted phenotype would help to map the location of acid adapted cells and tracking them over tumor progress and its therapy. We believe by designing new therapies against acid adapted cells as the most aggressive phenotype we can control the tumor growth. Eventually this can be used as combination therapy to cure cancer.
Results and discussion
Acid adaptation gives rise to more invasive phenotype
Cancer cells have to overcome acidosis as one of the first evolutionary barriers in their challenge to survive.16 Microenvironmental acidosis is not only highly selective in an evolutionary sense, it is also able to induce new aggressive phenotypes through promoting genomic instability.17 As a result, the early malignant cancers are a dynamically evolving school of cells living in their distinct acidic microhabitats that ensure the emergence of more aggressive phenotype as “pioneers.”18 To investigate this, we adapted cells at to growth at acidic pH (6.5) until their growth rates were equivalent to their parental cells grown at high pH (7.4). As shown in Figure 1A, acid adapted MCF-7 cells have different shape and size than their parental cells that they are evolved from. They are smaller and have a more mesenchymal shape, similar to aggressive triple negative cells, MDA-mb-231. Wound healing assays on acid adapted and non-adapted MCF-7 showed adaptation to acidosis make the cancer cells more proliferative and migratory (Fig. 1B). To validate the migratory phenotype of acid adapted cells we also applied a label-free migration assay using an Xcelligene® system (Fig. 1D), showing an increase in the migratory phenotype of acid-adapted cells. Acid adapted MCF-7 cancer cells are also spherogenic (Fig. 1C). They don’t need any matrix to assemble into spheres in a 3D culture, contrary to their parental cells that require matrix to form a sphere. These phenotypic differences imply a more tumorogenic phenotype of acid-adapted cells.
Figure 1.
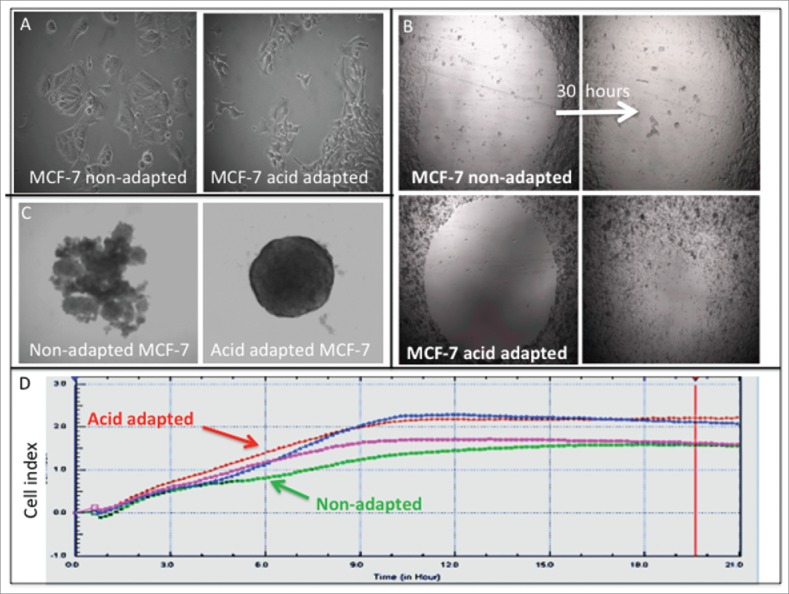
Acid adaptation changes the phenotype of cancer cells toward more aggressive one: (A) size and shape of NA and AA MCF-7 cells. (B) Wound healing assay shows acid adapted MCF_7 cells are more migratory than non-adapted cells. (C) Acid adapted MCF-7 cells are more amenable to spheroiogenic. (D) Migration assay using Xcelligene system confirms the migratory behavior of acid adapted MDA-mb-231 (red) cancer cells compared to non-adapted (green) one in low pH media (P < 0.05 from 3 hours on).
Animal model and 3D co-culture validate acid-induced emerging aggressive phenotype
To investigate this in vivo, we injected acid-adapted and non-adapted MDA-mb-231 cells into mice. We measured the tumor growth by caliper (Fig. 2A) and monitored the activity of the tumor using the cells luminescent activity, measured by Perkin Elmer IVIS-2000 (Fig. 2B). Acid-adapted cells formed bigger tumors in animals and grew faster, suggesting that a more aggressive phenotype emerged from acid adaptation as a response to a micro environmental force. Further we tested the local habitat of acid adapted cells in co-culture with their parental non-adapted cells in 3D spheroid model. We grew the spheres as hanging droplets and moved them to non-sticky ‘U’ shape wells filled with the acidic media (pH 6.5). Acid-adapted cells were labeled with RFP and non-adapted with GFP (Fig. 2C). Notably, over time, acid-adapted cells moved to the surface of the sphere, perhaps protecting the whole system from acid toxicity. These observations could be interpreted heuristically to suggest that acid-adapted cells can serve as defenders within their local society. This does not appear to be due to selective toxicity against non-adapted cells, as video (supplementary video 1) shows what appears to be active movement of the acid adapted cells toward the surface of the sphere.
Figure 2.
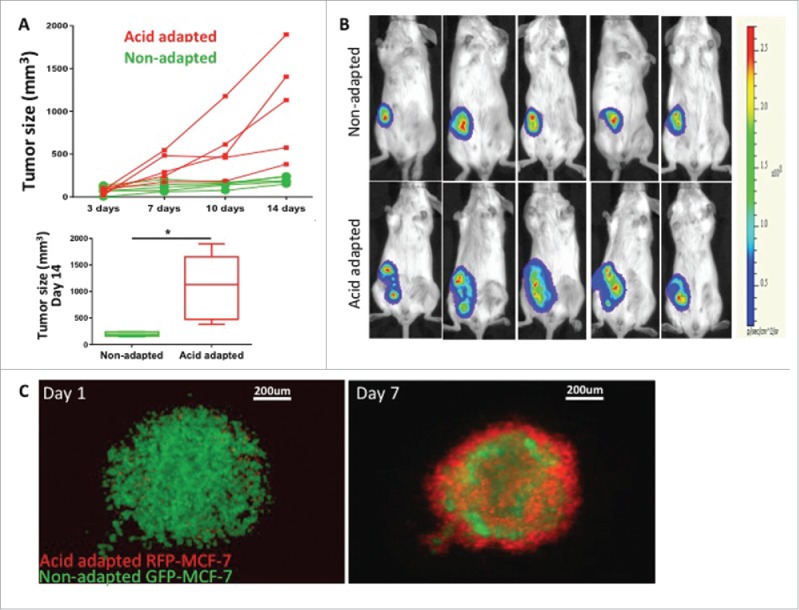
Acid adapted cells grow more rapidly in mouse xenografts. (A) Tumor size measured by caliper over 2 weeks (top) as well as statistical analysis of tumor size for day 14 (bottom) and (B) representative images of tumor bioluminescence after 14 d. The same number of acid adapted and non-adapted MDA-mb-231 cells were injected to nude mice. Acid adapted cells grew faster and make bigger tumors in vivo. (C) co-culture of acid adapted (red) and non-adapted (green) MCF-7 cells in a 3D spheroid model in acidic media (pH6.5). Acid adapted cells move to the edge of spheres, and may result in protecting the whole system from toxicity of acidic media.
Adaptation to local acidic habitat leads cancer cells to a “pioneer” phenotype
We believe evolution of cancer cells occurs on an adaptive landscape that is entirely local and tumor cells are responding to direct local microenvironmental forces and are not susceptible to systemic perturbations. Adaptation to these versatile local niches may also be connected to tumor heterogeneity. In solid tumors such as breast cancer, hyperplastic epithelial cells grow into ductal lumens pushing them further from the stromal vasculature, leading to selection for a glycolytic phenotype through hypoxia and consequently increased acidity that leads to acid adapted phenotype, as we showed previously for the cells at the center of DCIS samples.15 This acidic niche can be re-created in later stages through hard-wired glycolysis (Warburg effect) that generates acid-producing cancer cells. Notably, these appear to emerge at the leading edge of invading tumors.18 We showed this phenomenon in 3D cell culture above (Fig. 2C). To further examine this we also studied whole tumor mounts of breast cancer patients. This is shown in Figure 3 with staining of sequential cuts of the same tumor with CD34 as vasculature marker, CA9 as acid-producing phenotype marker, GLUT1 as glycolytic phenotype marker, and LAMP2 as acid-adapted phenotype marker. The co-localization of all the above-mentioned markers at the leading edge of this stage III tumor is consistent with the acid mediated invasion hypothesis showed also in Figure 2C. It appear that acid adaptation not only protects tumor cells from a harsh acidic environment, but also emboss them with some new characteristics that they can use to migrate and invade the adjacent tissues and eventually metastasize to farther locations.
Figure 3.
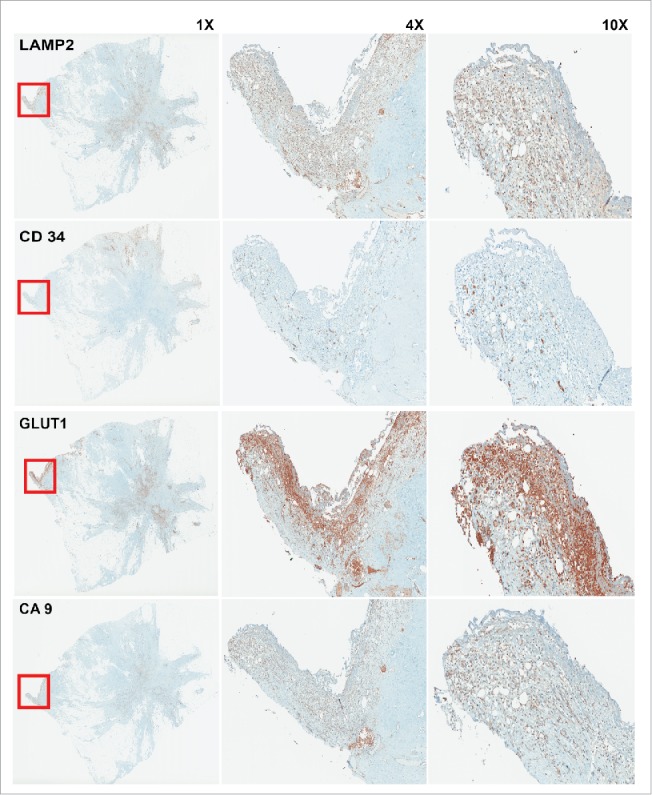
IHC staining of sequential cuts of patient sample. The red box shows the area with less vasculature marked by low CD34 staining. This area is hypoxic and acidic as depicted by GLUT1 and CA9 staining. As we see this area has highest expression of LAMP2 that is a marker for acid adapted cells. Acid adaptation plays a major role in acquiring new phenotype that make the adapted cells as pioneer cancer cells that can protect themselves and other local cancer cells and also lead the invasion and migration toward adjacent tissues.
Material and methods
Cell Culture and acid adaptation in vitro
MCF-7 and MDA-MB-231 cells were acquired from American Type Culture Collection (ATCC, Manassas, VA, 2007–2010) and were maintained in DMEM-F12 (Life Technologies) supplemented with 10% FBS (HyClone Laboratories). Growth medium was further supplemented with 25 mmol/L each of PIPES and HEPES and the pH adjusted to 7.4 or 6.7. Cells were tested for mycoplasma contamination and authenticated using short tandem repeat (STR) DNA typing according to ATCC's. To achieve acid adaptation cells were chronically cultured and passaged directly in pH 6.7 medium for approximately 3 months. Chronic low pH adapted cells underwent about 30 passages.
Transfection (GFP/RFP plasmids)
To establish stable cell lines, the MCF-7 and MDA-MB-231 cells were infected with Plasmids expressing RFP or GFP using Fugene 6 (Promega) at an early passage and were selected using 2 µg/ml puromycin (Sigma).
3D spheroid co-culture
Perfecta3®96-Well Hanging Drop Plates were used to grow the the primary spheres containing 10000 cells (5000 acid adapted MCF-7 RFP and 5000 non-adapted MCF-7 GFP). After 4 d the spheres were moved to non-adhesive U shape bottom 96 well plates containing low pH media and normal pH media as control. Evos microscope was used to image the sphere growth lively over one week. Data were analyzed in image J.
Tumor xenograft development and IVIS imaging
All animals were maintained in accordance with IACUC standards of care in pathogen free rooms, in the Moffitt Cancer Center and Research Institute (Tampa, FL) Vivarium. One week before inoculation with tumor cells, MDA-mb-231 cells (1 × 107) in the mammary fat pads, female nu/nu mice 6 to 8 weeks old (Charles River Laboratories) were placed in 2 cohorts per experiment. The tumor size was measured by a third person. Animal weights were measured and recorded twice weekly, and the overall health of each animal was noted to ensure timely endpoints within the experiment. Finally, animals were humanely euthanized and tumors were extracted and fixed in 10% formalin, paraffin embedded.
For the luciferase activity study, 5 × 106 MDA-mb-231/Luc cells were implanted into the right MFP of 6- to 8-week-old female nu/nu mice. Bioluminescence imaging was used to follow tumor formation, growth, and activity. Animals were anesthetized, and 300 μL of d-luciferin potassium salt (GoldBio) was introduced via intraperitoneal injection. Five minutes after the injection, a bioluminescence image was acquired using standard bioluminescence settings on the IVIS 200.
IHC of patient samples
Patients tumor blocks were collected from moffitt core anonymously. A 1:400 dilution of anti-LAMP2 (#ab37024, Abcam), anti-GLUT1, anti-CD34, and anti-CA9 (Abcam) were used as primary antibodies. Positive and negative controls were used. Normal placenta was used as a positive control for LAMP2, normal breast was used as a positive control for GLUT1, and normal kidney was used as a positive control for CA9. For the negative control, an adjacent section of the same tissue was stained without application of primary antibody and any stain pattern observed was considered as nonspecific binding of the secondary.
Statistical analysis
A 2-tailed unpaired Student t test was performed to compare of treated and non-treated groups of animals and also for tumors against normal cells to determine statistical significance. The significance level was set as P < 0.05.
Ethics statement
All procedures on animals were carried out in compliance with the Guide for the Care and Use of Laboratory Animal Resources (1996), National Research Council, and approved by the Institutional Animal Care and Use Committee, University of South Florida (IACUC# R4051).
Written informed consent was obtained via Institutional Review Board from the University of South Florida approval for the use of patient information for the publication of this report and any accompanying images. Patient samples were processed carefully based on IRB protocols (IRB# 16511).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Ibrahim-Hashim A, Wojtkowiak JW, de Lourdes Coelho Ribeiro M, Estrella V, Bailey KM, Cornnell HH, Gatenby RA, Gillies RJ. Free base lysine increases survival and reduces metastasis in prostate cancer model. J Cancer Sci Therapy. 2011;Suppl 1(4):S1–S5. doi: 10.4172/1948-5956.S1-004. PMID:24032073; h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, et al.. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60(24):7075--7083. [PubMed] [Google Scholar]
- [3].Gillies RJ, Gatenby RA. Hypoxia and adaptive landscapes in the evolution of carcinogenesis. Cancer Metastasis Rev. 2007;26(2):311--317. doi: 10.1007/s10555-007-9065-z. [DOI] [PubMed] [Google Scholar]
- [4].Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al.. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883--892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009--4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Baird RD, Caldas C. Genetic heterogeneity in breast cancer: the road to personalized medicine? BMC Med. 2013;11:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yachida S, Iacobuzio-Donahue CA. Evolution and dynamics of pancreatic cancer progression. Oncogene. 2013;32(45):5253--5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ercan D, Zejnullahu K, Yonesaka K, Xiao Y, Capelletti M, Rogers A, Lifshits E, Brown A, Lee C, Christensen JG, et al.. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29(16):2346--2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, Bailey K, Balagurunathan Y, Rothberg JM, Sloane BF, et al.. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013;73(5):1524--1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66(10):5216--5223. [DOI] [PubMed] [Google Scholar]
- [11].Gatenby RA, Gillies RJ, Brown JS. Of cancer and cave fish. Nat Rev Cancer. 2011;11(4):237--238. doi: 10.1038/nrc3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pilon-Thomas S, Kodumudi KN, El-Kenawi AE, Russell S, Weber AM, Luddy K, Damaghi M, Wojtkowiak JW, Mule JJ, Ibrahim-Hashim A, et al.. Neutralization of Tumor Acidity Improves Antitumor Responses to Immunotherapy. Cancer Res. 2016;76(6):1381--1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wojtkowiak JW, Rothberg JM, Kumar V, Schramm KJ, Haller E, Proemsey JB, Lloyd MC, Sloane BF, Gillies RJ. Chronic Autophagy Is a Cellular Adaptation to Tumor Acidic pH Microenvironments. Cancer Res. 2012;72(16):3938--3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10(9):623--635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- [15].Damaghi M, Tafreshi NK, Lloyd MC, Sprung R, Estrella V, Wojtkowiak JW, Morse DL, Koomen JM, Bui MM, Gatenby RA, et al.. Chronic acidosis in the tumour microenvironment selects for overexpression of LAMP2 in the plasma membrane. Nat Commun. 2015;6:8752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8(1):56--61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- [17].Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012;12(7):487--493. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lloyd MC, Cunningham JJ, Bui MM, Gillies RJ, Brown JS, Gatenby RA. Darwinian dynamics of intratumoral heterogeneity: not solely random mutations but also variable environmental selection forces. Cancer Res. 2016;76(11):3136–3144. doi: 10.1158/0008-5472. PMID:27009166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


