Abstract
Non-small cell lung cancer (NSCLC) is one of the most common malignancies worldwide, and it occurs at a higher frequency in males. HOXD-AS1, an important cancer-associated long noncoding RNA (lncRNA), contributes to the development and progression of several cancers. However, the exact roles of HOXD-AS1 in NSCLC progression are still unknown. Here, we investigated the underlying mechanisms of HOXD-AS1 in human NSCLC tissues. We found that lncRNA HOXD-AS1 was specifically upregulated (P<0.001) in NSCLC tissues and promoted cancer cell growth by targeting miR-147a. Moreover, HOXD-AS1 expression positively correlated with NSCLC clinical pathologic characteristics (tumor size, P=0.006; tumor stage, P=0.044; recurrence, P=0.031) and survival rate (P=0.003). HOXD-AS1 knockdown reduced proliferation and promoted apoptosis of NSCLC cells. The dual-luciferase reporter assay showed that HOXD-AS1 could negatively regulate the expression of miR-147a. miR-147a inhibition abrogated the effect of HOXD-AS1 knockdown on the proliferation and apoptosis of NSCLC cells. Furthermore, HOXD-AS1 positively regulated the expression of pRB (a tumor suppressor protein) in NSCLC cells. Taken together, our data indicated that HOXD-AS1 might be an oncogenic lncRNA that promotes proliferation of NSCLC and could be a therapeutic target in NSCLC.
Keywords: non-small cell lung cancer, HOXD-AS1, proliferation, miR-147a
Introduction
Lung cancer is a common malignancy, and it has the mortality of cancer-related deaths worldwide. Lung cancer can be classified into small cell lung cancer and non-small cell lung cancer (NSCLC) based on their degrees of differentiation and morphologic characteristics. Among all lung cancer patients, 85% are NSCLC.1 Accumulating evidence has shown that there are many important molecules and processes involved in the occurrence and development of NSCLC, such as the altered gene expression induced by epigenetic regulation.2,3 At present, the involvement of noncoding genes in NSCLC has been well documented, and they consist mainly of miRNAs. They can affect the stability of mRNAs and regulate both the transcription of mRNA and, subsequently, the expression of protein-encoding genes at the translational level. It has been shown that miRNAs can regulate various biologic processes and play a critical role in the development and metastasis of cancers.4,5 Long noncoding RNAs (lncRNAs) account for the greatest proportion of the mammalian gene transcriptome and are different from many protein-encoding genes or microRNAs. lncRNAs, still belonging to the “dark matter in genomics”, are not well studied to date, and attempts to understand their function and mechanism are underway.6,7
In recent years, several lncRNAs have been demonstrated to be directly involved in the process of tumor development and metastasis.8,9 HOTAIR is the first lncRNA proved to have a transregulatory effect on transcription. By mediating chromatin remodeling, binding to polycomb repressive complex 2 (PRC2) and histone demethylation complex 1 (LSD1), and mediating the binding of these two protein complexes to specific sites on the genome, HOTAIR allows lysine methylation on specific residues of histones, resulting in gene silencing.10 It has been found that the expression of HOTAIR in various tumor tissues, including lung cancer, is closely related to tumor metastasis, recurrence, and clinical prognosis.11–13 In this study, we focused on a novel lncRNA, HOXD Cluster Antisense RNA 1 (HOXD-AS1), which is encoded by a member of the same gene family that encodes HOTAIR (the HOX gene family). The HOX gene family, first discovered in the study of homeosis in Drosophila, contains a series of evolutionarily conserved genes and plays an important role in embryonic development.14 Human HOX genes can be divided into A, B, C, and D gene clusters, which are located on different chromosomes. Each gene cluster contains 9–11 genes.15 It was found that the mutation in HOX genes may cause developmental disorders and abnormal formation of tissues and organs. HOX gene mutations can even induce cell malignant transformation to form tumors.16,17 A total of 231 lncRNAs have been annotated in the four HOX gene clusters,18 including HOTAIR, which is located between HOXC 11 and HOXC 12 genes and is on the antisense strand. HOXD-AS1, another lncRNA that has been annotated, is located between the HOXD1 and HOSD3 genes and is also located on the antisense strand. In addition to their proximity on the genome, HOXD-AS1, like HOTAIR, also plays an important role in the occurrence and progression of tumors, especially in the regulation of tumor metastasis. A recent study demonstrated that HOXD-AS1 is upregulated in bladder cancer and may be involved in the apoptosis and metastasis of tumor cells.19 However, neither the role of HOXD-AS1 in NSCLC nor its underlying mechanism has been elucidated.
Studies have shown that 50% of miRNAs are located in the fragile sites or tumor-related gene regions, which are often amplified, deleted, or rearranged in cancer cells, suggesting that some miRNAs may act as oncogenes or tumor suppressor genes.20–23 miR-147a was cloned and characterized by Lagos-Quintana et al in 2002 from mouse spleen tissue. Its homologous sequence was also found in the human genome at 9q33.2 and was originally named miR-147.24 Another member of the family, miR-147b, was identified on 15q21.1 in 2011.25 The mature sequences of these two miRNAs only differ in two nucleotides at the 3′ end. Few studies on the functional mechanism of miR-147a have been reported to date. It is known that miR-147b is induced by Toll-like receptors and is involved in the regulation of inflammatory responses of murine macrophages.26
In this study, we found that the expression levels of the lncRNA HOXD-AS1 were upregulated in NSCLC clinical tissue samples. We also report an interaction between HOXD-AS1 and miR-147a. Our findings provide a novel understanding for NSCLC progression and the mechanism involved.
Materials and methods
Cell lines and clinical samples
Eighty-seven human NSCLC tissues and their corresponding noncancerous adjacent tissues were collected at the Affiliated Hospital of Nantong University. All of the patient materials were obtained with appropriate written informed consent, and this study was approved by the Clinical Research Ethics Committees of the Affiliated Hospital of Nantong University.
Human NSCLC cell lines, including A549, H1703, SK-MES-1, and NCI-H1299, were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 Medium (Hyclone, Beijing, China), supplemented with 10% fetal bovine serum (Hyclone) at 37°C in a 5% CO2 incubator.
Plasmid construction and cell transfection
Full-length fragments of HOXD-AS1 were subcloned into pcDNA3.1(+) vector (Thermo Fisher Scientific, Waltham, MA, USA) for HOXD-AS1 overexpression (forward primer: GGTCGACGTTTGTGCCGCGCGC; reverse primer: CGCGGCCGCTGACACTTTGAA). The miR-147a binding sequence in the HOXD-AS1 gene (wild-type HOXD-AS1) and the mutant seed region of HOXD-AS1 were obtained and cloned downstream of the luciferase gene in pGL3-control vector (Promega, Shanghai, China), yielding the plasmids HOXD-AS1-WT and HOXD-AS1-MUT, respectively. The primers for HOXD-AS-WT construction were: forward primer: GGTCGACTGATAGGGAGCTTGGTAGCTA and reverse primer: CGCGGCCGCTCTGAAGAAAGGGGTCAGG. The HOXD-AS-MUT fragment was generated by overlapping polymerase chain reaction (PCR; template primers: ATTTGTTGTTAAATTAAAACCTCT and AGAGGTTTTAATTTAACAACAAAT).
si-RNAs specifically targeting HOXD-AS1(si-HOXD-AS1) and the control si-RNA (si-NC) were synthesized by Ribobio (Guangzhou, China). The target sequence was 5′-GAAAGAAGGACCAAAGTAA-3′. An miR-147a mimic, an miR-147a inhibitor, and the corresponding controls were purchased from GenePharma (Shanghai, China). SK-MES-1 cells were transfected with pcDNA3.1-HOXD-AS1 for overexpression and A549 cells with si-HOXD-AS1 for downregulation. The miR-147a mimic and miR-147a inhibitor dual-luciferase assay plasmids were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer’s specifications.
RNA extraction and quantitative reverse-transcription PCR
Total RNA was isolated by Trizol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. Total RNA samples were reverse-transcribed with PrimeScript RT Reagent Kit (Thermo Fisher Scientific). Quantitative reverse-transcription PCR (qRT-PCR) assay was carried out for HOXD-AS1 and retinoblastoma protein (pRB) detection with SYBR Premix Ex Taq II (Takara Biotechnology, Dalian, China) and monitored with Roche Cobas® z480 (Roche Molecular Systems, Pleasanton, CA, USA). Reaction conditions were as follows: 95°C for 10 s, followed by 40 cycles of 95°C for 5 s, 60°C for 15 s, and 72°C for 30 s. For miR-147a detection, qRT-PCR was conducted using a TaqMan MicroRNA Assay (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. A comparative cycle threshold method was used to calculate fold change in gene expression. The HOXD-AS1 expression level was normalized to β-actin, and miR-147a expression was normalized to U6. All experiments were carried out in triplicate. The primers are listed as follows: HOXD-AS1 forward primer: 5′-GGCTCTTCCCTAATGTGTGG-3′ and reverse primer: 5′-CTCTGGTTGGGTGACTGGTT-3′; pRB forward primer: 5′-TCAGTTGGTCCTTCTCGGTC-3′ and reverse primer: 5′-TGTGAACATCGAATCATGGAA-3′. β-actin was used as an internal control. The primers for β-actin were 5′-AGCGAGCATCCCCCAAAGTT-3′ and 5′-GGGCACGAAGGCTCATCATT-3′.
Dual-luciferase reporter assay
The binding sites between HOXD-AS1 and miR-147a were predicted using DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web). HEK293T cells were placed on a 24-well plate and grown to 80% confluence. HOXD-AS1-WT and HOXD-AS1-MUT were cotransfected with 50 nM miR-147a mimic (or miR-NC) into HEK293T cells. Forty-eight hours after transfection, the luciferase activities were detected using the Dual-Luciferase® Reporter Assay System (Promega).
Cell proliferation assay
Cells were seeded into 96-well plates with 5×103 cells/well and cultured overnight. Cell proliferation was determined using a cell counting kit-8 (CCK-8; Dojindo Laboratories, Rockville, MD, USA), according to the manufacturer’s instructions. The absorbance value of each sample was spectrophotometrically determined at a wavelength of 450 nm.
Western blot analysis
Western blot experiments were used to measure the pRB expression level. The total protein from the cultured cells was extracted in cell lysis buffer (Boster, Wuhan, China) and quantified using the Bradford method. Twenty micrograms of protein was loaded and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After transferring to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA), the membrane was incubated overnight at 4°C with primary antibody against pRB (1:1,000; Proteintech, Rosemont, IL, USA). A primary antibody against β-actin (1:2,000, Proteintech) was used to detect the expression of β-actin (loading control). Then, the membranes were incubated with secondary antibody (1:2,000; Cell Signaling Technology, Danvers, MA, USA) for 2 h at room temperature. Protein bands were visualized using Electro-Chemi-Luminescence (ECL) (Tanon, Shanghai, China) and detected using BioImaging Systems (Tanon).
Cell cycle and apoptosis analysis
Cells were transfected for 48 h and fixed with 75% ethanol at 4°C overnight. The fixed cells were incubated with 50 μg/mL propidium iodide for 30 min in the dark at 37°C. Then, the cells were analyzed using a FACScan (Becton-Dickinson, Franklin Lakes, NJ, USA). The ModFit software (Becton-Dickinson) was used to quantify the cells in different phases of the cell cycle. For apoptosis analysis, dual staining using the fluorescein isothiocyanate Annexin V Apoptosis Detection Kit (Sigma-Aldrich Co., St Louis, MO, USA) was performed according to the manufacturer’s instructions.
Statistical analysis
SPSS software version 19.0 (IBM Corporation, Armonk, NY, USA) and GraphPad Prism 6 (GraphPad Software, Inc., San Diego, CA, USA) were used for statistical analysis. Experimental results are presented as mean ± SD. Comparisons between two groups were conducted using two-tailed Student’s t-test or chi-square test, and differences were considered statistically significant when P<0.05.
Results
HOXD-AS1 is upregulated in NSCLC tissues and predicts a poor outcome for NSCLC patients
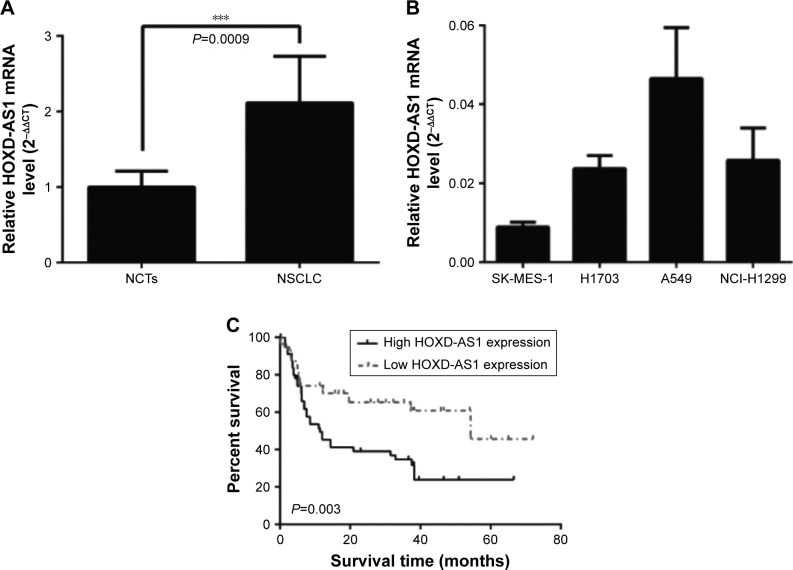
The levels of HOXD-AS1 were detected in 87 NSCLC tissues and their corresponding noncancerous tissues (NCTs) by qRT-PCR. The HOXD-AS1 expression was significantly upregulated in cancerous tissues compared with NCTs (2.1-fold average increase; P<0.001), as shown in Figure 1A. We investigated the levels of HOXD-AS1 expression in different NSCLC cell lines (A549, H1703, SK-MES-1, and NCI-H1299). HOXD-AS1 expression was much higher in A549 cells than in the other cell lines and was the lowest in SK-MES-1 cells (Figure 1B).
Figure 1.
HOXD-AS1 is overexpressed in NSCLC and correlates with patient outcome.
Notes: (A) qRT-PCR evaluation of HOXD-AS1 expression in NSCLC samples compared with NCTs. (B) Expression levels of HOXD-AS1 expression in different NSCLC cell lines, including A549, H1703, SK-MES-1, and NCI-H1299 cells. (C) Overall survival Kaplan–Meier analysis of NSCLC patients. ***P<0.001.
Abbreviations: HOXD-AS1, HOXD cluster antisense RNA 1; NCTs, noncancerous tissues; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
To correlate HOXD-AS1 expression with clinical pathologic features, patients were classified into a relatively high-HOXD-AS1 group and a relatively low group using the median expression level of HOXD-AS1 in NSCLC tissues (Table 1). HOXD-AS1 expression has highly significantly correlated with tumor size (P=0.006) and significantly correlated with TNM stage (P=0.044) and recurrence status (P=0.031). Kaplan–Meier analysis and log-rank test were used to evaluate the effects of HOXD-AS1 expression on overall survival. The results showed that patients with higher HOXD-AS1 expression had a significantly poorer prognosis than patients with lower HOXD-AS1 expression (P=0.003; Figure 1C).
Table 1.
Correlation of the expression of HOXD-AS1 with NSCLC clinical pathologic features
| Characteristic | Number of patients | HOXD-AS1 mRNA expression
|
||
|---|---|---|---|---|
| High | Low | P-value | ||
| Gender | 0.714 | |||
| Male | 50 | 29 | 21 | |
| Female | 37 | 20 | 17 | |
| Age (years) | 0.570 | |||
| <65 | 35 | 21 | 14 | |
| ≥65 | 52 | 28 | 24 | |
| Size of tumor (cm) | 0.006* | |||
| ≤ 3 | 45 | 19 | 26 | |
| >3 | 42 | 30 | 12 | |
| TNM stage | 0.044* | |||
| I | 14 | 4 | 10 | |
| II | 26 | 14 | 12 | |
| III | 47 | 31 | 16 | |
| Recurrence status | 0.031* | |||
| Positive | 48 | 32 | 16 | |
| Negative | 39 | 17 | 22 | |
Note:
Statistically significant difference (P<0.05).
Abbreviations: HOXD-AS1, HOXD cluster antisense RNA 1; NSCLC, non-small cell lung cancer.
HOXD-AS1 promotes NSCLC cell proliferation and cell cycle progression and suppresses apoptosis
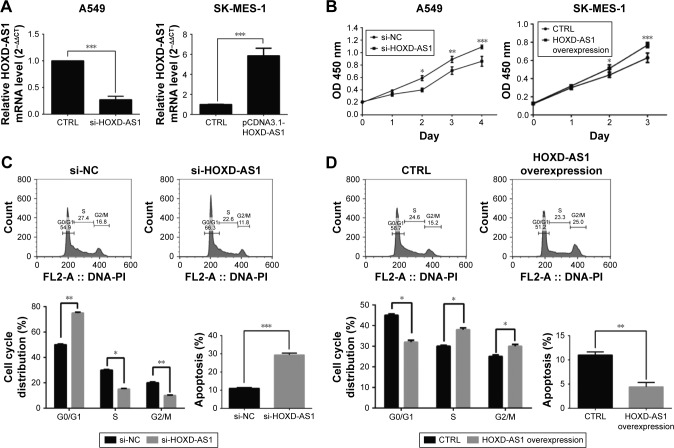
To investigate the function of HOXD-AS1 in the development of NSCLC, A549 and SK-MES-1 cells were used to establish cell lines with knockdown (26.1%, P<0.001) or overexpression (5.86-fold, P<0.001) of HOXD-AS1, respectively, and the efficiency was verified by qRT-PCR (Figure 2A). A CCK-8 assay was performed to explore the effect of HOXD-AS1 expression on the proliferation of NSCLC cells. HOXD-AS1 knockdown induced by si-HOXD-AS1 greatly inhibited cell proliferation in A549 (decreased by 27.1% in 2 days, P=0.031), and SK-MES-1 cells with HOXD-AS1 overexpression showed a significant increase in growth compared with the control group (increased by 1.52-fold in 2 days, P=0.028), as shown in Figure 2B.
Figure 2.
The expression of HOXD-AS1 affects NSCLC cell proliferation, cell cycle and apoptosis.
Notes: (A) qRT-PCR evaluation of HOXD-AS1 expression in HOXD-AS1–knockdown A549 and HOXD-AS1–overexpressing SK-MES-1 cells. (B) Cell proliferation assay was performed in HOXD-AS1–knockdown A549 and HOXD-AS1–overexpressing SK-MES-1 cells. Cell cycle and apoptosis analysis were performed in (C) HOXD-AS1–knockdown A549 and (D) HOXD-AS1–overexpression. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: CTRL, control; HOXD-AS1, HOXD cluster antisense RNA 1; NC, negative control; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; FL, fluorescence; PI, propidium iodide.
Flow cytometry analysis was used to detect cell cycle progression and apoptosis. Cell cycle analysis revealed that HOXD-AS1–knockdown cells were largely arrested at the G0/G1 phase (parental cells, 51.5% compared with HOXD-AS1–knockdown cells, 78.9%, P=0.0003) and showed fewer S-phase cells (parental cells, 23.4% compared with HOXD-AS1–knockdown cells, 14.9%; P=0.042) than the si-NC group, and the percentage of apoptotic cells in the HOXD-AS1–knockdown A549 cells was much higher than that in the control (parental cells, 21.3% compared to HOXD-AS1–knockdown cells, 11.3%), as shown in Figure 2C. Results also showed that overexpression of HOXD-AS1 in SK-MES-1 cells promoted cell cycle progression (G0/G1 phase, from 43.8% to 30.4%, P=0.027; S-phase, from 28.3% to 32.7%, P=0.033) and inhibited apoptosis (from 11.7% to 4.9%), as shown in Figure 2D. Taken together, these data indicated that HOXD-AS1 promotes cell proliferation and cell cycle progression and that it suppressed apoptosis in NSCLC.
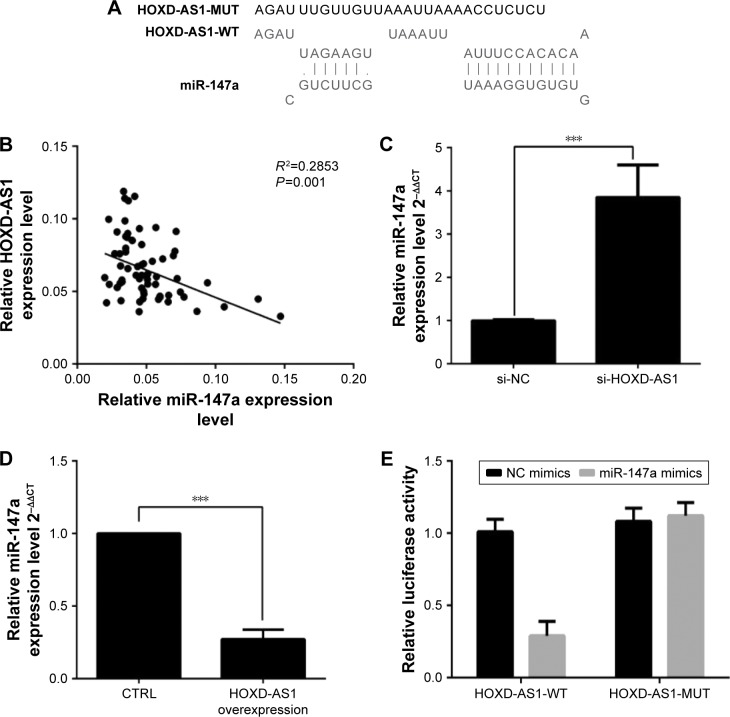
HOXD-AS1 negatively regulates the expression of miR-147a in NSCLC cells
To investigate further the mechanism by which HOXD-AS1 regulates NSCLC progression, we detected the association between miR-147a and HOXD-AS1. Using the DIANA tools, miR-147a was identified as potentially able to bind miRNAs with HOXD-AS1 (Figure 3A). We examined the potential correlation between the RNA expression levels of HOXD-AS1 and miR-147a, and a negative correlation between their expression levels was observed in clinical specimens (P=0.001), as shown in Figure 3B. To determine the effect of HOXD-AS1 on the expression of miR-147a, the expression levels of miR-147a were detected by qRT-PCR in the HOXD-AS1–knockdown A549 cells and SK-MES-1 cells with HOXD-AS1 overexpression. In the HOXD-AS1–knockdown A549 cells, the expression level of miR-147a was 3.8-fold greater than in the control cells (P<0.0001), as shown in Figure 3C. For SK-MES-1 cells with HOXD-AS1 overexpression, the miR-147a expression was significantly inhibited (decreased by 23.4%, P<0.0003), as shown in Figure 3D. The relationship between HOXD-AS1 and miR-147a was verified using a dual-luciferase reporter assay. The luciferase activity of the reporter HOXD-AS1-WT (HOXD-AS1 sequence with wild-type miR-147a binding site, which was cloned into the pGL3 vector) was reduced in cells transfected with the miR-147a mimic, but HOXD-AS1-MUT (loss of the miR-147a binding ability by incorporating a mutated miR-147a binding site) was completely refractory to miR-147a-induced luciferase reporter repression (Figure 3E), indicating that miR-147a binds to HOXD-AS1 in a sequence-specific manner.
Figure 3.
HOXD-AS1 negatively regulates the expression of miR-147a in NSCLC cells.
Notes: (A) Binding site was predicted between HOXD-AS1 and miR-147a using DIANA tools. (B) Correlation analysis was performed to detect the relationship between HOXD-AS1 and miR-147a in NSCLC tissues. The expression level of miR-147a was detected in (C) HOXD-AS1–knockdown A549 and (D) HOXD-AS1–overexpressing cells. (E) Luciferase activity of reporters containing HOXD-AS1-WT and HOXD-AS1-MUT sequences in 293T cells transfected with miR-147a mimic or control. ***P<0.001.
Abbreviations: HOXD-AS1, HOXD cluster antisense RNA 1; HOXD-AS1-MUT, HOXD-AS1 sequence with mutational miR-147a binding site, which was cloned into pGL3 vector; HOXD-AS1-WT, HOXD-AS1 sequence with miR-147a wild-type binding site, which was cloned into pGL3 vector; NC, negative control; NSCLC, non-small cell lung cancer.
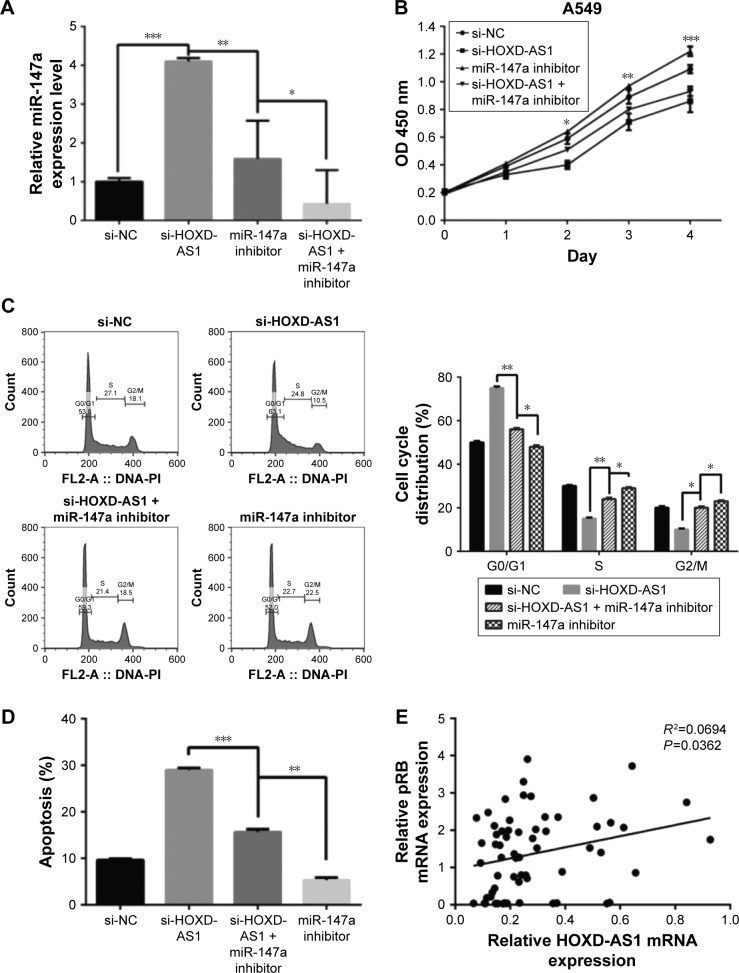
miR-147a inhibition abrogates the effect of HOXD-AS1 knockdown on NSCLC cell proliferation by regulating pRB expression
To further evaluate the effect of miR-147a’s regulation by HOXD-AS1 on NSCLC cell proliferation, an miR-147a inhibitor was transfected into HOXD-AS1–knockdown A549 cells or untreated A549 cells. The expression level of miR-147a was decreased in cells cotransfected with si-HOXD-AS1 and miR-147a inhibitor (compared with cells transfected only with miR-147a inhibitor, P=0.017), as observed in Figure 4A. The suppression of cell proliferation induced by HOXD-AS1 knockdown was abrogated by miR-147a inhibition in 2 days (HOXD-AS1–knockdown cells transfected with miR-147a inhibitor compared with parental cells transfected with miR-147a inhibitor, P=0.0008; Figure 4B). Similarly, miR-147a inhibition also reversed the effect of si-HOXD-AS1 on the cell cycle (decreased by 23.1% in G0/G1 phase, increased by 19.8% in S phase) and apoptosis (decreased by 37.8%). Moreover, proliferation and cell cycle progression were significantly promoted in A549 cells transfected with miR-147a inhibitor compared with cells cotransfected with si-HOXD-AS1 and miR-147a inhibitor; however, the proportion of apoptotic cells was reduced (Figure 4C and D).
Figure 4.
miR-147a inhibition abrogates the effect of HOXD-AS1 knockdown on NSCLC cell proliferation by regulating pRB expression.
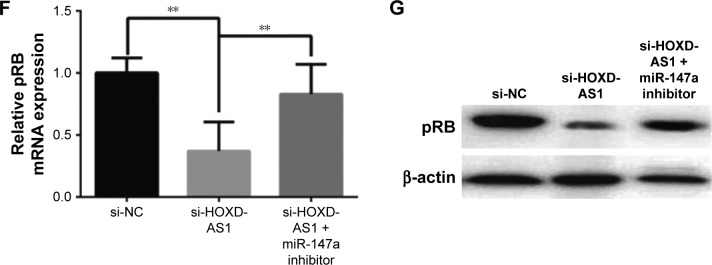
Notes: A549 cells were transfected with si-NC, si-HOXD-AS1, si-HOXD-AS1+miR-147a inhibitor, and miR-147a inhibitor. (A) The expression of miR-147a in these cells. (B) CCK-8 assay was performed to examine cell proliferation. (C) Cell cycle status was determined. (D) Cell apoptosis was detected by flow cytometry. (E) HOXD-AS1 positively correlated with pRB expression in NSCLC (GDS3627). mRNA (F) and protein (G) levels of pRB were examined in cells transfected with si-NC, si-HOXD-AS1, and si-HOXD-AS1+miR-147a inhibitor. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: CCK-8, cell counting kit-8; HOXD-AS1, HOXD cluster antisense RNA 1; NC, negative control; NSCLC, non-small cell lung cancer; pRB, retinoblastoma protein; FL, fluorescence; PI, propidium iodide.
It has been reported that the pRB is the direct target of miR-147a, which inhibits cell proliferation in NSCLC cells by downregulating cell cycle proteins.27 Thus, we explored the potential correlation between pRB and HOXD-AS1 expression. We analyzed the Gene Expression Omnibus (GEO) profile data (GDS3627) and found that the expression level of HOXD-AS1 was positively correlated with pRB in NSCLC (P=0.0362), as shown in Figure 4E. We also found that both the mRNA (decreased by 48.2%, P=0.007) and the protein levels of pRB were reduced in HOXD-AS1–knockdown A549 cells, and that the miR-147a inhibitor abrogated the downregulation of pRB that was induced by si-HOXD-AS1 (1.7-fold increase in mRNA level; Figure 4F and G).
Discussion
Accumulating evidence has shown that there are many important molecules involved in the occurrence and development process of NSCLC. Possible roles for lncRNAs in malignancy development have been proposed, since their biologic functions and molecular mechanisms remain largely unclear.6,28–30 In this study, we found that the expression levels of the lncRNA HOXD-AS1 were much higher in NSCLC tissues than in their corresponding NCTs. Furthermore, the expression level of HOXD-AS1 correlated with a poor prognosis for NSCLC patients and was negatively correlated with survival times. Functional and mechanistic studies revealed that HOXD-AS1 exerts its growth-promoting functions by acting as a competitive endogenous RNA (ceRNA) of miR-147a.
At present, a variety of lncRNAs have been identified as potential biomarkers in NSCLC. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is an lncRNA that is over 8,000 nt long. It is located at 1lq13.1 in the genome, and its upregulation can be seen in melanoma, prostate carcinoma, NSCLC, and other cancers.31–33 Expression of MALAT1 in NSCLC is both stage- and tissue specific. A significant rise in MALAT1 expression has been observed in tumor samples collected from patients with distant metastasis, and the expression is closely related to the TNM staging. Thus, MALAT1 can be used as a marker molecule for the early-stage diagnosis and prognosis of NSCLC.
The gene encoding HOXD-AS1 is located in the HOXD cluster, and its transcript is a novel lncRNA. HOX genes are key developmental regulators, and their aberrant expression is often associated with malignancy.34 The HOXD3 gene, adjacent to the HOXD-AS1 locus, has been reported to promote the metastatic potential of lung cancer cells.35 In this study, higher expression of HOXD-AS1 was observed in NSCLC tissues than in their corresponding NCTs. As HOXD-AS1 was expressed at the highest level in A549 cells and at the lowest level in SK-MES-1 cells of all the NSCLC cell lines used, we studied the effect of HOXD-AS1 on cell proliferation and apoptosis using HOXD-AS1–knockdown A549 cells and HOXD-AS1–overexpressing SK-MES-1 cells. The knockdown of HOXD-AS1 promotes NSCLC cell proliferation and cell cycle progression, but it suppresses apoptosis. As expected, HOXD-AS1 overexpression in SK-MES-1 showed the opposite effects, suggesting that HOXD-AS1 might play a crucial role in promoting proliferation and inhibiting apoptosis of NSCLC cells. More importantly, we found that NSCLC patients with high HOXD-AS1 expression levels had significantly lower survival rates compared to those with low HOXD-AS1, and HOXD-AS1 expression correlated with NSCLC clinical pathologic factors. Our results are consistent with previous reports showing that HOXD-AS1 could act as an oncogene. In breast cancer, overexpression of BRMS gene leads to downregulation of HOXD-AS1, revealing its potential oncogenic role.36 In neuroblastoma, HOXD-AS1 is upregulated and closely associated with progression and prognosis.37,38
Recent studies have described an intricate interaction among different types of RNAs, including mRNAs and ncRNAs (such as lncRNAs, pseudogenes, and circular RNAs). The lncRNAs with miRNA targeting sequences can bind with miRNAs and act as ceRNAs, thus inhibiting the regulation of target genes by miRNAs.28,29 These RNAs, just like sponges, decrease the biologic effects of multiple miRNAs, thereby removing suppression of their target genes and increasing the expression levels.39,40 We found that HOXD-AS1 could potentially bind to miR-147a and verified this with a dual-luciferase reporter assay. In addition, the expression of miR-147a was reduced in HOXD-AS1–overexpressing SK-MES-1 cells and increased in HOXD-AS1–knockdown A549 cells. Furthermore, a negative correlation between HOXD-AS1 and miR-147a expression was observed in NSCLC tissues. We also found that miR-147a inhibition reversed the effect of si-HOXD-AS1 on the cell cycle progression and apoptosis. An increasing number of lncRNAs are being reported to serve as ceRNA in NSCLC as well.41,42 The lncRNA MDl can bind with miR-133 and miR-135 to reduce the inhibition of translation of target genes and eventually contribute to increased levels of MAML1 and MEF2C proteins as downstream targets of the miRNAs.43 Nie et al reported that UCA1 was highly expressed in NSCLC tissues, and that patients with high expression of UCA1 had a poor prognosis. At the same time, they found that UCA1 could demonstrate its carcinogenic effects by interactions with miR-193a-3p.44
Hsa-miR-147a is closely related to miR-210, differing by one nucleotide in the seed region. It has been reported that miR-147a is upregulated in squamous cell carcinoma of the tongue, human gastric cancer, small cell lung cancer, and hepatocellular carcinoma (HCC).45–48 Like miR-210, miR-147a inhibits cell proliferation by downregulating cell cycle proteins such as pRB, CycB, CycA, and Cdk6.27,49,50 Here, we analyzed the GEO profile data (GDS3627) and found that the expression level of HOXD-AS1 positively correlated with pRB in NSCLC. In addition, we detected the regulation of pRB by HOXD-AS1 and found that the pRB expression was reduced in HOXD-AS1–knockdown A549 cells. miR-147a inhibitor abrogated the downregulation of pRB by si-HOXD-AS1. Thus, we suggest that miR-147a inhibition abrogates the effect of HOXD-AS1 knockdown on NSCLC cell proliferation by regulating the expression of the cell cycle protein pRB. However, some reports demonstrate that miR-147a affects cell development, migration, and invasion, but they do not report that it has an influence on cell proliferation.51–53
Taken together, this study demonstrated that HOXD-AS1 was highly expressed in NSCLC tissues. The expression of HOXD-AS1 positively correlated with clinical pathologic characteristics and survival rate in NSCLC. We also found that HOXD-AS1 promoted NSCLC cell proliferation by inhibiting miR-147a. These data provide new insights into NSCLC tumor progression and a novel promising predictive biomarker and potential therapeutic target for NSCLC.
Conclusion
We demonstrate that HOXD-AS1 promotes proliferation and inhibits apoptosis of NSCLC cells. The expression of HOXD-AS1 in NSCLC tumors also correlates with clinical pathologic features such as survival rate, tumor size, stage, and recurrence. We also found that HOXD-AS1 affects miR-147a and pRB expression in NSCLC cells. Our data suggest that HOXD-AS1 might be an oncogenic lncRNA and could be a therapeutic target in NSCLC.
Acknowledgments
This work was supported by the Ministry of Science and Technology of China (Grants 2014BAI02B01 and 2015BAI08B02), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant No 11KJB180010), and the Large Instruments Open Foundation of Nantong University (Grant No KFJN1701).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Liu X, Xu Y, Pang Z, et al. Knockdown of SUMO-activating enzyme subunit 2 (SAE2) suppresses cancer malignancy and enhances chemotherapy sensitivity in small cell lung cancer. J Hematol Oncol. 2015;8(1):67. doi: 10.1186/s13045-015-0164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan J, Zhang F, Sun C, et al. miR-134: A Human Cancer Suppressor? Mol Therapy Nucleic Acids. 2017;6:140–149. doi: 10.1016/j.omtn.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palanichamy JK, Rao DS. miRNA dysregulation in cancer: towards a mechanistic understanding. Front Genet. 2014;5:54. doi: 10.3389/fgene.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun C, Li S, Zhang F, et al. Hsa-miR-329 exerts tumor suppressor function through down-regulation of MET in non-small cell lung cancer. Oncotarget. 2016;7(16):21510–21526. doi: 10.18632/oncotarget.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22(1):1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34(2):613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 11.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Sun M, Lu K, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregulation of p21(WAF1/CIP1) expression. PLoS One. 2013;8(10):e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao W, Ribeiro Rde O, Liu D, et al. EZH2 promotes malignant behaviors via cell cycle dysregulation and its mRNA level associates with prognosis of patient with non-small cell lung cancer. PLoS One. 2012;7(12):e52984. doi: 10.1371/journal.pone.0052984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57(3):367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 15.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Scott MP. Vertebrate homeobox gene nomenclature. Cell. 1992;71(4):551–553. doi: 10.1016/0092-8674(92)90588-4. [DOI] [PubMed] [Google Scholar]
- 17.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78(2):191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 18.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Zhuang C, Liu Y, et al. Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer. J Exp Clin Cancer Res. 2016;35(1):99. doi: 10.1186/s13046-016-0372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Li Y, Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29(7):937–948. doi: 10.1038/onc.2009.406. [DOI] [PubMed] [Google Scholar]
- 21.Xi Y, Wang L, Sun C, Yang C, Zhang F, Li D. The novel miR-9501 inhibits cell proliferation, migration and activates apoptosis in non-small cell lung cancer. Med Oncol. 2016;33(11):124. doi: 10.1007/s12032-016-0837-6. [DOI] [PubMed] [Google Scholar]
- 22.Sun C, Li S, Zhang F, et al. The novel miR-9600 suppresses tumor progression and promotes paclitaxel sensitivity in non-small-cell lung cancer through altering STAT3 expression. Mol Ther Nucleic Acids. 2016;5(11):e387. doi: 10.1038/mtna.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun C, Huang C, Li S, et al. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget. 2016;7(7):8341–8359. doi: 10.18632/oncotarget.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 25.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci U S A. 2009;106(37):15819–15824. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertero T, Grosso S, Robbe-Sermesant K, et al. “Seed-Milarity” confers to hsa-miR-210 and hsa-miR-147b similar functional activity. PLoS One. 2012;7(9):e44919. doi: 10.1371/journal.pone.0044919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun K, Peng T, Chen Z, Song P, Zhou X. Long non-coding RNA LOC100129148 functions as an oncogene in human nasopharyngeal carcinoma by targeting miR-539-5p. Aging. 2017;9(3):999–1011. doi: 10.18632/aging.101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun C, Li S, Zhang F, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7(32):51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song P, Yin S. Long non-coding RNA EWSAT1 promotes human nasopharyngeal carcinoma cell growth in vitro by targeting miR-326/-330-5p. Aging. 2016;8(11):2948–2960. doi: 10.18632/aging.101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Y, Zhang X, Hao Y, Fang Z, He Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014;24(4):335–341. doi: 10.1097/CMR.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt LH, Gorlich D, Spieker T, et al. Prognostic impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non-small-cell lung cancer. J Thorac Oncol. 2014;9(9):1294–1304. doi: 10.1097/JTO.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 33.Ren S, Liu Y, Xu W, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190(6):2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki YJ, Hamada J, Tada M, et al. HOXD3 enhances motility and invasiveness through the TGF-beta-dependent and -independent pathways in A549 cells. Oncogene. 2002;21(5):798–808. doi: 10.1038/sj.onc.1205126. [DOI] [PubMed] [Google Scholar]
- 36.Lee DC, Kang YK, Kim WH, et al. Functional and clinical evidence for NDRG2 as a candidate suppressor of liver cancer metastasis. Cancer Res. 2008;68(11):4210–4220. doi: 10.1158/0008-5472.CAN-07-5040. [DOI] [PubMed] [Google Scholar]
- 37.Batagov AO, Yarmishyn AA, Jenjaroenpun P, Tan JZ, Nishida Y, Kurochkin IV. Role of genomic architecture in the expression dynamics of long noncoding RNAs during differentiation of human neuroblas-toma cells. BMC Syst Biol. 2013;7(Suppl 3):S11. doi: 10.1186/1752-0509-7-S3-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yarmishyn AA, Batagov AO, Tan JZ, et al. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics. 2014;15(Suppl 9):S7. doi: 10.1186/1471-2164-15-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CL, Tseng YW, Wu JC, et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015;44:71–81. doi: 10.1016/j.biomaterials.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Li P, Yang R, et al. microRNA-30b inhibits cell invasion and migration through targeting collagen triple helix repeat containing 1 in non-small cell lung cancer. Cancer Cell Int. 2015;15(1):85. doi: 10.1186/s12935-015-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A feed-forward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell. 2014;53(3):506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 45.Han ZB, Zhong L, Teng MJ, et al. Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol Oncol. 2012;6(4):445–457. doi: 10.1016/j.molonc.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14(9):2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 47.Yi Z, Fu Y, Ji R, Li R, Guan Z. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS One. 2012;7(8):e43184. doi: 10.1371/journal.pone.0043184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao Y, Suo AL, Li ZF, et al. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2(6):963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 49.Sun C, Li S, Li D. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget. 2016;7(24):35960–35978. doi: 10.18632/oncotarget.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun C, Liu Z, Li S, et al. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget. 2015;6(28):25533–25574. doi: 10.18632/oncotarget.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seok JK, Lee SH, Kim MJ, Lee YM. MicroRNA-382 induced by HIF-1alpha is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014;42(12):8062–8072. doi: 10.1093/nar/gku515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uhlmann S, Mannsperger H, Zhang JD, et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol. 2012;8(1):570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatura R, Buchholz M, Dickson D, et al. microRNA profiling: increased expression of miR-147a and miR-518e in progressive supranuclear palsy (PSP) Neurogenetics. 2016;17(3):165–171. doi: 10.1007/s10048-016-0480-6. [DOI] [PubMed] [Google Scholar]