Abstract
Arginylation is an emerging protein modification mediated by arginyltransferase ATE1, shown to regulate embryogenesis and actin cytoskeleton, however its functions in different physiological systems are not well understood. Here we analyzed the role of ATE1 in brain development and neuronal growth by producing a conditional mouse knockout with Ate1 deletion in the nervous system driven by Nestin promoter (Nes-Ate1 mice). These mice were weaker than wild type, resulting in low postnatal survival rates, and had abnormalities in the brain that suggested defects in neuronal migration. Cultured Ate1 knockout neurons showed a reduction in the neurite outgrowth and the levels of doublecortin and F-actin in the growth cones. In wild type, ATE1 prominently localized to the growth cones, in addition to the cell bodies. Examination of the Ate1 mRNA sequence reveals the existence of putative zipcode-binding sequences involved in mRNA targeting to the cell periphery and local translation at the growth cones. Fluorescence in situ hybridization showed that Ate1 mRNA localized to the tips of the growth cones, likely due to zipcode-mediated targeting, and this localization coincided with spots of localization of arginylated β-actin, which disappeared in the presence of protein synthesis inhibitors. We propose that zipcode-mediated co-targeting of Ate1 and β-actin mRNA leads to localized co-translational arginylation of β-actin that drives the growth cone migration and neurite outgrowth.
1. Introduction
Protein arginylation mediated by arginyltransferase ATE1 is an emerging regulatory modification that consists of posttranslational tRNA-mediated addition of arginine to proteins. Multiple prior studies demonstrated the essential role of arginylation in embryogenesis (Kwon et al., 2002), cell migration (Karakozova et al., 2006), and protein homeostasis (Kashina, 2014). Arginylation targets a large number of proteins in vivo, including some of the major components of the cytoskeleton (Saha and Kashina, 2011; Wong et al., 2007). Our prior data show that non-muscle β-actin is arginylated in migratory fibroblasts (Karakozova et al., 2006). Lack of arginylation has been linked to impairments in cell migration (Karakozova et al., 2006) and actin network maintenance (Saha et al., 2010), however it is not known whether these effects are global or locally targeted to the leading edge of the cell, and whether similar arginylation-dependent regulation also drives the migration of other cell types.
Multiple studies over the years have implicated arginylation in neuronal function (Galiano et al., 2016). It has been suggested that arginylation facilitates nerve regeneration after injury (Wang and Ingoglia, 1997) and, more recently, participates in neural tube closure (Kim et al., 2016). Despite these intriguing observations, no direct functional studies of protein arginylation in the brain and neurons have ever been conducted.
Here we used conditional mouse knockout model to address the role of protein arginylation in the brain. Our results demonstrate that lack of arginylation in the brain leads to a defect in neurite outgrowth, resulting in behavioral abnormalities and high rates of postnatal lethality in mice. We find that Ate1 mRNA contains a putative zipcode binding sequence that likely targets it for local synthesis at the neuronal growth cones. Both ATE1 and arginylated β-actin are localized at the growth cones, and lack of arginylation leads to a marked reduction in growth cone spreading, accompanied by the corresponding decrease in the actin polymer.
Our results suggest a novel mechanism that regulates neurite outgrowth during development via arginylation and potentially involves targeted cotranslational arginylation of β-actin in the developing growth cones.
2. Results
2.1. Mice lacking arginylation in the brain exhibit abnormalities at birth suggesting defects in neuronal migration
To test the role of arginylation in brain development, we produced a brain-specific Ate1 knockout mouse by crossing our existing Ate1flox/flox mouse line (with the first four critical exons of the Ate1 gene flanked by LoxP sites) with the commercially available mice expressing Cre recombinase under the brain-specific Nestin promoter that activates in mouse nervous system progenitor cells at E10.5 (Dahlstrand et al., 1995). In Nes-Cre mice the transgene expression can be detected in multiple structures throughout the body (Fig. S1), so their crossing into the Ate1flox/flox mouse line would drive Ate1 deletion in the nervous system.
Unlike the complete Ate1 knockout mice, which die at E12.5–E14.5 during development (Kwon et al., 2002), Nes-Ate1 mice developed to full term and were born at the expected ~ 25% ratio, with the body weight and appearance at birth indistinguishable from their wild type littermates. However, these newborn mice were visibly less active than wild type, easily pushed away by their littermates during feeding and showing no inclination to explore the environment within days after birth. These newborns exhibited dramatically reduced growth in the first days of postnatal life, likely due to their inability to compete for the mother's milk with wild type littermates. Without intervention, most of these mice died within the first two weeks. Keeping them alive required nutritional supplementation (yogurt drops) and extended time with the mother in the absence of wild type littermates, and with this kind of care Nes-Ate1 mice could survive to adulthood.
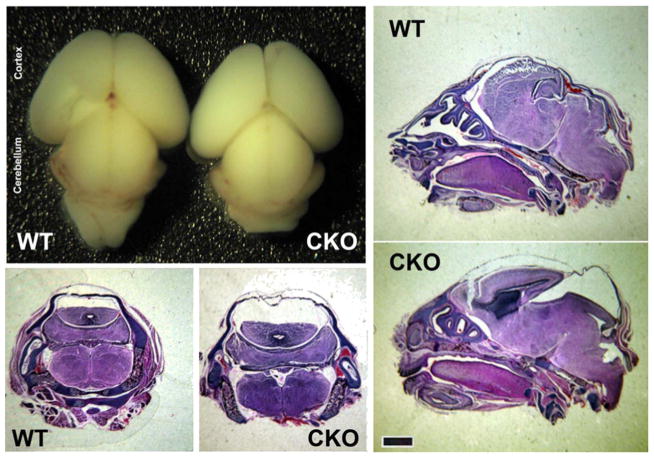
Brains from Nes-Ate1 neonates were similar to control in overall morphology and size (Fig. 1, top left), suggesting that the large-scale brain patterning was not affected by Ate1 knockout. However, sections through the whole head revealed that Nes-Ate1 neonates had a larger skull cavity compared to control (Fig. 1, bottom left and right panels), somewhat reminiscent of hydrocephalus. To test for hydrocephalus, we performed magnetic resonance imaging (MRI) on the fixed newborn pups, but saw no gross dilations in the ventricles (Fig. S2), suggesting that the skull cavities seen on paraffin-embedded sections are due to tissue collapse during flxation and processing rather than the presence of cavities at the start. This defect could potentially be associated with the previously observed abnormalities in the neural tube closure in Ate1 knockout (Kim et al., 2016), or with the reduction of the tissue stiffness due, e.g., to actin cytoskeleton defects previously reported in Ate1 knockout (Cornachione et al., 2014; Kurosaka et al., 2012; Leite et al., 2016; Saha et al., 2010).
Fig. 1. Brain-specific Ate1 knockout mice exhibit abnormalities at birth.
Whole brain images(top left), and H & E stained coronal (two bottom left panels) and sagittal (right panels) sections through the whole heads of wild type (WT) and Nes-Ate1 (CKO) mice. Scale bar, 1 mm.
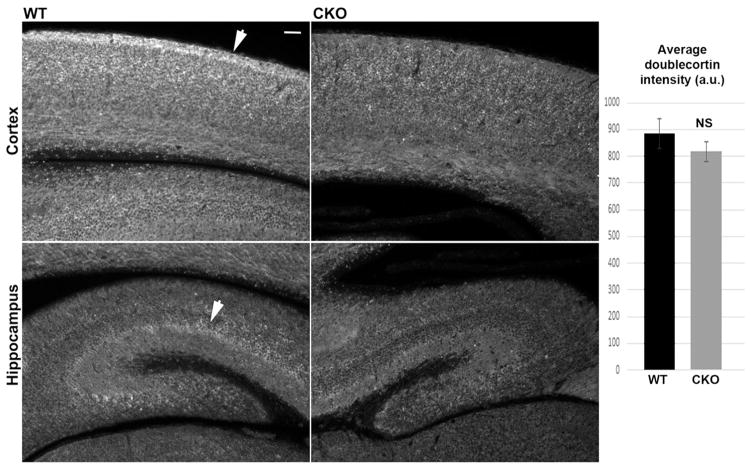
Since arginylation is known to regulate cell migration, we next assessed the overall distribution of doublecortin, a common marker for neurogenesis that supports neuronal migration and neurite outgrowth in embryogenesis and early postnatal development (Friocourt et al., 2003; Weimer and Anton, 2006). In sections of cortex and hippocampus, doublecortin staining showed abnormal distribution in Ate1 knockout, prominently diminished in neurite-rich areas on the outside of the cortex and in hippocampus layers (arrows in Fig. 2, left). At the same time, the overall levels of doublecortin remained unchanged (Fig. 2, right). In addition, no overall reduction in the levels of doublecortin mRNA, as well as several other cytoskeletal proteins typically associated with neuronal migration, was seen (Fig. S3), suggesting a specific defect in doublecortin distribution rather than systemic change in protein levels.
Fig. 2. Brain-specific Ate1 knockout mice have abnormal distribution of doublecortin in the brain.
Left, sections of the cortex of the neonatal wild type (WT) and Nes- Ate1 (CKO) brain stained with antibodies to doublecortin. Images were equalized by gray levels to enable direct comparisons of the differences in the staining patterns and intensity. Arrows indicate the doublecortin-enriched areas in the WT, which are prominently absent in CKO. Scale bar, 50 μm. Right, quantifications for thresholded intensities obtained from 12 difierent sections of 2 pairs of WT and CKO mice. Error bars represent SEM, n = 12. P = 0.34, determined by 2-tailed unpaired t-test.
2.2. Lack of arginylation causes defects in neurite outgrowth
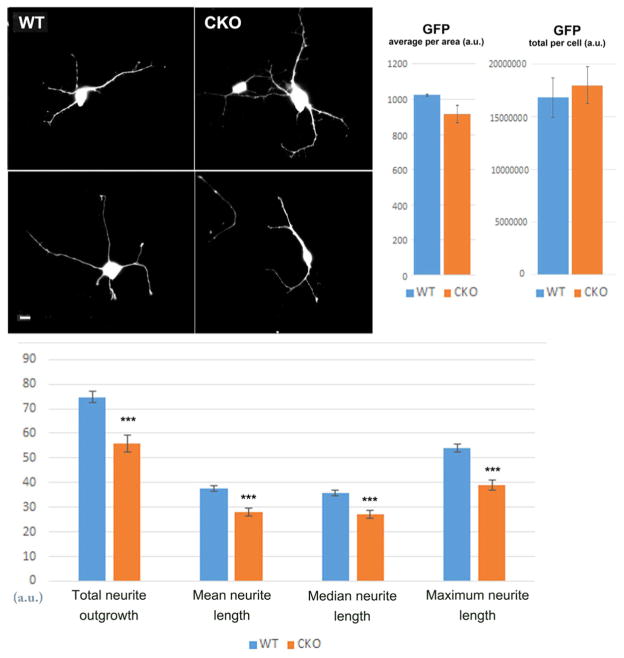
To further test the effects of Ate1 deletion on neurons, we isolated primary cortical and hippocampal neurons from WT and Nes-Ate1 neonates and examined their morphology after 3–7 days in culture. Overall, both types of neurons looked morphologically similar as they attached to the tissue culture dishes and developed a network of processes over time. To analyze their morphology in more detail, we transiently transfected cultured primary hippocampal neurons with GFP (to enable easy visualization of individual neurons), and used fluorescence microscopy to measure the total cell volume, neurite outgrowth and length. While the volume was similar in both types of neurons, suggesting an overall similar cell morphology and thickness, Ate1 knockout neurons had 20–25% lower total neurite length compared to wild type, indicating defects in neurite outgrowth (Fig. 3).
Fig. 3. Ate1 knockout neurons exhibit reduced neurite outgrowth.
Top, representative images of GFP-transfected wild type (WT) and Nes-Ate1 (CKO) cultured primary hippocampal neurons (left) and quantification of average and total GFP fluorescence per area (right). Scale bar, 20 μm. Bottom, quantification of neurite outgrowth from the images similar to those shown on top, using Metamorph Imaging Software. Error bars represent SEM, n = 401 neurites (WT) and 1608 neurites (CKO), quantified from 66 images of neuron cultures derived from 2 embryos for WT and 214 images derived from 4 embryos in CKO. *** indicates P value < 0.000003, determined by 2-tailed unpaired t-test.
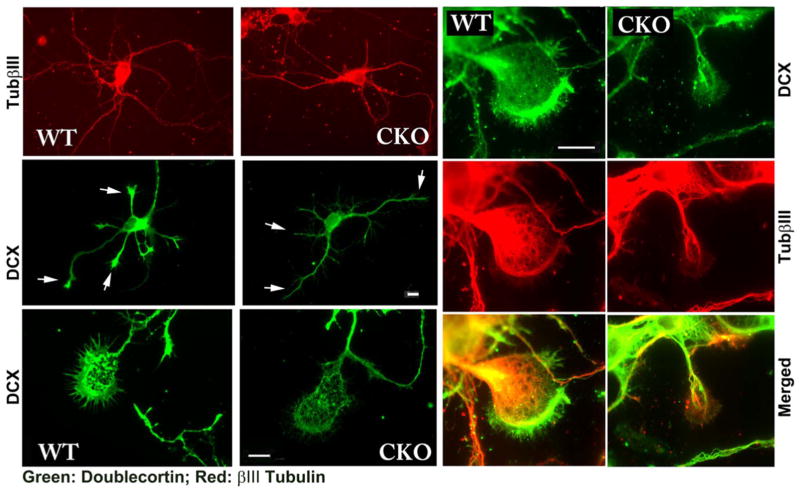
We next stained cultured neurons with antibodies to doublecortin, which promotes neurite outgrowth through association with the microtubule cytoskeleton and facilitating actin-microtubules cross talk (Coquelle et al., 2006; Jean et al., 2012; Lambert de Rouvroit and Goffinet, 2001; Tsukada et al., 2005). In wild type neurons, double-cortin localized throughout the cell body and processes and showed prominent staining at the tips of the growing neurites, where it is believed to play a role in growth cone guidance (Fig. 4). In Ate1 knockout neurons, doublecortin showed similar localization throughout the cell, but no prominent staining at the neurite tips. Since GFP distribution in GFP-transfected neurons suggested no apparent change in the thickness of neurite and growth cone area in Ate1 knockout (Fig. 3), we conclude that this decrease in doublecortin staining at the neurite tips is due to specific doublecortin redistribution rather than non-specific change in cell morphology. No corresponding changes in tubulin levels or distribution was observed (Fig. 4), indicating that this difference was not due to the perturbation of the microtubules.
Fig. 4. Ate1 knockout neurons have reduced doublecortin staining in the neurites and growth cones.
Representative immunofluorescence images of cultured primary cortical neurons from wild type (WT) and Nes-Ate1 (CKO) neonates, stained with antibodies to doublecortin (green) and beta III tubulin (red). Arrows in the right-hand middle panels indicate the distal areas of neurites showing prominent doublecortin staining in WT and a reduction in doublecortin staining in CKO. Scale bars, 20 μm, shown separately for the top four images on the left, bottom left pair, and six right-hand panels.
2.3. ATE1 is targeted to the tips of the growing neurites
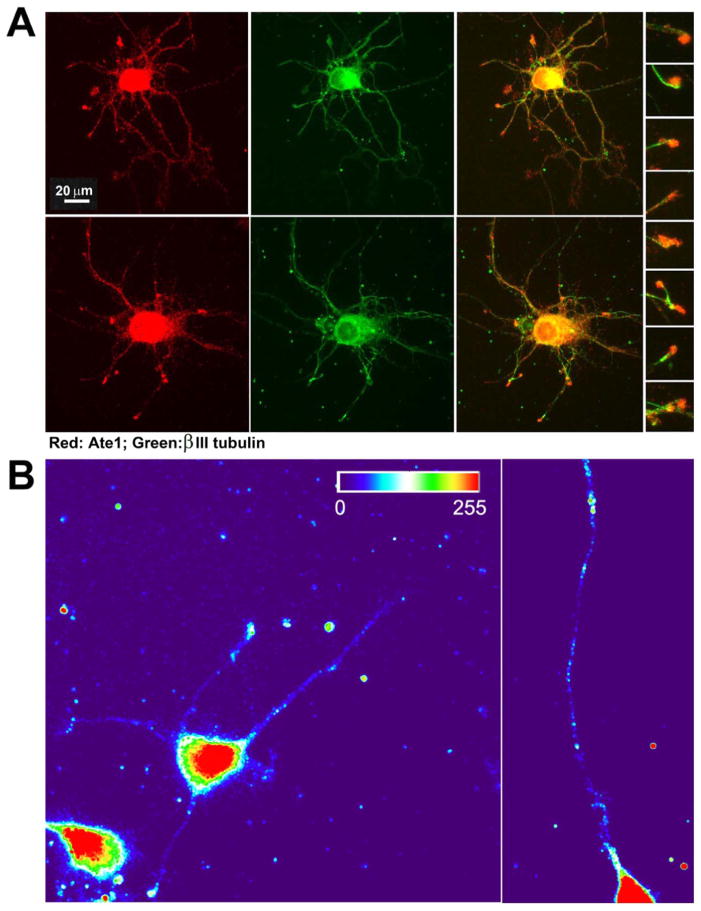
It has been previously shown that ATE1 facilitates migration of non-muscle cells (Karakozova et al., 2006; Kurosaka et al., 2010). Some of the prior studies reported a transient increase of the ATE1 signal at the edge of migratory fibroblasts (Kwon et al., 1999; Wang et al., 2011). Since neuronal growth cones share many similarities with migrating non-muscle cells, it is possible that ATE1 may act similarly in neurons, localizing at the neurite tips and facilitating growth cone migration. To test this, we stained cultured neurons with antibodies to ATE1. Consistent with prior observations, ATE1 in neurons localized diffusely throughout the cell bodies and neurites, and appeared to prominently target the neurite tips (Fig. 5A).
Fig. 5. ATE1 localizes at the tips of the growing neurites.
A. Representative images of primary cultured wild type mouse hippocampal neurons double-stained with antibodies to ATE1 (red) and beta III tubulin (green). The column of inset images on the right shows enlarged view of a set of growth cones taken from merged images to enable simultaneous view of ATE1 and beta III tubulin distribution. B. Heat maps of the fluorescence in situ hybridization (FISH) signal in wild type neurons using probes against Ate1 mRNA.
It has been previously found that many proteins essential for growth cone migration and guidance are targeted to the distal neural tips prior to their translation as zipcode-bound mRNA particles, which become unpackaged and locally translated during active migration of neurites (see, e.g., Tiruchinapalli et al., 2003; Yao et al., 2006). One such zipcode-targeted protein is β-actin, a prominent substrate of ATE1, whose arginylation is essential for cell migration in fibroblasts (Karakozova et al., 2006). To test the possibility that ATE1 localization to the growth cone tips may be zipcode-mediated, we first analyzed the mRNA sequences of all four ATE1 isoforms to search for the zipcode-binding consensus sequence (Chao et al., 2010; Patel et al., 2012), normally located in the 3′ UTR. Two out of four ATE1 isoforms (the more abundant and active ATE1-1 and ATE1-2 (Rai and Kashina, 2005)) contained a near-conventional zipcode-binding sequence that initiated in its 5′ UTR immediately upstream of the first ATG and spanned the first few codons of the coding sequence (Fig. S5), suggesting that ATE1-1 and ATE1-2 mRNA may be zipcode-localized. To test this, we performed fluorescence in situ hybridization (FISH) to detect Ate1 mRNA in the cultured neurons. This staining revealed that, in addition to the localization of Ate1 mRNA in the cell body, a subset of FISH signal was distributed throughout the neurites and prominently localized to the growth cones (Fig. 5B). This localization pattern strongly resembles that of other zipcode-localized mRNA in neurons reported in other studies, confirming that Ate1 mRNA is likely zipcode-localized.
Thus, ATE1 is targeted to the neuronal growth cones, most likely through zipcode-mediated localization of its mRNA.
2.4. Arginylated β-actin localizes to the tips of the growing neurites
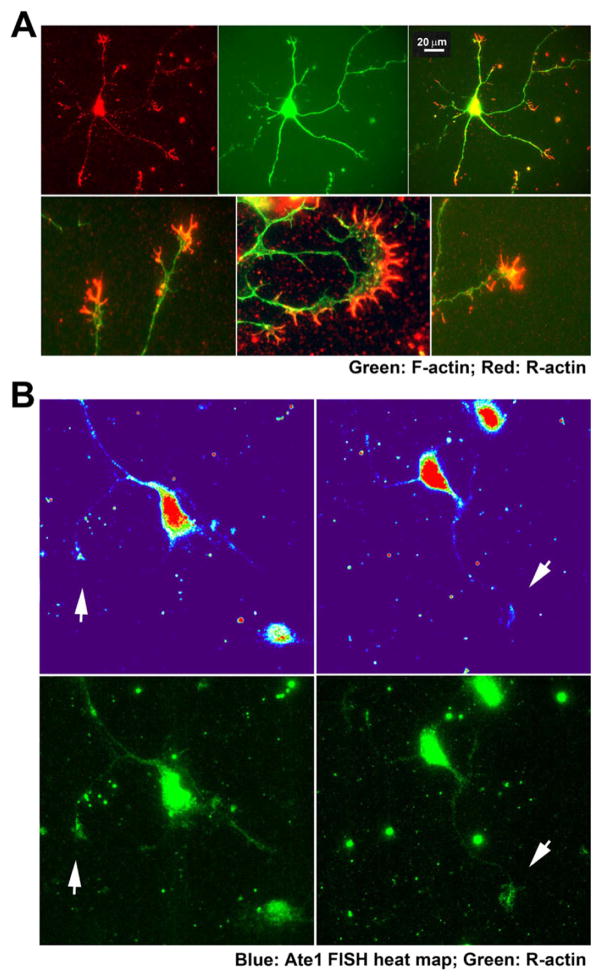
Our prior studies showed that cell migration in non-muscle cells is facilitated by arginylation of β-actin (Karakozova et al., 2006), a protein that is also known to localize to the neuronal growth cones through the mRNA/zipcode-mediated targeting. To analyze the distribution of arginylated β-actin in the neurons, we used arginylated β-actin antibody, recently developed by EMD Millipore. Staining with this antibody revealed a prominent increase of the arginylated β-actin in the growth cones, concentrated mostly inside the filopodia that facilitate the growth cone extension (Fig. 6A). This arginylated β-actin staining was far more pronounced in the growth cones than anywhere else in the neurites, suggesting that most of the actin arginylation is likely happening at these sites.
Fig. 6. Arginylated β-actin and Ate1 mRNA co-localize at the tips of the growth cones.
A, Representative images of primary cultured wild type mouse hippocampal neurons double-stained with antibodies to arginylated β-actin (red) and F-actin (stained with phalloidin-Alexa 488, green). The bottom row shows enlarged views of growth cones. B, Representative images of the neurons showing the heat map of Ate1 mRNA FISH signal (top) and staining with antibodies against arginylated β-actin (bottom). Arrows indicate points of colocalization of the two signals at the growth cones. Scale bar, 20 μm.
To test whether arginylated actin signal coincides with the distribution of Ate1 mRNA FISH signal, we performed a double staining for these two markers in the neurons. Under the harsh flxation conditions favorable for FISH, arginylated actin staining was less pronounced than during milder flxation used for neuronal staining in Fig. 6A, however it still localized mostly to the neurite tips and growth cones. Moreover, where seen, these patches of arginylated actin signal strongly coincided with those for the FISH signal (Fig. 6B). These data strongly suggest that ATE1 locally synthesized in the growth cones correlates with local β-actin arginylation.
2.5. ATE1 and arginylated actin localization at the growth cones depends on active translation
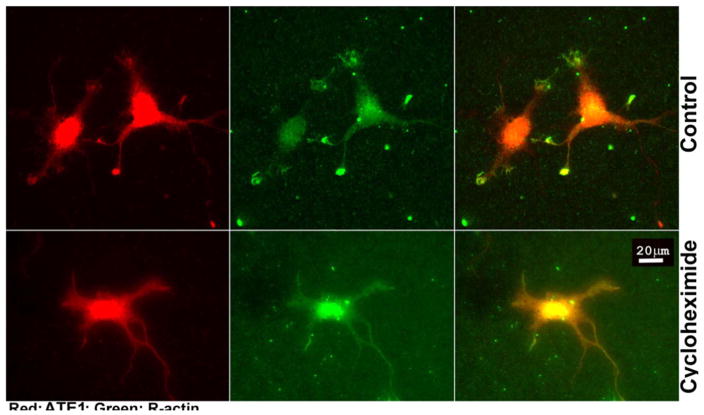
Our previous studies suggest that β-actin arginylation happens co-translationally and is regulated via its nucleotide coding sequence (Zhang et al., 2010). We also found that a subset of ATE1 in the cell is associated with the ribosomes (Wang et al., 2011). Given that both ATE1 and β-actin mRNA co-localize in the neuronal growth cones where we also detect the majority of actin arginylation, it appears likely that β-actin arginylation at the growth cones may be coupled to translation of both ATE1 and β-actin. If true, actin arginylation in the growth cone would not occur in the absence of active translation. To test this prediction, we compared the distribution of ATE1 and arginylated β-actin in control cultured neurons and neurons treated with the translation inhibitor cycloheximide. Remarkably, this treatment greatly reduced ATE1 staining at the tips of the neurites, while not affecting the staining and distribution of ATE1 in the rest of the neurons (Fig. 7, left). Moreover, this treatment completely abolished the increased arginylated actin signal at the neuronal growth cones (Fig. 7, middle and right). These results demonstrate that localization of both ATE1 and arginylated β-actin to the neuronal growth cones requires active translation.
Fig. 7. Localization of ATE1 and arginylated β-actin at the neuronal growth cones depends on active translation.
Representative images of control (top) and cycloheximide-treated (bottom) primary cultured mouse hippocampal neurons stained with antibodies to ATE1 (red) and arginylated β-actin (green). Scale bar, 20 μm.
2.6. Lack of arginylation affects the spreading of the growth cones
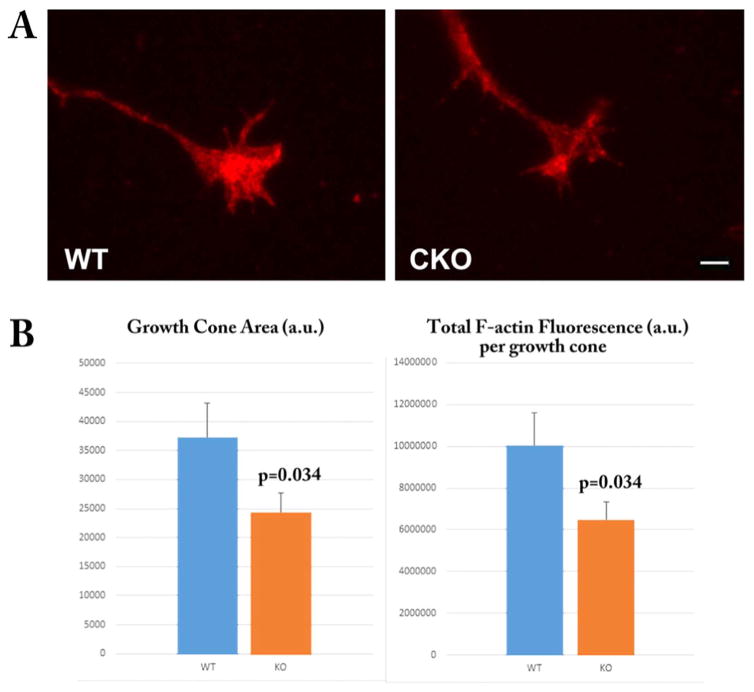
In our previous work we found that lack of arginylation results in reduced cell spreading and disorganization of the actin cytoskeleton (Karakozova et al., 2006; Saha et al., 2010). During neurite outgrowth, the growth cones depend on similar migratory mechanisms, suggesting that lack of arginylation may exert similar effects on their spreading and actin polymerization in the growth cones. To test this, we quantified the total growth cone area and the levels of fluorescent phalloidin staining (which reflects the actin polymer level) in the growth cones of WT and Ate1 knockout neurons (Fig. 8). We found that the growth cone area was prominently and significantly reduced in Ate1 knockout neurons compared to wild type, indicating the reduced spreading reminiscent of the defects seen in cultured fibroblasts. This defect could potentially underlie the impairment in the neurite outgrowth seen in Ate1 knockout neurons.
Fig. 8. Arginylation knockout leads to reduced growth cone area and reduced F-actin in the growth cones.
Top, representative images of wild type (WT) and Nes-Ate1 (CKO) growth cones stained with Rhodamine-Phalloidin to detect F-actin. Bottom, quantification of growth cone area and total and average F-actin fluorescence in the growth cones quantified from the images similar to those shown on top, using Metamorph Imaging Software. Error bars represent SEM, n = 50 (WT) and 46 (CKO). P-values were calculated using 1-tailed unpaired t-test. Scale bar, 20 μm.
3. Discussion
Our work for the first time addresses the role of ATE1 in brain development and neurite outgrowth. It has been previously found that arginylation is essential for normal development and functioning of the cardiovascular system (Kwon et al., 2002), neural crest morphogenesis (Kurosaka et al., 2010), and maintenance of skeletal muscle strength (Cornachione et al., 2014), however no study of ATE1 effect on the brain development and function has never been done. In this paper, we show that ATE1 drives neuritogenesis and facilitates neurite outgrowth, that lack of ATE1 leads to behavioral abnormalities and high rates of early postnatal lethality in mice, and that these effects are potentially mediated, at least in part, by zipcode-mediated co-targeting of Ate1 and β-actin mRNA to the neuronal growth cones, followed by localized arginylation of β-actin in the growth cones, likely by locally synthesized ATE1. While it is possible, and likely, that other ATE1 protein targets contribute to the overall mouse phenotype, this is the first indication that ATE1 can directly target a key component of neuronal migration with effects on the cellular level.
Our prior studies using cultured embryonic fibroblasts showed that lack of ATE1 leads to a decrease in cell's ability to spread on the substrate in culture (Karakozova et al., 2006; Saha et al., 2010). In agreement with this, Ate1 knockout neurons have a reduced outreach and neurite length, as well as a reduced area of growth cones compared to wild type, the parameters that define the neurons’ ability to spread. It appears likely that the underlying mechanisms for ATE1-dependent cell spreading may be similar in both cell types.
Our data show that Ate1 knockout is accompanied by abnormal distribution of doublecortin in the brain and its prominent reduction in the growth cone area. Doublecortin has been previously shown to bind microtubules and is proposed to mediate the actin-microtubule crosstalk during growth cone movement (Coquelle et al., 2006; Jean et al., 2012; Lambert de Rouvroit and Goffinet, 2001; Tsukada et al., 2005). Its altered distribution in Ate1 knockout is consistent with the previously proposed role of arginylation in actin cytoskeleton function and may reflect a directly or indirect link of arginylation in double-cortin-mediated migration of the growth cone via mechanisms that coordinate the action of actin and microtubules.
We previously found that the migratory fibroblasts depend on β-actin arginylation for their ability to form the leading edge lamellipodia (Karakozova et al., 2006) and to maintain directional migration (Kurosaka et al., 2010). Consistent with this, we find here that arginylated β-actin localizes to the leading edge in neuronal growth cones. Moreover, arginylated β-actin appears to target fllopodia, the structures essential for leading edge extension in neurons. While the exact effect of arginylation on actin properties is still under investigation, its role in polarized elongation of the actin network to propel lamellipodia movement in the growth cone is an exciting possibility to be studied.
It has been shown in multiple studies that directional migration in many cell types depends on zipcode-mediated targeting of mRNA to the cell leading edge, where its local translation is believed to facilitate the protruding behavior of the cell (Perycz et al., 2011; Yao et al., 2006). This mechanism is especially important in neurons, the specialized cells in which the leading edge areas in the growth cones are often located at a far great distances from the cell body, thus limiting the ability to transport newly synthesized proteins to the cell periphery as rapidly as needed. One of the most prominent proteins whose function in growth cone migration depends on this zipcode-mediated targeting is β-actin (Ross et al., 1997). Numerous studies show that this targeting is important for cell migration (Shestakova et al., 2001), but the exact underlying mechanism has not been clearly understood, given the fact that β-actin is overall highly abundant in cells regardless of the sites of its localization. Our findings that Ate1 is also zipcode-localized and that β-actin arginylation targets the growth cones opens up an exciting possibility that zipcode-mediated mRNA transport may facilitate actin arginylation during leading edge extension and directional migration.
We have previously shown that arginylation of β-actin is coupled to its fast translation rate, suggesting that actin arginylation is likely a co-translational event. Our current study supports this idea by demonstrating that actin arginylation at the neuronal growth cones requires active translation. It is attractive to think that zipcode-mediated co-targeting of β-actin and Ate1 mRNA and their colocalized translation in the growth cones are required to facilitate actin arginylation, required to facilitate actin's activity at the cell leading edge (Fig. 9). Elucidating the global implications of this actin coding mechanism in various cell types constitutes an exciting direction of further studies.
Fig. 9. Regulation of neurite outgrowth by localized co-translational arginylation of β-actin.
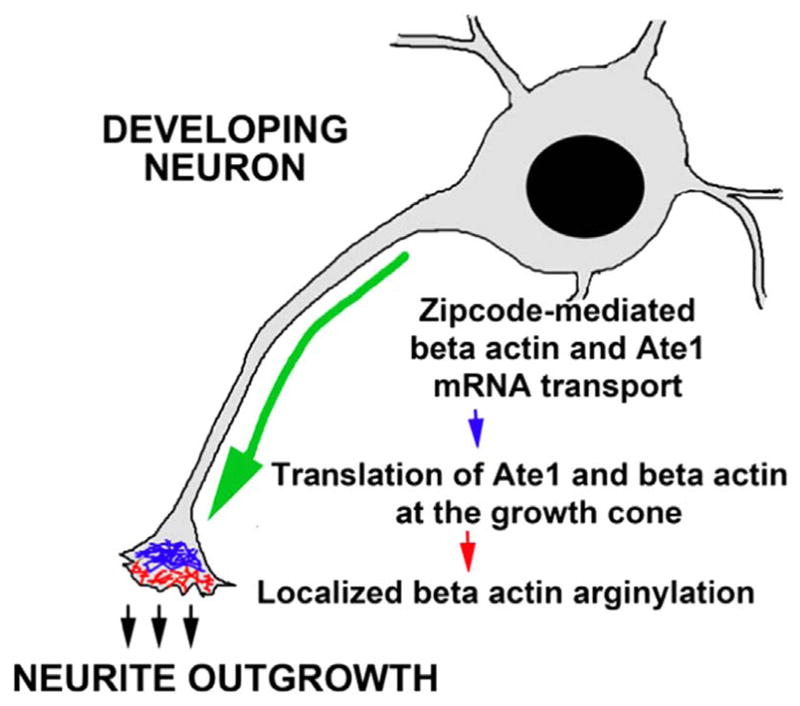
Zipcode-mediated transport of β-actin and Ate1 mRNA to the growth cones leads to their co-localized translation and subsequent co-translational arginylation of β-actin. We propose this to be the mechanism that facilitates the neurite outgrowth by maintain polarized actin elongation in the growth cones.
4. Materials and methods
4.1. Mice
Ate1-floxed mice, previously generated in our lab (Kurosaka et al., 2010; Leu et al., 2009) were crossed with commercially available mouse line expressing Cre recombinase under Nestin promoter (Jackson Laboratory strain B6.Cg-Tg(Nes-cre)1Kln/J). Mice were bred and maintained in a mixed C57Bl6/129CVJ background according to the proper animal protocols.
4.2. Antibodies
Rat monoclonal ATE1 antibody (EMD Millipore, MABS436) was previously described in Wang et al. (2011). Rabbit polyclonal antibody to arginylated actin (R-actin) was obtained from EMD Millipore (ABT264) as part of their product validation study. Batch 13 of the antibody, not currently licensed for commercial use, is shown in the majority of the study. Anti-Doublecortin antibody (ab18723 dilutions: 1:200 for immunofluorescence) was purchased from Abcam. Mouse monoclonal anti- β-III tubulin was from R & D Systems (MAB1195, dilutions: 1:100 for immunofluorescence). Rhodamine-phalloidin was purchased from Sigma and used at the working concentration of 100 nM.
4.3. R-actin antibody staining
Cultured neurons were fixed by incubation with 4% paraformaldehyde (PFA), washed twice with PBS, and permeabilized with ice-cold acetone for 5 min, followed by incubation for 30 min in a blocking solution (5% horse serum in PBS). Cells were incubated overnight at 4 °C or for 2 h at room temperature with primary antibodies against polyclonal anti-R-actin, washed with PBS, and incubated for 1 h at room temperature with Alexa Fluor 488-conjugated anti-rabbit antibody (Molecular Probes, Invitrogen) at 1:1000 dilution. After the staining, cover slips with cells were washed in PBS and mounted into ProLong Diamond anti-fade mounting media (Life Technologies).
4.4. Immunohistochemistry
Paraffin-embedded brain sections were deparaffinized with xylene, re-hydrated with sequential ethanol:water series (95:5, 80:20, 50:50 and 30:70), washed with water, boiled in 10 mM sodium citrate /0.05% Tween 20, pH 6.0 for 20 min for antigen retrieval, then blocked with 0.1 M Tris pH7.6 supplemented with 3% BSA for 1 h at room temperature, and treated with primary antibodies diluted in 0.1 M Tris pH 7.6 supplemented with 3% BSA overnight at 4 °C. After treatment with primary antibodies, samples were washed with 0.1 M Tris, treated with fluorescent dye-conjugated secondary antibodies for 2 h at room temperature, then stained with DAPI for 10 min. After triple washes with water, samples were embedded in Aqua Poly/Mount (Polysciences, Inc., 18606).
4.5. Neuron cultures
Primary cultures of neurons from wild type and Ate1 knockout P0 newborn mice were prepared as described in Nunez (2008) for the rat neurons, except that mice were used instead. Embryos were euthanized by decapitation and their brains were immediately dissected to isolate cortex and hippocampus. Neurons from both regions of the brain were dissociated in 0.25% pre-warmed trypsin, and plated at approximate density of 0.5 × 106 cells per 35 mm Glass Bottom Dishes (Matek Corportation, uncoated) pre-coated the day before the experiment with 0.1 mg/ml Poly-D-Lysine (Sigma). Cells were initially plated in Neurobasal medium (1× B27 from a 50× stock, 1× GlutaMAX from a 100× stock, 50 U/ml penicillin and 50 μg/ml streptomycin) containing 10% Fetal Bovine Serum (FBS), and 2–18 h after the plating the medium was changed to Neurobasal medium without serum for continued neuronal growth.
For treatment with translation inhibitors and R-actin antibody staining, 3-days-old cultures of primary mouse hippocampal neurons were obtained from the Neurons-R-Us facility at the University of Pennsylvania and either kept intact or treated with 5 μg/ml cycloheximide (or DMSO for control) for 2 h, followed by fixation with 4% PFA at room temperature for 30 min.
For GFP transfection experiments, primary neuron cultures 8–12 h after plating were treated with pEGFP-N2 vector (Clontech) in the presence of Lipofectamine 2000 reagent (ThermoFisher Scientific) according to the manufacturer's protocol. Cells were visualized approximately 3 days after transfection and individual GFP-expressing cells were photographed to examine the overall cell morphology and neurite outgrowth.
4.6. Fluorescence in situ hybridization
Ate1 mRNA probes (conjugated to Quasar 570 dye) were designed and purchased from LGC Biosearch Technologies and fluorescence in situ hybridization was carried out as per manufacturers’ protocol. Briefly, 3 day old mouse hippocampal neurons growing on glass coverslips were fixed in 4% (w/v) PFA at room temperature for 30 min followed by treatment with 70% alcohol at 4 °C for 1 h. Cells were incubated with 125 nM probes at 37 °C overnight. Cells were stained with DAPI (5 ng/ml) and mounted using Prolong Diamond (Life Technologies).
4.7. Imaging
All fluorescence images shown in the paper are wide-field images, acquired using Nikon fluorescence microscope and digital Hamamatsu Orca AG and Andor IXon Ultra EMCCD camera.
4.8. Image processing and quantification
In each experiment, the images were acquired with identical exposure for wild type and knockout, using a Hamamatsu Orca AG CCD camera (12-bit) or Andor IXon Ultra EMCCD camera (16-bit). For image quantification, the 12/16-bit raw gray level images (with the maximum gray value in the image well below the saturation limit of the detector) were thresholded to the identical gray scale values to remove the background for quantification. The thresholds were chosen manually as the maximally inclusive area covering the entire range of the detectable gray levels in a representative image and maintained constant for all the other wild type and knockout images in the set. Following thresholding, integrated morphometric measurement in Metamorph Imaging software was used to determine the total fluorescence signal in the image, quantified as total gray levels above threshold.
For presentation in the manuscript, the 12/16-bit raw images were converted to 8-bit images and re-scaled (with a gamma factor of 1) so that all images had the same lower and upper cut-off values. This form of processing does not alter the relative gray levels between images, while making the images suitable for display utilizing the maximum possible dynamic range. These identically re-scaled images were pseudo-colored using Metamorph “color combine” function and further processed in Adobe Photoshop, where the equally scaled wild type and knockout images were placed side by side into the same panel and uniform contrast was applied to the entire panel using the “levels” function in Adobe Photoshop.
For quantification of neurite length and total outgrowth, we used “neurite outgrowth” module of the Metamorph Imaging Software (Molecular Devices, Inc.) to analyze the thresholded images of GFP-transfected neurons. The growth cone area and intensity of different marker staining in the neurons and the growth cones was measured in thresholded images using the “integrated morphometry analysis” function of the Metamorph Imaging Software.
4.9. Gene and protein nomenclature
Throughout the manuscript, we used the Guidelines for Nomenclature of Genes, Genetic Markers, Alleles, and Mutations in Mouse and Rat found at http://www.informatics.jax.org/mgihome/nomen/gene.shtml
Following this nomenclature, gene and mRNA symbols were italicized, with only the first letter in upper-case (e.g., Ate1), and protein symbols were not italicized with all letters in upper-case (e.g., ATE1).
4.10. Statistics
P-values were determined by one-tailed Welch's t-test (for the RNAseq data in Fig. S3) and one-tailed and two-tailed unpaired Student's t-test (for all other figures, as indicated).
Supplementary Material
Acknowledgments
We thank Margaret Maronski (Neurons-R-Us facility at the University of Pennsylvania) for providing cultures of primary mouse hippocampal neurons for FISH and arginylated actin staining, University of Pennsylvania Small Animal Imaging facility for MRI, and Dr. Stephen Jones for helpful suggestions and critical reading of the manuscript. This work was supported by NIH grants GM104003 and GM117984.
The authors declare no competing financial interests.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ydbio.2017.08.027.
References
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle FM, Levy T, Bergmann S, Wolf SG, Bar-El D, Sapir T, Brody Y, Orr I, Barkai N, Eichele G, Reiner O. Common and divergent roles for members of the mouse DCX superfamily. Cell Cycle. 2006;5:976–983. doi: 10.4161/cc.5.9.2715. [DOI] [PubMed] [Google Scholar]
- Cornachione AS, Leite FS, Wang J, Leu NA, Kalganov A, Volgin D, Han X, Xu T, Cheng YS, Yates JR, 3rd, Rassier DE, Kashina A. Arginylation of myosin heavy chain regulates skeletal muscle strength. Cell Rep. 2014;8:470–476. doi: 10.1016/j.celrep.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrand J, Lardelli M, Lendahl U. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system brain. Res Dev Brain Res. 1995;84:109–129. doi: 10.1016/0165-3806(94)00162-s. [DOI] [PubMed] [Google Scholar]
- Friocourt G, Koulakoff A, Chafey P, Boucher D, Fauchereau F, Chelly J, Francis F. Doublecortin functions at the extremities of growing neuronal processes. Cereb Cortex. 2003;13:620–626. doi: 10.1093/cercor/13.6.620. [DOI] [PubMed] [Google Scholar]
- Galiano MR, Goitea VE, Hallak ME. Post-translational protein arginylation in the normal nervous system and in neurodegeneration. J Neurochem. 2016;138:506–517. doi: 10.1111/jnc.13708. [DOI] [PubMed] [Google Scholar]
- Jean DC, Baas PW, Black MM. A novel role for doublecortin and doublecortin-like kinase in regulating growth cone microtubules. Hum Mol Genet. 2012;21:5511–5527. doi: 10.1093/hmg/dds395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR, 3rd, Mogilner A, Zebroski H, Kashina A. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- Kashina A. Protein arginylation, a global biological regulator that targets actin cytoskeleton and the muscle. Anat Rec. 2014;297:1630–1636. doi: 10.1002/ar.22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim S, Lee JH, Kwon YT, Lee MJ. Ablation of Arg-tRNA-protein transferases results in defective neural tube development. BMB Rep. 2016;49:443–448. doi: 10.5483/BMBRep.2016.49.8.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka S, Leu NA, Zhang F, Bunte R, Saha S, Wang J, Guo C, He W, Kashina A. Arginylation-dependent neural crest cell migration is essential for mouse development. PLoS Genet. 2010;6:e1000878. doi: 10.1371/journal.pgen.1000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka S, Leu NA, Pavlov I, Han X, Ribeiro PA, Xu T, Bunte R, Saha S, Wang J, Cornachione A, Mai W, Yates JR, 3rd, Rassier DE, Kashina A. Arginylation regulates myofibrils to maintain heart function and prevent dilated cardiomyopathy. J Mol Cell Cardiol. 2012;53:333–341. doi: 10.1016/j.yjmcc.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Kashina AS, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Kashina AS, Davydov IV, Hu RG, An JY, Seo JW, Du F, Varshavsky A. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- Lambert de Rouvroit C, Goffinet AM. Neuronal migration. Mech Dev. 2001;105:47–56. doi: 10.1016/s0925-4773(01)00396-3. [DOI] [PubMed] [Google Scholar]
- Leite FS, Minozzo FC, Kalganov A, Cornachione AS, Cheng YS, Leu NA, Han X, Saripalli C, Yates JR, 3rd, Granzier H, Kashina AS, Rassier DE. Reduced passive force in skeletal muscles lacking protein arginylation. Am J Physiol Cell Physiol. 2016;310:C127–C135. doi: 10.1152/ajpcell.00269.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu NA, Kurosaka S, Kashina A. Conditional Tek promoter-driven deletion of arginyltransferase in the germ line causes defects in gametogenesis and early embryonic lethality in mice. PLoS One. 2009;4:e7734. doi: 10.1371/journal.pone.0007734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez J. Primary culture of hippocampal neurons from P0 newborn rats. J Vis Exp. 2008 doi: 10.3791/895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, Chao JA. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 2012;26:43–53. doi: 10.1101/gad.177428.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perycz M, Urbanska AS, Krawczyk PS, Parobczak K, Jaworski J. Zipcode binding protein 1 regulates the development of dendritic arbors in hippocampal neurons. J Neurosci: Off J Soc Neurosci. 2011;31:5271–5285. doi: 10.1523/JNEUROSCI.2387-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Kashina A. Identification of mammalian arginyltransferases that modify a specific subset of protein substrates. Proc Natl Acad Sci USA. 2005;102:10123–10128. doi: 10.1073/pnas.0504500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Kashina A. Posttranslational arginylation as a global biological regulator. Dev Biol. 2011;358:1–8. doi: 10.1016/j.ydbio.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Mundia MM, Zhang F, Demers RW, Korobova F, Svitkina T, Perieteanu AA, Dawson JF, Kashina A. Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Mol Biol Cell. 2010;21:1350–1361. doi: 10.1091/mbc.E09-09-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova EA, Singer RH, Condeelis J. The physiological significance of beta -actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci USA. 2001;98:7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent traficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci: Off J Soc Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Prokscha A, Ungewickell E, Eichele G. Doublecortin association with actin fllaments is regulated by neurabin II. J Biol Chem. 2005;280:11361–11368. doi: 10.1074/jbc.M405525200. [DOI] [PubMed] [Google Scholar]
- Wang J, Han X, Saha S, Xu T, Rai R, Zhang F, Wolf YI, Wolfson A, Yates JR, 3rd, Kashina A. Arginyltransferase is an ATP-independent self-regulating enzyme that forms distinct functional complexes in vivo. Chem Biol. 2011;18:121–130. doi: 10.1016/j.chembiol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YM, Ingoglia NA. N-terminal arginylation of sciatic nerve and brain proteins following injury. Neurochem Res. 1997;22:1453–1459. doi: 10.1023/a:1021998227237. [DOI] [PubMed] [Google Scholar]
- Weimer JM, Anton ES. Doubling up on microtubule stabilizers: synergistic functions of doublecortin-like kinase and doublecortin in the developing cerebral cortex. Neuron. 2006;49:3–4. doi: 10.1016/j.neuron.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Wong CCL, Xu T, Rai R, Bailey AO, Yates JR, Wolf YI, Zebroski H, Kashina A. Global analysis of posttranslational protein arginylation. PLoS Biol. 2007;5:e258. doi: 10.1371/journal.pbio.0050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Zhang F, Saha S, Shabalina SA, Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329:1534–1537. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.