Abstract
Development of vaccines against mosquito-borne Flaviviruses is complicated by the occurrence of antibody-dependent enhancement (ADE), which can increase disease severity. Long-term delivery of neutralizing antibodies (nAbs) has the potential to effectively block infection and represents an alternative to vaccination. The risk of ADE may be avoided by using prophylactic nAbs harboring amino acid mutations L234A and L235A (LALA) in the immunoglobulin G (IgG) constant region. Here, we used recombinant adeno-associated viruses (rAAVs) to deliver the anti-dengue virus 3 (DENV3) nAb P3D05. While the administration of rAAV-P3D05-rhesus immunoglobulin G1 (rhIgG1)-LALA to rhesus macaques engendered DENV3-neutralizing activity in serum, it did not prevent infection. The emergence of viremia following DENV3 challenge was delayed by 3–6 days in the rAAV-treated group, and replicating virus contained the envelope mutation K64R. This neutralization-resistant variant was also confirmed by virus outgrowth experiments in vitro. By delivering P3D05 with unmutated Fc sequences, we further demonstrated that DENV3 also evaded wild-type nAb prophylaxis, and serum viral loads appeared to be higher in the presence of low levels of unmutated P3D05-rhIgG1. Our study shows that a vectored approach for long-term delivery of nAbs with the LALA mutations is promising, but prophylaxis using a single nAb is likely insufficient at preventing DENV infection and replication.
Keywords: dengue virus, monoclonal antibody, neutralizing antibody, AAV, gene delivery
Vectored-delivery of virus-neutralizing antibodies could represent an alternative approach to vaccines in controlling viral replication. Magnani and colleagues show that despite sustained antibody transgene expression, dengue virus can evade single serotype-specific antibody gene-based prophylaxis by the emergence of a viral variant that is not sensitive to neutralization.
Introduction
The use of antibodies (Abs) is making a major impact on medicine and expanding our tools for the prevention and therapy of a variety of human viral pathogens.1 Prophylaxis using Abs is especially compelling for diseases that are not yet preventable by vaccination. For instance, HIV-specific neutralizing Abs (nAbs) are currently being tested for HIV treatment and prevention, National Library of Medicine: NCT02716675 and NCT02568215.2, 3, 4, 5, 6, 7 The Ebola epidemic has highlighted the utility of ZMapp—a cocktail of three anti-Ebola nAbs—during an outbreak.8, 9, 10, 11, 12 Administration of a single nAb up to 5 days post infection blocks the development of disease in Ebola-infected macaques.13 Like Ebola, there are a multitude of emergent viral diseases that could be prevented with passive Ab strategies. Recently, outbreaks of the dengue virus (DENV), Zika virus (ZIKV), yellow fever virus (YFV), and others have caused enormous morbidity, with billions of people at risk of infection.14, 15, 16 It is possible that the impact and magnitude of these outbreaks could have been reduced if Ab therapy and prophylaxis were available.
The main limitation of passive nAb administration is the short-lived immunity conferred to its recipients, due to nAb clearance. Attempts to solve this problem include Fc modifications to increase nAb half-lives or use of gene delivery methods to engender long-term activity.17, 18 Most promisingly, gene therapy has the potential to sustain protective nAb levels indefinitely. In fact, nonhuman primates that received recombinant adeno-associated viruses (rAAVs) generated circulating nAbs for months to years.18, 19, 20, 21 If AAV proves to be safe and effective in the delivery of nAbs, it will greatly enhance our options for intervention to prevent infection and disease.
The role of Abs in dengue infection, and possibly most flaviviral infections, is paradoxical.22 The natural immunity triggered by DENV infection is likely mediated by the induction of potent nAbs. However, it is well accepted that the presence of low levels of anti-DENV Abs can enhance viral replication and increase disease severity.23 Thus, even potent nAbs might enhance the infection if present at subneutralizing concentrations. Accordingly, a major concern of nAb-based strategies against DENV is to avoid antibody-dependent enhancement (ADE).23 Fortunately, enhancement of viral replication can be prevented in vitro and in vivo, by altering the amino acid residues that interact with Fc receptors (FcRs).24, 25 Several monoclonal Abs with different neutralization characteristics have been isolated.26, 27, 28, 29, 30 Some of these Abs can prevent viral replication in mouse models, when administered prior to infection.30, 31, 32 These studies highlight the concerns and potential of nAb-mediated prophylaxis against DENV.
Our long-term goal is to prevent flaviviral infections through vectored delivery of nAbs. In contrast to vaccination, we can selectively deliver nAbs in combinations that mediate potent neutralization without inducing cross-reactive non-neutralizing responses. Here, we have engineered rAAV vectors to deliver the serotype-specific anti-DENV3 nAb P3D05 in rhesus immunoglobulin G1 (rhIgG1) L234A and L235A (LALA) and wild-type (WT) versions. While six in eight macaques sustained high levels of P3D05 in serum for months, this single nAb was insufficient for preventing DENV3 infection after a subcutaneous challenge. Regardless of the Fc version, DENV3 replicated to high levels and an escape mutant was identified. Furthermore, we show that low levels of WT nAbs can lead to enhanced viral replication. Ultimately, our results indicate that a single nAb will be insufficient for protecting naive hosts from flaviviral infections.
Results
The human monoclonal nAb P3D05 binds and neutralizes DENV3, but does not bind nor neutralize DENV1, DENV2, or DENV4 (unpublished data). This nAb blocks DENV3 infection of Vero cells at low concentrations, in the range of the most potent DENV3-specific nAbs isolated to date (inhibitory concentration [IC50] = 0.03 μg mL−1). To investigate if P3D05 nAb prophylaxis can be used as an alternative to vaccination, we encoded the antibody chains in a rAAV vector (Figure 1A). rAAV particles carrying nAb-encoding constructs have been used successfully to express and sustain nAb levels for extended periods of time, prolonging the protective effects of the delivered nAbs.18, 19 We cloned the variable heavy (VH) and variable light (VL) chains from the P3D05 human nAb with rhesus constant heavy and kappa regions to create a chimeric human/rhesus immunoglobulin G1 (IgG1).21 We confirmed that this construct produced the bioactive P3D05 nAb when transfected into 293 cells (data not shown). A C9 tag was fused to the heavy chain C terminus to enable the quantification of the recombinant nAbs in vivo. To curb the theoretical risk of ADE, we engineered the rhIgG1 with a LALA double mutation, which greatly reduces Ab binding to Fc receptors.32, 33 We packaged the single-stranded recombinant genome into an rAAV serotype 1 (rAAV1) capsid, as described previously.20, 21, 34
Figure 1.
AAV-Delivered P3D05-rhIgG1-LALA Engenders Virus-Neutralization Activity in Serum
(A) The human VH and VL chains of the P3D05 nAb were fused to rhesus constant regions and linked into a single expression cassette with a F2A self-processing peptide. A LALA mutation was introduced in the rhIgG1 molecule, and a C9 tag was added at the C terminus of the heavy chain. (B) To test the efficacy of the AAV-based prophylaxis, we used the outlined experiment with two groups of four macaques (AAV recipients and no AAV controls). (C) Production of nAbs in serum was measured by a C9-ELISA. (D) We confirmed the presence of bioactive molecules in vivo by assessing the serum DENV3-neutralization titers in the AAV recipients.
We administered the rAAV vector encoding P3D05-rhIgG1-LALA to four rhesus macaques (group 1; Figure 1B). The rAAV (2.5 × 1012 vector genome DNA molecules kg−1) was delivered by four intramuscular injections. The concentrations of the P3D05 nAb in circulation increased quickly, reaching a peak of ∼40–80 μg mL−1 between days 21–29 post rAAV inoculation (Figure 1C). We did not measure host immune responses against the transgene or the vector. At 3 weeks post rAAV delivery, we observed an increased clearance of the circulating nAbs, which stabilized after the fifth week. One of the recipients (r13076) was remarkably different from the others in which the nAbs fell to undetectable levels by week 5. Based on these nAb concentrations, neutralization titers exceeded 1,000 times the nAb IC50. To confirm the biological activity of the recombinant nAbs, we tested the ability of nAb-containing serum to neutralize DENV3 (Figure 1D). At 3 weeks post rAAV delivery, the serum of the vector recipients had neutralization titers in the range of 1 in 2,934 to 1 in 7,969, as assessed by a Vero cell plaque reduction neutralization titer test (PRNT50). Thus, by using this intramuscular gene transfer strategy, we generated very high levels of DENV3-specific nAbs in serum up to the time of the challenge (D38 post rAAV).
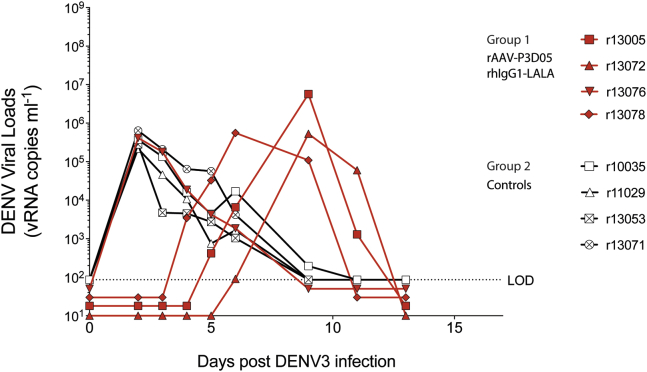
To investigate whether rAAV-encoded nAbs prevent infection, we challenged the four rAAV-treated and four control animals with DENV3, subcutaneously. The quantity of infecting DENV during a human exposure has not been established, but estimates using mosquito-transmission models of another flavivirus indicated that high doses of the related West Nile Virus are inoculated.35 We, therefore, used a challenge dose of 6 × 105 plaque forming units (PFUs). The nAb-treated animals appeared to control virus replication for the first few days post challenge. However, to our surprise, the virus rebounded at days 3–6 post challenge, even in the face of high nAb titers (Figure 2). In contrast to the other monkeys in group 1, animal r13076 had neither detectable levels of delivered nAbs nor detectable neutralizing activity in the serum at the time of the challenge (Figure 1D). Indeed, r13076 had identical DENV replication kinetics to those seen in control animals, consistent with the absence of nAbs in this animal. At D22 post challenge, the naive animals also developed anti-DENV responses, confirming viral infection (Figure 1D). Hence, we found evidence of viral replication in all animals.
Figure 2.
Viral Loads
After subcutaneous DENV3 challenge, viremia was measured by real-time qRT-PCR. Serum DENV3 vRNA (vRNA) levels from rAAV-P3D05-rhIgG1-LALA recipients (group 1) and control macaques (group 2) animals are shown in red and black, respectively. A dotted line indicates the limit of detection (LOD). Median viral loads below the LOD were staggered for visualization purposes only.
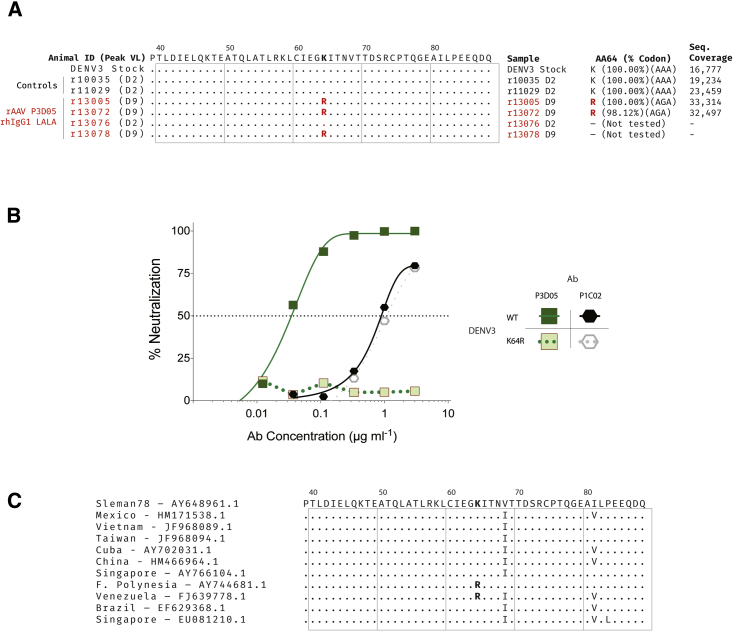
Except for animal r13076, which had no detectable nAbs, all of the other rAAV-treated animals sustained high levels of circulating nAbs for some time during the post challenge period (Figure 1C). It was unclear, therefore, how the virus replicated to such high levels in the presence of high concentrations of a potent nAb in serum. To investigate, we sequenced the viral envelope genes (M and E) at the peak replication time points (Figure 3). We found a single mutation (K64R) in the envelope protein E in all three AAV recipients that had measurable nAbs in serum (Figure 3A). The sequences of the viruses from the controls or r13076 (the animal with low nAb levels) were identical to the parental challenge strain. Deep sequencing of the envelope genes confirmed that the only site that differs among all the consensus sequences is in the E protein at amino acid position 64. Moreover, we observed limited within-sample amino acid diversity, with most of the minor variants being present at less than 2% despite high read coverage. Importantly, the K64R mutation was not identified in the challenge stock, even with read coverage of over 10,000×. We then cultured WT virus in the presence of the P3D05 nAb and observed that the same mutation arose in vitro. We next made K64R mutant virus stocks and demonstrated that this DENV3 mutant was resistant to the P3D05 nAb, but sensitive to neutralization by another nAb (Figure 3B). Interestingly, viruses with this mutation have been observed in clinical specimens from Venezuela and French Polynesia (Figure 3C). In summary, these data demonstrated that the P3D05-rhIgG1-LALA nAb selected for escape variants in vivo.
Figure 3.
Neutralization-Resistant DENV Mutants Emerge In Vivo and In Vitro
(A) We sequenced the M and E genes of circulating viruses at peak replication post challenge. A single mutation, K64R, was identified in all rAAV recipients with measurable nAb titers at the time of the challenge. The frequency of the primary codon at position 64 in each sample and respective sequencing coverage is also shown. (B) DENV3 with the same K64R mutation was generated after in vitro serial passage in media supplemented with P3D05. The DENV3 K64R virus was resistant to neutralization by P3D05. (C) Alignments of E gene sequences from DENV3 isolates.
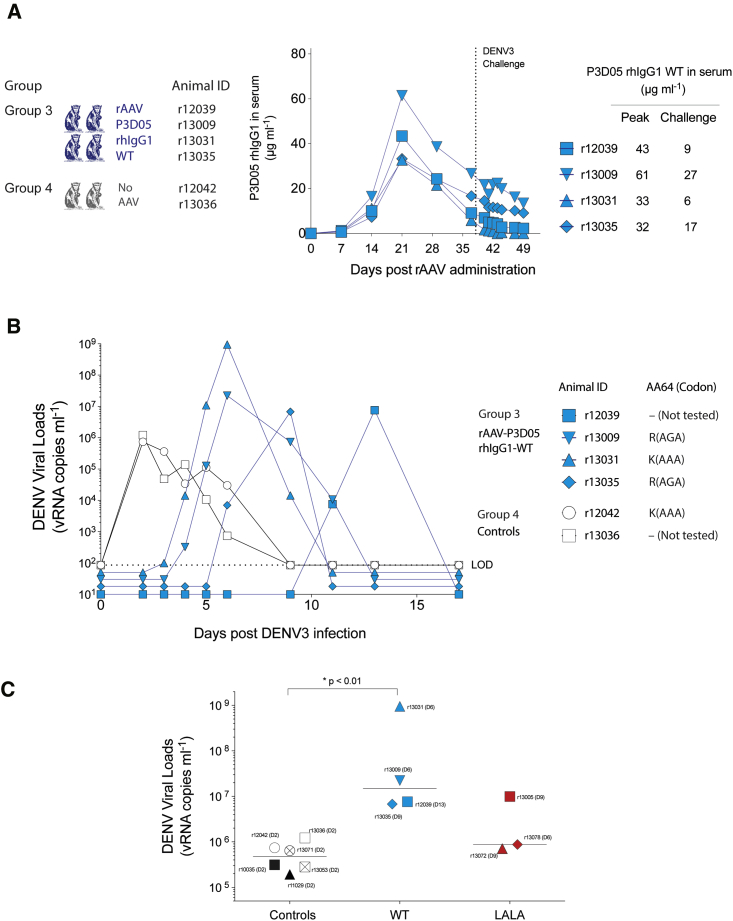
The introduction of the LALA mutation is known to abrogate several potentially important Ab effector mechanisms, including complement binding and antibody-dependent cell cytotoxicity (ADCC).33 It is possible, therefore, that nAbs with this mutation could be less efficient in clearing the virus, especially after cell infection. Thus, in the absence of infected cell clearance mechanisms, any infection event could lead to uncontrolled viral replication and de novo generation of escape variants. To test the hypothesis that Ab effector mechanisms are required for nAb-mediated protection against DENV3, we reengineered the vectors to express P3D05 as a WT rhIgG1 (Figure 4). After rAAV administration, as expected, we detected nAbs in the sera of all four treated macaques (group 3). Similar to the previous experiment, one animal, r13031, had lower levels of nAbs at the time of the challenge (Figure 4A). Irrespective of the nAb titers, the challenge virus infected all macaques (Figure 4B). The emergence of detectable viral loads in the treated animals was delayed as compared to the untreated animals. Strikingly, the rAAV-P3D05-rhIgG1-WT recipients had DENV3 peak viral loads (median 1.50 × 107 viral RNA [vRNA] copies mL−1; range from 6.77 × 106 to 9.49 × 108 vRNA copies mL−1) that were much higher than those that were measured in the controls (median 4.77 × 105 vRNA copies mL−1; range from 1.93 × 105 to 1.22 × 106 vRNA copies mL−1) and the rAAV-P3D05-rhIgG1-LALA recipients (median 8.76 × 105 vRNA copies mL−1; range from 7.08 × 105 to 9.93 × 106) (Figure 4C). The macaque with the lowest nAb levels (r13031) had viral levels exceeding those of the controls by over 1,000-fold (9.49 × 108 vRNA copies mL−1 of r13031 versus the median of 8.76 × 105 vRNA copies mL−1 of controls). Remarkably, the virus from r13031 contained no envelope mutations, while DENV3 with the same K64R escape mutation identified previously replicated in the other rAAV-treated animals. Of note, the serum collection frequency was reduced after day 6 post challenge because of the small size of the animals. It is possible, therefore, that we missed the peak of viral replication in this animal. In summary, these data are consistent with classic ADE, suggesting that the high virus replication observed in r13031 was indeed facilitated by this animal’s subneutralizing levels of WT rhIgG1 nAb. Therefore, delivery of a potent rhIgG1 nAb was not sufficient to prevent infection post DENV3 challenge, regardless which IgG1 Fc version was used.
Figure 4.
Prophylaxis with WT P3D05 Does Not Prevent Viremia and Enhances Viral Replication
(A) We reengineered the rAAV constructs to express a WT-P3D05-rhIgG1 molecule and monitored nAb production in vivo with ELISA to the C9 tag. (B) Four rAAV-P3D05-rhIgG1-WT recipients and two controls were challenged with DENV3 at 37 days post rAAV inoculation. Serum DENV3 vRNA levels from rAAV-P3D05-rhIgG1-WT recipients (group 3) and control macaques (group 4) animals are shown in blue and black, respectively. A dotted line indicates the limit of detection (LOD). Median viral loads below the LOD were staggered for visualization purposes only. (C) Mean peak viral loads of no AAV (controls), rAAV-P3D05-rhIgG1-WT (WT), and rAAV-P3D05-rhIgG1-LALA (LALA). The animal r13076 did not have measurable nAbs at the time of the challenge and was excluded from this analysis. *Mann-Whitney nonparametric rank test.
Discussion
The recent emergence of ZIKV in the Americas resulted in thousands of newborns with severe brain damage and demonstrated that we remain unprepared for emerging outbreaks.36, 37 ADE and cross-reactivity are among the main challenges for the quick development of safe vaccines for flaviviruses.22, 23, 38 Other prophylactic approaches could be helpful in the control of these viruses. In the experiments described here, we evaluated protection mediated by a vector-delivered human nAb against a flavivirus using a nonhuman primate model of infection. We achieved and sustained high levels of the potent P3D05 nAb in circulation for extended periods of time. However, despite the presence of high titers of neutralizing activity conferred by a single nAb, this was not sufficient to protect from infection.
A few, but important, caveats were noticed during our experiments. First, challenge dose and route may be important determinants of the outcome of any preventive or therapeutic flavivirus experiment.39, 40 We used the subcutaneous route to deliver DENV3, at a dose likely to exceed the minimal infectious dose, which has not yet been determined. The virus dose delivered during natural exposure is difficult to measure and is thought to be affected by the levels of viral replication in the mosquito, the extent of probing activity, and the number of bites.40 While a smaller inoculum might have been easier to be protected against, the best estimates indicate that substantially higher doses of flaviviruses can be delivered by a mosquito after a blood meal.35 Thus, nAb treatments should be capable to neutralize large doses of virus.
High levels of neutralizing activity in the serum did not translate to protection from a challenge. In vitro virus neutralization of Vero cell infection is the most commonly used assay to estimate flavivirus-neutralizing antibody (nAb) capability.41, 42 Given that Vero-based assays often fail to predict protection against dengue infection,43, 44 it is possible that this test might not recapitulate essential aspects of DENV infection in vivo. The limitations of this in vitro neutralization assay include the existence of viruses in different maturation states and use of several receptors to achieve infection.41, 42, 43
Our study clearly shows that prophylaxis with a single potent nAb against DENV3 can be evaded by the emergence of circulating viral variants that are not sensitive to neutralization. In the presence of high nAb titers, the replicating virus contained a single mutation that provided neutralization resistance in all animals. Acute escape from Abs in vivo, in a DENV macaque model, has already been reported using a chimpanzee monoclonal nAb with defective FcR binding.45 Our findings, therefore, confirm the notion that flaviviruses can escape from single nAbs both in vivo and in vitro. The underlying mechanism of the generation of the K64R escape mutation remains unknown. It is possible that underrepresented Ab-resistant viral species were present in the challenge stock, despite being undetectable by deep sequencing. Intriguingly, the macaques were aviremic for up to 10 days, before DENV3 could be detected in the circulation. A possible interpretation of the delayed replication kinetics is that the escape mutation was generated de novo. This explanation would be consistent with the fact that AAV-treated animals had different delays in viral replication. In this alternative scenario, either the large dose of infecting virus was sufficient to bypass the nAbs and infect a small number of cells, or the virus remained compartmentalized in sites protected from neutralization. Thus, an initial failure to sterilize the incoming virus could result in productively infected cells and enable viral replication, thereby spawning escape mutants.
The introduction of the LALA mutation is known to abrogate ADCC.33 Consequently, after cells became infected in rAAV-P3D05-rhIgG1-LALA-treated animals, there may have been no specific anti-viral response that might have limited the generation of de novo virus production. We, therefore, reasoned that these effector mechanisms against infected cells may be necessary for achieving sterilizing protection against viral challenge. However, the use of WT nAb sequences did not result in protection or reduced viral replication. On the contrary, DENV replication in the presence of low WT nAb amounts was enhanced. Despite the high viral loads in these animals, no obvious clinical manifestation was noticed. A large hematoma developed in animal r13009 at 37 days post challenge (unpublished data), possibly associated with blood collection. These data highlight the real risk of ADE after therapy with WT nAbs.
In summary, our results point to the unfortunate, but likely scenario that a single potent nAb will not be sufficient for preventing or treating DENV infections. Furthermore, our data confirm that even a potent WT nAb is capable of enhancing infection. These findings could have profound implications not only for DENV, but other flaviviruses as well.46, 47, 48 To ultimately succeed, a nAb-based therapy for a flavivirus must address viral escape and ADE. Of note, prevention of dengue infection with monoclonal Abs (mAbs) was previously achieved in immunodeficient mouse models of dengue.30, 31, 32 However, murine models are artificially constrained by the reduced permissiveness of mouse cells, and the extent that these findings will be translatable to humans remains unclear.39 In contrast, nonhuman primate (NHP) models have been useful in predicting human clinical trial outcomes during dengue vaccine development.39 The failure of the P3D05 nAb to prevent infection could be associated with the nature of this particular nAb and its targeted epitope. However, nonhuman primate studies that used a single nAb-prophylaxis strategy using a different nAb also resulted in viremia and seroconversion.45, 49 Additionally, nonhuman primate models of Ab-enhanced DENV infection show that, under certain conditions, the administered WT Abs can increase peak viremia.50, 51 The outlook for nAb-based prevention and therapies is, nonetheless, still promising. In a recent study, the administration of high doses of a single nAb failed to prevent viremia and seroconversion, but similar levels of polyclonal Abs isolated from sera succeeded.49 It is possible then that combinations of several nAbs that do not select for escape mutations in vitro will be sufficient to control viral replication. In addition, the recent isolation of potent and serotype-cross-reactive nAbs expands the prophylaxis options to different conserved epitopes.26, 30 Finally, we have described here the use of rAAVs for long-term sustained expression of nAbs in nonhuman primates. Indeed, we confirmed sustained production of recombinant Abs (rAbs) even at day 499 post rAAV administration (r13005 42 μg mL−1; r13072 46 μg mL−1; r13076 17 μg mL−1; and r13078 76 μg mL−1). rAAV-delivered nAbs is an innovative and promising approach to provide immunity against diseases for which there are complications that might be caused by vaccines. Thus, the vectored-delivery of several potent nAbs and the long-term maintenance of high serum levels of these nAbs could represent an alternative approach to vaccines in controlling viral replication.
Materials and Methods
Ethics Statement
The Indian rhesus macaques (Macaca mulatta) utilized in this study were housed at the Wisconsin National Primate Research Center (WNPRC). The WNPRC is accredited by the American Association of Accreditation of Laboratory Animal Care (Animal Welfare Assurance No. A3368-01). All animals were cared for in accordance with the guidelines of the Weatherall Report, the Animal Welfare Act, and the NIH for care and use of laboratory animals.52 Animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Wisconsin (IACUC; protocol G00734). Procedures were performed to ensure that discomfort was limited to that unavoidable in the conduct of the research plan. Sedatives were applied as necessary for blood and tissue collections and, analgesics were used when determined appropriate by veterinary medical staff. Animals were anesthetized during experimental procedures with ketamine (up to 15 mg kg−1 intramuscularly) or up to 7 mg kg−1 ketamine and up to 0.03 mg kg−1 dexmedetomidine via the intramuscular route. The macaques in this study were managed according to the animal husbandry program of the WNPRC, which aims at providing consistent and excellent care to nonhuman primates at the center. This program is employed by the Colony Management Unit and is based on the laws, regulations, and guidelines promulgated by the United States Department of Agriculture (e.g., the Animal Welfare Act and its regulations, and the Animal Care Policy Manual), Institute for Laboratory Animal Research (e.g., Guide for the Care and Use of Laboratory Animals, 8th edition), Public Health Service, National Research Council, Centers for Disease Control, and the Association for Assessment and Accreditation of Laboratory Animal Care International. The nutritional plan utilized by the WNPRC is based on recommendations published by the National Research Council. Specifically, macaques were fed twice daily with 2050 Teklad Global 20% Protein Primate Diet and food intake was closely monitored by animal research technicians. This diet was also supplemented with a variety of fruits, vegetables, and other edible objects as part of the environmental enrichment program established by the Behavioral Management Unit. Paired/grouped animals exhibiting stereotypical and/or incompatible behaviors were reported to the Behavioral Management staff and managed accordingly. All primary enclosures (i.e., stationary cages, mobile racks, and pens) and animal rooms were cleaned daily with water and sanitized at least once every 2 weeks. Lights were on a 12:12 diurnal schedule. Animals had access to more than one category of enrichment at WNPRC. The IACUC proposal included a written scientific justification for any exclusions from some parts of the enrichment plan. Research-related exemptions are reviewed at least annually. This was a nonterminal experiment, so no euthanasia was performed. All animals were returned to the WNPRC’s colony after study completion.
Animals
This was a nonterminal experiment, with the hypothesis that rAAV-delivered nAbs are sufficient to prevent DENV viremia after the challenge. Animals were prescreened based on their serological status, and only animals free of serum nAbs against AAV1 were selected. All 14 animals were returned to the WNPRC’s colony after study completion.
Vector Construction and rAAV Preparation
In order to encode P3D05 in a single AAV, we based our constructs on plasmids previously optimized for expression of nAbs in rhesus macaques.18, 20, 21 We synthesized genes encoding the nAbs expression cassette, flanked by the AAV’s inverted terminal repeats (ITRs). A cytomegalovirus (CMV) promoter was used to express the antibody (Figure 1A). The 3′ UTR contained a polyadenylation signal from SV40. To confirm that all expression elements were present in the final plasmid preparations, we digested each construct with PvuII with NcoI; SpeI with AflII; PvuII with AgeI; EcoRI with NotII; and SmaI. The synthetic nucleic acid was packaged into AAV-1 using previously described methods.20, 21, 34 To confirm the expression of the Ab product encoded on our AAV construct, we have transfected HEK293T/17 cells (American Type Culture Collection [ATCC]) with the rAAV-DNA encoding P3D05 and measured the production of rAb using an ELISA against the C9 tag and the neutralization potency of the rAb.
rAAV and DENV Administration
A total of 14 Indian rhesus macaques (eight males and six females) were used to test our rAAV-mediated prophylaxis strategy. These macaques were separated into four groups, as follows: group 1 (rAAV-P3D05-rhIgG1-LALA, n = 4); group 2 (no AAV controls, n = 4); group 3 (rAAV-P3D05-rhIgG1-WT, n = 4); and group 4 (no AAV controls, n = 2). Recombinant AAV diluted in RPMI was administered to macaques weighing 2–5 kg in four 0.5 mL intramuscular injections in the quadriceps and biceps muscles. Each animal received 2.5 × 1012 viral genome copies kg−1. To measure antibody titers generated prior to the challenge, we collected sera in the weeks 0, 1, 2, 3, 4, and 5 post rAAV. Animals were challenged with 6 × 105 PFUs of recombinant DENV3 (AY648961.1; lot: DEN3 Sleman/78 1A1 TD3+V2+C6/36p1 1/14/2008),53 delivered subcutaneously. After the challenge, we collected serum at the indicated time points to measure DENV viremia and seroconversion.
PRNT
PRNTs were conducted as previously described.54 Briefly, serum samples were serially diluted in Opti-MEM (Thermo Fisher Scientific) supplemented with 2% human serum albumin, 2% fetal bovine serum (FBS), and gentamicin. DENV3 was diluted to a final concentration of ∼500–1,000 PFU mL−1 in the same diluent added to equal volumes of the diluted sample. The virus/serum mixture was incubated at 37°C for 30 min. Cell culture medium was removed from 90% confluent monolayer cultures of Vero cells (ATCC) on 24-well plates and 100 μL of the virus/Ab mixture was transferred onto duplicate cell monolayers. Cell monolayers were incubated for 60 min at 37°C and overlaid with 1% methylcellulose in Opti-MEM (Thermo Fisher Scientific) supplemented with 2% FBS 2 mM glutamine + 50 μg mL−1 gentamicin. Samples were incubated at 37°C for 4 days, after which plaques were visualized by immunoperoxidase staining and a 50% plaque-reduction neutralization titer was calculated.
ELISA
The production of rAb in sera was quantitated in a rC9 ELISA assay. In brief, 96-well ELISA plates were coated overnight with 2 μg mL−1 of the mouse-anti-Rhodopsin (C9) monoclonal Ab clone 1D4 (EMD Millipore) diluted in PBS. Each plate was washed five times with PBS-Tween20, and the wells were blocked with 5% skimmed milk in PBS for 1 hr at 37°C. Subsequently, the plate was washed with PBS and serum samples were added to designated wells. After 1 hr incubation at 37°C, the plate was washed and detection was carried out using a mouse anti-monkey IgG-horseradish peroxidase (HRP) clone SB108a (SouthernBiotech), which was added to all wells at a dilution of 1:2,000. Following a 1 hr incubation at 37°C, the plate was washed and developed with 3,3’,5,5’-tetramethylbenzidine (TMB) substrate at room temperature for 3–4 min. Reaction was then stopped with the TMB stop solution, and absorbance was read at 450 nm.
Viral Loads
Real time quantitative reverse-transcription PCR (qRT-PCR) was used for the measurement of viral loads in sera, based on a previously validated assay.55, 56 In brief, RNA was extracted from 140 μL of frozen sera using the QIAamp vRNA Mini Kit (QIAGEN). The total nucleic acid was eluted in two centrifugation steps with 40 μL of buffer AVE each. A real-time qRT-PCR reaction was then carried out with 20 μL of samples and 10 μL of primer, probes, and TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems). We used precombined probe and primers (500 nM primers and 250 nM probe; Integrated DNA Technologies). The primer and probe sequences were modified to match sequences to the DENV3 Sleman/78 isolate (AY648961.1) and were as follows: DENV3 P1 5′GGACTGGACACACGCACCCA3′; P2 5′AATGTCTCTACCTTCTCGACCTGTCT3′; and Probe 5′ACCTGGATGTCGGCTGAAGGAGCTTG3′. The probe was double quenched (ZEN/Iowa Black FQ) and labeled with the carboxyfluorescein (FAM) dye (Integrated DNA Technologies). The PCR conditions consisted of a reverse transcription step at 50°C for 30 min, followed by a step of 95°C for 10 min, then 40 cycles at 95°C for 15 s, and 60°C for 1 min. Three-fold serial dilutions of total DENV3 RNA (Vircell; lot: 15MBC057001) starting at 800 copies μL−1 were used as standards, and results were reported as the equivalent vRNA genomes per milliliter.
In Vitro Escape Assay
Vero cells (160,000 cells per well) were infected with virus in the presence or absence of P3D05. DENV3-Sleman/78 was used at a final MOI of approximately 0.3 virions per well. The purified mAb P3D05 was added to a final concentration of 1 μg mL−1. Virus with antibody or virus and media mixtures were incubated for 1 hr at 37°C before cell adsorption. Cells were infected (1 hr) and then washed and incubated in media with or without 1 μg mL−1 P3D05 supplementation. On the fourth day post infection, 50 μL of the cell-free supernatant was used to infect a new batch of cells. This process was repeated until cytopathic effect was clearly observed. From each passage, the remainder of the supernatant was frozen in aliquots for vRNA sequencing.
Envelope Sequencing
To sequence the envelope from serum virus, we amplified a 1,857 bp segment containing the M and E genes in a one-step Superscript III RT-PCR reaction (Life Technologies) using the following primers: DV3 620 5′GAAGACATTGACTGCTGGTGCAAC3′ and DV3 2476 5′GACGAAAATTCCACTTCCACATTTGAGTTC3′. The amplified DNA was then sequenced in multiple reactions of 500 bp each, with 2–3× coverage of the whole segment via Sanger sequencing. The same procedure was used to generate amplicons for Illumina deep sequencing. Illumina library construction was performed using Nextera XT (Illumina) according to manufacturer’s protocol. Sequencing was performed on the Illumina MiSeq Platform, generating 250-bp paired-end reads. De novo genome assembly and variant calling were performed using VICUNA and V-Phaser 2, respectively.57, 58
Author Contributions
Conceptualization, D.M.M., R.C.D., E.G.K., and D.I.W.; Writing - Original Draft, D.M.M. and D.I.W.; Writing - Review & Editing, D.M.M., R.C.D., D.R.B., and D.I.W.; Investigation, D.M.M., M.J.R., V.K.B., M.J.G., N.P.-L., H.S.M., A.D., L.G.-N., Q.S., R.M.N., M.P., E.G.R., and S.S.W.; Resources, C.G.T.S., E.G.K., G.G., and D.I.W.; Funding Acquisition, G.G. and D.I.W.; Methodology, D.M.M., M.A.M., J.M.M.-N., S.P.F., and S.S.W.; Project Administration, D.M.M. and D.I.W.; and Supervision, D.M.M., T.M.A., S.S.W., D.R.B., G.G., R.C.D., E.G.K., and D.I.W.
Conflicts of Interest
The authors declare no financial conflicts of interest.
Acknowledgments
This work was supported by NIH grant 4P01AI094420-05, the Wallace H. Coulter Center for Translational Research at the University of Miami, and the Miami Clinical and Translational Science Institute (CTSI). This work was in part supported by NIH grant P51OD011106 to the WNPRC at the University of Wisconsin-Madison. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We would like to thank Kim Weisgrau, Jessica Furlott, Nicholas Pomplun, and Logan Vosler for technical support and Eric Peterson, Kristin Crosno, and Wendy Newton for providing care of the rhesus macaques used in this experiment. We also thank the veterinary and animal care staff for their contribution to this study.
References
- 1.Qiu X., Wong G., Audet J., Bello A., Fernando L., Alimonti J.B., Fausther-Bovendo H., Wei H., Aviles J., Hiatt E. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Institute of Allergy and Infectious Diseases (NIAID). (2016). Evaluating the safety and efficacy of the VRC01 antibody in reducing acquisition of HIV-1 infection among men and transgender persons who have sex with men. https://clinicaltrials.gov/ct2/show/NCT02716675. NLM Identifier: NCT02716675.
- 3.National Institute of Allergy and Infectious Diseases (NIAID). (2016). Evaluating the safety and efficacy of the VRC01 antibody in reducing acquisition of HIV-1 infection in women. https://clinicaltrials.gov/ct2/show/NCT02568215. NLM Identifier: NCT02568215.
- 4.Ledgerwood J.E., Coates E.E., Yamshchikov G., Saunders J.G., Holman L., Enama M.E., DeZure A., Lynch R.M., Gordon I., Plummer S., VRC 602 Study Team Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin. Exp. Immunol. 2015;182:289–301. doi: 10.1111/cei.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheid J.F., Horwitz J.A., Bar-On Y., Kreider E.F., Lu C.L., Lorenzi J.C., Feldmann A., Braunschweig M., Nogueira L., Oliveira T. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature. 2016;535:556–560. doi: 10.1038/nature18929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caskey M., Klein F., Nussenzweig M.C. Broadly neutralizing antibodies for HIV-1 prevention or immunotherapy. N. Engl. J. Med. 2016;375:2019–2021. doi: 10.1056/NEJMp1613362. [DOI] [PubMed] [Google Scholar]
- 7.Caskey M., Schoofs T., Gruell H., Settler A., Karagounis T., Kreider E.F., Murrell B., Pfeifer N., Nogueira L., Oliveira T.Y. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 2017;23:185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haque A., Hober D., Blondiaux J. Addressing therapeutic options for Ebola virus infection in current and future outbreaks. Antimicrob. Agents Chemother. 2015;59:5892–5902. doi: 10.1128/AAC.01105-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuyama W., Marzi A., Nanbo A., Haddock E., Maruyama J., Miyamoto H., Igarashi M., Yoshida R., Noyori O., Feldmann H., Takada A. Discovery of an antibody for pan-ebolavirus therapy. Sci. Rep. 2016;6:20514. doi: 10.1038/srep20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu X., Audet J., Lv M., He S., Wong G., Wei H., Luo L., Fernando L., Kroeker A., Bovendo H.F. Two-mAb cocktail protects macaques against the Makona variant of Ebola virus. Sci. Transl. Med. 2016;8:329ra33. doi: 10.1126/scitranslmed.aad9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey R.T., Jr., Dodd L., Proschan M.A., Neaton J., Neuhaus Nordwall J., Koopmeiners J.S., Beigel J., Tierney J., Lane H.C., Fauci A.S., PREVAIL II Writing Group. Multi-National PREVAIL II Study Team A randomized, controlled trial of ZMapp for Ebola virus infection. N. Engl. J. Med. 2016;375:1448–1456. doi: 10.1056/NEJMoa1604330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dörnemann J., Burzio C., Ronsse A., Sprecher A., De Clerck H., Van Herp M., Kolié M.C., Yosifiva V., Caluwaerts S., McElroy A.K., Antierens A. First newborn baby to receive experimental therapies survives Ebola virus disease. J. Infect. Dis. 2017;215:171–174. doi: 10.1093/infdis/jiw493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corti D., Misasi J., Mulangu S., Stanley D.A., Kanekiyo M., Wollen S., Ploquin A., Doria-Rose N.A., Staupe R.P., Bailey M. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- 14.Smith D.R. Waiting in the wings: The potential of mosquito transmitted flaviviruses to emerge. Crit. Rev. Microbiol. 2017;43:405–422. doi: 10.1080/1040841X.2016.1230974. [DOI] [PubMed] [Google Scholar]
- 15.Tam C.C., Khan M.S., Legido-Quigley H. Where economics and epidemics collide: migrant workers and emerging infections. Lancet. 2016;388:1374–1376. doi: 10.1016/S0140-6736(16)31645-2. [DOI] [PubMed] [Google Scholar]
- 16.Barrett A.D. Yellow fever in Angola and beyond--the problem of vaccine supply and demand. N. Engl. J. Med. 2016;375:301–303. doi: 10.1056/NEJMp1606997. [DOI] [PubMed] [Google Scholar]
- 17.Ko S.Y., Pegu A., Rudicell R.S., Yang Z.Y., Joyce M.G., Chen X., Wang K., Bao S., Kraemer T.D., Rath T. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature. 2014;514:642–645. doi: 10.1038/nature13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs S.P., Desrosiers R.C. Promise and problems associated with the use of recombinant AAV for the delivery of anti-HIV antibodies. Mol. Ther. Methods Clin. Dev. 2016;3:16068. doi: 10.1038/mtm.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson P.R., Schnepp B.C., Zhang J., Connell M.J., Greene S.M., Yuste E., Desrosiers R.C., Clark K.R. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 2009;15:901–906. doi: 10.1038/nm.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs S.P., Martinez-Navio J.M., Piatak M., Jr., Lifson J.D., Gao G., Desrosiers R.C. AAV-delivered antibody mediates significant protective effects against SIVmac239 challenge in the absence of neutralizing activity. PLoS Pathog. 2015;11:e1005090. doi: 10.1371/journal.ppat.1005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs S.P., Martinez-Navio J.M., Gao G., Desrosiers R.C. Recombinant AAV vectors for enhanced expression of authentic IgG. PLoS ONE. 2016;11:e0158009. doi: 10.1371/journal.pone.0158009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acosta E.G., Bartenschlager R. Paradoxical role of antibodies in dengue virus infections: considerations for prophylactic vaccine development. Expert Rev. Vaccines. 2016;15:467–482. doi: 10.1586/14760584.2016.1121814. [DOI] [PubMed] [Google Scholar]
- 23.Wang T.T., Sewatanon J., Memoli M.J., Wrammert J., Bournazos S., Bhaumik S.K., Pinsky B.A., Chokephaibulkit K., Onlamoon N., Pattanapanyasat K. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science. 2017;355:395–398. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramadhany R., Hirai I., Sasaki T., Ono K., Ramasoota P., Ikuta K., Kurosu T. Antibody with an engineered Fc region as a therapeutic agent against dengue virus infection. Antiviral Res. 2015;124:61–68. doi: 10.1016/j.antiviral.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Lai C.J., Goncalvez A.P., Men R., Wernly C., Donau O., Engle R.E., Purcell R.H. Epitope determinants of a chimpanzee dengue virus type 4 (DENV-4)-neutralizing antibody and protection against DENV-4 challenge in mice and rhesus monkeys by passively transferred humanized antibody. J. Virol. 2007;81:12766–12774. doi: 10.1128/JVI.01420-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dejnirattisai W., Wongwiwat W., Supasa S., Zhang X., Dai X., Rouvinski A., Jumnainsong A., Edwards C., Quyen N.T., Duangchinda T. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai W.-Y., Lai C.-Y., Wu Y.-C., Lin H.-E., Edwards C., Jumnainsong A., Kliks S., Halstead S., Mongkolsapaya J., Screaton G.R., Wang W.K. High-avidity and potently neutralizing cross-reactive human monoclonal antibodies derived from secondary dengue virus infection. J. Virol. 2013;87:12562–12575. doi: 10.1128/JVI.00871-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith S.A., de Alwis A.R., Kose N., Harris E., Ibarra K.D., Kahle K.M., Pfaff J.M., Xiang X., Doranz B.J., de Silva A.M. The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. mBio. 2013;4:1–12. doi: 10.1128/mBio.00873-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Priyamvada L., Cho A., Onlamoon N., Zheng N.Y., Huang M., Kovalenkov Y., Chokephaibulkit K., Angkasekwinai N., Pattanapanyasat K., Ahmed R. B cell responses during secondary dengue virus infection are dominated by highly cross-reactive, memory-derived plasmablasts. J. Virol. 2016;90:5574–5585. doi: 10.1128/JVI.03203-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu M., Zuest R., Velumani S., Tukijan F., Toh Y.X., Appanna R., Tan E.W., Cerny D., MacAry P., Wang C.-I., Fink K. A potent neutralizing antibody with therapeutic potential against all four serotypes of dengue virus. npj. Vaccines (Basel) 2017;2:2. doi: 10.1038/s41541-016-0003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beltramello M., Williams K.L., Simmons C.P., Macagno A., Simonelli L., Quyen N.T., Sukupolvi-Petty S., Navarro-Sanchez E., Young P.R., de Silva A.M. The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams K.L., Sukupolvi-Petty S., Beltramello M., Johnson S., Sallusto F., Lanzavecchia A., Diamond M.S., Harris E. Therapeutic efficacy of antibodies lacking Fcγ receptor binding against lethal dengue virus infection is due to neutralizing potency and blocking of enhancing antibodies [corrected] PLoS Pathog. 2013;9:e1003157. doi: 10.1371/journal.ppat.1003157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hessell A.J., Hangartner L., Hunter M., Havenith C.E., Beurskens F.J., Bakker J.M., Lanigan C.M., Landucci G., Forthal D.N., Parren P.W. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 34.Gao, G., and Sena-Esteves, M. (2012). Introducing genes into mammalian cells: viral vectors. In Molecular Cloning, Vol 2: A Laboratory Manual, M.R. Green and J. Sambrook, eds. (Cold Spring Harbor Laboratory Press), pp 1209–1313. [DOI] [PubMed]
- 35.Styer L.M., Kent K.A., Albright R.G., Bennett C.J., Kramer L.D., Bernard K.A. Mosquitoes inoculate high doses of West Nile virus as they probe and feed on live hosts. PLoS Pathog. 2007;3:1262–1270. doi: 10.1371/journal.ppat.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brasil P., Pereira J.P., Jr., Moreira M.E., Ribeiro Nogueira R.M., Damasceno L., Wakimoto M., Rabello R.S., Valderramos S.G., Halai U.A., Salles T.S. Zika virus infection in pregnant women in Rio de Janeiro. N. Engl. J. Med. 2016;375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honein M.A., Dawson A.L., Petersen E.E., Jones A.M., Lee E.H., Yazdy M.M., Ahmad N., Macdonald J., Evert N., Bingham A., US Zika Pregnancy Registry Collaboration Birth defects among fetuses and infants of US women with evidence of possible Zika virus infection during pregnancy. JAMA. 2017;317:59–68. doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- 38.Dejnirattisai W., Supasa P., Wongwiwat W., Rouvinski A., Barba-Spaeth G., Duangchinda T., Sakuntabhai A., Cao-Lormeau V.M., Malasit P., Rey F.A. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zompi S., Harris E. Animal models of dengue virus infection. Viruses. 2012;4:62–82. doi: 10.3390/v4010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mores C.N., Christofferson R.C., Davidson S.A. The role of the mosquito in a dengue human infection model. J. Infect. Dis. 2014;209(Suppl 2):S71–S78. doi: 10.1093/infdis/jiu110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas S.J., Nisalak A., Anderson K.B., Libraty D.H., Kalayanarooj S., Vaughn D.W., Putnak R., Gibbons R.V., Jarman R., Endy T.P. Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: How alterations in assay conditions impact performance. Am. J. Trop. Med. Hyg. 2009;81:825–833. doi: 10.4269/ajtmh.2009.08-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timiryasova T.M., Bonaparte M.I., Luo P., Zedar R., Hu B.T., Hildreth S.W. Optimization and validation of a plaque reduction neutralization test for the detection of neutralizing antibodies to four serotypes of dengue virus used in support of dengue vaccine development. Am. J. Trop. Med. Hyg. 2013;88:962–970. doi: 10.4269/ajtmh.12-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Byers A.M., Broder R., Haupfear K., Timiryasova T.M., Hu B.T., Boaz M., Warren W.L., Jackson N., Moser J.M., Guy B. Influence of FcγRIIa-expressing cells on the assessment of neutralizing and enhancing serum antibodies elicited by a live-attenuated tetravalent dengue vaccine. Open Forum Infect. Dis. 2015;2:ofv172. doi: 10.1093/ofid/ofv172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirivichayakul C., Sabchareon A., Limkittikul K., Yoksan S. Plaque reduction neutralization antibody test does not accurately predict protection against dengue infection in Ratchaburi cohort, Thailand. Virol. J. 2014;11:48. doi: 10.1186/1743-422X-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goncalvez A.P., Purcell R.H., Lai C.J. Epitope determinants of a chimpanzee Fab antibody that efficiently cross-neutralizes dengue type 1 and type 2 viruses map to inside and in close proximity to fusion loop of the dengue type 2 virus envelope glycoprotein. J. Virol. 2004;78:12919–12928. doi: 10.1128/JVI.78.23.12919-12928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond M.S. Progress on the development of therapeutics against West Nile virus. Antiviral Res. 2009;83:214–227. doi: 10.1016/j.antiviral.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diamond M.S., Pierson T.C., Fremont D.H. The structural immunology of antibody protection against West Nile virus. Immunol. Rev. 2008;225:212–225. doi: 10.1111/j.1600-065X.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roehrig J.T., Staudinger L.A., Hunt A.R., Mathews J.H., Blair C.D. Antibody prophylaxis and therapy for flavivirus encephalitis infections. Ann. N Y Acad. Sci. 2001;951:286–297. doi: 10.1111/j.1749-6632.2001.tb02704.x. [DOI] [PubMed] [Google Scholar]
- 49.Simmons M., Putnak R., Sun P., Burgess T., Marasco W.A. Antibody prophylaxis against dengue virus 2 infection in non-human primates. Am. J. Trop. Med. Hyg. 2016;95:1148–1156. doi: 10.4269/ajtmh.16-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halstead S.B. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J. Infect. Dis. 1979;140:527–533. doi: 10.1093/infdis/140.4.527. [DOI] [PubMed] [Google Scholar]
- 51.Goncalvez A.P., Engle R.E., St Claire M., Purcell R.H., Lai C.J. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc. Natl. Acad. Sci. USA. 2007;104:9422–9427. doi: 10.1073/pnas.0703498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.National Research Council (U.S.) Committee for the Update of the Guide for the Care and Use of Laboratory Animals, Institute for Laboratory Animal Research (U.S.), and National Academies Press (U.S.). (2011). Guide for the Care and Use of Laboratory Animals. 8th edition (National Academies Press) pp xxv, 220.
- 53.Blaney J.E., Jr., Matro J.M., Murphy B.R., Whitehead S.S. Recombinant, live-attenuated tetravalent dengue virus vaccine formulations induce a balanced, broad, and protective neutralizing antibody response against each of the four serotypes in rhesus monkeys. J. Virol. 2005;79:5516–5528. doi: 10.1128/JVI.79.9.5516-5528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durbin A.P., Karron R.A., Sun W., Vaughn D.W., Reynolds M.J., Perreault J.R., Thumar B., Men R., Lai C.J., Elkins W.R. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3′-untranslated region. Am. J. Trop. Med. Hyg. 2001;65:405–413. doi: 10.4269/ajtmh.2001.65.405. [DOI] [PubMed] [Google Scholar]
- 55.Santiago G.A., Vergne E., Quiles Y., Cosme J., Vazquez J., Medina J.F., Medina F., Colón C., Margolis H., Muñoz-Jordán J.L. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl. Trop. Dis. 2013;7:e2311. doi: 10.1371/journal.pntd.0002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson B.W., Russell B.J., Lanciotti R.S. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 2005;43:4977–4983. doi: 10.1128/JCM.43.10.4977-4983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X., Charlebois P., Macalalad A., Henn M.R., Zody M.C. V-Phaser 2: variant inference for viral populations. BMC Genomics. 2013;14:674. doi: 10.1186/1471-2164-14-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X., Charlebois P., Gnerre S., Coole M.G., Lennon N.J., Levin J.Z., Qu J., Ryan E.M., Zody M.C., Henn M.R. De novo assembly of highly diverse viral populations. BMC Genomics. 2012;13:475. doi: 10.1186/1471-2164-13-475. [DOI] [PMC free article] [PubMed] [Google Scholar]