Abstract
Ex vivo gene therapy using lentiviral vectors (LVs) is a proven approach to treat and potentially cure many hematologic disorders and malignancies but remains stymied by cumbersome, cost-prohibitive, and scale-limited production processes that cannot meet the demands of current clinical protocols for widespread clinical utilization. However, limitations in LV manufacture coupled with inefficient transduction protocols requiring significant excess amounts of vector currently limit widespread implementation. Herein, we describe a microfluidic, mass transport-based approach that overcomes the diffusion limitations of current transduction platforms to enhance LV gene transfer kinetics and efficiency. This novel ex vivo LV transduction platform is flexible in design, easy to use, scalable, and compatible with standard cell transduction reagents and LV preparations. Using hematopoietic cell lines, primary human T cells, primary hematopoietic stem and progenitor cells (HSPCs) of both murine (Sca-1+) and human (CD34+) origin, microfluidic transduction using clinically processed LVs occurs up to 5-fold faster and requires as little as one-twentieth of LV. As an in vivo validation of the microfluidic-based transduction technology, HSPC gene therapy was performed in hemophilia A mice using limiting amounts of LV. Compared to the standard static well-based transduction protocols, only animals transplanted with microfluidic-transduced cells displayed clotting levels restored to normal.
Keywords: microfluidics, lentivirus, gene therapy, hemophilia, CAR-T cells, hematopoietic stem cells, cell therapy, cell manufacturing
Graphical Abstract

Leveraging mass transport-based principles of their microfluidic transduction system, Tran et al. demonstrate enhanced gene transfer efficiency by reducing the required amounts of virus and transduction times for adequate gene transfer in primary human T cells and hematopoietic stem cells while also increasing in vivo gene expression in animal models.
Introduction
Clinical trials targeting hematopoietic stem and progenitor cells (HSPCs) and T cells for treatment of genetic disorders and cancers are finding increasing success using HIV-derived lentiviral vectors (LVs).1, 2, 3, 4 Additionally, the improved safety and efficacy of current self-inactivating LVs provides an attractive option for gene therapy.5, 6 Despite these advances, LV manufacture remains inefficient, scale limited, and expensive.7 Moreover, lengthy ex vivo cell transduction protocols require large quantities of LVs to achieve the desired clinical outcomes.8, 9 Ongoing clinical studies expose the technological limitations and highlight the need for improved clinical LV transduction efficiency, as entire LV production runs may only supply sufficient material to dose a few patients. To address these issues, previous attempts to improve transduction using polycations,10 spinoculation,11 recombinant fibronectin fragment coatings (e.g., RetroNectin [RN]),12 or biological adjuvants13 have been explored with mixed success. High-throughput screening has also identified potential transduction-enhancing reagents, although further testing and characterization are required.14 These problematically high vector requirements needed for sufficient levels of gene transfer are due in part to the large minimum working volumes of standard transduction systems (e.g., culture plates, flasks, and bags). Moreover, the MOI, a commonly used parameter representing the ratio of viral particles to target cells for a given transduction, has consistently proven to be unreliable to predict dosages of LV necessary for therapeutic benefit.15, 16 Instead, LV concentration by volume [(v/v)%] is emerging as a means to describe approaches that maximize the amount of virus used without inducing vector-related cytotoxicity and nutrient deprivation.17, 18 In clinical protocols, these concentrations correspond to an MOI as high as 100 with several rounds of transduction, despite theoretical models based on a Poisson distribution that suggest that an MOI of 2 is sufficient to enable ∼87% of the cells to interact with at least one infectious unit.
Taking all of the above into account, we hypothesized that this disparity is due in part to a lack of efficient transport of virus to cells. With a half-life on the order of hours at 37°C, LV particles may only travel an average distance of ∼600 μm from their starting point through Brownian motion.19, 20 However, transductions conducted in the aforementioned standard cell culture vessels must traverse distances on the order of millimeters to centimeters to reach cells, suggesting that most of the virus is wasted due to insufficient transport before viral degradation. By spatially constraining cells and virus to small volumes over large surface areas and microscale heights, microfluidic systems eliminate the biotransport issues associated with standard cell culture systems. Using immortalized cell lines that are generally permissive to viral transduction, we (1) established a microfluidic transduction system, (2) determined which parameters were critical for enhancing gene transfer, and (3) explored the feasibility of scaling up toward clinically appropriate cell numbers. Furthermore, we tested the microfluidic transduction system on primary human and murine cells for ex vivo gene therapy applications for hemophilia A, which has proven to be a challenging clinical target to efficiently transduce the number of cells needed to effectively achieve curative plasma coagulation factor VIII (fVIII) activity levels.21
Microfluidics have enabled miniaturization of biological assays and processes by reducing reagent consumption and leveraging microscale fluidic physics.22 By investigating factors such as MOI, LV (v/v)% concentration, transduction height (diffusion distance), and transduction times in the microfluidic system compared to well plates requiring larger working volumes, we determined that microfluidic devices could be leveraged to create a more efficient transduction platform for gene therapy that significantly reduces the amount of virus required for efficient gene transfer. Furthermore, we demonstrated that physical factors contribute largely to the low transduction efficiencies observed in primary cells, as use of the microfluidic system significantly reduced virus usage and improved transduction kinetics. This work demonstrates that microfluidics have the potential to enable the scale up and commercialization of lentiviral-based gene and cell therapies.
Results
We first explored the microfluidics parameter space relevant to LV transduction efficiency using vesicular stomatitis virus G glycoprotein (VSV-G) pseudotyped LVs encoding a GFP reporter transgene for transduction of hematopoietic cell lines. Head-to-head comparisons of microfluidic and well-plate controls were conducted such that the basal surface area, cell numbers, transduction times, and amounts of LV used were maintained constant unless otherwise noted. Clinical transduction protocols maximize the amount of LV in the parameter space afforded by low cellular toxicity. The downside to this strategy is the significant LV waste resident in the culture dead volume (Figure 1A). Standard systems all require relatively large working volumes to mitigate evaporation and overcome surface tension to fully wet the surface. Our design relies on micron-scale heights to overcome diffusion limitations and minimize working volumes while maintaining large surface areas that mirror that of standard 96- and 6-well plates to accommodate clinically relevant target cell numbers (Figure 1B). As a result, the minimized volumes allow high LV concentrations to be achieved without having to increase the amount of LV used in the microfluidics.
Figure 1.
Microfluidics Are an Enabling Platform for More Efficient LV Transduction
(A) Microfluidics bring LV particles within closer proximity to target cells without requiring large quantities that would otherwise be wasted due to their short half-life. (B) Example of microfluidic devices that accommodate 106 (top) and 105 (bottom) cells with a surface area equivalent to a 6-well plate and a 96-well plate, respectively. The scale bar represents 1 cm. (C) Transductions using the same amount of GFP-LV and Jurkat cells (constant MOI) for various volumes/fluid heights reveal a height range (<100 μm) in which diffusion no longer limits vector integration as indicated by the plateau (n = 2–4). Data represent means ± SD. (D) Jurkat transductions comparing utilization efficiencies for a constant GFP-LV (v/v)% concentration at various transduction volumes demonstrates that minimizing total volume more efficiently utilizes available LV (n = 2–4). Data represent means ± SD. (E) A constant ratio of cells and virus (MOI of 1) loaded into 106 and 107 scale microfluidics for 5 hr produces nearly identical transduction of Jurkat cells with a GFP-LV. Data represent mean ± SD. Insets are photos of different versions of microfluidics used for each condition. The scale bar represents 3 cm.
Decreased Diffusion Lengths in Microfluidics Enhance Transduction Efficiency at Multiple Scales
Initial studies utilized Jurkat and K562 cell lines as surrogates for primary human T cells and HSPCs, respectively. Previous studies have shown that keeping the number of infectious units constant while minimizing the total transduction volume results in greater gene transfer.23 Given microfabrication constraints and typical cell sizes, we used small-scale microfluidics with varying heights from 30 to 457.5 μm, corresponding to total volumes of 1–15 μL. These were compared to 96-well-plate transductions with volumes of 50–200 μL, corresponding to fluid heights of 1,555–6,220 μm. Surface area was kept constant in both the microfluidics and wells to ensure that the observed results were due only to differences in height.
For identical MOIs, GFP expression increased with decreasing fluid height in microfluidic transductions due to the increased LV (v/v)% concentration. Transduction reached a plateau for fluid heights of 90.2 μm or less, indicating that LV transport was no longer diffusion limited because associated increases in LV concentration did not further increase transduction (Figure 1C). This highlights that microfluidics enabled transductions to operate in a range where MOI was relevant and unencumbered by diffusion limitations. In the absence of diffusion limitations, estimations for vector integration based on MOI may be derived from a Poisson distribution. Noting that some cells will be infected multiple times while others remain uninfected, the Poisson distribution describes the MOI-dependent probability of interaction between the vector and cells.24 To assess the efficiency of our system, we defined a utilization efficiency: the ratio of theoretical infectious units required to achieve the observed transduction based on a Poisson distribution of viral infectivity to the number of infectious units input to the system. The derivation of this equation is provided in Equation S1. Evaluating the utilization efficiency revealed that even for a short transduction at a constant 10% LV (v/v)% concentration, smaller diffusion lengths (fluid height) achieved only within the microfluidic regime resulted in more efficient uptake of LV, while the 96-well transductions all utilized <10% of the available LV (Figure 1D). Diffusion length is also critical for the scalability of the microfluidic. When cells in a larger microfluidic with a 107-cell capacity were transduced, transduction was identical to a smaller-scale microfluidic with a 106-cell capacity for a constant MOI and equivalent height despite the order of magnitude difference in targeted cells (Figure 1E). Taken together, these results show that geometrically constraining the system in which transduction occurs increases gene transfer events in a scalable manner.
Factors That Increase Transduction in Standard Systems Are Amplified in Microfluidics
We then used a microfluidic system accommodating 106 cells for head-to-head comparisons with six-well plates in which the number of infectious units, cell numbers, and transduction times were kept constant between the two systems. Cell density, which corresponds to the number of targets for LV particles to infect, was determined to affect utilization efficiency proportionally in both systems. Due to the increased mass transport and LV (v/v)% concentration in the microfluidic, greater LV availability resulted in increased utilization efficiency for increasing target cell numbers. According to first-order mass action kinetics, the rate of vector adsorption is proportional to the vector concentration and the target bed cell density (i.e., the probability of infection proportionally increases) (Figure 2A). This result is in agreement with previous studies that used smaller quantities of adherent NIH 3T3 cells.16
Figure 2.
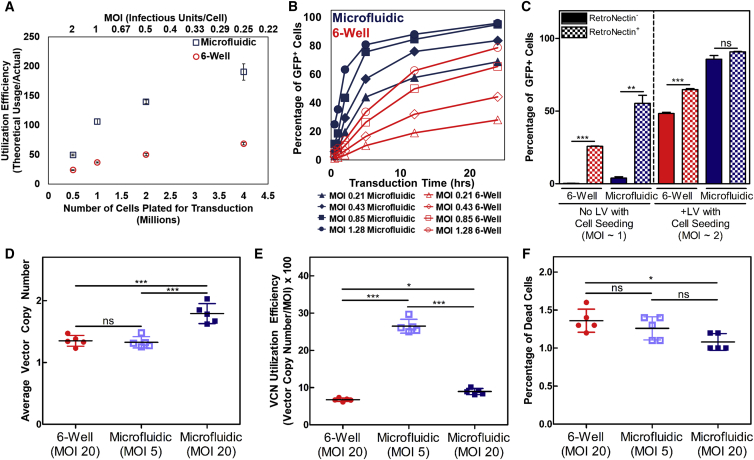
Factors That Readily Increase Transduction Are Amplified in Microfluidic Systems and Are Optimized for Primary Human T Cell Transductions
(A) Greater utilization of GFP-LV can be achieved in microfluidics by increasing the number of Jurkat cells targeted because virus-cell interactions are enhanced (n = 3). Data represent mean ± SD. (B) Assessment of Jurkat transduction kinetics between six-well plates and microfluidic systems at increasing MOIs using a GFP-LV. Both time and LV savings are possible using microfluidics. (C) RN coating can efficiently immobilize infectious particles in microfluidics compared to six-well plates. While RN significantly improves transduction in six-well plates, microfluidics marginally benefit from RN coating due to the high efficiency inherent in microfluidics (n = 3). Data represent mean ± SD. **p < 0.01, ***p < 0.001 (unpaired Student’s t test with Welch’s correction for unequal variances). (D and E) Microfluidic systems enable vector savings or increased gene transfer using a clinically processed fVIII-LV in primary human T cells as shown by VCN analysis (D) and VCN utilization efficiency (E). (F) Cell viability is not negatively affected by microfluidic transductions. Transduction time = 12 hr. (n = 5). Data represent means ± SD. *p < 0.05, ***p < 0.001 (one-way ANOVA). ns, not significant.
Beyond virus savings, ex vivo culture sensitivity of the target cell and transduction time are also important parameters constrained by LV half-life. Therefore, we assessed transduction kinetics for various MOIs between the two systems (Figure 2B). Increasing the number of infectious units (and thus the MOI) improved transduction in both systems, which further highlights the concentration dependence of LV transduction. However, the microfluidic still outperformed the six-well plate in all head-to-head comparisons. Higher transduction in the six-well plate was only achieved by using 6-fold more virus than the microfluidic and for transduction times greater than 12 hr. The viral supernatant collected at each time point and used for secondary transductions on naive Jurkat cells also confirmed that less LV was wasted in the microfluidic (Figure S1A). Despite the small quantities of LV available at a lower MOI, the number of cells transduced resulted in utilization efficiencies greater than 100%. Since the LV used is replication incompetent, this result indicates that initial titers may be underestimated by 2- to 5-fold due to the diffusion-limited well-based nature and variability in the titration process, which has been addressed elsewhere24 (Figure S1B). Furthermore, in silico modeling (COMSOL) utilizing the governing diffusion equation for the transport of a dilute species (LV) and a surface reaction (LV binding to cells) was consistent with our experimental results (Figure S1C), indicating that the microfluidic enhanced transduction primarily via physical mechanisms. However, the number of infectious units input to the simulation needed to be increased by 3-fold to more closely approximate the results observed in vitro, further highlighting the underestimation of infectious titer when relying on diffusion-limited systems for titration. COMSOL model parameters are displayed in Table S1.25
Existing Transduction-Enhancing Compounds Extend the Capabilities of Microfluidics
Although previous reports indicate that RN, one of the few clinically approved transduction-enhancing compounds, may be optimized for adsorption to plastics such as polystyrene, cyclo-olefin, polyethylene, and Teflon,26 no characterization for other materials such as polydimethylsiloxane (PDMS), the most commonly utilized polymer for soft lithography and microfluidics, has yet been assessed. To that end, we aimed to determine microfluidic compatibility with RN, which has the benefit of binding cells and virus for co-localization, although varying efficacy with different viral vectors and envelope proteins has been reported.12, 27 For our purposes, we sought to elucidate (1) the cell immobilization efficiency of RN coatings, which enables perfusion of nutrients in the microfluidic system; and (2) the extent of transduction in enhancement RN can provide in microfluidic transductions. We optimized the surface concentration of RN to efficiently immobilize non-adherent cell types while maintaining the capability to effectively bind virus without requiring trypsinization for cell removal (R.T. and W.C.L., unpublished data).
To determine whether cells could be immobilized in the microfluidic for perfusion, we compared the RN coating of the microfluidic system with polystyrene six-well plates following the product manual at our optimized surface concentration. After cells were seeded and allowed a brief incubation period to adhere to the RN, the microfluidics were flushed at 18.4 μL/min for 1 hr to remove non-immobilized or loosely bound cells, while six-well plates were gently pipetted to achieve a similar effect; counts of collected cells from flushing are shown (Figure S2A). In separate studies, we applied two standard RN-mediated infection protocols to the microfluidic system: (1) the RN-bound virus infection method, where vector is pre-adsorbed onto an RN-coated plate followed by loading of cells after washing away unbound vector; and (2) the supernatant infection method, in which cells and vector are mixed together and loaded onto an RN-coated plate. We sought to systematically determine the efficacy of both methods, comparing the microfluidic to the six-well plate. Microfluidics and six-well plates were loaded with vector for 1 hr at 37°C in both bare and RN-coated microfluidics to determine the efficiency of the RN-bound virus infection method. Cells were then loaded into all conditions with either vector-free or vector-containing media to assess the supernatant infection method. RN coating is shown to clearly bind virus and with greater efficiency in the microfluidic, as seen by the greater transduction in microfluidics where cells were seeded without additional vector (Figure 2C). However, cells that were loaded with vector into microfluidics produced similar levels of transduction regardless of RN coating, while an ∼34% increase was observed in six-well plate transductions with an RN coating. In general, cells were retrieved from both the microfluidics and six-well plates with ≥85% efficiency regardless of RN coating (Figure S2B). Cells that were expanded from each condition also demonstrated similar fold increases, indicating that cell proliferation was not altered (Figure S2C). Overall, these results demonstrate that microfluidics serve as a versatile platform that can effectively utilize existing transduction-enhancing compounds to synergistically maximize efficiency and maintain cell viability.
Microfluidics Reduce LV Requirements in Primary Human T Cells and CD34+ HSPCs
To further demonstrate microfluidic efficacy in more clinically relevant cells, we transduced primary human T cells with clinically processed GFP-LVs and a high-titer fVIII-LV. Here we utilized an adapted protocol giving all cells identical conditioning so that any differences in transduction were solely due to the selected transduction platform.28 Primary human T cells were transduced with a GFP-LV to determine optimal transduction times. An alternative utilization efficiency parameter was defined, dividing the measured average vector copy number (VCN) by the MOI (Equation S2) to obtain the ratio of the integrated vector copies to the amount of infectious units used for transduction. This parameter was used to compare different conditions across 4- and 12-hr transduction times. The VCN, VCN utilization efficiency, percentage of GFP+ cells, and percentage of dead cells as quantified by eFluor 780 viability staining are displayed in Figure S3.
Separate 12-hr transductions using an fVIII-LV resulted in significantly greater VCNs (p < 0.001) for microfluidic transductions utilizing equal amounts of LV compared to six-well plates. Concurrent microfluidic transductions utilizing 4-fold less LV yielded equal VCNs as the six-well plates, indicating that current production levels of genetically modified T cells may potentially be obtained with 4-fold less LV simply by using microfluidics (Figure 2D). Assessing the VCN utilization efficiency of these transductions also revealed that dosing a lower MOI in microfluidics was the most efficient, indicating that transduction may further be optimized to reduce wasted LV (Figure 2E). Cell death was also minimized, with all conditions resulting in <2% positive staining for dead cells (Figure 2F).
Primary CD34+ HSPCs, one of the other major cell targets in gene therapy, are notoriously difficult to transduce. We compared an optimized well-plate transduction protocol mimicking clinical conditions to an adapted microfluidic transduction protocol using a clinically processed fVIII-LV. To accommodate a higher MOI in the microfluidic, LV was pre-adsorbed onto an RN coating for 2 hr prior to the loading of cells and additional LV at a 30% (v/v)% concentration. Average VCNs per cell transduced using microfluidics were not statistically different from those in the well plates despite using half the amount of virus in half the transduction time (Figure 3A), thus resulting in a higher VCN utilization efficiency (Figure 3B). Cell viability was also maintained across all conditions (Figure 3C) and colony-forming units (CFUs) from harvested cells plated in methylcellulose media were comparable between well-plate and microfluidic transductions (Figure 3D). Thus, microfluidics are able to improve transduction efficiency in multiple clinically relevant primary human cell types without compromising cell viability or stem cell characteristics.
Figure 3.
CD34+ Cells Are More Efficiently Transduced in Microfluidic Systems
(A) VCN analysis demonstrates that transduction times and fVIII-LV usage are reduced by at least 50% using microfluidics. (B) Microfluidic systems enable greater VCN utilization efficiency compared to 24-well transductions. (C) Cell viability as measured by the trypan exclusion assay is not reduced from microfluidic systems. (D) Stem cell properties are not negatively impacted by microfluidic systems, as demonstrated by the human colony-forming assay (n = 3). Data represent means ± SD. *p < 0.05 (one-way ANOVA). Tdx, transduced; ns, not significant.
Microfluidic-Transduced HSPCs Correct Hemophilia A in a Murine Model
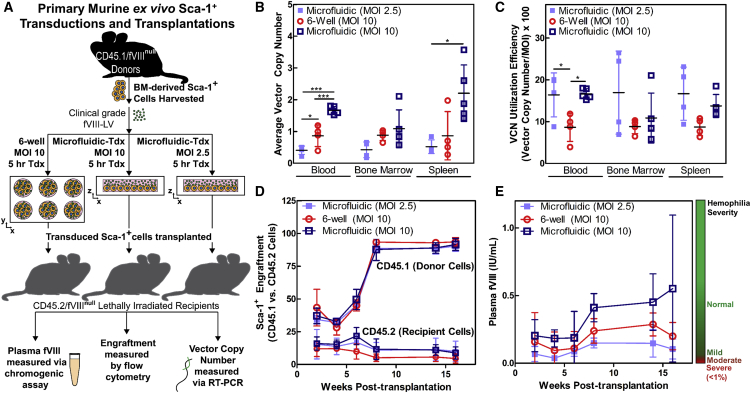
Although cell viability did not appear to suffer from microfluidic transduction in vitro, we tested the in vivo efficacy and safety of the microfluidic system in a hemophilia A murine model (fVIII-deficient mice) (Figure 4A). Transductions of Sca-1+ cells isolated from CD45.1+ donors were performed using highly concentrated, clinically processed fVIII-encoding LV in microfluidics and six-well plates for 5 hr at an MOI of 10. An additional group of cells was transduced in microfluidics using 4-fold fewer infectious units (MOI of 2.5). From the pool of transduced cells, 1 million CD45.1+ cells from each group were then transplanted into a corresponding lethally irradiated CD45.2+ fVIII-deficient recipient. Plasma fVIII levels were measured via a chromogenic fVIII assay and engraftment was assessed via flow cytometry by monitoring populations of CD45.1+ cells in the recipient mice every 2 weeks until 8 weeks post-transplantation when fVIII levels stabilized. The average VCN and the VCN utilization efficiency of blood, bone marrow, and spleen cells were obtained after 16 weeks (Figures 4B and 4C). Microfluidic transductions at an MOI of 10 yielded the highest VCN in all tissues and exhibited no statistical differences in engraftment (Figure 4D). Three of five mice transplanted with microfluidic-transduced cells at an MOI of 10 produced plasma fVIII levels within the normal range compared to mild hemophilia levels in those transplanted with the same number of infectious units in a six-well plate or microfluidic-transduced cells at an MOI of 2.5 despite the relatively low dosages of LV and short transduction times (Figure 4E). Therefore, microfluidics demonstrated pre-clinical safety and efficacy with the potential to reduce both transduction times and vector usage.
Figure 4.
Transplantation of Microfluidic-Transduced Cells Results in Greater Rates of Gene Transfer and Higher fVIII Production in a Hemophilia A Animal Model
(A) Pre-clinical simulation of murine gene therapy for hemophilia A using a clinically processed fVIII-LV. Microfluidic transductions are compared to an equivalent standard protocol using a six-well plate. (B and C) Microfluidic systems improve gene transfer efficiency, as indicated by VCN analysis (B) as well as VCN utilization efficiency (C) across various mouse tissues (n = 5 mice, MOI of 10, microfluidic; n = 4 mice, MOI of 10, six-well plate; and n = 4 mice, MOI of 2.5, microfluidic). Data represent means ± SD. *p < 0.05 and ***p < 0.001 (one-way ANOVA of individual tissue groups). (D) High levels of donor (CD45.1+) cell engraftment were maintained across all conditions. (E) Only mice transplanted with microfluidic-transduced Sca-1+ cells at an MOI of 10 were able to restore plasma fVIII levels to normal. Time course of averaged plasma fVIII levels for each group of mice. Tdx, transduced. Data represent mean ± SD.
Discussion
In the current study, we translated state-of-the-art transduction protocols to a scalable microfluidic platform enabling significant reduction in LV requirements and transduction times by leveraging micron-scale heights to overcome diffusion limitations of current systems. Transduction efficiency can be further maximized by incorporating existing transduction-enhancing methods such as RN, polycations, and biological adjuvants. Previous studies have shown that transduction of adherent cell types using adenoviral and retroviral vectors can be enhanced with various perfusion regimes, although this becomes less practical as the system is scaled up.29, 30 Although flow rates >18.4 μL/min can be used without inducing significant shear stress on cells due to the large channel widths used, flow rates should be minimized to keep cells immobilized on the RN coatings when necessary. Perfusion as a means of virus delivery to cells in our larger-scale microfluidics would artificially recreate the issue of limiting virus-cell contact, since the majority of the virus would reside within tubing or syringes before finally moving through the microfluidic. As such, we believe that the microfluidic system is best suited for perfusion of nutrients instead of virus. RN, which is a costly clinical reagent, is shown to be effective at immobilizing non-adherent cells and binding virus in the microfluidic system even when using 5-fold less than the recommended amount (Figure S2A). Furthermore, the microfluidic system enhances efficiency such that RN coatings do not confer much benefit unless the RN-bound virus infection method is combined with the supernatant infection method (Figure 2C). However, the benefit of using RN in microfluidics is apparent for conditions where the constrained geometry would otherwise limit the amount of infectious units or nutrients provided to cells. Only VSV-G pseudotyped LVs were assessed in this study, although the physics-based mechanism of action should enable a wide variety of vector envelopes and cell types to be compatible with microfluidic transductions, as demonstrated by the increased transduction in primary human and murine cells. Overall, the microfluidic platform outperforms current clinical transduction platforms in all aspects studied.
Human Jurkat and K562 hematopoietic cell lines as well as primary mouse and human HSPCs and T cells were efficiently transduced in the microfluidic system. Previous studies and current practices highlight the importance of minimizing the total volume of transduction for efficient gene transfer, but practical limits of conventional cell culture platforms do not allow further reduction of these transduction volumes.23, 31 Alternatively, novel micro- and nano-well arrays developed for stem cell aggregate formation and single cell processing, respectively, may provide potential methods of physically constraining the transduction space to enhance efficiency.31, 32, 33 However, the translation of these formats to clinical-scale gene therapy applications is not as straightforward due to limitations in fabrication, loading, and removal of cells. Conventional microfluidics are also typically designed to minimize volume, cell numbers, or cell residence time. Rather than adapt existing formats toward gene therapy applications, we designed a microfluidic platform specifically for transduction with a focus on cell capacity, scalability, loading, immobilization, and retrieval. Our most current device design is able to accommodate 107 cells (Figure 1E) while maintaining the dimensions necessary to operate in this optimal range, although further scale up to 108–109 cells required for clinical-scale transductions1 is readily achievable by linking the devices in parallel (R.T., unpublished data). Within the assortment of microscale heights explored, we identified a range (≤∼100 μm) in which diffusion limitations are surpassed, as indicated by the plateau in transduction efficiency observed in Figure 1D. Despite having increased vector concentration at shorter heights, transduction efficiencies in this range did not increase because the amount of infectious units became the limiting factor rather than the vector concentration. In this regime where the amount of infectious units and number of target cells are the critical variables, the theoretical transduction governed by the MOI and Poisson distribution better correlates with the actual observed gene transfer, unlike conventional systems where a large MOI is required for the desired result.
While cell viability has not been an issue in any of the in vitro or in vivo studies we performed to date, gas exchange and nutrient replenishment may potentially be problematic for future scale up toward 108–109 cells. The geometric constraints of the space in which transduction occurs in microfluidics can limit the amount of nutrients and gas available to cells, although studies encapsulating single cells in picoliter droplets have demonstrated that high cell viability is preserved despite reduced volumes.34 These concerns are further mitigated by the significantly shortened transduction times made possible in the microfluidic system, which have not resulted in altered cell physiology or significantly higher cell death for the cell lines or with primary human cells used in this work (Figures 2F, 3C, 3D, S2C, and S3D). Most notably, the long-term repopulating capability of mouse HSPCs was not negatively affected. Although mouse HSPCs are more easily transduced than human cells, we have demonstrated that microfluidics also improve transduction of human CD34+ cells without affecting viability or colony-forming potential when compared to optimized well transductions. These results are encouraging in moving toward transplantation into a humanized mouse system to demonstrate true clinical efficacy, as we believe that the duration of LV-RN adsorption and transduction times can be further optimized. Such experiments would provide valuable insight into our system, since the CD68 promoter of the fVIII-LV used in our primary human cell studies was limited to expression of fVIII only in cells of the myeloid lineage. Therefore, transduced T cells of the lymphoid lineage and undifferentiated CD34+ cells did not produce detectable levels of fVIII, although our animal studies demonstrated that the VCN is proportional to fVIII production, which suggests that the VCN is an adequate readout for transduction efficiency in our primary human cell studies.
Based on our current data and our experience with pre-clinical development of a CD34+ fVIII-LV gene therapy product candidate (G.D., H.T.C., and C.B.D., unpublished data), we estimate that an order-of-magnitude reduction in vector costs per patient may be attainable by using microfluidics. Enhanced transduction in the microfluidic system was shown to be independent of the device fabrication method. Therefore, manufacture of microfluidic systems that meet common good manufacturing practices (cGMP) standards should be feasible using methods such as injection molding or hot embossing of polystyrene or other thermoplastics. Furthermore, integration into automated cell processing systems adapted for gene therapy applications have the potential to significantly decrease costs further.35 Previous studies have also estimated that variable costs in genetically modified hematopoietic stem cell products can theoretically be reduced as the number of products generated increases.36, 37 Incorporating the microfluidic system into a standard cell manufacturing process would increase production output for a given amount of viral vector, which is a significant cost driver due to the lengthy, expensive, and scale-limited manufacturing process. When combined with other cost savings from reduced cytokine and reagent usage typically required for primary cell culture, the microfluidic platform represents an exceptional tool for cell manufacturing for clinical gene therapy. With clinical gene therapy rapidly advancing with definite evidence of success and licensed products, associated advances in vector manufacturing and utilization will be essential to routine clinical implementation and globalization.
Materials and Methods
Cell Lines, Media, and LV
K562 and Jurkat cells (ATCC) were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), l-glutamine, 25 mM HEPES, and 1% penicillin/streptomycin (Pen/Strep). GFP and fVIII-LV were supplied by Expression Therapeutics and were prepared by a commercial manufacturing organization for clinical development of a CD34+ fVIII-LV gene therapy product candidate (G.D., H.T.C., and C.B.D., unpublished data). Infectious particle titers of the GFP-LV were determined from 293FT cell line transductions (8.9 × 107 TU/mL) and that of the fVIII-LV was determined from 293T/17 cell line transductions (4.82 × 108 TU/mL). All LVs tested were replication incompetent and pseudotyped with the VSV-G envelope. Separate authentication or mycoplasma testing was not performed on cell lines.
Microfluidics Fabrication
All devices were designed to have an equivalent surface area to a comparable polystyrene well plate. Small-scale (105 cell) microfluidics had a matching surface area with individual wells of a 96-well plate (0.32 cm2), while all large-scale (106-cell) microfluidics had a matching surface area with individual wells of a six-well plate (9.5 cm2). Microfluidics used for CD34+ transductions had a matching surface area with individual wells of a 12-well plate (3.8 cm2). Scaled up 107 cell microfluidics had a surface area of approximately 51.5 cm2 with a total volume of approximately 670 μL.
Small-scale devices with volumes ranging from 1 to 15 μL and 92 μL large-scale devices were made using traditional soft lithography as described elsewhere.38 Briefly, a thick layer of SU-8 photoresist (MicroChem) was spincoated onto a silicon wafer to achieve the desired height of the channel. The photoresist was baked according to the manufacturer guidelines to drive out the solvent. The device pattern was then crosslinked into the photoresist by exposing the coated wafer to UV light through a photomask containing the desired pattern. Non-crosslinked photoresist was removed by treatment with SU-8 developer. The completed mold was treated with hexamethyldisilazane (Sigma-Aldrich) overnight by evaporation onto the surface before casting PDMS (Sylgard 184; Dow Corning).
Master molds for 200 μL and scaled-up 107 microfluidics were made from computer numerical control (CNC) micromilling of aluminum. PDMS was then cast onto the mold and peeled off to obtain the negative pattern of the channel. Channel dimensions were measured using a Dektak 150 profilometer (Veeco Instruments). Inlet and outlet ports were punched before plasma or corona treatment of the PDMS for covalent bonding to a flat PDMS slab, sealing the channel.
Microfluidics were made using double-sided silicone adhesive transfer tape (3M) to assess transduction kinetics and CD34+ cell transductions. Channel patterns were drawn using AutoCAD software (Autodesk). The channels were then cut from the double-sided silicone adhesive using a Silhouette craft cutter (Silhouette America). The double-sided silicone adhesive containing the channel pattern was then applied to a 150 mm polystyrene petri dish, followed by a flat slab of PDMS with inlet and outlet ports pre-punched to seal the channel. The resulting devices were 48 μL in volume with a height of approximately 50 μm for the transduction kinetics microfluidics and approximately 100 μm for the CD34+ microfluidics.
Cell Line Transduction
Prior to transduction, cells were centrifuged at 200 × g for 10 min to remove growth media. Cells were then re-suspended in fresh growth media containing 8 μg/mL polybrene and clinical-grade GFP-LV ranging from 0.1 to 89.9 μL. All microfluidic transductions were conducted simultaneously in direct comparisons with well-plate controls containing the same amount of LV for the same transduction times ranging from 0.5 to 24 hr unless otherwise noted. Small-scale direct comparisons were conducted in a 96-well plate with a total volume of 50–200 μL. All large-scale direct comparisons were conducted in a six-well plate with a total volume of 1 mL. Small-scale transductions targeted 70,000 cells, while large-scale transductions targeted 500,000–4,000,000 cells. Prior to adding cells and LV, the devices were placed under a vacuum for at least 10 min to facilitate uniform loading and removal of air bubbles.
RN Coating
Devices were coated overnight with 10 μg RN (1.05 μg/cm2; Takara Bio) the day before transduction. Polystyrene six-well plates were also coated with the same amount for comparison to the standard. BSA (2% in 1× Dulbecco’s PBS [DPBS]) was incubated in the devices and six-well plates for 30 min at room temperature to block non-specific binding. The blocking solution was flushed out with 1× DPBS and aspirated from the devices and six-well plates prior to use. To assess the efficacy of the RN-bound virus infection method, 22.5 μL vector stock (∼2 × 106 transducing units [TU]) was pre-adsorbed onto RN-coated microfluidics and six-well plates for 1 hr at 37°C before cells were seeded. Bare microfluidics and six-well plates were also loaded with vector stock as negative controls. Following the pre-adsorption phase, 2 million cells were loaded into microfluidics and six-well plates with either vector-free or vector-containing media (additional 22.5 μL vector stock added) to assess the supernatant infection method.
Cell Collection
After each specified transduction time, devices were moderately tapped along the underside of the channel to release cells from the surface. Channels were then flushed with 1 mL 1× DPBS in small-scale devices or 5–6 mL 1× DPBS in large-scale devices following directly into 15 mL conical tubes. 1× DPBS was added to the well controls to achieve comparable final volumes and pipet mixed for cell removal. Due to the reduced RN surface concentration, cells did not require trypsinization for removal. All cells were then centrifuged at 200 × g for 10 min. The viral supernatant was removed, and cells were plated with fresh growth medium.
Assessment of GFP-LV Transduction Efficiency
Cells were maintained in culture following removal from the devices and wells for at least 72 hr before assessing GFP expression of transduced cells with a BD C6 Accuri flow cytometer.
Primary Human T Cell Culture
Frozen human pan T cells were purchased from AllCells. Cells were thawed following the AllCells thawing protocol and allowed to recover in RPMI 1640 media supplemented with 10% FBS, l-glutamine, 25 mM HEPES, and 1% Pen/Strep for 24 hr. CD3- and CD28-coated beads (Miltenyi Biotec) were then added to the cells at a 1:1 bead/cell ratio with 100 IU/mL human IL-2 (IL-2; PeproTech) for T cell activation. The cells and beads were then transferred to a six-well plate at a density of 1–2 × 106 cells/mL per cm2. Cells were ready for transduction after a 24-hr activation period.
Primary Human T Cell Transduction
200 μL devices and six-well plates were coated with 20 μg (2.10 μg/cm2) RN using the process described above. Each microfluidic and six-well plate was loaded with 2 million cells with beads, 60 μL GFP-LV, 6 μg/mL Polybrene, and 100 IU/mL human IL-2.
T Cell Transduction Assessment
Cells that were transduced with the GFP-LV were stained with eFluor 780 viability dye (eBioscience) at least 72 hr post-transduction. T cell purity was confirmed with a CD3-V450 (500A2) antibody (BD Biosciences). Transduction efficiencies are determined as the percentage of GFP+ live cells as obtained with a BD LSR II flow cytometer. Cell samples were blinded to the operator before processing for the VCN. Genomic DNA was harvested from cells using the DNeasy blood and tissue kit (QIAGEN). Real-time qPCR was used to determine the VCN. fVIII-transduced cells received identical processing, without GFP assessment.
Primary Human CD34+ HSPC Cell Culture
Frozen human mobilized peripheral blood (mPB) CD34+ cells were purchased from AllCells. Upon receipt, mPB CD34+ cells were thawed and counted prior to assessment of viability via trypan blue dye exclusion. For transduction, mPB CD34+ cells were re-plated at a cell density of 1 × 106 cells/mL in RN-coated plates in stem cell growth media (SCGM) containing 300 ng/mL recombinant human Flt-3 Ligand (rhFlt-3L), 300 ng/mL recombinant human stem cell factor (rhSCF), 100 ng/mL recombinant human thrombopoietin (rhTPO), and 60 ng/mL recombinant human IL-3 (rhIL-3) and stimulated overnight at 37°C/5% CO2. SCGM, rhFlt-3L, rhSCF, rhTPO, and rhIL-3 were all purchased from CellGenix.
Primary Human CD34+ Transduction with fVIII-LV
After overnight stimulation, transduction was initiated by removing 50% of the conditioned medium, pelleting any cells in suspension, re-suspending in SCGM containing cytokine mix and LV at an MOI of 50, and adding the resuspended mixture back to cells on RN-coated 24-well plates. Similarly, cells were pelleted, re-suspended in SCGM containing cytokine mix and LV at MOI 25, and loaded into RN-coated microfluidics. Cells were transduced for 24 hr at 37°C /5% CO2 in the 24-well plates while cells were transduced for 6 hr at 37°C/5% CO2, followed by an additional 6 hr after loading fresh cytokine mix-containing SCGM to replenish nutrients (total transduction time: 12 hr). After 12 hr of transduction, cells were removed from the microfluidics via DPBS flushing, washed to remove unbound LV, and plated in fresh SCGM containing cytokine mix for an additional 12 hr. Cells transduced in 24-well plates were similarly removed after 24 hr of transduction and washed to remove unbound LV. At the end of the transduction period, both 24-well and microfluidic-transduced cells were plated for CFU analysis.
CFU Analysis
Clonogenic potential post-transduction was assessed by a colony-forming cell (CFC) assay and enumeration of individual CFUs compared to mock (untransduced) cells. Briefly, a 10× solution of cells was mixed with MethoCult H4434 Classic medium (StemCell Technologies) and dispensed into dishes using a syringe attached to a blunt-end needle. Plated cells were incubated for 10 days at 37°C/5% CO2, at which point the individual colonies (CFUs) were counted manually on an inverted microscope. A total of 1,000 cells per well were plated for each assay and each assay was performed in duplicate.
VCN Analysis
After CFU enumeration, each assay well was re-suspended in DPBS to dissolve methylcellulose and replicates combined. Cells were pelleted, washed once with DPBS, and stored at −20°C for up to 1 month for genomic DNA isolation. Genomic DNA from washed cell pellets was isolated using the DNeasy Blood & Tissue Kit (QIAGEN) following the manufacturer’s protocol for “cells in culture.” Genomic DNA was quantitated using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific) and equal amounts of genomic DNA were used for SYBR Green-based real-time qPCR analysis using transgene-specific primers and compared to a plasmid standard curve of known quantities.
Animal Studies
All animal studies and procedures were performed at Emory University under the guidelines set by the Emory University Institutional Animal Care and Use Committee. C57BL/6 Exon-16 (E16)-disrupted hemophilia A mice were used in all animal experiments. Generation of this murine model for hemophilia A was described previously.39 For transduction efficiency studies, both male and female mice aged between 8 and 10 weeks were used. No randomization was carried out to determine animal groups. Based on statistical power estimates (alpha = 0.05, power = 0.8, sigma = 0.3) assuming fVIII expression levels drawn from previous animal studies (G.D., J.E.S., H.T.C., and C.B.D., unpublished data), a minimum of three mice per group would be needed to determine differences. Total animal numbers for each group are given.
Murine Sca-1+ Cell Isolation, Transduction, Transplantation, and Analysis
Bone marrow was isolated from cleaned hind leg femurs and tibias of C57BL/6 E16 hemophilia A mice with the CD45.1 allele. Sca-1+ cells were incubated with biotin anti-Sca-1 antibody followed by anti-biotin microbeads (Miltenyi Biotec) and passed through a magnetic separation column for positive selection. Isolated cells were then cultured overnight at a density of 2 × 106 cells/mL in StemPro media supplemented with stem cell factor (100 ng/mL), murine IL-3 (20 ng/mL), human IL-11 (100 ng/mL), human Flt-3 ligand (100 ng/mL), StemPro nutrient supplement (40×), l-glutamine (100×), and Pen/Strep (100×).
On the day of transduction, 2 million cells were loaded into each bare microfluidic and six-well plate with 8 μg/mL Polybrene as described above with a clinical-grade fVIII-LV for 5 hr. Cells were then collected, washed, and re-suspended in fresh 1× DPBS for transplant.
At least 4 hr prior to transplantation, a separate cohort of 8- to 10-week-old C56BL/6 E16 hemophilia A mice with the CD45.2 allele were given two doses of lethal irradiation at 5.5 Gy (total 11 Gy) 4 hr apart. After transduction and washing of LV from the donor cells, 1,000,000 cells were transplanted into each recipient mouse via retro-orbital injection.
Mice were bled every 2 weeks for the first 8 weeks via tail vein microsampling. Flow cytometry was run on collected blood cells and stained for CD45.1 and CD45.2 to assess engraftment. The following antibody-fluorophore conjugates were used for flow cytometry: CD45.2-APC (558702), Gr1-APC-Cy7 (557661), Mac1-APC-Cy7 (557657), CD45.1-PE (553776), CD3e-V450 (560801), and CD45R/B220-PE-Cy7 (552772) (BD Biosciences). Complete blood counts were also conducted to monitor populations of various white blood cells, including lymphocytes, monocytes, and granulocytes. Plasma was isolated and used to measure fVIII plasma levels via a commercially available chromogenic substrate assay (Chromogenix Coatest SP FVIII; diaPharma). After 16 weeks, mice were euthanized, and cells from blood, spleen, and bone marrow were harvested for RT-PCR to quantify the LV copy number. The chromogenic fVIII assay and VCN analysis were conducted as previously described.40 The operator was blinded to the cell samples before processing for VCN.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software). All data are represented as means ± SD. Multiple groups were compared using one-way ANOVA followed by Bonferroni post hoc analysis. Normal distributions were tested with the Shapiro-Wilk test for normality and equal variances were tested with the Levene test for equality of variances at the 0.05 level.
Author Contributions
R.T. and D.R.M. designed the microfluidic devices. R.T., G.D., C.B.D, H.T.S., and W.A.L. designed the experiments. R.T., G.D., J.E.S., A.M.L., Y.Q., Y.S., and W.C.L. performed the experiments and analyzed the data. H.A. and O.B. created the COMSOL model. R.T. drafted the manuscript. J.M.LD, H.T.S., C.B.D., and W.A.L. conceived the project and edited the manuscript.
Conflicts of Interest
C.B.D., W.A.L., D.R.M., H.T.S., and R.T. are inventors on a patent application describing microfluidic lentiviral transduction technology filed by Emory University/Children’s Healthcare of Atlanta and the Georgia Institute of Technology. C.B.D. and H.T.S. are co-founders of Expression Therapeutics, LLC, and own equity in the company. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict of interest policies.
Acknowledgments
Financial support was provided by the NIH (R01-HL121264 to W.A.L.) and a research partnership between Children’s Healthcare of Atlanta and the Georgia Institute of Technology. This work was performed in part at the Georgia Tech Institute for Electronics and Nanotechnology, a member of the National Nanotechnology Coordinated Infrastructure, which is supported by the National Science Foundation (grant ECCS-1542174).
Footnotes
Supplemental Information includes three figures, one table, and two equations and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.07.002.
Contributor Information
Christopher B. Doering, Email: cdoerin@emory.edu.
Wilbur A. Lam, Email: wilbur.lam@emory.edu.
Supplemental Information
References
- 1.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Veres G., Schmidt M., Kutschera I., Vidaud M., Abel U., Dal-Cortivo L., Caccavelli L. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 3.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zufferey R., Dull T., Mandel R.J., Bukovsky A., Quiroz D., Naldini L., Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naldini L. Lentiviruses as gene transfer agents for delivery to non-dividing cells. Curr. Opin. Biotechnol. 1998;9:457–463. doi: 10.1016/s0958-1669(98)80029-3. [DOI] [PubMed] [Google Scholar]
- 7.Ausubel L.J., Hall C., Sharma A., Shakeley R., Lopez P., Quezada V., Couture S., Laderman K., McMahon R., Huang P. Production of CGMP-grade lentiviral vectors. Bioprocess Int. 2012;10:32–43. [PMC free article] [PubMed] [Google Scholar]
- 8.Millington M., Arndt A., Boyd M., Applegate T., Shen S. Towards a clinically relevant lentiviral transduction protocol for primary human CD34 hematopoietic stem/progenitor cells. PLoS ONE. 2009;4:e6461. doi: 10.1371/journal.pone.0006461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scaramuzza S., Biasco L., Ripamonti A., Castiello M.C., Loperfido M., Draghici E., Hernandez R.J., Benedicenti F., Radrizzani M., Salomoni M. Preclinical safety and efficacy of human CD34(+) cells transduced with lentiviral vector for the treatment of Wiskott-Aldrich syndrome. Mol. Ther. 2013;21:175–184. doi: 10.1038/mt.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis H.E., Morgan J.R., Yarmush M.L. Polybrene increases retrovirus gene transfer efficiency by enhancing receptor-independent virus adsorption on target cell membranes. Biophys. Chem. 2002;97:159–172. doi: 10.1016/s0301-4622(02)00057-1. [DOI] [PubMed] [Google Scholar]
- 11.O’Doherty U., Swiggard W.J., Malim M.H. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee H.-J., Lee Y.-S., Kim H.-S., Kim Y.-K., Kim J.-H., Jeon S.-H., Lee H.W., Kim S., Miyoshi H., Chung H.M., Kim D.K. Retronectin enhances lentivirus-mediated gene delivery into hematopoietic progenitor cells. Biologicals. 2009;37:203–209. doi: 10.1016/j.biologicals.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Wang C.X., Sather B.D., Wang X., Adair J., Khan I., Singh S., Lang S., Adams A., Curinga G., Kiem H.P. Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. 2014;124:913–923. doi: 10.1182/blood-2013-12-546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston J.M., Denning G., Moot R., Whitehead D., Shields J., Le Doux J.M., Doering C.B., Spencer H.T. High-throughput screening identifies compounds that enhance lentiviral transduction. Gene Ther. 2014;21:1008–1020. doi: 10.1038/gt.2014.80. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B., Metharom P., Jullie H., Ellem K.A.O., Cleghorn G., West M.J., Wei M.Q. The significance of controlled conditions in lentiviral vector titration and in the use of multiplicity of infection (MOI) for predicting gene transfer events. Genet. Vaccines Ther. 2004;2:6. doi: 10.1186/1479-0556-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreadis S., Lavery T., Davis H.E., Le Doux J.M., Yarmush M.L., Morgan J.R. Toward a more accurate quantitation of the activity of recombinant retroviruses: alternatives to titer and multiplicity of infection. J. Virol. 2000;74:1258–1266. doi: 10.1128/jvi.74.3.1258-1266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas D.L., Case S.S., Crooks G.M., Kohn D.B. Critical factors influencing stable transduction of human CD34(+) cells with HIV-1-derived lentiviral vectors. Mol. Ther. 2000;2:71–80. doi: 10.1006/mthe.2000.0094. [DOI] [PubMed] [Google Scholar]
- 18.Sutton K.S., Dasgupta A., McCarty D., Doering C.B., Spencer H.T. Bioengineering and serum free expansion of blood-derived γδ T cells. Cytotherapy. 2016;18:881–892. doi: 10.1016/j.jcyt.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Chuck A.S., Clarke M.F., Palsson B.O. Retroviral infection is limited by Brownian motion. Hum. Gene Ther. 1996;7:1527–1534. doi: 10.1089/hum.1996.7.13-1527. [DOI] [PubMed] [Google Scholar]
- 20.Higashikawa F., Chang L. Kinetic analyses of stability of simple and complex retroviral vectors. Virology. 2001;280:124–131. doi: 10.1006/viro.2000.0743. [DOI] [PubMed] [Google Scholar]
- 21.Rogers G.L., Herzog R.W. Gene therapy for hemophilia. Front. Biosci. (Landmark Ed.) 2015;20:556–603. doi: 10.2741/4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beebe D.J., Mensing G.A., Walker G.M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 23.Morgan J.R., LeDoux J.M., Snow R.G., Tompkins R.G., Yarmush M.L. Retrovirus infection: effect of time and target cell number. J. Virol. 1995;69:6994–7000. doi: 10.1128/jvi.69.11.6994-7000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shabram P., Aguilar-Cordova E. Multiplicity of infection/multiplicity of confusion. Mol. Ther. 2000;2:420–421. doi: 10.1006/mthe.2000.0212. [DOI] [PubMed] [Google Scholar]
- 25.Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dodo K., Chono H., Saito N., Tanaka Y., Tahara K., Nukaya I., Mineno J. An efficient large-scale retroviral transduction method involving preloading the vector into a RetroNectin-coated bag with low-temperature shaking. PLoS ONE. 2014;9:e86275. doi: 10.1371/journal.pone.0086275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandrin V., Boson B., Salmon P., Gay W., Nègre D., Le Grand R., Trono D., Cosset F.L. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood. 2002;100:823–832. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- 28.Bilal M.Y., Vacaflores A., Houtman J.C.D. Optimization of methods for the genetic modification of human T cells. Immunol. Cell Biol. 2015;93:896–908. doi: 10.1038/icb.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luni C., Michielin F., Barzon L., Calabrò V., Elvassore N. Stochastic model-assisted development of efficient low-dose viral transduction in microfluidics. Biophys. J. 2013;104:934–942. doi: 10.1016/j.bpj.2012.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cimetta E., Franzoso M., Trevisan M., Serena E., Zambon A., Giulitti S., Barzon L., Elvassore N. Microfluidic-driven viral infection on cell cultures: theoretical and experimental study. Biomicrofluidics. 2012;6:24127–2412712. doi: 10.1063/1.4723853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gierahn T.M., Wadsworth M.H., 2nd, Hughes T.K., Bryson B.D., Butler A., Satija R., Fortune S., Love J.C., Shalek A.K. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong Y., Ogunniyi A.O., Love J.C. Massively parallel detection of gene expression in single cells using subnanolitre wells. Lab Chip. 2010;10:2334–2337. doi: 10.1039/c004847j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moeller H.-C., Mian M.K., Shrivastava S., Chung B.G., Khademhosseini A. A microwell array system for stem cell culture. Biomaterials. 2008;29:752–763. doi: 10.1016/j.biomaterials.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen F., Zhan Y., Geng T., Lian H., Xu P., Lu C. Chemical transfection of cells in picoliter aqueous droplets in fluorocarbon oil. Anal. Chem. 2011;83:8816–8820. doi: 10.1021/ac2022794. [DOI] [PubMed] [Google Scholar]
- 35.Abou-El-Enein M., Bauer G., Reinke P. The business case for cell and gene therapies. Nat. Biotechnol. 2014;32:1192–1193. doi: 10.1038/nbt.3084. [DOI] [PubMed] [Google Scholar]
- 36.Abou-El-Enein M., Römhild A., Kaiser D., Beier C., Bauer G., Volk H.-D., Reinke P. Good Manufacturing Practices (GMP) manufacturing of advanced therapy medicinal products: a novel tailored model for optimizing performance and estimating costs. Cytotherapy. 2013;15:362–383. doi: 10.1016/j.jcyt.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Abou-El-Enein M., Bauer G., Medcalf N., Volk H.-D., Reinke P. Putting a price tag on novel autologous cellular therapies. Cytotherapy. 2016;18:1056–1061. doi: 10.1016/j.jcyt.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 38.del Campo A., Greiner C. Su-8: a photoresist for high-aspect-ratio and 3d submicron lithography. J. Micromech. Microeng. 2007;17:R81. [Google Scholar]
- 39.Zakas P.M., Brown H.C., Knight K., Meeks S.L., Spencer H.T., Gaucher E.A., Doering C.B. Enhancing the pharmaceutical properties of protein drugs by ancestral sequence reconstruction. Nat. Biotechnol. 2017;35:35–37. doi: 10.1038/nbt.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston J.M., Denning G., Doering C.B., Spencer H.T. Generation of an optimized lentiviral vector encoding a high-expression factor VIII transgene for gene therapy of hemophilia A. Gene Ther. 2013;20:607–615. doi: 10.1038/gt.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.