Highlights
-
•
Portrait of somatic alterations in HPV-negative early stage tongue tumors with tobacco signature.
-
•
Upregulation of genes related to EMT pathway identified by transcriptome analysis.
-
•
MMP10 could be a candidate prognostic biomarker to stratify patients who develop metastases.
Keywords: HPV-negative early stage tongue cancer, Tobacco/nut chewers, Whole exome and transcriptome sequencing, Nodal metastases, Matrix metalloproteinases
Abstract
Objectives
Nodal metastases status among early stage tongue squamous cell cancer patients plays a decisive role in the choice of treatment, wherein about 70% patients can be spared from surgery with an accurate prediction of negative pathological lymph node status. This underscores an unmet need for prognostic biomarkers to stratify the patients who are likely to develop metastases.
Materials and methods
We performed high throughput sequencing of fifty four samples derived from HPV negative early stage tongue cancer patients habitual of chewing betel nuts, areca nuts, lime or tobacco using whole exome (n = 47) and transcriptome (n = 17) sequencing that were analyzed using in-house computational tools. Additionally, gene expression meta-analyses were carried out for 253 tongue cancer samples. The candidate genes were validated using qPCR and immuno-histochemical analysis in an extended set of 50 early primary tongue cancer samples.
Results and conclusion
Somatic analysis revealed a classical tobacco mutational signature C:G > A:T transversion in 53% patients that were mutated in TP53, NOTCH1, CDKN2A, HRAS, USP6, PIK3CA, CASP8, FAT1, APC, and JAK1. Similarly, significant gains at genomic locus 11q13.3 (CCND1, FGF19, ORAOV1, FADD), 5p15.33 (SHANK2, MMP16, TERT), and 8q24.3 (BOP1); and, losses at 5q22.2 (APC), 6q25.3 (GTF2H2) and 5q13.2 (SMN1) were observed in these samples. Furthermore, an integrated gene-expression analysis of 253 tongue tumors suggested an upregulation of metastases-related pathways and over-expression of MMP10 in 48% tumors that may be crucial to predict nodal metastases in early tongue cancer patients. In overall, we present the first descriptive portrait of somatic alterations underlying the genome of tobacco/nut chewing HPV-negative early tongue cancer, and identify MMP10 as a potential prognostic biomarker to stratify those likely to develop metastases.
Introduction
Tongue cancer is the most predominant form of oral cancer in developed countries with a varying incidence in developing countries [1]. The major etiological factors associated with tongue cancer include tobacco related products, alcohol and human papilloma virus (HPV) infections [2]. These factors lend to variability across populations, particularly in the Indian subcontinent wherein chewing betel-quid comprising betel leaf (Piper betel), areca nut (Areca catechu) and slaked lime (predominantly calcium hydroxide) is a part of the tradition [3]. While tobacco usage shows a 5–25-fold increased risk of cancer [4], HPV infection defines clinical and molecularly distinct subgroups of head and neck squamous cell carcinoma (HNSCC) patients [5]. Such as, HPV-negative tumors are driven by amplification at 11q13, EGFR and FGFR loci; focal deletions at NSD1, FAT1, NOTCH1, SMAD4 and CDKN2A loci; and, point mutations in TP53, CDKN2A, FAT1, PIK3CA, NOTCH1, KMT2D, and NSD1 [6], [7]. On the other hand, HPV-positive tumors harbor TRAF3, ATM deletion, E2F1 amplification, FGFR2/3 and KRAS mutations.
Another unique feature of tongue squamous cell carcinoma (TSCC) compared to other subsites of oral cancer is the occurrence of nodal metastases, observed in 27–40% of early stage (pT1 or pT2) patients. Neck dissection among them adds to morbidity and poor survival due to disease recurrence [8], [9], [10], [11]. Although poor prognostic indicators for TSCC such as occult node positivity, tumor depth, lymphovascular invasion and perineural invasion are well defined, there’s an unmet need for reliable and robust prognostic biomarkers to stratify the patients who are likely to have an adverse clinical outcome [11], [12]. Interestingly, most of the genomic analysis studies involving HPV negative TSCC have been restricted to advanced stage samples (pT3–pT4), while genomic alterations underlying HPV negative early tongue tumor genome remains largely unexplored. In the present study, we present a portrait of somatic alterations in HPV negative early tongue cancer (pT1–pT2) using integrative genomic approach to identify marker to stratify those likely to develop metastases.
Material and methods
Sample selection and patients details
The sample set and study protocol were approved by (ACTREC-TMC) institutional Internal Review Board. All the tissue samples used under study have been taken after obtaining informed consent from patients. Primary tongue tumors were staged as T1 (measuring ≤2 cm) or T2 (measuring >2 cm but <4 cm) as per AJCC (American Joint Committee on Cancer)/UICC (Union for International Cancer Control) TNM classification (7th edition) system and primary tumors with early stage (T1 and T2) were included in this study. Samples were duly verified by two independent reviewers for histological examinations such as normal sample verification, percent tumor nuclei and lymph node metastasis. The tumor sample with concordant histopathological diagnosis by both reviewers was included in the study. Tumor with at least >50% tumor nuclei was used for data analysis. Clinical, histological and etiological features were collected along with follow-up data for recurrence (Supplementary Table 1). None of the samples showed the presence of HPV using HPVDetector and PCR-based validation using the MY09/11 method as described previously [13], [14].
Exome capture and NGS DNA sequencing
Exome capture and sequencing were performed as described previously [14]. Briefly, TruSeq Exome Enrichment kit (Illumina) and NimbleGen SeqCap EZ Exome Library v3.0 were used to capture ∼62 Mb region of human genome comprising of ∼201,121 exons representing ∼20,974 gene sequences, including 5′UTR, 3′UTR, microRNAs and other non-coding RNA.
Somatic variant analysis, functional annotations and prioritization
The variant analysis was performed as described previously [14] and detailed in supplementary material and methods. MutSigCV v2.0 [15] and IntOgen [16] were used for identification of the significantly mutated gene and p value ≤ 0.05 was considered as the threshold for significance, as described earlier [17], [18]. Since our dataset was inherently not suitable for above tools due to a limited number of samples (n = 25), we have also performed extensive functional prediction tool based analysis for non-synonymous variants using nine different tools (detailed in supplementary material and methods). The total number of identified somatic substitutions in exome sequencing were extracted from the MutSigCV output and were processed to calculate the number and frequency distribution of various transitions and transversions.
Somatic copy number analysis from exome sequencing data and qPCR validation
DNA copy number analysis of exome sequencing data was performed as described previously [14] and detailed in supplementary material and methods. Genes with Segments-of-Gain-Or-Loss (SGOL) score ≥4 were defined as amplified genes and ≤−2 as deleted genes. The validation of somatic copy number changes was performed as described previously [14]. Details of the primers used for copy number study are provided in Supplementary Table S15.
Transcriptome sequencing and data analysis to identify expressed genes
Transcriptome libraries for sequencing were constructed according to the TruSeq RNA library protocol (Illumina) outlined in TruSeq RNA Sample Prep (Illumina) performed as described previously [14] and detailed in supplementary material and methods. Transcriptome data analysis was performed using previously published a protocol for transcriptome sequencing data analysis [19]. First, to identify the bona fide expressed transcripts, we filtered all the transcripts which were lowly expressed (≤0.1 log 10 (RSEM + 1)) for each sample; second, transcript expressed in 10% of samples was considered as a candidate expressed gene in tongue tumor tissue. A list of 16,525 transcripts identified to be expressed in TSCC tumors were used to filter mutation and DNA copy number changes in this study.
Gene expression dataset meta-analysis and RT-qPCR validation
TSCC gene expression profiling studies were identified by searching Gene Expression Omnibus (GEO) database [20] using keyword ‘tongue cancer’, ‘tongue tumor’. The TCGA-HNSCC dataset were downloaded from Cancer Genome Browser [21] and tongue sub-site data was extracted for the analysis. The criteria for selection of data set included fresh frozen tumor samples with corresponding normal sample and studies with >10 patient samples in the cohort. Studies involving cell lines, <10 samples, and non-human tissue samples were excluded. Raw data from 4 GEO microarray based data (GSE34105, GSE13601, GSE9844, and GSE31056) was analyzed using BRB array tool [22]. Briefly, non-variable genes were excluded from the analysis based on the log expression variation filter (variance of a gene across the arrays) followed by a class comparison of samples based on normal verses tumor comparison. Genes were considered as differentially regulated if a gene followed 1.5 <fold change <−1.5 filter along with p-value <0.05. Gene set enrichment analysis (GSEA) was performed selecting KEGG gene set in MSigDB [23] to identify underlying biological processes and pathways. The validation of gene expression was performed using quantitative reverse transcriptase PCR analysis as described previously [24] [14]. The primer sequences for genes are provided in Supplementary Table S16.
Immunohistochemical analysis
Immunohistochemical staining, was performed with the help of pathologist as described previously [14] and is detailed in supplementary material and methods.
Statistical analysis
The clinicopathologic association analysis was performed using IBM SPSS statistics software version 2. Test for overlap significance for a number of genes overlap for copy number changes different studies and databases were carried out using previously described method (http://nemates.org/MA/progs/representation.stats.html). The mutual exclusivity and co-occurrence analysis were performed using Gitools [25]. The significant differences between selected two groups were estimated using Unpaired Student t-test and P-value <0.05 was considered as a threshold for statistical significance.
Results
Patient details
Fifty-seven patient-matched normal early tongue cancer patient tumor were analyzed for somatic mutations, copy number changes, and differential expression by whole exome and transcriptome sequencing approach (Supplementary Fig. S1a, b). The clinicopathological details for fifty-seven in the cohort are detailed in Supplementary Table S1. In brief, our cohort comprised of 72% male; 61% tobacco users; 80% chewers of either betel, tobacco, areca, or lime; and, 28% smokers, with a median age of 45 years (range 25–72 years). 56% of all patients with pT1 and pT2 staged tongue tumors were lymph node positive (n = 32) at the time of registration. Primary treatment modality for all the patients was surgery followed by radiation and chemotherapy alone or in combination with chemo-radiation therapy. None of the patients were positive for HPV infection as reported previously [13], [14]. Forty patient follow-up data was available and median survival duration for the cohort was 29 months (range 2–42 months). During this time period, there were 9 recurrences, 6 metastasis and 8 fatal events.
Somatic variants in HPV negative early tongue squamous cell carcinoma
We performed whole exome sequencing of forty seven samples including early TSCC tumor (n = 24) and matched normal (n = 23) samples. We captured ∼62 Mb of coding genome at a median coverage of 97x for tumor samples and 103x coverage for normal samples. Somatic variants were called using Mutect [26] and GATK algorithm [27]. We identified a total of 2969 somatic mutations across 19 TSCC patients (5 patients were excluded from further analysis due to low coverage and/or poor correlation with their matched normal), which included 1693 missense, 60 nonsense, 972 silent, 124 splice site as well as 120 indels. The aggregated non-silent mutation rate across the dataset was 4.12 per Mb, consistent with the literature [28], [29]. The sequencing statistics and somatic mutational features are provided in Supplementary Tables S2–S3 and Supplementary Fig. 2a–e.
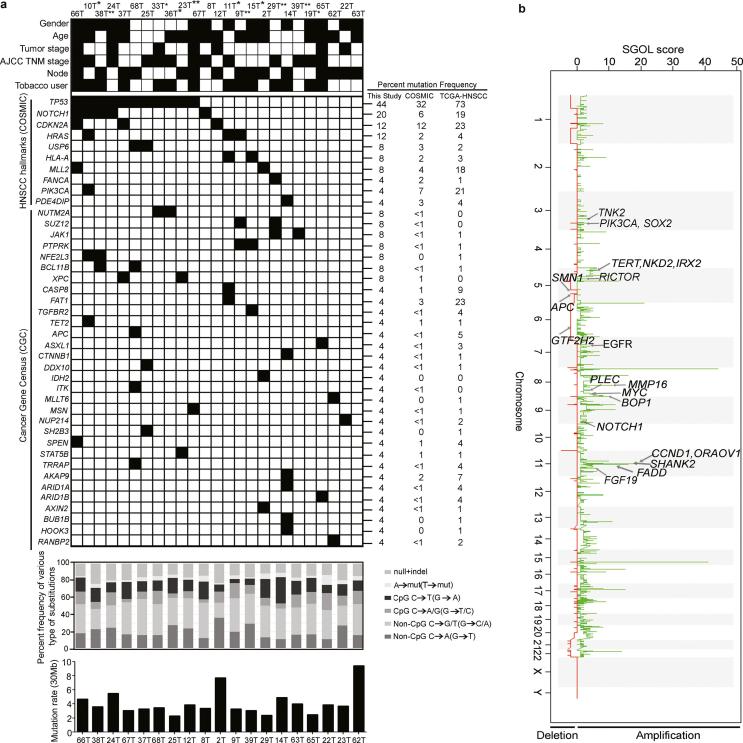
863 of 1693 non-silent somatic variants across 767 genes were predicted to be deleterious (Supplementary Table S4). Further posterior filtering of these variants was performed by restricting to 33 genes found to be significantly mutated using IntOgen (Q-value ≤ 0.05) as potential driver variants (Supplementary Table S5) [16]. Among the HNSCC hallmark genes reported in COSMIC database, we observed recurrent mutations in TP53 (44%), NOTCH1 (20%), CDKN2A (12%), HRAS (12%), USP6 (8%); while, mutations in FANCA, HLA-A, PIK3CA, KMT2D and PDE4DIP were observed as non-recurrent (Fig. 1a). Overall the frequency of mutations observed in the hallmark genes were consistent with COSMIC and TCGA HNSCC data with altered frequency for TP53 and NOTCH1, but consistent with reports from ICGC-India (Gingivo-buccal) [28], tongue subsite from India and Asia [29], [30] (Supplementary Table S6). A known mutational signature feature induced by tobacco C:G > A:T transversion was found to be represented in high fraction (53%) (Supplementary Fig. 2b Fig. 2b), which is much higher than observed in various cancers (15–26%) not associated with tobacco [31] and consistent with reports in HNSCC [6], [28].
Fig. 1.
Identification of somatic mutations and DNA copy number changes in HPV-negative early stage TSCC tumors. (a) Mutational features of 25 early tongue squamous carcinoma samples: 19 of 24 whole exome (5 samples excluded due to low quality reads) and 6 of 11 whole transcriptome sequencing (excluding 5 common samples). Samples ID’s with asterisk (**) denotes samples with exome and transcriptome sequencing; (*) samples with transcriptome sequencing alone. Different clinicopathological factors such as; gender, age, tumor stage, AJCC TNM stage and lymph node metastasis status and etiological factors such as tobacco users are shown for each patient. The black solid boxes denotes gender: male, age: >45 years, tumor stage: pT1, AJCC-TNM stage: Stage I–II, nodal status: positive and tobacco habit. The white boxes denote gender: female, age: <45 years, tumor stage: pT2, AJCC-TNM stage: Stage III–IV, nodal status: negative and without tobacco habit. Grey filled boxes denotes no information available. The ten HNSCC hallmark genes and cancer gene census (COSMIC) found to be mutated in data, is arranged in decreasing order of percent frequency. Black filled box denotes presence of a somatic mutation in the patient. Mutation frequencies for the hallmark and cancer census genes observed in this study (n = 25), COSMIC-HNSCC (n ≥ 500) and TCGA-HNSCC (n = 279) samples. The substitution frequencies spectrum for each patient for whole exome sequencing data is shown. Percent frequency of various types of SNVs and indels are shown. Different types of substitutions shown by different shades. Somatic non-silent mutation rate/30 Mb derived from whole exome sequencing data for each tumor is shown. (b) Somatic DNA copy number changes identified using Exome sequencing data. Somatic DNA copy number gains and losses were generated using Segments-of-Gain-Or-Loss (SGOL) scores across 23 TSCC patients. SGOL score is plotted (horizontal axis) for DNA copy number gains (green) and losses (red) are plotted as a function of distance along with human genome (vertical axis). Representative amplified and deleted regions are annotated for HNSCC-associated genes and denoted by an arrow. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Somatic copy number alterations derived from whole exome sequencing data
We used Control-FREEC [32] and cghMCR package to identify genomic regions harboring statistically significant copy number gains and losses relative to normal tissues. 440 amplified and 2275 deleted regions were identified across 23 TSCC patients (Supplementary Tables S7, S8). 18 genes exhibited copy number greater than three (Fig. 1b). Among most frequently amplified regions include 11q13.3 (CCND1, FGF19, ORAOV1, FADD); 8q21.3 (MMP16), and genes BOP1 (8q24.3), EGFR (33%); RICTOR (33%), PLEC (33%), TERT (26%), TNK2 (26%), PIK3CA and SOX2 (22%) MYC (14%) and NOTCH1 (14%). Comparative analysis of amplified and deleted gene with previous HNSCC including tongue cancer studies [28], TCGA-HNSCC [6], Vettore et al. [30] and PanCancer [33] revealed statistically significant overlap in the number of genes (Supplementary Table S9).
Additionally, we validated copy number changes in hallmark genes using qPCR (Supplementary Fig. 3). We observed significant co-amplification of CCND1, FGF19, ORAOV1, FADD (P-value < 0.01, Co-occurrence); PIK3CA and SOX2 (P-value < 0.001) in this study and TCGA-tongue tumors, which contains genes implicated in cell cycle, cell death/NF-kB pathway and, consistent with previously described in HPV-negative HNSCC tumors [6], [7] (Supplementary Fig. 4a). Interestingly, EGFR amplification was significantly mutually exclusive to CCND1, FGF19, ORAOV1 and FADD amplification (P-value < 0.01, MutEx) including TCGA-tongue cohort (Supplementary Fig. 4a,c) [6], suggesting unique molecular features associated in our study cohort. These novel genetic associations could serve as pathognomonic alterations in HPV-negative early TSCC tumors.
Whole transcriptome sequencing data identify upregulation of metastases related pathway in early tongue cancer
We performed whole transcriptome sequencing of five adjacent normal, twelve tumor tissue samples to generate an average of 25 and 34 million paired end reads, respectively, that clustered distinctly as shown in Supplementary Fig. 5a–c. Cufflinks [34], a transcript assembler, was used to perform reference guided assembly of the transcripts with an average expression of 11,824 (SD ± 606) genes per sample ≥ 1 FPKM. Of 17 samples that were whole transcriptome sequenced, one normal showed poor distribution of reads (Supplementary Fig. 5a) and tumor samples were misclassified (Supplementary Fig. 5b), were removed them from differential gene expression analysis resulting in 4 normal samples and 11 tumor tissues as shown in Fig. 2a. To identify the differentially expressed genes (DEGs) in whole transcriptome dataset, we used Cuffmerge and Cuffdiff method [34] and applied P-value ≤ 0.05 (Unpaired student-t-test) and log fold change 2 as a cut-off to identify 739 significantly differentially expressed genes (Fig. 2a; Supplementary Table S10).
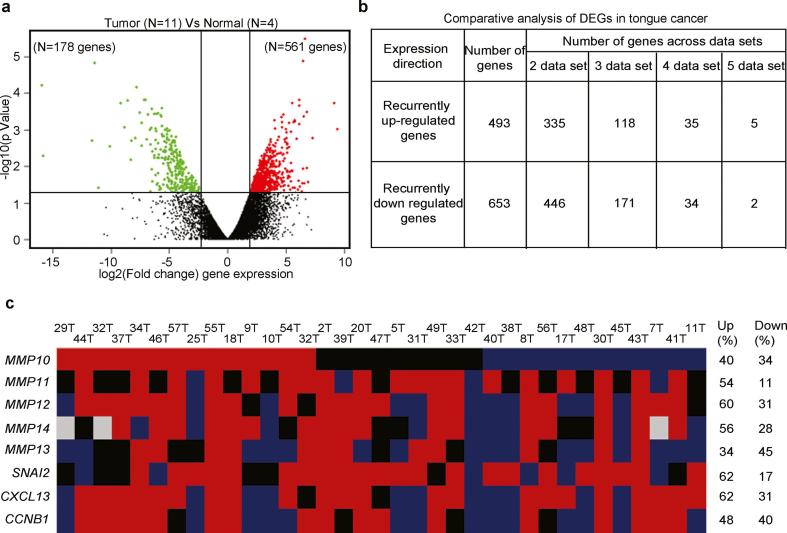
Fig. 2.
Differential expression profile of tongue squamous cell carcinoma using mRNA sequencing and meta-analysis identifies MMP10 up regulation. Differential expression analysis to identify the distinct gene expression profile of tongue tumors. (a) Volcano plot representation of differentially expressed in between early tongue tumors and adjacent normal tongue tissues. The red and green dots denote the up-regulated and down-regulated differentially expressed genes with P value <0.05 and fold changes ≥2 or ≤−2 for, respectively. (b) The tabular representation of a number of genes overlapped in tongue cancer across different studies. (c) Schematic representation of commonly up-regulated genes qRT-PCR validation in a cohort of 35 paired tongue tumor samples. The Red denotes up-regulation, blue as downregulation, black as basal expression and gray color; experiment could not be done or results could not be acquired. The ≥2 mean fold change is for up-regulation, ≤0.5 mean fold change for down-regulation and in between 1.99 and 0.501 mean fold change as a basal level expression compared to the adjacent normal tissue sample. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Further, to determine whether gene expression profile in this study was in agreement with the previous studies, we performed a systematic meta-analysis of 4 GEO (microarray) and TCGA (transcriptome sequencing) datasets comprising of 243 tongue tumors and 79 adjacent normal tissue samples expression profile using BRB array toolkit [22] (Supplementary Table S11) and fold change 1.5 and P-value ≤ 0.05 was applied as a cutoff to identify the DEGs in each dataset. We identified an average 1281 (SD ± 719) genes to be significantly differentially expressed, where average 619 (SD ± 364) and 662 (SD ± 447) genes showing up and down regulation, respectively (Supplementary Table S11). In overall, the average number of up-regulated genes were comparable with the study based on our cohort (Supplementary Table S11). To identify the commonly deregulated genes across dataset we performed recurrence based comparative analysis across dataset and observed 1146 genes to be deregulated in two or more number of datasets (Supplementary Tables S12 and S13). Among the 1146 deregulated genes, 493 and 653 were showing common upregulation and down-regulation in ≥2 datasets (including this study) in the meta-analysis (Supplementary Fig. 6a). Interestingly, we observed significant overlap i.e. 39% (196/493) up-regulated genes (P-value < 0.0001); and 20% (133/653) down regulated genes overlap (P-value < 0.0001) with recurrently up-regulated genes in previous datasets (Fig. 2b). Next, to gain broader insight into biological processes related to the commonly DEGs in tongue cancer, we performed gene set enrichment analysis against KEGG gene sets using MSigDB [23]. For the up-regulated genes, significantly enriched KEGG gene sets include several pathways involved in tumor cells metastasis process consistent with previous reports in HNSCC and tongue cancer [35] (Supplementary Fig. 6b, left panel). The down regulated genes, significantly enriched KEGG gene sets include pathways implicated in detoxification of carcinogenic compounds and environmental toxins such as drug metabolism consistent with previous reports in HNSCC, including tongue tumors [35] (Supplementary Fig. 6b, right panel). Interestingly, Arachidonic acid metabolism pathway was previously shown to be down regulated and inactivated via somatic mutations in Indian Gingivobuccal cancer patients [36], suggesting its possible tumor suppressive role via downregulation in tongue cancer patients in this study.
Upregulation of MMP10 and other MMPs in early stage tongue primary tumors
Several matrix metalloproteinase (MMPs) family genes were among the highly up-regulated genes across ≥3 dataset (Supplementary Table S15). While the role of MMP1, 3, 7 and 9 have previously been described in head and neck cancer [37], [38], [39], we set to ask if remaining 5 of 9 matrix metalloproteinases (MMP10, 11, 12, 13 and 14) along with 3 hallmark genes CXCL13, CCNB1 and SNAI2, as described in Table S12— also play a role in head and neck cancer. The real-time PCR based validations were performed across an additional set of 35 primary paired normal tongue tumors samples. The validated genes were ranked based on their differential expression (Fig. 2c and Fig. 3a). MMP10 differential expression was most significant (p < 0.0001) that was further validated histochemically (Fig. 3b–d). Incidentally, MMP10 is known to be involved in promoting metastases [38] and inflammation [40] in other cancers.
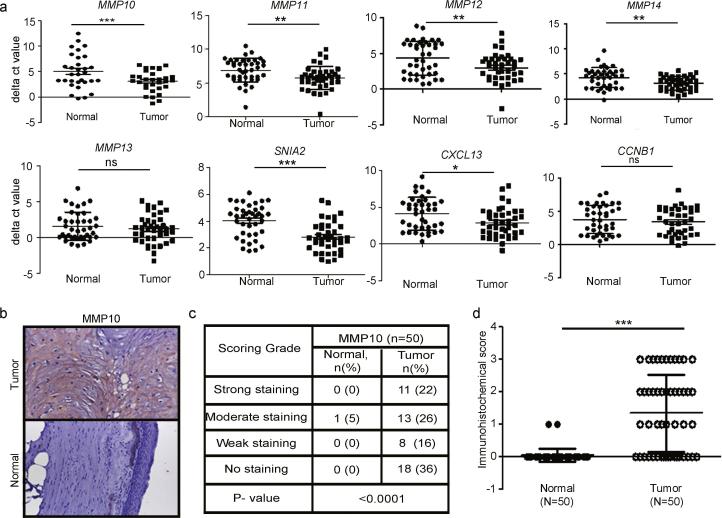
Fig. 3.
qRT-PCR validation of MMPs and immunohistochemical analysis of MMP10 in early stage tongue cancer. (a) qRT-PCR analysis of MMP11, MMP12, MMP13, MMP14, CXCL13, CCNB1, and SNAI2 transcript expression in paired normal early tongue tumors (n = 35). Dot plot representation of ΔCt value distribution and its significance between normal and tumors tongue tissue samples for MMP10. Each dot represents the average normalized ΔCt value of a gene in a single sample. Median with interquartile range is shown for each gene for normal and tumor samples. P-value is denoted as *; P < 0.01, **; P < 0.001, ***; P < 0.0001. (b) Representative IHC stained photomicrographs of tongue tumors and normal samples. The brown color indicates positive expression of MMP10 protein. (c) The tabular representation of different immunohistochemical scores grades of MMP10 in early tongue tumors. P-value is denoted as ***; P < 0.0001. The P-value was calculated by Mann-Whitney U test using GraphPad Prism 5 program and P-value ≤0.05 was considered as a threshold for statistical significance. (d) Dot plot representation of immunohistochemical score of MMP10 expression in tongue tumors and adjacent normal tissues (n = 50). Each dot represents that final IHC score for each sample and median with interquartile range is shown. Median with interquartile range is shown for MMP10 protein expression in normal and tumor samples. P-value is denoted as ***; P < 0.0001.
Next, we performed immunohistochemical based validation of MMP10 expression in 50 primary early stage paired normal tongue tumors. In the adjacent normal samples, positive MMP10 staining was not observed, whereas, positive cytoplasmic staining of MMP10 was detected in 32/50 (64%) of tongue cancer patients tumors (Fig. 3b), consistent with the previous report in HNSCC tumors [35]. About 48% of primary tongue tumors displayed strong or moderate immunostaining of MMP10 protein, whereas 62% tongue tumors showed weak or no staining (Fig. 3c). In overall, statistically significant differences in immunohistochemical scores were observed in tumors as compared to adjacent normal tissues (P-value < 0.0001, unpaired student-t-test) (Fig. 3d) and MMP10 protein was up-regulated in a large proportion of primary tongue tumors.
Clinical correlation with genetic alterations in early tongue cancer
The cohort did not reveal any significant association between clinical features such as age, gender, tumor stage, American Joint committee on Cancer (AJCC) TNM stage, nodal status, smoking, alcohol, tobacco usages with mutations in HNSCC hallmark gene; TP53, NOTCH1, CDKN2A, CASP8, HRAS and PIK3CA (Supplementary Table S14). However, we observed 3 of 3 patients with HRAS mutation were tobacco chewers, which is consistent with previous reports in Indian oral cancer patients [41]. As shown in Table 1, an association of MMP10 transcript and protein expression analyzed in 35 and 43 primary TSCC patients showed a marginal significance with tobacco habit (P-value = 0.057). Survival data of these patients were far from maturity. Thus, analysis of GEO gene expression dataset of HNSCC (GSE2837) based on MMP10 transcript expression performed showed poor survival in the cancer patients with high MMP10 expression, similar to as observed in breast cancer (GSE2990), lung cancer (GSE31210; 11117), liposarcomas (GSE30929), and colorectal cancer (GSE12945) using PrognoScan [42] and PROGgene [43] (Fig. 4). The association of MMP10 expression with poor survival in early TSCC remains to be verified in a larger sample set.
Table 1.
Association of clinical features of early stage TSCC patients with MMP10 transcript and protein level expression.
| Clinicopathological features | MMP10 expression, number (%), along the column |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | N |
MMP10 transcript expression (n = 35) |
P value* | N | MMP10 protein expression (n = 43) |
P value* | |||
| Basal or Down | Up-regulated | Negative | Positive | ||||||
| Age | >45 years | 15 | 9 (41%) | 6 (46%) | 1 | 21 | 12 (50%) | 9 (47%) | 1 |
| <45 years | 20 | 13 (59%) | 7 (54%) | 22 | 12 (50%) | 10 (53%) | |||
| Gender | Male | 25 | 17 (77%) | 8 (62%) | 0.444 | 30 | 18 (75%) | 12 (63%) | 0.509 |
| Female | 10 | 5 (23%) | 5 (38%) | 13 | 6 (25%) | 7 (37%) | |||
| AJCC TNM stage | I–II | 13 | 7 (32%) | 6 (46%) | 0.48 | 16 | 8 (33%) | 8 (42%) | 0.752 |
| III–IVA | 22 | 15 (68%) | 7 (54%) | 27 | 16 (67%) | 11 (58%) | |||
| Node negative | 16 | 7 (54%) | 6 (46%) | 16 | 8 (34%) | 8 (42%) | |||
| Smoking | Smoker | 12 | 9 (41%) | 3 (23%) | 0.463 | 16 | 8 (33%) | 8 (42%) | 0.752 |
| Non-smoker | 23 | 13 (59%) | 10 (77%) | 27 | 16 (67%) | 11 (58%) | |||
| Alcohol | Yes | 8 | 5 (23%) | 3 (23%) | 1 | 14 | 8 (33%) | 6 (32%) | 1 |
| No | 27 | 17 (77%) | 10 (77%) | 29 | 16 (67%) | 13 (68%) | |||
| Tobacco | Yes | 24 | 18 (82%) | 6 (46%) | 0.057 | 28 | 14 (58%) | 14 (74%) | 0.349 |
| No | 11 | 4 (18%) | 7 (54%) | 15 | 10 (42%) | 5 (27%) | |||
N: Number of patients.
Chi-square t-test.
Fig. 4.
Kaplan-Meier survival curve disease-free survival and overall survival in various cancer types based on the level of MMP10 gene expression. Kaplan-Meier plot of DFS and OS analysis in HNSCC studies based on low and high MMP10 gene expression. The red and green lines denote high and low MMP10 expressing patient’s survival, respectively. The number of samples and median survival in each group is denoted. The log-rank test was applied to accesses the statistical differences in median survival and P-value ≤ 0.05 was considered as statistical significance. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
Here, we describe the landscape of genomic alterations in a unique set of early staged HPV-negative tobacco or nut chewing tongue cancer patients, using whole exome sequencing and transcriptome sequencing. While lack of survival data is a major limitation of the study, several lines of distinct features underlie this study attributing to unique etiology, subsite, and specific population, which has been previously described for HNSCC [7].
Firstly, the mutational profile of large fraction of patients display high frequency (53%) of C:G > A:T transversion in exome sequencing data—a hallmark of smokeless tobacco usage—reflecting tobacco as the most predominant etiological agent; which is considerably higher than observed (15–26%) in various non-tobacco associated cancer types [31]. We also observed an enriched fraction of C > T transition and C > G transversion, consistent with previous report in gingiva-buccal (ICGC-India) and tongue tumors with tobacco chewing habit [28], [29]. The C > G transversions are known to be caused by tobacco due to reactive oxygen species (8-oxoguanine lesions) and or APOBEC family of cytidine deaminases genes overactivity induced by deamination of 5-methyl-cytosine to uracil in CpG island as described previously. Moreover, the predominance of G > A transition in TP53 gene in this study (57%; 4/7) is consistent with previous reports in betel quid and tobacco chewing associated oral squamous cell carcinoma tumors from Indian population [44], [45].
Secondly, recent reports suggest the presence of low frequency (∼5%) of RAS mutations in tongue tumors [29], [30]. However, we observed 12% HRAS mutations, which though were all tobacco chewers, consistent with previous reports from the Indian population [41]. Similarly, unlike previously reported, inactivating and low-frequency mutation in NOTCH1 in HNSCC [6], [28], [46], [47], most of the mutations were missense, consistent with a recent report in the Asian population and our report [14]. However, consistent with previous reports, frequent copy number alterations including gains at 5p, 8q, 20q, 22q and 11q and losses at 1p, 5p, 6q, 7p and 21q [6], [28], [29], [30] were significantly represented. Moreover, deleterious somatic variants in HNSCC hallmark genes: TP53, NOTCH1, CDKN2A, CASP8, PIK3CA, USP6, MLL2, HLA-A, FANCA, PDE4DIP, and FAT1 were also identified [29], [30]. Furthermore, significantly co-occurring alterations in FADD CCND1, FGF19, and ORAOV1 (P < 0.0001) were found to occur mutually exclusive with EGFR amplification among HPV-negative early TSCC tumors, as previously described in other cancers [6], [7]. Interestingly, EGFR and CCND1 oncogenic events are known to act via a common RAS-MAPK Kinase pathway to promote cell cycle and known to act as a driver of oral cancer tumorigenesis [48], [49], [50]. The mutual exclusivity of EGFR and CCND1 amplification suggests activation of a common downstream signalling pathway in different TSCC patients via diverse genetic alterations. Thus this finding may affect the benefit of common downstream inhibitor, such as MAPK inhibitor, to a broader spectrum of patients.
Thirdly, differential gene expression analysis showed significant up-regulation of gene-sets primarily involved in epithelial to mesenchymal transition (EMT) processes, corroborating with known occult lymph node metastasis and invasive behaviour of early stage tongue tumors [51]. Furthermore, meta-analysis approaches of gene expression studies lead to a precise estimation of recurrently expressed genes across data set. The overexpression of MMPs has been known to be involved in ECM degradation thereby facilitating the process of tumor invasion and metastasis leading to an aggressive course of disease in HNSCC patients [37]. qRT-PCR validation across 35 paired normal early stage primary tumors for up-regulated genes (MMP10, MMP11, MMP12, MMP14, CXCL13, CCNB1, SNIA2) showed significant up-regulation in tumors suggesting reliability of genes identified from this study. Of the multiple MMPs found to be up-regulated, immunohistochemical analysis of MMP10 in 50 paired normal early stage primary tumors showed significant up-regulation of protein expression in primary tumors owing its possible role in early stage progression as described in other cancer types as a potential prognostic biomarker to stratify those likely to develop metastases [52], [53]. However, insights about their specific role await validation in a larger independent cohort with survival followed by functional analysis.
Acknowledgment
All members of the Dutt laboratory for critically reviewing the manuscript. Sandor Proteomics Pvt. Ltd., Hyderabad, India and Medgenome Labs Pvt. Ltd, Bengaluru, India, for providing Exome and Transcriptome library preparation services. We acknowledge BTIS facility, funded by the Department of Biotechnology (DBT), Govt. of India, at ACTREC. A.D. is supported by an Intermediate Fellowship from the Wellcome Trust/DBT India Alliance (IA/I/11/2500278), by a grant from DBT (BT/PR2372/AGR/36/696/2011), and intramural grants (IRB project 92 and 55). P.U. is supported by a senior research fellowship from CSIR. N.G. is supported by a junior research fellowship from Tata Memorial hospital. S.D. and A.J. are supported by a junior research fellowship from ACTREC. B. D. is supported by a junior research fellowship from CSIR. P.C. supported by a senior research fellowship from ACTREC. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
Authors contributions
Conception and design: Pawan Upadhyay and Amit Dutt.
Development of methodology: Pawan Upadhyay, Nilesh Gardi, Sanket Desai, Pratik Chandrani and Amit Dutt: Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Pawan Upadhyay, Nilesh Gardi, Sanket Desai, Pratik Chandrani, Bhasker Dharavath, Asim Joshi, Priynacka Arora, Munita Bal and Sudhir Nair.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Pawan Upadhyay, Nilesh Gardi, Sanket Desai, Pratik Chandrani and Amit Dutt.
Writing, review, and/or revision of the manuscript: Pawan Upadhyay and Amit Dutt.
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Sudhir Nair and team.
Study supervision: Amit Dutt.
Conflict of interest
The authors declare no competing financial interests.
Footnotes
The raw sequence data has been deposited at the ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), hosted by the European Bioinformatics Institute (EBI), under the following accession number: E-MTAB-4654: Whole transcriptome tissue samples, E-MTAB-4653: Whole exome tumor tissue samples, E-MTAB-4618: Whole exome normal tissue samples. Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.oraloncology.2017.08.003.
Appendix A. Supplementary material
References
- 1.Majchrzak E., Szybiak B., Wegner A., Pienkowski P., Pazdrowski J., Luczewski L. Oral cavity and oropharyngeal squamous cell carcinoma in young adults: a review of the literature. Radiol Oncol. 2014;48:1–10. doi: 10.2478/raon-2013-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garnaes E., Kiss K., Andersen L., Therkildsen M.H., Franzmann M.B., Filtenborg-Barnkob B. Increasing incidence of base of tongue cancers from 2000 to 2010 due to HPV: the largest demographic study of 210 Danish patients. Brit J Cancer. 2015;113:131–134. doi: 10.1038/bjc.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datta S., Chaturvedi P., Mishra A., Pawar P. A review of Indian literature for association of smokeless tobacco with malignant and premalignant diseases of head and neck region. Indian J Cancer. 2014;51:200–208. doi: 10.4103/0019-509X.146713. [DOI] [PubMed] [Google Scholar]
- 4.Wyss A., Hashibe M., Chuang S.C., Lee Y.C., Zhang Z.F., Yu G.P. Cigarette, cigar, and pipe smoking and the risk of head and neck cancers: pooled analysis in the international head and neck cancer epidemiology consortium. Am J Epidemiol. 2013;178:679–690. doi: 10.1093/aje/kwt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Rorke M.A., Ellison M.V., Murray L.J., Moran M., James J., Anderson L.A. Human papillomavirus related head and neck cancer survival: a systematic review and meta-analysis. Oral Oncol. 2012;48:1191–1201. doi: 10.1016/j.oraloncology.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seiwert T.Y., Zuo Z., Keck M.K., Khattri A., Pedamallu C.S., Stricker T. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res: Off J Am Assoc Cancer Res. 2015;21:632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharyya N., Fried M.P. Benchmarks for mortality, morbidity, and length of stay for head and neck surgical procedures. Arch Tolaryngol Head Neck Surg. 2001;127:127–132. doi: 10.1001/archotol.127.2.127. [DOI] [PubMed] [Google Scholar]
- 9.Kapoor C., Vaidya S., Wadhwan V., Malik S. Lymph node metastasis: a bearing on prognosis in squamous cell carcinoma. Indian J Cancer. 2015;52:417–424. doi: 10.4103/0019-509X.176750. [DOI] [PubMed] [Google Scholar]
- 10.Thiagarajan S., Nair S., Nair D., Chaturvedi P., Kane S.V., Agarwal J.P. Predictors of prognosis for squamous cell carcinoma of oral tongue. J Surg Oncol. 2014;109:639–644. doi: 10.1002/jso.23583. [DOI] [PubMed] [Google Scholar]
- 11.D'Cruz A.K., Vaish R., Kapre N., Dandekar M., Gupta S., Hawaldar R. Elective versus therapeutic neck dissection in node-negative oral cancer. New Engl J Med. 2015;373:521–529. doi: 10.1056/NEJMoa1506007. [DOI] [PubMed] [Google Scholar]
- 12.Ziober A.F., D'Alessandro L., Ziober B.L. Is gene expression profiling of head and neck cancers ready for the clinic? Biomark Med. 2010;4:571–580. doi: 10.2217/bmm.10.71. [DOI] [PubMed] [Google Scholar]
- 13.Chandrani P., Kulkarni V., Iyer P., Upadhyay P., Chaubal R., Das P. NGS-based approach to determine the presence of HPV and their sites of integration in human cancer genome. Brit J Cancer. 2015;112:1958–1965. doi: 10.1038/bjc.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Upadhyay P, Nair S, Kaur E, Aich J, Dani P, Sethunath V, et al. Notch pathway activation is essential for maintenance of stem-like cells in early tongue cancer. Oncotarget 2016. [DOI] [PMC free article] [PubMed]
- 15.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Perez A., Perez-Llamas C., Deu-Pons J., Tamborero D., Schroeder M.P., Jene-Sanz A. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10:1081–1082. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Upadhyay P, Gardi N, Desai S∗, Sahoo B, Singh A, Togar T, Iyer P, Prasad R, Chandrani P, Gupta S, Dutt A. TMC-SNPdb: an Indian germline variant database derived from whole exome sequences. Database. Oxford;2016. [DOI] [PMC free article] [PubMed]
- 18.Chandrani P., Prabhash K., Prasad R., Sethunath V., Ranjan M., Iyer P. Drug-sensitive FGFR3 mutations in lung adenocarcinoma. Anna Oncol: Off J Euro Soc Med Oncol. 2017;28:597–603. doi: 10.1093/annonc/mdw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman M., Craft B., Swatloski T., Cline M., Morozova O., Diekhans M. The UCSC cancer genomics browser: update 2015. Nucleic Acids Res. 2015;43:D812–D817. doi: 10.1093/nar/gku1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon R., Lam A., Li M.C., Ngan M., Menenzes S., Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Informatics. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choughule A., Sharma R., Trivedi V., Thavamani A., Noronha V., Joshi A. Coexistence of KRAS mutation with mutant but not wild-type EGFR predicts response to tyrosine-kinase inhibitors in human lung cancer. Brit J Cancer. 2014;111:2203–2204. doi: 10.1038/bjc.2014.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gagliardi A.R., Brouwers M.C., Bhattacharyya O.K., Guideline Implementation R. Application N. A framework of the desirable features of guideline implementation tools (GItools): Delphi survey and assessment of GItools. Implement Sci. 2014;9:98. doi: 10.1186/s13012-014-0098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.India Project Team of the International Cancer Genome c. Mutational landscape of gingivo–buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat commun 2013;4:2873. [DOI] [PMC free article] [PubMed]
- 29.Krishnan N, Gupta S, Palve V, Varghese L, Pattnaik S, Jain P, et al. Integrated analysis of oral tongue squamous cell carcinoma identifies key variants and pathways linked to risk habits, HPV, Clinical Parameters and Tumor Recurrence. F1000Res. 2015;4:1215. [DOI] [PMC free article] [PubMed]
- 30.Vettore A.L., Ramnarayanan K., Poore G., Lim K., Ong C.K., Huang K.K. Mutational landscapes of tongue carcinoma reveal recurrent mutations in genes of therapeutic and prognostic relevance. Genome Med. 2015;7:98. doi: 10.1186/s13073-015-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartzentruber J., Korshunov A., Liu X.Y., Jones D.T., Pfaff E., Jacob K. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 32.Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., Schleiermacher G. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., McGee J., Chen X., Doman T.N., Gong X., Zhang Y. Identification of druggable cancer driver genes amplified across TCGA datasets. PLoS ONE. 2014;9:e98293. doi: 10.1371/journal.pone.0098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thangaraj S.V., Shyamsundar V., Krishnamurthy A., Ramani P., Ganesan K., Muthuswami M. Molecular portrait of oral tongue squamous cell carcinoma shown by integrative meta-analysis of expression profiles with validations. PLoS ONE. 2016;11:e0156582. doi: 10.1371/journal.pone.0156582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biswas NK, Das S, Maitra A, Sarin R, Majumder PP. Somatic mutations in arachidonic acid metabolism pathway genes enhance oral cancer post-treatment disease-free survival. Nat commun 2014;5:5835. [DOI] [PubMed]
- 37.Iizuka S., Ishimaru N., Kudo Y. Matrix metalloproteinases: the gene expression signatures of head and neck cancer progression. Cancers. 2014;6:396–415. doi: 10.3390/cancers6010396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessenbrock K., Plaks V., Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overall C.M., Kleifeld O. Tumour microenvironment – opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 40.Murray M.Y., Birkland T.P., Howe J.D., Rowan A.D., Fidock M., Parks W.C. Macrophage migration and invasion is regulated by MMP10 expression. PLoS ONE. 2013;8:e63555. doi: 10.1371/journal.pone.0063555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sathyan K.M., Nalinakumari K.R., Kannan S. H-Ras mutation modulates the expression of major cell cycle regulatory proteins and disease prognosis in oral carcinoma. Mod Pathol. 2007;20:1141–1148. doi: 10.1038/modpathol.3800948. [DOI] [PubMed] [Google Scholar]
- 42.Mizuno H., Kitada K., Nakai K., Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genom. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goswami C.P., Nakshatri H. PROGgene: gene expression based survival analysis web application for multiple cancers. J Clin Bioinform. 2013;3:22. doi: 10.1186/2043-9113-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saranath D., Tandle A.T., Teni T.R., Dedhia P.M., Borges A.M., Parikh D. P53 inactivation in chewing tobacco-induced oral cancers and leukoplakias from India. Oral Oncol. 1999;35:242–250. [PubMed] [Google Scholar]
- 45.Kannan K., Munirajan A.K., Krishnamurthy J., Bhuvarahamurthy V., Mohanprasad B.K., Panishankar K.H. Low incidence of p53 mutations in betel quid and tobacco chewing-associated oral squamous carcinoma from India. Int J Oncol. 1999;15:1133–1136. doi: 10.3892/ijo.15.6.1133. [DOI] [PubMed] [Google Scholar]
- 46.Stransky N., Egloff A.M., Tward A.D., Kostic A.D., Cibulskis K., Sivachenko A. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agrawal N., Frederick M.J., Pickering C.R., Bettegowda C., Chang K., Li R.J. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsui I.F., Poh C.F., Garnis C., Rosin M.P., Zhang L., Lam W.L. Multiple pathways in the FGF signaling network are frequently deregulated by gene amplification in oral dysplasias. Int J Cancer. 2009;125:2219–2228. doi: 10.1002/ijc.24611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michikawa C., Uzawa N., Sato H., Ohyama Y., Okada N., Amagasa T. Epidermal growth factor receptor gene copy number aberration at the primary tumour is significantly associated with extracapsular spread in oral cancer. Brit J Cancer. 2011;104:850–855. doi: 10.1038/bjc.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi K.-I., Uzawa N., Myo K., Amagasa T. Simultaneous assessment of cyclin D1 and epidermal growth factor receptor gene copy number for prognostic factor in oral squamous cell carcinomas. Oral Sci Int. 2009;6:8–20. [Google Scholar]
- 51.Rickman D.S., Millon R., De Reynies A., Thomas E., Wasylyk C., Muller D. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27:6607–6622. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 52.Liu H., Qin Y.R., Bi J., Guo A., Fu L., Guan X.Y. Overexpression of matrix metalloproteinase 10 is associated with poor survival in patients with early stage of esophageal squamous cell carcinoma. Dis Esophagus: Off J Int Soc Dis Esophagus. 2012;25:656–663. doi: 10.1111/j.1442-2050.2011.01284.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhang G., Miyake M., Lawton A., Goodison S., Rosser C.J. Matrix metalloproteinase-10 promotes tumor progression through regulation of angiogenic and apoptotic pathways in cervical tumors. BMC Cancer. 2014;14:310. doi: 10.1186/1471-2407-14-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.