Introduction
Gastric adenocarcinoma is the third leading cause of cancer-related death worldwide, accounting for more than 720,000 deaths annually [1]. The strongest known risk factor for this devastating disease is infection with Helicobacter pylori, which drives the development of premalignant lesions (such as gastric atrophy, intestinal metaplasia, and dysplasia) that can lead to gastric cancer (Fig 1). However, although H. pylori is the most common bacterial infection worldwide and colonizes greater than 50% of the global population, only 1%–3% of infected individuals ever develop gastric cancer.
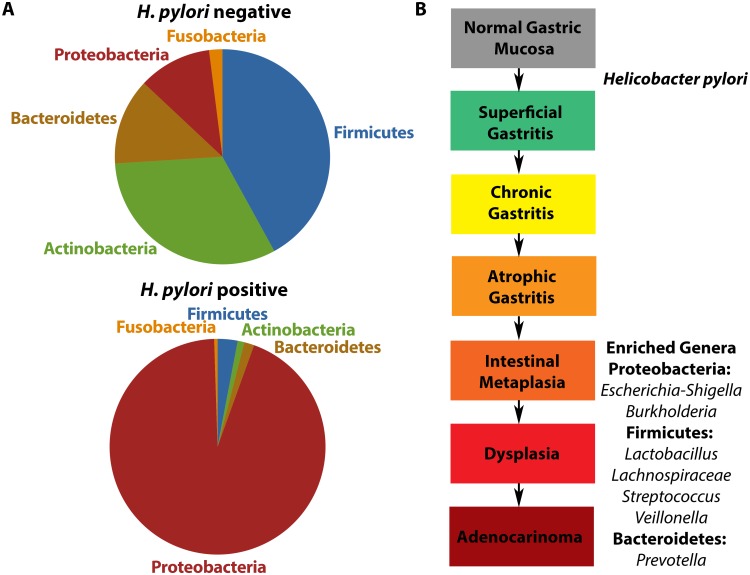
Fig 1. Alterations in the gastric microbiota following Helicobacter pylori infection and gastric disease progression.
(A) Schematic representation of the predominant phyla of the gastric microbiota based on H. pylori infection status. H. pylori-negative individuals harbor a microbiota that is more complex and highly diverse compared to H. pylori-positive individuals. (B) Schematic representation of the predominant genera at different stages within the gastric carcinogenesis cascade. Following infection with H. pylori, Proteobacteria and specifically H. pylori dominate the gastric microbiota. This leads to the development of chronic gastritis. In the later stages of the disease, ranging from intestinal metaplasia to gastric adenocarcinoma, a number of genera are enriched. These include Escherichia-Shigella and Burkholderia within the Proteobacteria phylum; Lactobacillus, Lachnospiraceae, Streptococcus, and Veillonella within the Firmicutes phylum; and Prevotella within the Bacteroidetes phylum.
Drivers of susceptibility to gastric carcinogenesis include H. pylori strain-specific virulence determinants, host constituents, and environmental factors. Along with these elements, the microbiota of the stomach may also influence the development of gastric malignancies. The acidic environment of the stomach in conjunction with low levels of cultured bacteria from this site previously led to assumptions that the gastric niche was not able to support a diverse microbial community. However, recent advances in DNA sequencing of conserved ribosomal RNA genes, phylogenetic analyses, and computational methods have uncovered a complex microbiota within the human stomach with the potential for disease induction [2].
The gut microbiota and dysbiosis
The human gut microbiota is critical for maintenance of human health and plays an integral role in energy metabolism, absorption of nutrients, and defense against invading pathogens [3–5]. However, this microbiota exists within a delicate balance that, if altered, becomes dysbiotic and contributes to aberrant proinflammatory immune responses, susceptibility to invading pathogens, and initiation of disease processes, including cancer [6]. Dysbiosis contributes to the pathogenesis of gastrointestinal carcinomas in the esophagus [7] and colon [8, 9], and specific bacterial species are associated with the development of colorectal cancer (Fusobacteria nucleatum and Escherichia coli) [10, 11] and gastric cancer (H. pylori) [12].
The relationship between specific microbial pathogens and carcinogenesis has been the subject of extensive investigation, and historically, the majority of research has focused on individual pathogens, such as H. pylori, and their ability to initiate and perpetuate disease. Advances in sequencing technology have greatly enhanced the ability of scientists to identify additional microbial species that may be potentially associated with various disease states, such as cancer, although establishing cause versus effect presents multiple challenges. Therefore, understanding how dysbiosis impacts aberrant host inflammatory responses and downstream carcinogenic cascades will be critical to accurately define the role of niche-specific microbiota in oncogenesis.
H. pylori, the human gastric microbiota, and gastric cancer
A first step in establishing causation is to take inventory of a particular resident microbial population, and several investigations have focused on defining microbial communities within the human stomach and the interactions of these populations with H. pylori (Fig 1A). Numerous groups have used PCR- and sequencing-based approaches to demonstrate that H. pylori-negative individuals harbor a highly diverse gastric microbiota dominated by 5 predominant phyla: Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, and Fusobacteria [13–15]. In contrast, among H. pylori-positive subjects, H. pylori is the single most abundant bacterium present in the stomach and accounts for between 72% and 97% of all sequence reads [13, 14].
These initial studies primarily focused on defining the composition of the gastric microbiome stratified by H. pylori infection status but not disease diagnosis. Subsequent human cross-sectional studies have compared the gastric microbiota in patients with pathologic outcomes that span the gastric carcinogenesis cascade (Fig 1B). One study demonstrated that H. pylori was present at relatively low abundance in patients with advanced premalignant lesions and that the microbiota of patients with gastric cancer were dominated by species of Lactobacillus, Streptococcus, Veillonella, and Prevotella [16]. Another study demonstrated a steady decrease in bacterial diversity of the gastric microbiota, with an increasing abundance of Lactobacillus and Lachnospiraceae in patients progressing along the carcinogenic cascade [17]. The increase in these genera validates other studies that demonstrated similar increases in the abundances of Lactobacilli [16] and Lachnospiraceae [18, 19] in tissue samples from gastric cancer patients. The abundance of Lactobacillus, Lachnospiraceae, Escherichia-Shigella, Nitrospirae, and Burkholderia is also enriched when gastric cancer patients are compared to controls [19], supporting previous findings that Lactobacillus and Lachnospiraceae are present at higher abundance in gastric cancer [16–18, 20] and that Escherichia-Shigella is enriched in patients with colorectal cancer [21]. It is important to note that these studies only identified genetic evidence of bacteria and in-depth studies to assess viability of these organisms have not been performed. However, these results raise an intriguing hypothesis, namely that gastric colonization by non-H. pylori bacteria, many of which also colonize the intestine, could impact the risk for gastric cancer.
Most bacteria cannot survive in the acidic environment of the stomach. However, it has been well established that, in a subset of persons, infection with H. pylori leads to achlorhydria and decreased acid secretion. Thus, long-term H. pylori colonization and neutralization of the gastric environment may directly contribute to alterations in the gastric microbiota. There are also clinical studies that support this concept, namely that patients treated with acid-suppressive drugs, such as proton pump inhibitors, exhibit a significant increase in the burden of non-H. pylori bacteria within the stomach. Of interest, this increase correlates with increased inflammatory responses, suggesting that non-H. pylori bacteria that colonize an achlorhydric stomach may have the capacity to promote inflammation that could potentially facilitate the progression to cancer [22, 23]. However, definitive evidence for this requires careful interventional studies that have yet to be performed.
Although these studies demonstrate associations between the human gastric microbiota and H. pylori infection (as well as various H. pylori-induced pathologies), they do not directly differentiate cause from effect, primarily due to their cross-sectional study designs. One longitudinal study that supported the role of non-H. pylori species in the development of cancer assessed the effects of H. pylori eradication therapy on gastric cancer incidence over a 15-year time period [24]. Despite only a 47% eradication rate for H. pylori, there were similar reductions in the incidence of gastric cancer among subjects who received antibiotics and were unsuccessfully eradicated compared to those who remained H. pylori-free [24]. These results suggest that bacteria in addition to H. pylori may have been affected by antibiotics, which may have contributed to attenuated rates of gastric cancer. There are also computational biology studies that support these concepts. Using a computerized search algorithm designed to identify the presence of bacterial DNA within interrogated known cancer genomes, these investigators determined that the type of cancer that harbored the second highest number of bacterial DNA sequences was gastric adenocarcinoma. However, the most common type of integrated bacterial DNA was not H. pylori but was instead Pseudomonas [25].
The effect of the gastric microbiota on H. pylori-induced gastric inflammation and cancer in rodents
The ability to establish causality is greatly enhanced by carefully controlled and manipulatable animal model systems. Inbred mice with defined genotypes are commonly used to study the effects of H. pylori infection on gastric diseases such as cancer. Of interest, the gastric microbiota of C57BL/6 mice is dominated by the same predominant phyla that have been reported in humans: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria [26]. Longitudinal studies in mice have provided more direct evidence of the contribution of non-H. pylori species to H. pylori-induced gastric carcinogenesis. For example, 1 study demonstrated that INS-GAS mice harboring a complex microbiota developed gastric cancer within 7 months following H. pylori infection, whereas the development of gastric cancer was markedly prolonged in germ-free mice that were monocolonized by H. pylori [27]. Following H. pylori infection, there was an overall decrease in gastric microbial diversity [27], similar to that observed in human populations, but there were no significant differences in the intestinal microbiota among any of the groups. These observations were studied in greater depth in an INS-GAS H. pylori mono-associated germ-free mouse model, where the addition of a restricted microflora accelerated the development of gastric cancer in conjunction with H. pylori [28]. Specifically, germ-free INS-GAS mice supplemented with a gastric and intestinal microbiota containing only 3 species of commensal intestinal bacteria (ASF356 Clostridium species, ASF361 Lactobacillus murinus, and ASF519 Bacteroides species) were sufficient to promote gastric neoplasia in H. pylori-infected INS-GAS mice to the same extent as observed in H. pylori-infected INS-GAS mice harboring a complex microbiota [28]. Importantly, these genera are also enriched in the stomachs of patients that develop premalignant and malignant lesions. Further supporting the concept of a contributory role of the gastric microbiota in promoting disease have been interventions with antibiotic therapy, which were shown to delay the onset of gastric cancer in INS-GAS mice in a manner that was not dependent on the presence of H. pylori [29]. Collectively, these results suggest that non-H. pylori bacteria can colonize the stomach and may represent an additional modifier of gastric cancer risk, particularly among H. pylori-infected individuals.
In addition to the stomach, bacteria within other microbial niches may exert a role in modulating H. pylori-induced gastric inflammatory responses. Two studies have shown that precolonization with intestinal Helicobacters (H. bilis, H. hepaticus, and H. muridarum) can either increase or decrease the severity of gastric inflammation induced by H. pylori by altering T-regulatory cell responses [30, 31]. Another study demonstrated that H. pylori per se is present within the intestine in a coccoid form and that the interaction between phagocytes and H. pylori within intestinal Peyer’s patches plays a critical role in modifying the intensity of H. pylori-induced gastritis [32]. However, other studies have shown that the microflora within the stomach can accelerate the progression of gastric cancer in the presence of H. pylori and do so with no differences detected in the composition of the intestinal microbiota [27, 28]. Addressing whether the resident intestinal microbiome directly contributes to the pathophysiology of H. pylori-induced gastric diseases is an avenue that requires further investigation, and it is important to consider that the effects of bacteria and microbial communities in the intestine and the stomach on gastric pathophysiology may not be mutually exclusive.
Conclusions
Evidence that the host microbiota specifically functions to promote health and prevent disease and that dysbiosis contributes to inflammation, susceptibility to pathogens, and diseases (including cancer) is undisputed [3]. As a result, the concept of specific microorganisms solely driving cancer initiation and progression may need to be modified in certain circumstances. Although great advances have been made in understanding the complex interplay between the gastric microbiota and H. pylori in the development of gastric inflammation and cancer, detailed studies are still needed in well-defined human populations to compare differences in the microbiota of H. pylori-infected persons with and without neoplastic lesions. Cross-sectional studies can provide initial insights into microbial associations with cancer; however, reverse effects are a concern, as it is difficult to discern whether carcinogenesis leads to changes in the local microenvironment that creates a new niche for microbes or whether alterations in the microbial population or its functions contribute to carcinogenesis [33]. Due to the acidic nature of the stomach, most bacteria cannot survive in this environment. However, infection with H. pylori leads to achlorhydria of the stomach in a subset of colonized persons; thus, long-term H. pylori colonization and neutralization of the gastric environment may directly contribute to alterations in the gastric microbiota.
Since the gastric microbiota is more austere in terms of microbial breadth and depth compared to the intestinal microbiota, future studies should focus on assessing whether the composition of the gastric microbiome in different anatomical regions of the stomach exerts differential effects on cancer risk. This could be done through site-specific topographical mapping of the microbiota in the presence or absence of H. pylori as well as assessing differences in relation to different disease states along the gastric carcinogenesis cascade. Clearly, longitudinal studies that utilize sequential sampling to elucidate the temporal nature of microbial associations with premalignant lesions are needed. Details regarding patient populations, including age, gender, diet, and other comorbidities need to be assessed and compared in a rigorous fashion to discern whether any of these variables affect the potential for the gastric microbiome to influence disease. Since studies of the gastric microbiota have largely focused on bacterial communities, more in depth studies elucidating effects of other microorganisms that potentially populate the stomach in addition to bacteria, including fungi, protists, archaea, and viruses, are needed to fully characterize the gastric microbiome and its relationship with cancer risk. Furthermore, to more definitively determine cause versus effect, studies may also need to incorporate humanized mouse models to discern effects of the human gastric microbiome on disease. As we begin to understand and elucidate the specific role of the gastric microbiota and its effects on human health and disease, studies of these microbial populations in innovative systems will likely yield translational opportunities to reduce gastric cancer morbidity and mortality by improving screening, prevention, and treatment. It is tempting to speculate that future studies will identify specific combinatorial populations of bacteria that are predictive of pathologic outcomes, yielding strategies to manipulate the microbiota to ultimately prevent disease.
Funding Statement
The authors would like to acknowledge support from the following NIH grants: R01CA077955 (RMP), R01DK058587 (RMP), P01CA116087 (RMP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210 . [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–10. doi: 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–14. doi: 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–7. doi: 10.1126/science.1223813 . [DOI] [PubMed] [Google Scholar]
- 5.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–30. doi: 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13(11):800–12. doi: 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22(22):5574–81. doi: 10.1158/1078-0432.CCR-16-1786 . [DOI] [PubMed] [Google Scholar]
- 8.Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C, et al. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol. 2014;20(4):908–22. doi: 10.3748/wjg.v20.i4.908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15(3):317–28. doi: 10.1016/j.chom.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014;20(4):859–67. doi: 10.1158/1078-0432.CCR-13-1343 . [DOI] [PubMed] [Google Scholar]
- 12.Wang LL, Yu XJ, Zhan SH, Jia SJ, Tian ZB, Dong QJ. Participation of microbiota in the development of gastric cancer. World J Gastroenterol. 2014;20(17):4948–52. doi: 10.3748/wjg.v20.i17.4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103(3):732–7. doi: 10.1073/pnas.0506655103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3(7):e2836 doi: 10.1371/journal.pone.0002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5(4):574–9. doi: 10.1038/ismej.2010.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009;58(Pt 4):509–16. doi: 10.1099/jmm.0.007302-0 . [DOI] [PubMed] [Google Scholar]
- 17.Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep. 2014;4:4202 doi: 10.1038/srep04202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley DR, Sieber KB, Robinson KM, White JR, Ganesan A, Nourbakhsh S, et al. Bacteria-human somatic cell lateral gene transfer is enriched in cancer samples. PLoS Comput Biol. 2013;9(6):e1003107 doi: 10.1371/journal.pcbi.1003107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. 2016;28(3):261–6. doi: 10.1097/MEG.0000000000000542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19(6):407–16. doi: 10.1111/hel.12145 . [DOI] [PubMed] [Google Scholar]
- 21.Leung A, Tsoi H, Yu J. Fusobacterium and Escherichia: models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev Gastroenterol Hepatol. 2015;9(5):651–7. doi: 10.1586/17474124.2015.1001745 . [DOI] [PubMed] [Google Scholar]
- 22.Mowat C, Williams C, Gillen D, Hossack M, Gilmour D, Carswell A, et al. Omeprazole, Helicobacter pylori status, and alterations in the intragastric milieu facilitating bacterial N-nitrosation. Gastroenterology. 2000;119(2):339–47. . [DOI] [PubMed] [Google Scholar]
- 23.Sanduleanu S, Jonkers D, De Bruine A, Hameeteman W, Stockbrugger RW. Non-Helicobacter pylori bacterial flora during acid-suppressive therapy: differential findings in gastric juice and gastric mucosa. Aliment Pharmacol Ther. 2001;15(3):379–88. . [DOI] [PubMed] [Google Scholar]
- 24.Ma JL, Zhang L, Brown LM, Li JY, Shen L, Pan KF, et al. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality. J Natl Cancer Inst. 2012;104(6):488–92. doi: 10.1093/jnci/djs003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson KM, Crabtree J, Mattick JS, Anderson KE, Dunning Hotopp JC. Distinguishing potential bacteria-tumor associations from contamination in a secondary data analysis of public cancer genome sequence data. Microbiome. 2017;5(1):9 doi: 10.1186/s40168-016-0224-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolig AS, Cech C, Ahler E, Carter JE, Ottemann KM. The degree of Helicobacter pylori-triggered inflammation is manipulated by preinfection host microbiota. Infect Immun. 2013;81(5):1382–9. doi: 10.1128/IAI.00044-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140(1):210–20. Epub 2010/10/19. doi: 10.1053/j.gastro.2010.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63(1):54–63. doi: 10.1136/gutjnl-2013-305178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee CW, Rickman B, Rogers AB, Muthupalani S, Takaishi S, Yang P, et al. Combination of sulindac and antimicrobial eradication of Helicobacter pylori prevents progression of gastric cancer in hypergastrinemic INS-GAS mice. Cancer Res. 2009;69(20):8166–74. doi: 10.1158/0008-5472.CAN-08-3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemke LB, Ge Z, Whary MT, Feng Y, Rogers AB, Muthupalani S, et al. Concurrent Helicobacter bilis infection in C57BL/6 mice attenuates proinflammatory H. pylori-induced gastric pathology. Infect Immun. 2009;77(5):2147–58. doi: 10.1128/IAI.01395-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge Z, Feng Y, Muthupalani S, Eurell LL, Taylor NS, Whary MT, et al. Coinfection with Enterohepatic Helicobacter species can ameliorate or promote Helicobacter pylori-induced gastric pathology in C57BL/6 mice. Infect Immun. 2011;79(10):3861–71. doi: 10.1128/IAI.05357-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai S, Mimuro H, Yamada T, Baba Y, Moro K, Nochi T, et al. Role of Peyer's patches in the induction of Helicobacter pylori-induced gastritis. Proc Natl Acad Sci U S A. 2007;104(21):8971–6. Epub 2007/05/16. doi: 10.1073/pnas.0609014104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. Br J Cancer. 2016;114(3):237–42. doi: 10.1038/bjc.2015.465 [DOI] [PMC free article] [PubMed] [Google Scholar]