Abstract
Pulmonary emphysema overlaps considerably with chronic obstructive pulmonary disease (COPD), and is traditionally subcategorized into three subtypes: centrilobular emphysema (CLE), panlobular emphysema (PLE) and paraseptal emphysema (PSE). Automated classification methods based on supervised learning are generally based upon the current definition of emphysema subtypes, while unsupervised learning of texture patterns enables the objective discovery of possible new radiological emphysema subtypes. In this work, we use a variant of the Latent Dirichlet Allocation (LDA) model to discover lung macroscopic patterns (LMPs) in an unsupervised way from lung regions that encode emphysematous areas. We evaluate the possible utility of the LMPs as potential novel emphysema subtypes via measuring their level of reproducibility when varying the learning set and by their ability to predict traditional radiological emphysema subtypes. Experimental results show that our algorithm can discover highly reproducible LMPs, that predict traditional emphysema subtypes.
Index Terms: CT, Lung, Emphysema, COPD, LDA, unsupervised learning, texture, classification
1. INTRODUCTION
In computer vision, topic models are popular tools capable of explaining complex macro-structures contained in an image via the use of “visual words” (i.e. visual primitives) [1, 2]. Topics (e.g., a human face) are defined via the learning of common co-occurence of visual words (e.g., two eyes, a nose, and a mouth) in documents (e.g., a set of portrait images). Visual topics convey very rich structural information that is able to extract the essence of image data content toward image interpretation and detection of more abstract visual concepts. Visual topics have been applied to medical image analysis for image-type classification and image retrieval [3]. Discovery of topics may also benefit disease subtypes discovery [4], and disease phenotyping.
In the context of radiological emphysema subtypes, which were initially defined at autopsy and have poor inter-rater agreement even in expert hands [5], we propose a learning framework to discover in a set of training CT scans (analogous to documents) some lung macroscopic patterns (LMPs, analogous to topics) of lung texture prototypes (LTPs, analogous to visual words) to encode emphysema subtypes. As introduced in [6], topics are learned with a probabilistic “mixed-membership” model called the Latent Dirichlet Allocation (LDA), which enables multiple topics per document. We assume that each subject is a mixture of multiple emphysema subtypes that can be explained by LMPs, and each LMP is closely related to a specific set and proportion of LTPs.
2. METHOD
This section is organized as follows: 1) Preprocessing (lung and emphysema segmentation and LTP labeling); 2) Unsupervised discovery of LMPs; and 3) Evaluation metrics of LMPs.
2.1. Emphysema Segmentation and LTP labeling
Following our previous works on quantifying emphysema subtypes on CT scans [7, 8] we pre-analyze each CT scan and generate two masks: the emphysema mask within the lung via HMMF regularized segmentation [7], and a LTP label mask, via assignment of 3D patches to the most similar LTP within a set of 100 LTPs generated using texton features [8].
2.2. Unsupervised Lung Macro Pattern (LMP) discovery
We assume that our dataset of M CT scans (documents) is generated from Nlmp LMPs (topics) using Nltp LTPs (words). LTP label is the observed variable, while the structure of the LMPs and their occurence in a CT scan are the two hidden (unobserved) variables. Discovering of the LMP topics is solved as the maximization of the posterior probabilities of the hidden variables given the observations. LDA is used as the probabilistic generative process of the observed data. Concretely, a LMP i is equipped with a probability distribution φ over the LTPs, using a Dirichlet with parameter β: φi ~ Dir(β) and a CT scan j is equipped with a probability distribution θ of LMPs, using a Dirichlet with parameter α: θj ~ Dir(α). α and β are Dirichlet prior hyperparameters.
The joint distribution of observed LTP and hidden LMP random variables is written as [6]:
| (1) |
where LTPj,t is the observed value of LTP t in document j. φi, θj and LMPj,t are hidden variables to be inferred. The generative LDA process is described as follows:
For a LMP i, a multinomial parameter φi is sampled from Dirichlet prior φi ~ Dir(β);
For a scan j, a multinomial parameter θj over the Nlmp LMPs is sampled from Dirichlet prior θj ~ Dir(α).
For a LTP t in scan j, its LMP is sampled from discrete distribution LMPj,t ~ Multinomia(θj).
The value LTPj,t of LTP t in scan j is sampled from the distribution of LMPj,t, LTPj,t~Multinomia(φLMPj,t).
Learning these various distributions (the set of LMPs and their associated LTP probabilities) is a Bayesian inference problem. As in [9], we used the Gibbs Sampling approach to maximize Eq. 1.
2.2.1. Localized LDA model
We perform LMP learning on local regions of interests rather than the whole lung. Each scan is quantized into a M* × Nltp-dimensional vector, where M* is the number of (local) document-level regions of interest (DROI) per scan. We chose overlapping DROIs of size between 1.5 and 4 times the patch size used for LTP labeling (PROI = 25×25×25 mm), and extracted an average of 50 DROIs per scan to achieve a balance between lung volume coverage and computational complexity of LDA. To infer emphysema-like topics, we only use the DROIs which have at least 1% of overlap with the HMMF emphysema mask for generating the LMPs.
2.2.2. Setting the number of LMPs
A common problem with LDA-based topic discovery models is that they do not guarantee global optimality, which can lead to instability and lack of reproducibility. To tackle this problem, we follow [10] which improved LDA for “community detection in networks” using the Infomap [11] graph partition to initialize the number of topics. Infomap is able to find stable clusters given a similarity graph using information compression technique. In contrast to standard unsupervised clustering algorithms, Infomap does not require the user to guess the number of clusters.
After extracting training ROIs at a given DROI size, we use Infomap to set Nlmp as follows: For each LTP, we generate the histogram of their frequency of occurrence over all the DROIs. We calculate the histogram intersections [12] as the measure of similarity of each pair of LTPs that co-appear in at least one DROI. To enforce sparsity in the similarity graph, we threshold similarity values below a threshold T. We then used Infomap to infer Nlmp based on this similarity graph. This sets the number of topics to discover and best guess of word composition of each topic. We then refine the estimation of the topics solving the LDA using Gibbs Sampling [9] to get the final LMPs. The hyper-parameters α and β were empirically set to 0.5 and 0.01. The threshold T is tested from 0 to 1 with 0.1 increments and we retain the maximal obtained value Nlmp to initialize Gibbs Sampling.
2.3. Evaluation metrics of LMPs
We evaluate the discovered LMPs based on their reproducibility over training sets and association with traditional radiological subtypes.
Given two sets of LMPs (topics) C and C′ learned on two distinct training sets, we propose the following metric to measure their reproducibility R:
| (2) |
where sim(Ck, C′πk) measures the similarity between LMPs Ck and and π denotes the permutation of indices leading to optimal matching of compared LMPs, using the Hungarian method [13]. We define two metrics for the similarity measurement, simA for measuring common LTP components in LMPs Ck and , and simB for measuring overlap of LMP labeling on a common test set, as follows:
| (3) |
where 1 is a column vector with all 1s (Ck1 = sum(Ck)).
| (4) |
where I is the 0–1 loss function, and LCk denotes LMP label masks using the LMPs Ck.
Noting MROI the ROIs used to label CT scans with LMPs, their size should not exceed the size of DROI and not be smaller than the size of PROI. We tested MROI of size between 1 and 3 times the PROI size. For each MROI, we get a normalized histogram of LTPs, and calculate its similarity with the LMPs composition using Euclidean distance. Each MROI is assigned to the LMP with the highest similarity.
The association of LMPs with traditional emphysema subtypes is measured as the intraclass correlation (ICC) of the percentage of emphysema subtypes (CLE, PLE, PSE) and non-emphysema (NE) between the visually assessed ground-truth reported in [5] and the predicted values from LMPs using a constrained multivariate regression [8].
3. EXPERIMENTS
Quality of generated topics was measured via: (1) measure of reproducibility of LMPs learned in two distinct training sets. (2) visual inspection of LMP samples; (3) Ability of LMPs to predict the traditional radiological emphysema subtypes. We performed two sets of experiments which are now described.
3.1. Synthetic Data
We first tested our algorithm on synthetic data generated as follows:
Generate a set of M =300 documents with N = 1000 words per document. There are Nltp=100 vocabularies.
Choose θi ~Dir(α), where i ∈{1, …, M} and Dir(α) is a Dirichlet distribution with α= 0.1;
Choose φk~Dir(β), where k∈{1, …,Nlmp} and β=0.01;
Use φ to generate a set of K = 3, 5, 10 topics made of vocabularies.
-
Use θi to generate a set of M = 300 documents with N words per document. Generation of the synthetic data is performed as follows: For each word position i, j, where j ∈ {1, …, N}, and i ∈ {1, …, M}:
Choose a topic zi,j ~ Multinomial(θi);
Choose a word wi,j ~ Multinomial(φzi,j).
We measured the similarity of the composition of the topics discovered by the LDA algorithm (using the known number of topics and random initialization) and the ground truth synthetic topics using the similarity metric R (using simA). The results for RK where K is the number of topics to discover, are: R3 = 1, R5 = 1, R10 = 0.99.
3.2. MESA COPD Study
We used N=203 out of 317 CT scans from the MESA COPD Study [5] by excluding the scans without emphysema. MESA COPD Study were acquired at full inspiration with either a Siemens 64-slice scanner or a GE 64-slice scanner, and reconstructed using B35/Standard kernels with axial resolutions within the range [0.58, 0.88]mm, and 0.625mm slice thickness. All CT scans were visually labeled by expert radiologists in [5] into global extents of each of the three traditional radiological emphysema subtypes over the lung volume.
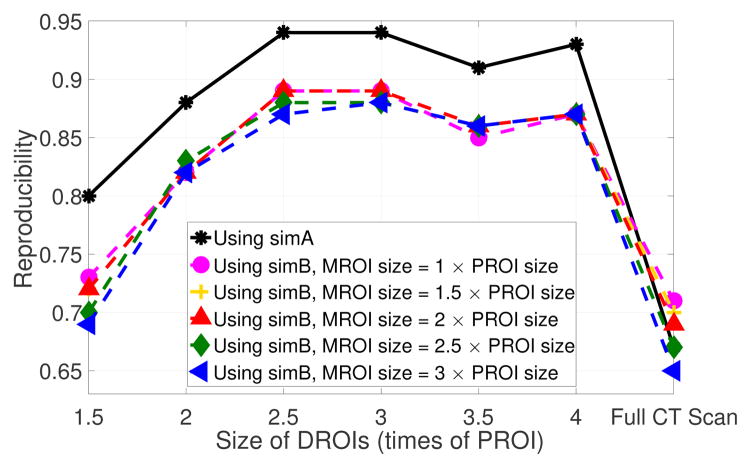
We split the study into three parts with 68, 68, 67 CT scans each, using the first two subsets as independent training sets and the third as a test set. We trained on the first two sets to generate two sets of LMPs and obtained Nlmp= [10, 8, 6, 6, 6, 6, 7] and Nlmp=[9, 7, 6, 6, 6, 6, 6] LMPs in each training run, using DROI sizes of [1.5,2,2.5,3,3.5,4,∞] times the LTP labeling patch ROI (PROI) size and where the ∞ means that the full lung volume field was used. Reproducibility of the learned LMPs is illustrated in Fig. 1. From the results using simA, we can see that highest reproducibility is achieved for the learned LMPs with DROI size (62.5 × 62.5 × 62.5)mm and (75 × 75 × 75)mm. In general, the LMPs learned on the full scan were not as good as those learned on DROIs. This verifies that spatial information is helpful for learning LMPs. From the results using simB, reproducibility follows a similar trend as with simA in terms of DROI size effect: the highest reproducibility is achieved with DROI size (62.5 × 62.5 × 62.5)mm and (75 × 75 × 75)mm. On the other hand, the size of MROI has no significant effect on the reproducibility of LMP labeling.
Fig. 1.
Reproducibility of LMPs with different DORIs sizes, and the reproducibility of labeling MROIs with different DROIs sizes in the validation set (N=67).
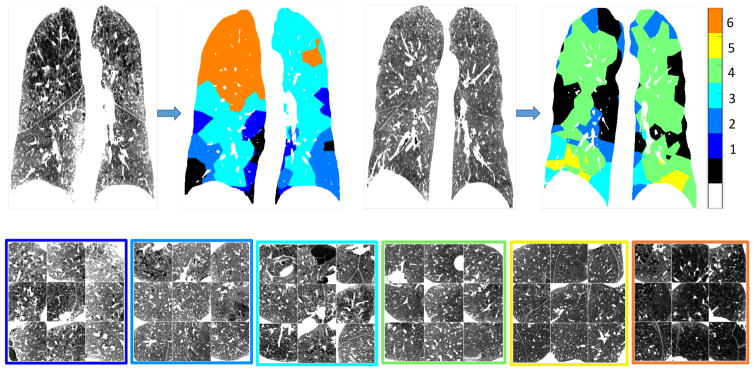
For visual illustration, we show in Fig. 2 some randomly selected MROIs (size = 75×75×75 mm) for each of the six LMPs learned from the first training set with DROI size=75×75×75 mm. These illustrations show that the MROIs are generally homogeneous within a LMP, and visually distinct between LMPs.
Fig. 2.
Examples of LMPs from 1 to 6. In the legend, ‘white’ is the background, ‘black’ is the area not covered by emphysema mask, and the other 6 colors are the 6 LMPs. Each MROI has the size of (75 × 75 × 75)mm.
Finally, we used the six generated LMPs, from the first training set with DROI size=75 × 75 × 75 mm, to predict the traditional radiological emphysema subtypes using a constrained multivariate regression [8]. We conducted three-fold cross-validations on the whole cohort to evaluate the ICC against visual labeling by radiologists, and the results are reported in Table 1. The 6-dimensional LMPs predicted traditional radiological subtypes better than the radiologist interrater reproducibility for CLE, PLE and NE. For PSE, however, the ICC of LMPs was not satisfactory. To investigate this issue, we augmented the LMPs by visually picking up the two LTPs within the disease LTPs (most occuring in disease subjects) that looked the most like PSE. Adding these 2 PSE-like LTPs to the LMPs corresponds to forcing the topic discovery process to consider these two LTPs as being topics on their own. The resultant ICC for PSE increased to 64%.
Table 1.
ICC of predicted of visual radiological subtypes. Three-fold cross validation on MESA COPD Study with 203 subjects.
| Method | CLE | PLE | PSE | NE |
|---|---|---|---|---|
| LMPs | 0.85 | 0.65 | 0.23 | 0.89 |
| Augmented LMPs | 0.86 | 0.65 | 0.64 | 0.89 |
| LTPs | 0.92 | 0.72 | 0.69 | 0.95 |
| Inter-rater | 0.74 | 0.59 | 0.67 | 0.76 |
4. DISCUSSION & CONCLUSION
In this paper, we have shown that topic discovery via Infomap and LDA can generate up to six highly reproducible emphysema-specific lung macroscopic patterns (LMPs) from a series of 100 pre-learned lung texture prototypes (LTPs), which are associated to traditional radiological emphysema subtypes. The PSE emphysema subtype was not properly discovered by the algorithm but easily added to the LMPs via picking up two visually-compatible LTPs. In future work, we will investigate how to add constraints on the LDA topic discovery process to preserve rare but important LTPs, such as PSE. In the longer term, we will also explore whether LMPs can be used to guide the discovery of novel clinical emphysema subtypes.
Acknowledgments
Funding provided by NIH/NHLBI R01-HL121270, R01- HL077612, RC1-HL100543, R01-HL093081 and N01-HC095159 through N01-HC-95169, UL1-RR-024156 and UL1-RR-025005.
References
- 1.Wang Hongxing, Zhao Gangqiang, Yuan Junsong. Visual pattern discovery in image and video data: a brief survey. Wiley Interdisc Rew.: Data Mining and Knowledge Discovery. 2014;4(1):24–37. [Google Scholar]
- 2.Zhang Quanshi, Song Xuan, Shao Xiaowei, Zhao Huijing, Shibasaki Ryosuke. Object discovery: Soft attributed graph mining. IEEE Trans Pattern Anal Mach Intell. 2016;38(3):532–545. doi: 10.1109/TPAMI.2015.2456892. [DOI] [PubMed] [Google Scholar]
- 3.Hofmanninger Johannes, Krenn Markus, Holzer Markus, Schlegl Thomas, Prosch Helmut, Langs Georg. Unsupervised identification of clinically relevant clusters in routine imaging data. MICCAI. 2016:192–200. [Google Scholar]
- 4.Binder Polina, Batmanghelich Nematollah Kayhan, Estépar Raúl San José, Golland Polina. Unsupervised discovery of emphysema subtypes in a large clinical cohort. MICCAI workshop on MLMI. 2016:180–187. doi: 10.1007/978-3-319-47157-0_22. [DOI] [PMC free article] [PubMed]
- 5.Smith Benjamin M, Austin John HM, Newell John D, D’Souza Belinda M, Rozenshtein Anna, Hoffman Eric A, Ahmed Firas, Graham Barr R. Pulmonary emphysema subtypes on computed tomography: the mesa copd study. The American journal of medicine. 2014;127(1):94–e7. doi: 10.1016/j.amjmed.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blei David M, Ng Andrew Y, Jordan Michael I. Latent dirichlet allocation. NIPS. 2001:601–608. [Google Scholar]
- 7.Häme Yrjö, Angelini Elsa D, Hoffman Eric A, Graham Barr R, Laine Andrew F. Adaptive quantification and longitudinal analysis of pulmonary emphysema with a hidden markov measure field model. IEEE transactions on Medical Imaging. 2014;33(7):1527–1540. doi: 10.1109/TMI.2014.2317520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Jie, Angelini Elsa D, Smith Benjamin M, Austin John HM, Hoffman Eric A, Bluemke David A, Graham Barr R, Laine Andrew F. Explaining radiological emphysema subtypes with unsupervised texture prototypes: Mesa copd study. MICCAI workshop on Medical Computer Vision: Algorithms for Big Data; 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths Thomas L, Steyvers Mark. Finding scientific topics. Proceedings of the National academy of Sciences. 2004;101(suppl 1):5228–5235. doi: 10.1073/pnas.0307752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancichinetti Andrea, Irmak Sirer M, Wang Jane X, Acuña Daniel E, Körding Konrad P, Nunes Amaral Luís A. A high-reproducibility and high-accuracy method for automated topic classification. CoRR. 2014 abs/1402.0422. [Google Scholar]
- 11.Rosvall Martin, Bergstrom Carl T. Maps of random walks on complex networks reveal community structure. Proceedings of the National Academy of Sciences. 2008;105(4):1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharjee Pijush Kanti. Integrating pixel cluster indexing, histogram intersection and discrete wavelet transform methods for color images content based image retrieval system. International Journal of Computer and Electrical Engineering. 2010;2(2):345. [Google Scholar]
- 13.Kuhn Harold W. 50 Years of Integer Programming 1958–2008. Springer; 2010. The hungarian method for the assignment problem; pp. 29–47. [Google Scholar]