SUMMARY
Targeted cancer therapies that use genetics are successful, but principles for selectively targeting tumor metabolism that is also dependent on the environment remain unknown. We now show that differences in rate-controlling enzymes during the Warburg Effect (WE), the most prominent hallmark of cancer cell metabolism, can be used to predict a response to targeting glucose metabolism. We establish a natural product, koningic acid (KA), to be a selective inhibitor of GAPDH, an enzyme we characterize to have differential control properties over metabolism during the WE. With machine learning and integrated pharmacogenomics and metabolomics, we demonstrate that KA efficacy is not determined by the status of individual genes, but by the quantitative extent of the WE leading to a therapeutic window in vivo. Thus, the basis of targeting of the WE can be encoded by molecular principles that extend beyond the status of individual genes.
eTOC Blurb

Liberti et al. use metabolic control analysis and multi-omics approaches to show that the enzyme GAPDH is rate limiting for the Warburg effect in cancer cells. They identify a therapeutic window where partial GAPDH inhibition is more selective for highly glycolytic tumors, highlighting how metabolism is an integral part of precision medicine.
INTRODUCTION
The general approach in cancer research is to define and then exploit the molecular requirements that distinguish tumor from normal. Targeted cancer therapies have used genomic alterations and differences in gene expression as biomarkers to predict therapeutic efficacy with great success. This strategy is currently the major paradigm within the current concept of precision medicine with each one’s disease having a specific molecular profile that determines the appropriate course of action (Collins and Varmus, 2015). In contrast, the determinants of therapies that target metabolism for the treatment of complex diseases, such as diabetes, do not depend on the status of a single gene because the principles that determine a specific response to alterations in metabolism are multifaceted convolutions of both genetic and environmental factors (Vander Heiden and DeBerardinis, 2017). Furthermore, predictive models for targeting cancer metabolism have not yet been successfully developed from the mutational or expression status of individual genes (Vander Heiden and DeBerardinis, 2017). Therefore, substantial challenges, especially regarding the appropriate populations that should be investigated, have limited advances in therapies that target cancer metabolism (Garber, 2016).
The most prominent hallmark of cancer metabolism is the Warburg Effect (WE) or aerobic glycolysis, which is defined by the increased uptake of glucose and incomplete oxidation or fermentation to lactate in the presence of oxygen. The WE has been extensively studied over the years and is routinely exploited clinically as a diagnostic of tumor burden with predictive features of cancer outcome (Liberti and Locasale, 2016; Vander Heiden, 2013). There are a number of strategies that have been proposed to target the WE including targeting oncogenes and tumor suppressor genes that interact with glucose metabolism and the specific isoforms of glycolytic enzymes that are expressed in cancer (Hay, 2016). However, full target inhibition, and thus complete ablation of an enzyme involved in metabolizing glucose, encounters limiting toxicity as glucose metabolism is required in nearly every mammalian tissue. Therefore, partial inhibition of glycolytic enzyme activity with selectivity against tumors is needed. Whether this specificity is possible and the strategy needed to define it is unknown.
An alternative approach to developing a predictive model for targeting cancer metabolism is to exploit differential properties of network activity (e.g. metabolic flux). In this scenario, the glycolytic pathway would be affected by a perturbation to one of its enzymes during conditions that promote the WE and the same perturbation would leave glucose metabolism lesser affected in conditions of normal physiology. This concept of synthetic lethality has been developed in cancer biology by identifying genetic events that create liabilities but a lesser-studied, if not novel approach to identify these vulnerabilities, is to use the quantitative properties encoded in the kinetics and thermodynamics of the metabolic network that are different during the WE. It is known that the level of control that a metabolic enzyme exerts on a metabolic flux depends on properties other than the expression or activity of the given enzyme. Thus, a change in enzyme activity could result in a different effect on metabolic output depending on other properties of the metabolic network. To our knowledge, this concept has not been explored in cancer therapy.
There are generally thought to be three rate-controlling enzymes in glycolysis: hexokinase, phosphofructokinase, and pyruvate kinase (Lehninger et al., 2013). However metabolic control analysis (MCA) (Fell, 1992), a quantitative approach to evaluate the amount of enzyme activity that is required to alter the overall output of a pathway, has suggested that the rate-control may be different during the WE (Shestov et al., 2014). Furthermore, MCA has proven invaluable in designing and controlling metabolic pathways for biotechnology applications (Bowden, 1999), but has not to our knowledge been explored in biomedical settings. We therefore reasoned that MCA, by identifying differences in biochemical regulation occurring at the systems or network level of the glycolytic pathway (i.e. flux control), could allow for selective targeting of the WE, thus identifying predictors for therapeutic response that are encoded within metabolism. In the present study, we sought to test this hypothesis.
RESULTS
Thermodynamic and kinetic analysis of rate-control in glycolysis
Based on our previously published mathematical model of glycolysis (Shestov et al., 2014) we performed metabolic control analysis (Heinrich and Schuster, 1996) on lactate production flux and evaluated thermodynamics for each step in glycolysis by calculating flux control coefficients (FCCs) and reaction free energies (ΔGs) (Figure 1A). While these models have limitations due to our incomplete knowledge of glycolysis, they can allow for discovery with further appropriate validation. Steps with higher FCCs and lower ΔGs are known to exert stronger control over flux through the pathway (Noor et al., 2014). We compared the FCCs of each enzymatic step in glycolysis to their ΔGs (Figure 1B). Two of the canonical rate-limiting enzymes of glycolysis, hexokinase (HK) and phosphofructokinase (PFK) exhibited high FCCs and low ΔGs as expected. However, a third glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), also displayed a high FCC and low ΔG, indicating that GAPDH also may be a rate-controlling enzyme of glycolysis in some settings.
Figure 1. Thermodynamic and kinetic analysis of rate-control in glycolysis.
(A) Glycolysis pathway with metabolites (black) and enzymes (blue). Model for flux control coefficients (FCCs) and reaction free energy (ΔG) values. f= flux.
(B) Median FCCs vs. median ΔGs of glycolytic enzymes. HK, hexokinase; PFK, phosphofructokinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
(C) Schematic showing flux inputs and outputs during Non-Warburg Effect and Warburg Effect conditions.
(D) Relationship between FCC values of GAPDH, HK, and PFK and extent of the Warburg Effect defined as ratio of lactate production flux to oxidative phosphorylation flux.
(E) Schematic demonstrating the high rate-limiting effect of GAPDH over cells carrying out the Warburg Effect compared to those undergoing oxidative phosphorylation. GA3P, glyceraldehyde-3-phosphate; 1,3-BPG, 1,3-bisphosphoglycerate.
To study relationship between the WE and control of glycolysis, we tuned the model to exhibit varying extents of the WE by altering activities of glycolytic enzymes with the highest variation in protein abundance (Figure 1C). Although HK, PFK, and GAPDH all exhibit considerable control over flux in glycolysis, only the value of GAPDH FCC increases during the WE (Figure 1D). Therefore, inhibiting GAPDH activity exerts larger reduction in glycolytic flux in cells undergoing a higher degree of the WE than in cells with lower glycolytic activity (Figure 1E), thus providing a rationale for exploring the control of glycolytic rate by GAPDH as a target against the WE.
Comparative metabolomics nominates a specific GAPDH inhibitor
We next evaluated the cytotoxic activities of several putative GAPDH inhibitors including arsenate, arsenic trioxide, 3-bromopyruvate, iodoacetate, and koningic acid (KA) (Campbell-Burk et al., 1987; Dai et al., 1999; Endo et al., 1985; Ganapathy-Kanniappan et al., 2009; Németi and Gregus, 2005), and found substantial variability (Figures S1A) indicating that they have different pharmacological activities and possibly act by different mechanisms. To further investigate this likely possibility, we generated dose-dependent metabolite profiles in HCT116 cells comprising over 350 metabolites and found that each profile exhibited differences in the levels of glycolytic metabolites and related pathways (Figures 2A–2E). However, with the exception of KA, all other profiles exhibited many other perturbations to metabolism as well. These effects were quantified using a network-based pathway analysis (Methods) that revealed glycolysis as the highest scoring pathway only in KA-treated cells (Figure 2F, 2G). Consistent with this finding, a docking analysis indicates that KA can directly bind to the active site of human GAPDH (Figure 2H).
Figure 2. Comparative metabolomics nominates a specific GAPDH inhibitor.
(A–E) Clustered heatmaps with pathway and dose annotations of dose-dependent global metabolic responses to putative GAPDH inhibitors.
(A) Arsenate (As2O4).
(B) Arsenic trioxide (As2O3).
(C) 3-bromopyruvate (3BP).
(D) Iodoacetate (IA).
(E) Koningic acid (KA).
(F) Network-based pathway analysis for each compound.
(G) Pathway analysis showing top 4 highest scoring pathways in response to KA.
(H) KA docking analysis to GAPDH active site Cys152.
At the concentration corresponding to the IC50 value of KA (Figure S1A), GAPDH activity decreased compared to vehicle (Figure S1B). To examine how KA-induced decreases in GAPDH activity exert cytotoxic effects, we evaluated the response at the IC50 using a volcano plot (Figure S1C), and confirmed that glycolytic metabolites were predominantly affected with an accumulation of glycolytic intermediates upstream of GAPDH occurring in a dose-dependent manner (Figure S1D). To analyze the consequences of glucose metabolism imbalance, we considered a kinetic flux profiling experiment (Methods) using uniformly labeled (U-13C)-glucose in HCT116 cells treated at the IC50 over 4 hours. As expected, a reduction of 13C-lactate in KA treated cells was observed, indicating decreased flux through the glycolytic pathway (Figure S1E). Relative levels of pentose phosphate pathway metabolites increased in the presence of KA and flux was disrupted (Figures S1F, S1G), as was flux into glycerol metabolism (Figure S1H). In addition, KA also reduces fluxes in pathways branching from glycolysis downstream of the GAPDH step, including de novo serine (Figure S1I), entry into the tricarboxylic acid cycle (Figures S1J), and palmitate, the product of de novo lipogenesis (Figure S1K). Dose-dependent changes in energy status (Figures S1L-S1N) and redox status (Figure S1O–S1T), both essential functions of glucose metabolism, were also affected. Together these findings indicate that KA induces global alterations in the metabolic network most consistent with direct targeting of GAPDH with a reduction in glycolytic rate and cytotoxicity likely due to the simultaneous effects on numerous metabolic pathways and functions.
Expression of a fungal-derived KA-resistant GAPDH allele renders human cells completely resistant to KA and reverses their metabolic profile
Notably, the cytotoxic effects of targeting metabolism can often be rescued by supplementation of nutrients that restore the defective metabolic functions, as is the case of metformin and mitochondrial metabolism (Liu et al., 2016). We thus attempted to restore cell viability by modulating nutrient availability, but were unable to rescue cytotoxicity induced by KA (Figures S2A-S2E). To determine whether other mechanisms and thus possible off-target effects may account for KA cytotoxicity, we considered alternative approaches to determine whether GAPDH is the mechanistic target of KA. Notably, KA is a natural product obtained from the Trichoderma fungus, which thrives in anaerobic environments rich in sugar. When encountering microbes that compete for its carbon source, it secretes KA as an antibiotic to eliminate these organisms (Sakai et al., 1990; Watanabe et al., 1993), while expressing a resistant allele of GAPDH (T. koningii KAr-GAPDH) (Figure 3A). Thus, we cloned the T. koningii KAr-GAPDH and expressed it in human HEK293T and HCT116 cells. After verifying that human cells can express T. koningii KAr-GAPDH (Figure 3B, S2F), we observed that HEK293T cells expressing T. koningii KAr-GAPDH exhibited complete cell viability (Figure 3C) and HCT116 cells expressing T. koningii KAr-GAPDH exhibited almost complete viability (Figure S2G) after treatment with 0–200μM KA. These results further demonstrated the specificity of KA towards GAPDH. In addition, while T. koningii KAr-GAPDH displays similarity to the active site of GAPDH with conservation of the reactive cysteine, it exhibits evolutionary divergence (Figures S2H-S2J) from mammalian GAPDH suggesting that acquiring resistance by mutating individual GAPDH residues is difficult.
Figure 3. Expression of a fungal-derived KA-resistant GAPDH allele renders human cells completely resistant to KA and reverses their metabolic profile.
(A) Schematic showing expression of a resistant allele of GAPDH by Trichoderma virens.
(B) Immunoblotting of parental, EV or KAr-GAPDH expressing HEK293T cells.
(C) Cell viability of HEK293T cells expressing KAr-GAPDH or EV (top left). Representative images of well (bottom left, top right) KAr-GAPDH or EV expressing cells treated with vehicle (0 μM) or KA (200μM).
(D) Volcano plots showing metabolite profiles of HEK293T cells expressing EV compared to those expressing KAr-GAPDH after treatment with DMSO or 5μM KA. Log2 fold change versus −log10 (p-value). Dotted lines along x-axis represent ±log2(2) fold change and dotted line along y-axis represents −log10(0.05). Glycolysis metabolites shown as red points. All other metabolites are black points.
(E) Glycolytic metabolite levels
(F) Pentose Phosphate Pathway levels.
(G) TCA cycle metabolite levels.
G6P, Glucose-6-Phoshate; F 1,6-BP, Fructose 1,6-Bisphosphate; DHAP, Dihydroxyacetone Phosphate; 3PG, 3-Phosphoglycerate; PEP, Phosphoenolpyruvate; S7P, Sedoheptulose-7-Phosphate; E4P, Erythrose-4-Phosphate; R5P, Ribose-5-Phosphate.
All data are represented as mean ±SEM from n = 3 biological replicates unless otherwise noted.
Since expression of T. koningii KAr-GAPDH successfully rescued cell viability in human cells treated with KA, we considered whether changes in metabolism observed in human cells treated with KA can be reversed upon KAr-GAPDH expression. After KA treatment, marked differences in metabolism in empty vector (EV) expressing cells were observed that were completely absent in T. koningii KAr-GAPDH-expressing cells (Figures 3D, S2K-S2L) and manifested in differential changes in the levels of glycolytic intermediates (Figure 3E, S2M), PPP (Figure 3F), and the TCA cycle (Figure 3G, S2N). Together, these data confirm that the mechanistic target of KA is indeed GAPDH in part by establishing that all disruptions to metabolism are ablated when cells are engineered to be resistant to KA by expressing a resistant allele of GAPDH.
The cytotoxic response to KA treatment is heterogeneous
We next measured the response to KA across a collection of 60 cancer cell lines from diverse tissue and genetic origins. At 10μM, there was a broad, but heterogeneous response to KA (Figure 4A) consistent with measured values of the IC50 for each line (Figure S3A). Consistent with earlier findings, HCT116 cells were only moderately sensitive to KA, therefore requiring a higher concentration of KA to trigger a disruption in the metabolic network. We identified three of the most resistant cell lines to KA as MCF-7, UACC-257, and NCI-H226 and their corresponding sensitive cell lines to KA as BT-549, SK-MEL-28, and NCI-H522 based on matching tissue type and subjected them to further analysis. Analysis of the cell lines considered showed that no single tissue type was more sensitive or resistant to KA (Figures S3B-S3D). We assessed whether inhibition of GAPDH activity accounts for the variability in cell line responses and found that GAPDH activity in response to KA treatment revealed little differences in the change in enzyme activity across both sensitive and resistant cells given KA at the same dose (Figures 4B, 4C). Thus, resistance to KA does not occur due to the inability to effectively inhibit GAPDH, rather it appears to occur by another mode of action.
Figure 4. The cytotoxic response to KA treatment is heterogeneous.
(A) Waterfall plot showing the difference in response of KA to 60 independent cell lines treated with vehicle (0.01% DMSO) or 10μM KA. Representative KA-resistant cell lines (Red, *) and KA-sensitive cell lines (Green, *).
(B) Relative GAPDH activity in representative KA-sensitive and resistant cell lines in response to vehicle (DMSO) or KA. SK-MEL-28 and UACC-257 were treated with vehicle or 1μM KA; NCI-H522 and NCI-H226 were treated with vehicle or 0.4μM KA; BT-549 and MCF-7 were treated with 0.7μM KA (n=2).
(C) Pearson correlation of KA IC50 values for KA-sensitive and resistant cell lines versus percent of GAPDH activity.
(D) Volcano plots showing metabolite profiles of breast cancer cell lines after treatment with DMSO or 90μM KA. Log2 fold change versus −log10 (p-value). Dotted lines along x-axis represent ±log2(2) fold change and dotted line along y-axis represents −log10(0.05). Glycolysis metabolites shown as red points. All other metabolites are black points.
(E) Melanoma cell lines as in (D).
(F) Non-small cell lung cancer cell lines as in (D).
(G) Kinetic flux profiling for lactate labeling from 13-C-glucose.
(H) Change in lactate flux in response to KA based on fold changes vs. relative fluxes of the vehicle group from 13C-lactate enrichment from U-13C-glucose.
(I) Glycolysis profiles for KA-sensitive and resistant breast, melanoma, and NSCLC cell lines treated with vehicle or their respective KA IC50 concentrations.
S7P, sedoheptulose-7-phosphate; F 1,6-BP, fructose 1,6-bisphosphate; G6P, glyceraldehyde-6-phosphate; DHAP, dihydroxyacetone phosphate; X5P, xylulose-5-phosphate; PEP, phosphoenolpyruvate; UDP-D-glucose, uridine diphosphate-D-glucose.
All data are represented as mean ±SEM from n = 3 biological replicates unless otherwise noted.
To determine whether nutrient supplementation could rescue cell viability in sensitive cells, we added serine and lactate, pyruvate, lactate, or 3PG to cells upon KA treatment (Figure S3E). Consistent with the findings in HCT116 cells (Figure S2E), nutrient supplementation was unable to rescue cell cytotoxicity. To determine whether differences in how KA affects metabolism contributes to the response, we generated metabolite profiles and found that sensitive cell lines revealed signatures nearly identical to those observed in previously characterized cell lines (Figures 2–3) whereas resistant cell lines did not show any significant metabolite alterations (Figures 4D–4F) as was also observed with kinetic flux profiling (Figures 4G–4H, S3F-S3H). Consistently, an accumulation of upper glycolytic intermediates and altered flux was observed only in sensitive cells (Figure 4I, S3I). Branching pathways were also affected whereby glucose flux decreased through TCA cycle intermediates and serine in a representative sensitive cell line (Figures S3J-S3M). Not surprisingly, overall TCA cycle metabolite levels remained unchanged, likely attributed to non-glucose carbon sources fueling the TCA cycle (Figure S3N). The metabolic changes observed in sensitive cells was confirmed using a pathway analysis and hierarchal clustering (Figures S4A-S4D). All together, these data strongly suggest that the heterogeneous response to KA observed across 60 cell lines can be attributed to the specific control that GAPDH exerts over the glycolytic pathway in certain cells, independent of the extent of enzyme inhibition and tissue type.
A multi-omics analysis reveals that only the extent of the WE predicts KA response
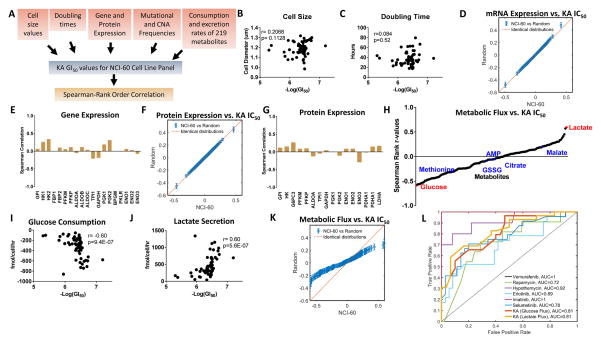
We next sought to determine if any molecular features predict KA sensitivity or response. We correlated KA IC50 values with multiple layers of physical and molecular information including cell size, doubling time, genetic status, mRNA expression, protein expression, and metabolic uptake and excretion rates that are available in 59 of the 60 cell lines (Dolfi et al., 2013; Jain et al., 2012) (Figure 5A). Neither cell size nor doubling time correlated with response to KA (Figures 5B–5C) nor did mutational status of several commonly mutated oncogenes and tumor suppressor genes including KRAS, PIK3CA, MYC, and TP53 (Figure S5A). Neither gene set enrichment analysis (GSEA) nor glycolytic enzyme expression reveal any significant association of any known gene sets to KA response (Figures 5D–5E, S5B) as was also observed when analyzing relationships with protein expression (Figures 5F, 5G). Strikingly, a comparison of metabolic uptake and excretion rates (Figure 5H) showed that glucose uptake (Spearman correlation, r = 0.60, p = 9.4 × 10−7) (Figure 5I) and lactate excretion (Spearman correlation, r = 0.60, p = 5.6 × 10−7) (Figure 5J), two parameters that quantitatively define the WE, had the highest Spearman correlation coefficients and were predictive of KA response. Moreover, we found that metabolic fluxes are significantly more correlated to KA IC50 values than to the random distribution (Figure 5K), which is not the case for mRNA and protein which appear random (Figures 5D, 5F). Therefore, of the roughly 50,000 molecular and physical variables surveyed, only glucose uptake and lactate excretion (i.e. the WE) were predictive of KA response allowing us to conclude that KA, and thus partial inhibition of GAPDH, specifically targets the WE.
Figure 5. A multi-omics analysis reveals that only the extent of the WE predicts KA response.
(A) Schematic workflow of spearman rank correlation calculations from metabolite consumption and excretion rates, cell size, doubling time, mutation and copy number alteration (CNA) frequencies, and gene and protein expression patterns with KA IC50 response across NCI-60 cell line collection.
(B) Cell size correlation with KA IC50 response across NCI-60 cell line panel.
(C) Doubling time correlation as in (B).
(D) Quantile-Quantile (Q-Q) plot comparing the quantiles from the correlation between mRNA expression levels and KA IC50 values in NCI-60 cell line screen with random values.
(E) Glycolysis gene expression correlation as in (B).
(F) Q-Q plot for protein expression levels as in (D).
(G) Glycolysis protein expression correlation as in (B).
(H) Correlations of uptake and secretion rates of metabolites.
(I) Glucose uptake correlation as in (B).
(J) Lactate secretion correlation as in (B).
(K) Q-Q plot for metabolic flux as in (D).
(L) Receiver-operating characteristic (ROC) curves of standard cancer therapies with specific targets and/or biomarkers. Area under the curve (AUC) assesses biomarker accuracy.
To address the diagnostic performance of glucose uptake and lactate secretion flux as predictive measurements of KA sensitivity, we generated receiver-operating characteristic (ROC) curves and calculated the area under the curve (AUC) (Figure 5L). Notably, we found that both glucose and lactate flux are strong predictors of KA response with an AUC=0.81, with prediction accuracy comparable to six representative targeted cancer therapies with known biomarkers. These data strongly support the clinical applicability and feasibility of using glycolytic flux, which is readily measured clinically with imaging, as predictors of KA efficacy.
KA is bioavailable and induces dynamic changes to glycolysis in vivo
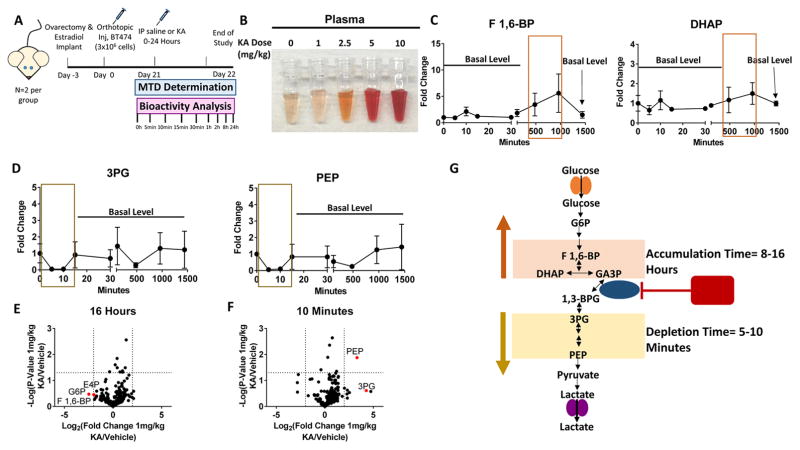
Thus far, we have established that in culture, the WE can be selectively targeted by exploiting metabolic control analysis and flux control through GAPDH. However, the effects on normal cells and whether a therapeutic window can be achieved in vivo are unclear. To begin to address these remaining questions, we first evaluated the effects of GAPDH inhibition in CD8+ cells since these cells are important in immune therapy settings. We found that KA does not have significant effects on proliferation, viability, or activation markers in activated CD8+ cells at lower concentrations (Figures S6A-S6I). We next evaluated the sensitivity of the highly glycolytic (Timmerman et al., 2013) estrogen-responsive breast cancer cell line, BT-474 to KA (IC50 =1μM) (Figure S6J). At KA concentrations corresponding to the IC50, GAPDH activity was reduced and glycolysis was altered (Figures S6K-S6M). Next, ovariectomized nu/nu mice were implanted subcutaneously with a slow-releasing 17β-estradiol pellet and BT-474 cells were injected orthotopically into the mammary fat pad (Methods). Tumors were established prior to toxicology studies (Figure 6A). 1 mg/kg KA was determined to be the maximum tolerated dose (MTD) based upon behavioral monitoring and adverse events at higher doses (hemolysis, hematuria, and anemia) (Figure 6B). To determine whether KA induces changes in glycolysis, we sacrificed mice over a time course of up to 24 hours and used metabolite profiling to examine the levels of glycolytic intermediates in upper glycolysis at different endpoints. Strikingly, fructose 1,6-bisphosphate (F1,6-BP) accumulated in tumors between 8–16 hours of treatment with 1 mg/kg KA, as did dihydroxyacetone phosphate (DHAP), albeit to a lesser extent (Figure 6C). Relative levels of 3-phosphoglycerate (3PG) and phosphoenolpyruvate (PEP), showed a decrease after 5–10 minutes of KA treatment, and recovered thereafter (Figure 6D). After 16 hours, upper glycolytic and PPP metabolites had the largest fold change increase (Figure 6E). At 10 minutes, PEP was significantly lower compared to all other detected metabolites, while 3PG had a similar trend (Figure 6F). These results indicate a dynamic effect on glycolysis whereby upper glycolytic metabolites accumulate within hours of KA treatment and lower glycolytic intermediates deplete within minutes, but then recover, likely due to a contribution of non-glucose carbon sources (Figure 6G). These data confirm that KA is bioavailable, tolerable at certain doses, and induces acute and dynamic changes in the glycolytic network in tumors.
Figure 6. KA is bioavailable and induces dynamic changes in glycolysis in vivo.
(A) Schematic of timeline and treatment regimen of E2 treated female nu/nu mice injected with BT-474 cells orthotopically. These mice were treated with either saline or 1mg/kg, 2.5mg/kg, 5mg/kg, or 10mg/kg to identify the maximum tolerated dose (MTD) for 24 hours.
(B) Plasma from MTD analysis for doses of 0–10mg/kg KA.
(C) Levels of upper glycolytic intermediates from 0–24 hours (n=2 per group).
(D) Levels of lower glycolytic intermediates as in (C).
(E) Volcano plot showing metabolic profile at 16 hour time-point. Log2 fold change versus −log10 (p-value). Dotted lines along x-axis represent ±log2(2) fold change and dotted line along y-axis represents −log10(0.05). Glycolysis and related metabolites shown as red points. All other metabolites are black points (n=2).
(F) 10 minute time-point as in (E).
(G) Schematic showing the dynamic response KA treatment.
G6P, glucose-6-phosphate; F 1,6-BP, fructose 1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GA3P, glyceraldehyde-3-phosphate; 1,3-BPG, 1,3-bisphosphoglycerate; 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate.
All data are represented as mean ±SEM from biological replicates.
A therapeutic window for precise targeting of the WE with KA can be achieved in vivo
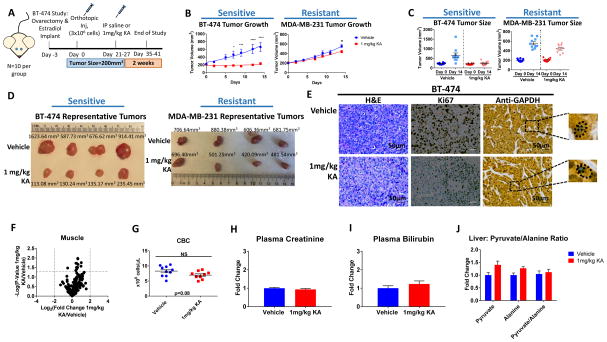
Given the bioavailability and tolerability of KA, we identified a resistant cell line to KA, MDA-MB-231 (Figures S6N-S6O) and carried out metabolite profiling in the presence of KA, in which no metabolic changes were observed in response to KA treatment (Figures S6P-S6Q). We next established two orthotopic breast cancer models in which mice were injected with either sensitive BT-474 cells or resistant MDA-MB-231 cells into the mammary fat pad and were treated daily with 1mg/kg KA, monitoring body weight and tumor growth for 14 days (Figure 7A). We determined that the circulating plasma concentration was 0.7μM KA (Figure S7A–S7C). No significant changes in body weight were observed in BT-474 tumor bearing mice (p=0.37, two-tailed student’s t-test) or MDA-MB-231 tumor bearing mice (p=0.86, two-tailed student’s t-test) indicative of drug tolerability (Figure S7D). Importantly, tumor growth in KA treated BT-474 tumor bearing mice was substantially suppressed (p<0.0001, two-way ANOVA) whereas tumor growth in KA treated MDA-MB-231 cells was only marginally suppressed (p=0.02, two-way ANOVA) over the course of 2 weeks (Figures 7B–7D). In addition, tumor morphology was markedly different in KA treated mice and cell proliferation was significantly decreased (p=4.0 × 10−5, unpaired multiple-t-test) (Figures 7E, S7E). GAPDH is suspected to have nuclear functions in cells (Barber et al., 2005; Jung et al., 2014), but remained localized to the cytoplasm, indicative of its role in glycolysis as its primary mode of action for tumor suppression upon KA treatment (Figure 7E).
Figure 7. A therapeutic window for precise targeting of the WE with KA can be achieved in vivo.
(A) Schematic of timeline and treatment of E2 treated female nu/nu mice injected with KA-sensitive BT-474 cells and female nu/nu mice injected with KA-resistant MDA-MB-231 cells orthotopically and treated with either saline or 1mg/kg KA.
(B) Average tumor volume (mm3) over 14 days in mice injected with BT-474 or MDA-MB-231 cells treated with vehicle or 1mg/kg KA (n=10 per group).
(C) Tumor volume of each individual mouse from day 0 and day 14 in the BT-474 and MDA-MB-231 tumor models (n=10).
(D) Representative BT-474 and MDA-MB-231 tumors from vehicle treated and 1mg/kg KA treated E2 treated female nu/nu mice on day 14.
(E) Representative hematoxylin and eosin (H&E) and immunohistochemical (IHC) staining of serial sections from nu/nu mice injected with BT-474 cells treated with vehicle or 1mg/kg KA.
(F) Volcano plot showing metabolic profile from skeletal muscle treated with vehicle or 1mg/kg KA from BT-474 tumor model. Log2 fold change versus −log10 (p-value). Dotted lines along x-axis represent ±log2(2) fold change and dotted line along y-axis represents −log10(0.05). Significantly different metabolites shown as red points. All other metabolites are black points (n=3).
(G) Complete blood count (CBC) for BT-474 mice treated with vehicle or 1mg/kg KA after 14 days (n=9 for vehicle group; n=10 for treatment group).
(H) Relative levels of creatinine from plasma (n=10 per group).
(I) Relative levels of bilirubin from plasma (n=10 per group).
(J) Pyruvate, alanine, and pyruvate to alanine ratio in liver (n=10 per group).
Data are represented as mean ±SEM, biological replicates.
*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 as determined by two-way ANOVA.
Given the efficacy of KA and issues of toxicity that typically limit further clinical evaluation of glycolysis inhibitors (Raez et al., 2013), we examined the effects on metabolism in healthy tissue. Metabolomics in skeletal muscle showed no differences upon KA treatment (Figures 7F, S7F). Metabolomics on whole blood showed as expected, altered glycolysis (Figure S7G), but no substantial differences in complete blood count (CBC) in BT-474 tumor bearing mice (p=0.08, two-tailed student’s t-test) or in MDA-MB-231 tumor bearing mice (p=0.41, two-tailed student’s t-test) (Figure 7G and S7H). Creatinine (p=0.35, two-tailed student’s t-test) (Figure 7H), bilirubin (p=0.34, two-tailed student’s t-test) (Figure 7I), and alanine aminotransferase (ALT) activity (p=0.95, two-way ANOVA) (Figure 7J), markers of renal and hepatic toxicity, showed no differences. Energy charges (ATP/ADP) in BT-474 tumor, skeletal muscle, blood, and liver, also showed no significant changes (Figures S7I-S7L). Similarly, oxidized to reduced glutathione ratios were not altered (Figure S7M). While no changes were observed in energy metabolites in tumors, it is possible that non-glucose courses are compensating for glycolysis inhibition at longer times given changes in energy metabolites in cell culture (Figure S1L–S1T). Lastly, we determined the levels of pyruvate and lactate from multiple tissues as a readout of gluconeogenesis. We found that in KA-treated BT-474 tumor bearing mice, pyruvate and lactate were elevated in tumors (Figure S7N), but remained mostly unchanged in liver, plasma, and muscle (Figures S7O-S7Q). These data show that KA exerts bioactivity mainly in the tumors, while sparing normal tissues.
DISCUSSION
Targeting the WE by affecting GAPDH
There have been numerous efforts to target what have been considered to be rate-limiting steps in glycolysis (Galluzzi et al., 2013; Hay, 2016; Vander Heiden, 2013). Most notably PKM2, an isoform of pyruvate kinase, has been proposed to be limiting for the WE and has been the focus of many drug development efforts. However, targeting it successfully has been challenging with numerous conflicting findings (Israelsen et al., 2013). Our results suggest that GAPDH appears to be the step in glycolysis whose regulatory properties are most different during the WE. We were able to demonstrate a remarkable selectivity towards the WE by targeting GAPDH with KA as was concluded by evaluating the determinants of its efficacy in over sixty cell lines and in vivo. Notably, this selectivity to KA treatment was manifested across the diverse cell and tissue types of an entire mammal. Importantly, despite glycolysis being required for nearly every cell in the animal, little to no whole body metabolic changes were observed at efficacious doses. Thus partial inhibition of GAPDH while exhibiting selective toxicity in tumors could be tolerated in most healthy cells.
We also determined that this compound, KA, is a candidate for further pre-clinical analysis since its cytotoxicity appears to occur specifically by acting on GAPDH. We found that, when KA is effective, it exerts numerous simultaneous metabolic disturbances across many pathways that are each individually considered to be targets in cancer (Locasale, 2013; Svensson et al., 2016; Weinberg and Chandel, 2015), which may be attractive for therapy.
A natural product with the WE as the mechanistic target
Other successful cancer therapeutics such as rapamycin have been isolated originally as natural products for their antibiotic activity (Vezina et al., 1975). Rapamycin was later shown to have rich underlying biology after identifying its mechanistic target, TOR. Our study provides another example of a natural product with antibiotic activity and rich underlying mammalian biology (Liberti and Locasale, 2016) as its mechanistic target (i.e. the WE). It is likely that in addition to being an agent with possible therapeutic value, KA will serve as a probe to more specifically explore the WE and delineate its physiological and pathophysiological functions.
Metabolic control analysis and principles for targeting cancer metabolism
Our findings illustrate how specific control of metabolic flux by an enzyme in a defined metabolic state can be exploited for selective targeting of that metabolic status. While metabolic control analysis (MCA) is widely used in broad applications such as the metabolic engineering of biofuels or antibiotic production in microbes (Bowden, 1999), our studies show that these concepts can be more broadly applicable to address biomedical questions. It should be noted that there are caveats to using MCA in that it depends on the selection and analysis of parameters and the boundary of the specific metabolic network model, and other assumptions. In addition, the assumption that the metabolic system is in steady-state only partially resembles a real system. Nonetheless, we were able to show that MCA could reveal network properties of glucose metabolism specific to the WE. Importantly, we were able to use MCA to define pharmacological interventions that specifically disrupt metabolic pathways important in neoplastic settings, but render healthy tissue largely unaffected. This concept was developed by first demonstrating that during the WE, a hallmark of cancer metabolism, the rate-controlling steps in glycolysis are different than in fully oxidative energy metabolism. In this case, the effect of a change in GAPDH activity is amplified under WE conditions as reflected by the drastic changes in glycolytic flux in KA-sensitive cells, but not KA-resistant cells (despite similar degrees of GAPDH inhibition). This differential notably is a systems level property of how the metabolic network is configured and is independent of the specific behavior of any particular enzyme outside of the context of how the flux through the metabolic network is configured.
Comparative metabolomics to define signatures of drug response
Altogether this study consists of a wealth of metabolomics data with over 500 metabolite profiles generated throughout the course of this study. Although many of these compounds are commonly used as GAPDH inhibitors in the literature, we found that their metabolome-wide responses were markedly different. Using a global metabolite profile as a handle on cellular physiology, these differences in responses could be assessed and used for subsequent molecular characterization as when we expressed a resistant allele of GAPDH from Trichoderma virens in human cells. Given the advent of this technology, we are hopeful that this resource and framework will allow for similar in-depth characterization when evaluating other pharmacological or genetic perturbations to metabolism. Such resources comparable in scope have been documented for yeast as in a recent study that evaluated the amino acid profiles of a yeast deletion library (Mulleder et al., 2016). This current analysis enables characterization of a phenotypic response in mammalian systems.
Metabolism and precision medicine
Therapeutic applications to cancer metabolism have often been limited by defining the appropriate contexts in which a given agent may be effective. Unlike cases where genetic status determines the success or failure of a given therapy, therapies that affect metabolism may have more complicated biomarkers, if any. In fact, recent studies have suggested that environmental factors such as tissue type or nutritional status have a larger influence on cancer cell metabolism (Davidson et al., 2016; Hensley et al., 2016; Mayers et al., 2016; Yuneva et al., 2012) than the genetic lesions that have been thought to determine the extent of the WE (Vander Heiden et al., 2009). Therefore, the metabolic state of the tumor is a complex function of genetic, protein and environmental status. Consistent with these findings, we remarkably found that of over 50,000 variables surveyed including the mutational status of oncogenes that are known to influence the WE, only a specific configuration of metabolism involving the extent of glucose uptake and lactate secretion was predictive of the outcome of a treatment with an agent that targets a glycolytic enzyme. Thus, this study provides one of the first examples of a targeted agent whose biomarker cannot be defined by genetic status, but by the value of a specific metabolic flux. Since metabolite measurements are routinely used clinically as diagnostics and FDG-PET uptake, which is a surrogate for glycolytic rate, as a standard approach to monitor tumor progression, our findings could be straightforwardly employed in the clinic. Importantly, this study demonstrates that a complete understanding of pharmacogenomics (Iorio et al., 2016) that uses multi-omics data to predict drug responses also likely requires information encoded at the level of metabolites and metabolic fluxes.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by, the corresponding author Jason W. Locasale (jason.locasale@duke.edu)
Experimental Model and Subject Details
Cell Culture
All cells were cultured in full media containing RPMI-1640 (Gibco), 10% heat-inactivated fetal bovine serum (FBS), 100U/mL penicillin, and 100mg/ml streptomycin. All cell lines were obtained from the American Tissue Culture Collection (ATCC), except UACC-57, which was obtained from the National Cancer Institute (NCI) at the National Institutes of Health (Bethesda, MD, USA). Cells were cultured in a 37°C, 5% CO2 atmosphere. For koningic acid (KA) dose response curves, either KA (Adipogen, #AG-N2-0118-M001; Isolated In-House) solubilized in water was added to the media at the respective concentrations or 0.01% water for the vehicle. At the start of each experiment, cells were seeded at a density of 1×106 cells for 10cm plates for protein collection, 3×105 cells/well in a 6-well plate for metabolite collection, and 5×103 cells/well in a 96-well plate for cell viability and activity assays. Cells were allowed to adhere for 24 hours.
CD8 Activation
CD8+ cells were isolated from the spleens of 8–12 week old C57BL/6J mice (The Jackson Laboratory, 000664) fed standard chow maintained under IACUC approved protocols. CD8+ cells isolated to greater than 95% purity by CD8+ negative isolation kit (Miltenyi, #130-095-236). CD8+ cells were stained using the proliferative dye Cell Trace Violet (LifeTech, #C34557) before stimulation as per manufacturer’s protocol. Cells were stimulated on 5 μg/mL anti-CD3/CD28-coated plates (ebioscience, aCD3 #16-0031-85; aCD28 #16-0281-85) in RPMI 1640 media containing IL-2 (‘activated’, eBioscience, #14-8021-64, 10 ng/mL), or in tissue-culture treated plates in media containing IL-7 (‘naive’, Peprotech, #217-17), 1 ng/mL) for up to 7 days. Drug was added at the beginning of activation. Cells were removed at each day described for viability (Propidium Iodide, Sigma-Aldritch, #P4864), cell counts, proliferation, and/or surface markers of activation. Cell surface markers used were CD8-APC (ebioscience, #17-0081-82), CD62L-APC (ebioscience, #17-0621-82), CD44-PE (ebioscience, #12-0441-82), and CD62L-FITC (ebioscience, #11-0621-81). At day 4 and 7, cells were removed for intracellular granzyme B PE stain (ebioscience, #12-8898-80). All data were acquired in triplicate on a MacsQuant Analyzer (Miltenyi Biotec) and analyzed using FlowJo V10 (TreeStar software).
Animal Experiments
All animal experiments were performed in compliance with guidelines of the Animal Welfare Act and the guide for the care and use of laboratory animals following protocols approved by the Duke University Institutional Animal Care and Use Committee (IACUC) protocol number A011-16-01. For BT-474 tumor bearing mice, 6-week old female Foxn1nu mice (The Jackson Laboratory, 007850) were ovariectomized and subcutaneously implanted at surgery with 0.72 mg/60 day estradiol pellets (Innovative Research of America, #SE-121). Septra antibiotic treatment (Hi-Tech Pharmacal, 50383-823-16) (drinking water) was initiated at this time and continued throughout the study. Two days later, 3×106 BT-474 breast cancer cells (mixed 1:1 with Matrigel) (Corning, #354234) were injected orthotopically into the mammary fat pad. For MDA-MB-231 tumor bearing mice, 6-week old female nu/nu mice (internal breeding colony) were injected orthotopicallly with 3×106 MDA-MB-231 breast cancer cells (mixed 1:1 with Matrigel) into the mammary fat pad. Tumor volume, animal body weight, and behavior were evaluated 3 times weekly until tumors reached 200mm3 volume. At that point, BT-474 tumor bearing mice were randomized to the following treatment groups for daily intraperitoneal (IP) injections to determine the maximum tolerated dose (MTD): vehicle (5% DMSO/95% sterile saline), 1 mg/kg KA, 2.5 mg/kg KA, 5 mg/kg KA, and 10 mg/kg KA. MDA-MB-231 tumor bearing mice were randomized to vehicle (5% DMSO/95% sterile saline) and 1 mg/kg KA. KA was resuspended in DMSO prior to dilution in saline immediately prior to use. Mice were evaluated during treatment daily for behavior and 3X weekly for weight loss. Mice receiving >1 mg/kg KA were euthanized 24–48 hours after administration due to human endpoints (behavior, hematuria, and anemia), and 1 mg/kg KA was determined to be the MTD. Tumor measurements were taken three times weekly for 2 weeks. For efficacy studies, after 2 weeks of treatment, the mice were anesthetized with isoflurane and euthanized by cervical dislocation 30 minutes after the final administration of KA prior to harvest of tumors and tissues for analysis. For pharmacokinetics analysis, mice were treated with vehicle or 1 mg/kg KA from 0–24 hours, and anesthetized and euthanized at different time points by the same methods already described, followed by tumor and tissue collection. For metabolomics, tumors, tissue, and plasma were flash frozen in liquid nitrogen. For immunohistochemistry, tumors and tissue were fixed in 10% formalin. For complete blood count (CBC) analysis, whole blood was mixed with 0.9% sodium chloride (1:200) and cells were counted using a hemocytometer.
Method Details
Drug Treatments
For all cell lines, IC50 values were measured by seeding 5×103 cells/well in triplicate in a 96-well plate and allowed to adhere for 24 hours. The following day, 0–500μM concentrations of either vehicle (DMSO), arsenate (As2O4) (Sigma-Aldrich, #A6756), arsenic trioxide (As2O3) (Sigma-Aldrich, #17971), 3-bromopyruvate (3BP) (Sigma-Aldrich, #16490), iodoacetate (IA) (Sigma-Aldrich, #I2512), or koningic acid (KA) were added. After 24 hours, cell viability assays were carried out using MTT as previously described. IC50 values of drug concentrations were used unless otherwise stated.
Cell Viability Assays
For all cell lines, 5×103 cells/well were seeded in triplicate a 96-well plate and allowed to adhere for 24 hours. The following day, vehicle or KA was added to each well at the respective concentrations. After 24 hours, the media was aspirated and replaced with 100 μl phenol-red free RPMI-1640 (Gibco) and 12mM 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium (MTT) (Thermo Fisher Scientific, #M6494) was added to the cells. After 4 hours, the media containing MTT was aspirated and 50 μl DMSO was added to dissolve the formazan and read at 540nm. For HEK293T cells, 100μl of SDS-HCl solution was added to each well and incubated for 4 hours at 37°C, followed by absorbance reading at 570nm.
Nutrient Supplementation in Media
For HCT116 and KA-sensitive cell lines, 5×103 cells/well were seeded in triplicate with complete RPMI-1640 media in 96-well plates and allowed to adhere for 24 hours. The following day, the respective treatment media was added in the absence or presence of KA at the cell lines’ IC50. MTT assays were carried out as previously described. All treatment media used were as follows: Low Glucose Media: RPMI-1640 with 0.5mM glucose (VWR, #97061-168), 10% heat-inactivated FBS, 100 U/ml penicillin and 100 μg/ml streptomycin with or without 3mM N-acetyl-cysteine (NAC) (Sigma-Aldrich, #A9165); Pyruvate Media: RPMI-1640 with 5mM pyruvate (Santa Cruz Biotechnology, #sc-208397), 10% heat-inactivated FBS, 100 U/ml penicillin and 100 μg/ml streptomycin with or without 3mM NAC; Lactate Media: RPMI-1640 with 5mM lactate (Santa Cruz, Biotechnology, #sc-253582), 10% heat-inactivated FBS, 100 U/ml penicillin and 100 μg/ml streptomycin with or without 3mM NAC; Serine Media: RPMI-1640 with 2mM additional serine (Amresco, #1B1103), 10% heat-inactivated FBS, 100 U/ml penicillin and 100 μg/ml streptomycin with or without 3mM NAC; Serine and Lactate Media: RPMI-1640 with 2mM additional serine, 5mM lactate, 10% heat-inactivated FBS, 100 U/ml penicillin and 100 μg/ml streptomycin; 3-phosphoglycerate (3PG) Media: RPMI-1640 with 2mM 3PG (Sigma-Aldrich, #P8877), 10% heat-inactivated FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. Cells were grown at 37 °C with 5% CO 2.
Tumor Cell Line Sensitivity Analysis
One-dose and five-dose screens were carried out by the Developmental Therapeutics Program (DTP) at the National Cancer Institute at National Institutes of Health (Bethesda, MD USA) on a panel of 59 independent cancer cell lines (Shoemaker, 2006). For the one-dose screen, 10μM KA or 0.01% DMSO (Vehicle) was used and for the five-dose screen, concentrations of 0–100μM KA or vehicle was used. The tumor cell lines were all grown in RPMI-1640 media containing 5% FBS and 2mM L-glutamine. Cells were seeded at 5,000 to 40,000 cells/well, depending on cell line doubling time, in 96-well plates containing 100μl media and incubated at 37°C, 5% CO2 for 24 hours before addition of KA. After 24 hours, two plates of each cell line were fixed in situ with TCA, to represent a measurement of the population of each cell line at the time of treatment. Following addition of vehicle or KA, the plates were incubated for 48 hours at 37°C, 5% CO2. For adherent cells, cold TCA was added to fix the cells in situ and incubated for 60 minutes at 4°C. The supernatant was discarded and the plates were washed five times with tap water and left to air dry. Sulforhodamine (SRB) solution at 0.4% (w/v) in 1% acetic acid was added to each well, and plates were incubated for 10 minutes at room temperature. After staining, unbound dye was removed by washing five times with 1% acetic acid that the plates were left to air dry. Bound stain was solubilized with 10 mM trizma base, and the absorbance was read at 515nm. For suspension cells, the methodology remained the same except the assay was terminated by fixing settled cells at the bottom of the wells by adding 80% TCA. Time zero (Tz), control growth (C), treatment growth (Ti) with the drug at one and five concentration levels, percent growth inhibition (GI), and lethal concentrations (LC) were recorded. Data for the BT-474 cell line was added to the one-dose screen and five-dose screen.
GAPDH Activity Assay
Using a GAPDH Activity Assay Kit (BioVision, #K680), all cells were seeded at 1×106 cells per 10cm plate with either 0.01% DMSO (Vehicle) or KA. After 24 hours, cells were lysed, NADH standard curve was made, and cells were measured at 450 nm in kinetic mode for 60 minutes at 37°C according to the manufacturer’s instructions.
Lentiviral Transfection and Transduction
KAr-GAPDH cDNA was made by de novo gene synthesis (Origene) and subcloned into pLenti-C-Myc-DDK-IRES-Puro Expression Vector (KAr-GAPDH) (Origene/Blue Heron). HEK293T cells were seeded at 1×106 cells per 10 cm plate in DMEM (Gibco) supplemented with 10% Heat-Inactivated FBS, 100 U/mL penicillin, and 100 mg/ml streptomycin and allowed to adhere and reach 70% confluency. After, 6 μg KAr-GAPDH, (Origene/Blue Heron, cDNA: D14519.1) or empty vector (EV) (Origene, #PS100069), 4 μg PsPAX2 packaging vector (Addgene, #12260), and 2 μg PMD2.G envelope expressing plasmid (Addgene, #12259) were diluted into 600μl jetPRIME buffer (Polyplus Transfection, #114-07) and vortexed. Next, 24μl jetPRIME transfection reagent (Polyplus Transfection, #114-07) was added to the mixture, vortexed for 10 seconds, and centrifuged for 30 seconds. The mixture was left to incubate for 10 minutes at room temperature. Next, 600 μl of the transfection mix was added to each plate dropwise into serum containing DMEM, evenly distributed, and incubated at 37°C, 5% CO2. The transfection medium was replaced with fresh medium after 4 hours and returned to the incubator for 24 hours. Virus was collected and filtered with 0.4 μm filter (VWR International). HCT116 cells were seeded at 50% confluency per 10 cm plate in RPMI-1640 (Gibco) supplemented with 10% Heat-Inactivated FBS, 100 U/mL penicillin and 100 mg/ml streptomycin and allowed to adhere overnight. The next day, virus-rich media was added to cells (1:1 with fresh RPMI-1640 media) for 24 hours. After, virus-rich media was removed and cells were plated with fresh RPMI-1640 media. HEK293T and HCT116 cells were incubated with 1 μg/ml puromycin for 48 hours and EV and KAr-GAPDH expressing HEK293T and HCT116 cells were verified by western blotting.
Immunoblotting
Samples were homogenized in 200 μl 1X RIPA buffer (VWR International) supplemented with 100 μM phenylmethylsulfonyl fluoride (PMSF), 2 μg/μl aprotinin, 5 μg/ml pepstatin, 1X phosphatase inhibitor cocktail, and 2 mM Dithiothreitol (DTT). And centrifuged at 14,000 rpm for 30 minutes at 4°C. The supernatant was transferred to a clean tube and a Bradford Assay (Bio-Rad) was carried out to quantify protein concentration. Protein samples were loaded onto TGX stain-free precast gels (Bio-Rad) and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked in 5% dry non-fat milk in TBST and incubated with anti-GAPDH (Abcam, #ab9485) 1:2500 in 5% BSA in TBST and anti-Actin 1:2000 (Thermo Scientific, #MA5-15739) in 5% dry non-fat milk in TBST. Horseradish peroxidase-conjugated anti-mouse (Rockland, #610-1302) and anti-rabbit (Rockland, #611-1302), 1:2000, were used as secondary antibodies. Chemiluminescent signals were detected with Clarity Western ECL Detection Kit (Bio-Rad, #1705061) and imaged using the ChemiDoc Touch Imaging System (Bio-Rad).
Culture and Isolation of Koningic Acid from Trichoderma virens
Trichoderma virens (G-4) (ATCC, MYA-297) was cultured aerobically at 26.8°C for 5 days on potato dextrose agar (Fisher scientific, #DF0013-17-6) plates (25 cm). Tween-20 (0.1% in H2O, 15 mL) was added to each plate and spores were scraped off the surface of the agar and fungal tissue using a cell spreader. The concentration of spore suspensions was determined by counting spores using a hemacytometer. Then, 1 L of liquid medium in a 2 L Erlenmeyer flask was inoculated with 1×106 spores mL−1. The culture broth consisted of malt extract medium containing 3% malt extract (BD Biosciences, #211677), 2% glucose (VWR, #97061-168), and 0.1% peptone (BD Biosciences, #211677). Liquid cultures were placed at 25°C with shaking at 200 rpm for 7 days.
After 7 days, liquid fungal cultures including fungal tissue and media were frozen using a dry ice acetone bath, and lyophilized. The lyophilized residues were extracted with 500 mL of methanol (Fisher Scientific, #A452) (containing 0.2 % acetic acid (Fisher Scientific, #A38-500)) for 3.5 hours with vigorously stirring. Extracts were filtered over cotton, evaporated to dryness, and stored in 8 mL vials.
Isolation and Purification of Koningic Acid
Methanol extracts derived from 1 L cultures were fractionated using large-scale reverse-phase flash chromatography on a Teledyne ISCO CombiFlash chromatography system with a Teledyne C18 gold (100 gram) column with acetonitrile (organic phase) and 0.1 % acetic acid in water (aqueous phase) as solvents at a flow rate of 60 mL/min. A linear ramp from 0 % organic to 100 % organic over 30 min was used and fractions containing KA were collected, evaporated to dryness, and transferred to 8 mL glass vials. Then, KA was dissolved in a minimal volume of methanol and further purified via semi-preparative HPLC using an Agilent XDB C-18 column (25 cm × 10 mm, 5 μm particle diameter) acetonitrile (organic phase) and 0.1 % acetic acid in water (aqueous phase) as solvents at a flow rate of 3.6 mL/min. A solvent gradient scheme was used, starting at 5 % organic for 3 min, followed by a linear increase to 100 % organic over 27 min, holding at 100 % organic for 5 min, then decreasing back to 5 % organic over 0.1 min, and holding at 5 % organic for the final 4.9 min, for a total of 40 min. Fractions containing pure KA were collected, evaporated to dryness, and transferred to 8 mL glass vials and stored at -20 °C.
U-13C-Glucose Stable Isotope Labeling
All cells (300,000 cells/well) were plated in a 6-well plate and allowed to adhere to the plate for 24 hours. Cells were then treated with either vehicle or KA for 6 hours, then replaced with RPMI-1640 media containing 11mM U13C-glucose (Cambridge Isotope Laboratories, Inc., #CLM-1396) and vehicle or KA for 0–24 hours. Metabolites were then extracted.
Metabolite Extraction
Metabolite extraction and subsequent Liquid-Chromatography coupled to High Resolution Mass Spectrometry (LC-HRMS) for polar metabolites of each cell line was carried out using a Q Exactive Plus as previously described (Liu et al., 2014a; Liu et al., 2014b). For culture from adherent cell lines, media was quickly aspirated. Next, 1 mL of extraction solvent (80% methanol/water) cooled to -80°C was added immediately to each well and the plates were then transferred to -80°C for 15 minutes. After, the plates were removed and the cells were scraped into the extraction solvent on dry ice. For tissue, the sample was homogenized in liquid nitrogen and then 5 to 10 mg was weighed in a new Eppendorf tube. Ice-cold extraction solvent (200 μl) was added to each tissue sample and homogenized using a tissue homogenizer. The homogenate was incubated on ice for 10 minutes. For plasma or media, 20 μl was transferred to a new Eppendorf tube containing 80 μl HPLC grade water. Next, 400 μl of ice-cold methanol was added to the sample for a final methanol concentration of 80% (v/v). Samples were incubated on ice for 10 minutes. For absolute quantitation of KA in plasma, 0–10μM KA standards were added to extraction solvent before centrifugation. All metabolite extractions were centrifuged at 20,000 g at 4°C for 10 minutes. Finally, the solvent in each sample was evaporated using a speed vacuum for metabolite analysis. For polar metabolite analysis, the cell metabolite extract was first dissolved in 15 μl water, followed by dilution with 15 μl methanol/acetonitrile (1:1 v/v) (optima LC-MS grade, Fisher Scientific, methanol, #A456; acetonitrile, #A955). Samples were centrifuged at 20,000 g for 10 minutes at 4°C and the supernatants were transferred to LC vials. The injection volume for polar metabolite analysis was 5 μl.
Liquid Chromatography
An XBridge amide column (100 × 2.1 mm i.d., 3.5 μm; Waters) was employed on a Dionex (Ultimate 3000 UHPLC) for compound separation at room temperature. Mobile phase A is water with 5mM Ammonium Acetate, pH 6.9, and mobile phase B is 100% Acetonitrile. The gradient is linear as follows: 0 minutes, 85% B; 1.5 minutes, 85% B; 5.5 minutes, 35% B; 10 minutes, 35% B; 10.5 minutes, 35% B; 10.6 minutes, 10% B; 12.5 minutes, 10% B; 13.5 minutes, 85% B; and 20 minutes, 85% B. The follow rate was 0.15 ml/min from 0 to 5.5 minutes, 0.17 ml/min from 6.9 to 10.5 minutes, 0.3 ml/min from 10.6 to 17.9 minutes, and 0.15 ml/min from 18 to 20 minutes. All solvents are LC-MS grade and purchased from Fisher Scientific.
Mass Spectrometry
The Q Exactive Plus MS (Thermo Scientific) is equipped with a heated electrospray ionization probe (HESI) and the relevant parameters are as listed: evaporation temperature, 120°C; sheath gas, 30; auxillary gas, 10; sweep gas, 3; spray voltage, 3.6 kV for positive mode and 2.5 kV for negative mode. Capillary temperature was set at 320°C, and S lens was 55. A full scan range from 70 to 900 (m/z) was used. The resolution was set at 70,000. The maximum injection time was 200 ms. Automated gain control (AGC) was targeted at 3×106 ions. For targeted MS/MS analysis of KA, the isolation width of the precursor ion was set at 1.5 (m/z), high energy collision dissociation (HCD) was 35%, and max IT was 100 ms. The resolution and AGC were 70,000 and 200,000 respectively.
Peak Extraction and Data Analysis
Raw data collected from LC-Q Exactive Plus MS was processed on Sieve 2.0 (Thermo Scientific, https://www.thermofisher.com/order/catalog/product/IQLAAEGABSFAHSMAPV). Peak alignment and detection were performed according to the protocol described by Thermo Scientific. For a targeted metabolite analysis, the method “peak alignment and frame extraction” was applied. An input file of theoretical m/z and detected retention time of 197 known metabolites was used for targeted metabolite analysis with data collected in positive mode, while a separate input file of 262 metabolites was used for negative mode. m/z width was set to 10 ppm. The output file including detected m/z and relative intensity in different samples was obtained after data processing. If the lowest integrated mass spectrometer signal (MS intensity) was less than 1000 and the highest signal was less than 10,000, then this metabolite was considered below the detection limit and excluded for further data analysis. If the lowest signal was less than 1000, but the highest signal was more than 10,000, then a value of 1000 was imputed for the lowest signals. Mass isotopomer distributions (MID) were calculated and samples were normalized by comparing the ratio of glucose-derived labeled metabolites to unlabeled metabolites within each sample. Quantitation and statistics were calculated using Microsoft Excel and GraphPad Prism 6.0.
Kinetic flux profiling
The time-dependent lactate labeling pattern was modeled as with the following equation:
In which [X*] is the concentration of labeled lactate, XT is the total concentration (both labeled and unlabeled) of lactate, fX is the lactate production flux. This model was fit to lactate MIDs using the fit() function in MATLAB to determine relative lactate production fluxes. Relative lactate pool sizes were estimated from MS signal intensities.
Thermodynamic and Kinetic Analysis
Mathematical model of glycolysis downloaded from the BioModels Database repository (EMBL-EBI, http://www.ebi.ac.uk/biomodels-main/, MODEL1504010000) was first translated to C++ code using the SBML translator module in the Systems Biology Workbench (http://sbw.sourceforge.net/) (Sauro et al., 2003) then simulated using the ODE solver gsl_odeiv2_step_msbdf in the GNU Scientific Library (Free Software Foundation, http://www.gnu.org/software/gsl/). FCC of an enzymatic step was computed by replacing Vmax of the enzyme with 1.01Vmax and Vmax/1.01 while keeping all other parameters fixed, computing the corresponding steady state lactate fluxes (fLac), then estimating FCC accordingly: . ΔGs were calculated from standard reaction free 2 log1.01 energies and steady state concentrations of the metabolites. Extent of the Warburg Effect was tuned by increasing Vmaxs of the glycolytic enzymes GLUT, HK, PFK, PGK, MCT from 0.1 to 10 folds of their original values and decreasing Vmax of PDH from 10 to 0.1 folds of its original value simultaneously. Extent of the Warburg Effect was quantified by ratio of lactate production flux to oxidative phosphorylation flux.
Integrative Analysis of Drug Response and Multi-omics Data
All correlations were carried out using pearson or spearman correlations to GI50 values of KA to each of the 59 cell lines tested from the NCI-60 cell line panel using GraphPad Prism 6.0 or R Statistical Programming (The R Foundation, https://www.r-project.org/). Metabolic consumption and excretion rates from NCI-60 cell line panel were acquired from Jain et al. 2012 (Jain et al., 2012). Cell size and doubling time were acquired from Dolfi et al. 2013 (Dolfi et al., 2013). Gene expression analyses were completed by manually dividing the 59 cell lines tested with KA into two groups (KA-resistant and KA-sensitive) based on the distribution of GI50 values. Gene set enrichment analysis (GSEA) was then performed (The Broad Institute, http://software.broadinstitute.org/gsea/index.jsp) using all of the genes for differential expression analysis between KA-sensitive and KA-resistant cell lines. For protein expression analyses the efficacy of KA (in terms of GI50) were plotted against the absolute protein quantification of glycolytic genes in the NCI-60 cell line panel. Protein quantification data was obtained from Gholami et al. 2013 (Gholami et al., 2013) and genes were filtered and mapped to their KEGG biochemical pathways as previously described in Madhukar et al. 2015 (Madhukar et al., 2015) P-values, R2, Pearson correlation values were calculated using the cor.test function within R. Mutation and copy number alterations (CNAs) were obtained from cBioPortal (http://www.cbioportal.org) for Cancer Genomics. Random distributions for the Q-Q plots were generated by permuting the original data 1000 times and calculating Spearman correlation coefficients accordingly. Error bars on the Q-Q plots represent for standard deviation of the quantiles of the 1000 random distributions. Drug activity z-scores of the targeted therapies calculated from their GI50 values on the NCI-60 cell line panel were available from CellMiner (National Cancer Institute, https://discover.nci.nih.gov/cellminer/). ROC curves were created for the targeted therapies using the perfcurve() function in MATLAB (MathWorks, Inc, https://www.mathworks.com/product/matlab.html), by labeling the cell lines according to presence of the genomic feature targeted and using drug activities as predictive scores. For KA, cell lines were labeled ‘high’ or ‘low’ based on whether the glucose or lactate flux in this cell line is higher or lower than the median value among all cell lines and ROC curves were created similarly.
Koningic Acid Docking to GAPDH
The virtual compound screening ZINC Database (http://zinc.docking.org) was used to obtain a ready-to-dock format (mol2) of KA (ZINC 15272438)(Irwin and Shoichet, 2005; Irwin et al., 2012). Next, docking between KA and the human GAPDH structure was performed using molecular docking on the Swiss Dock server using default settings (http://www.swissdock.ch) (Grosdidier et al., 2011). Since previous studies have already shown that the epoxide group of KA interacts with GAPDH active site cysteine 152 via alkylation, the highest scoring binding model with this interaction was used to infer the drug docking (Sakai et al., 1988; Sakai et al., 1991). The 3-D structure of the docked model was analyzed using UCSF Chimera 1.10.2 (http://www.rbvi.ucsf.edu/chimera) (Pettersen et al., 2004). Amino acid hydrophobicity was assigned to the structure as an attribute in Chimera with a pre-defined hydrophobicity scale (Kyte and Doolittle, 1982).
GAPDH Protein Multiple Sequence Alignment Comparisons
All multiple sequence alignments were carried out using Clustal Omega https://www.ebi.ac.uk/Tools/msa/clustalo) (Sievers et al., 2011). For phylogenetic analyses, outputs from Clustal Omega were used and inputted into Clustal W2-Phylogeny (https://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/) (Larkin et al., 2007) to generate a phylogenetic tree from the homology alignment. Percent Identity Matrices were calculated based on the data output from Clustal W2-Phylogeny.
Analysis of Metabolomics Data
GENE-E and Morpheus software were used for hierarchal clustering and heatmap generation (The Broad Institute, https://software.broadinstitute.org/GENE-E/index.html). For hierarchal clustering, spearman correlation parameters were implemented for row and column parameters, with the exception of putative GAPDH drug response data, in which hierarchal clustering for row parameters only was used. Quantile normalization was used to normalize the data, with the exception of glycolysis data in HCT116 cells, in which and log2 normalization was used.
For pathway enrichment analyses, MetaboAnalyst 3.0 was used (http://www.metaboanalyst.ca/faces/home.xhtml) (Xia et al., 2015). To do this, HMDB IDs from the metabolites that were significantly enriched in their respective treatment groups (greater than ±log2(2) fold change with a p<0.05) were inputted. The pathway library that was chosen was Homo sapiens and Fishers’ Exact test was the method employed for over-representation analysis. For pathway topology analysis, relative betweenness centrality was chosen as the node importance measure.
Paraffin Embedding and Immunohistochemistry
All paraffin embedding, tissue sectioning, and immunohistochemical (IHC) staining was carried out by Duke University Pathology Research Histology and Immunohistochemistry Laboratory shared resource facility. For tissue embedding and sectioning, previously established protocols were followed (Fischer et al., 2007). For hematoxylin and eosin (H&E) staining of tissue sections, standard protocols were used (Fischer et al., 2008). For Ki67 staining for cell proliferation, Ki67 was detected by biotinylated goat anti-rabbit 1:300 (Thermo Scientific,# BA-1000) and the ABC Elite (Vector, #PK-7100). For GAPDH staining to detect GAPDH localization, GAPDH was detected by the anti-rabbit HQ 1:200 (Abcam, #760-4815) and the anti-HQ system (Roche/Ventana, #760-4820). All slides were observed and photographed under the microscope.
To quantify Ki-67 staining, ImageJ (National Institute of Health, https://imagej.nih.gov/ij/index.html) was used. To do this, a threshold was set (8-Bit Auto Threshold) until all the stained areas were selected. Next, specific parameters were chosen for measurements (area, min & max gray value, area fraction, limit to threshold). After, intensity measurements were performed and output was area. The areas were compared to the background to achieve percentage of staining.
Quantification and Statistical Analysis
Unless otherwise noted, all error bars were reported ±SEM. with n=3 independent biological measurements and statistical tests resulting in p-value computations were computed using a student’s t-test two tailed. Tumor volumes in the animal studies were analyzed by 2-way ANOVA of time matched values, followed by Bonferroni multiple comparison test. All statistics were computed using GraphPad Prism 6 (GraphPad, https://www.graphpad.com/scientific-software/prism).
Data and Software Availability
Metabolomics data have been deposited in Mendeley Data and are accessible through http://dx.doi.org/10.17632/wmk2prwynj.1.
Supplementary Material
HIGHLIGHTS.
Flux control analysis reveals GAPDH as rate-limiting during the Warburg Effect (WE)
Koningic acid (KA) is nominated and validated as a specific GAPDH inhibitor
Machine learning shows that the quantitative extent of the WE predicts KA response
Partial GAPDH inhibition is tolerable and selective for highly glycolytic tumors
Acknowledgments
We thank the members of the Locasale laboratory for their comments and advice. We also thank Alexander P. Yllanes for help with the mouse studies. Support from the National Institutes of Health (T32GM007273 (MVL), R01CA193256 (JWL), R00CA168997 (JWL), R01CA174643 (DPD), T32GM008500 (JAB)), the National Science Foundation (MVL), the Sloan Foundation (MVL), the International Life Sciences Institute (JWL), and the Canadian Institutes of Health Research (X.G.) are gratefully acknowledged. A provisional patent related to this work has been filed.
Footnotes
Author Contributions
Conceptualization, M.V.L. and J.W.L.; Animal Experiments, S.E.W., D.P.M., M.V.L., X.G., and R.B.; Koningic Acid Isolation and Purification, M.V.L., J.A.B. and F.C.S.; Metabolomics, M.V.L. and X.L.; Data Analysis, M.V.L, M.M., M.O.J., J.C.R., Z.D., N.S.M., O.E., I.I.C.; Mathematical Modeling, Z.D., A.A.S.; All Other Experiments, M.V.L.; Writing, M.V.L. and J.W.L.; Supervision, M.V.L., and J.W.L.
Conflicts of Interest
The authors declare no conflicts of interest at this time.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiological genomics. 2005;21:389–395. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- Bergmann FT, Sauro HM. SBW - A modular framework for systems biology. Proceedings of the 2006 Winter Simulation Conference; 2006. p. 1637. [Google Scholar]
- Bowden AC. Metabolic control analysis in biotechnology and medicine. Nat Biotechnol. 1999;17:641–643. doi: 10.1038/10854. [DOI] [PubMed] [Google Scholar]
- Campbell-Burk S, Jones K, Shulman R. Phosphorus-31 NMR saturation-transfer measurements in Saccharomyces cerevisiae: characterization of phosphate exchange reactions by iodoacetate and antimycin A inhibition. Biochemistry. 1987;26:7483–7492. doi: 10.1021/bi00397a043. [DOI] [PubMed] [Google Scholar]
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Weinberg RS, Waxman S, Jing Y. Malignant cells can be sensitized to undergo growth inhibition and apoptosis by arsenic trioxide through modulation of the glutathione redox system. Blood. 1999;93:268–277. [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfi SC, Chan LLY, Qiu J, Tedeschi PM, Bertino JR, Hirshfield KM, Oltvai ZN, Vazquez A. The metabolic demands of cancer cells are coupled to their size and protein synthesis rates. Cancer & metabolism. 2013;1:1. doi: 10.1186/2049-3002-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Hasumi K, Sakai K, Kanbe T. Specific inhibition of glyceraldehyde-3-phosphate dehydrogenase by koningic acid (heptelidic acid) The Journal of antibiotics. 1985;38:920–925. doi: 10.7164/antibiotics.38.920. [DOI] [PubMed] [Google Scholar]
- Fell DA. Metabolic control analysis: a survey of its theoretical and experimental development. Biochem J. 1992;286(Pt 2):313–330. doi: 10.1042/bj2860313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AH, Jacobson KA, Rose J, Zeller R. Paraffin embedding tissue samples for sectioning. CSH protocols. 2007;2008 doi: 10.1101/pdb.prot4989. pdb. prot4989-pdb. prot4989. [DOI] [PubMed] [Google Scholar]
- Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harbor Protocols. 2008;2008 doi: 10.1101/pdb.prot4986. pdb. prot4986. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nature reviews Drug discovery. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- Ganapathy-Kanniappan S, Geschwind JFH, Kunjithapatham R, Buijs M, Vossen JA, Tchernyshyov I, Cole RN, Syed LH, Rao PP, Ota S. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is pyruvylated during 3-bromopyruvate mediated cancer cell death. Anticancer research. 2009;29:4909–4918. [PMC free article] [PubMed] [Google Scholar]
- Garber K. Cancer anabolic metabolism inhibitors move into clinic. Nature Research. 2016 doi: 10.1038/nbt0816-794. [DOI] [PubMed] [Google Scholar]
- Gholami AM, Hahne H, Wu ZX, Auer FJ, Meng C, Wilhelm M, Kuster B. Global Proteome Analysis of the NCI-60 Cell Line Panel. Cell Rep. 2013;4:609–620. doi: 10.1016/j.celrep.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic acids research. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nature Reviews Cancer. 2016 doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich R, Schuster S. The regulation of cellular systems. New York: Chapman & Hall; 1996. [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Goncalves E, Barthorpe S, Lightfoot H, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK. ZINC-a free database of commercially available compounds for virtual screening. Journal of chemical information and modeling. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. Journal of chemical information and modeling. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DW, Kim WH, Seo S, Oh E, Yim SH, Ha HH, Chang YT, Williams DR. Chemical Targeting of GAPDH Moonlighting Function in Cancer Cells Reveals Its Role in Tubulin Regulation. Chemistry & biology. 2014;21:1533–1545. doi: 10.1016/j.chembiol.2014.08.017. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of molecular biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. Clustal W and Clustal X version 2.0. bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM. Lehninger principles of biochemistry. 6. New York: W.H. Freeman; 2013. [Google Scholar]
- Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends in biochemical sciences. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Romero IL, Litchfield LM, Lengyel E, Locasale JW. Metformin Targets Central Carbon Metabolism and Reveals Mitochondrial Requirements in Human Cancers. Cell Metabolism. 2016 doi: 10.1016/j.cmet.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ser Z, Cluntun AA, Mentch SJ, Locasale JW. A strategy for sensitive, large scale quantitative metabolomics. Journal of visualized experiments: JoVE. 2014a doi: 10.3791/51358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ser Z, Locasale JW. Development and quantitative evaluation of a high-resolution metabolomics technology. Analytical chemistry. 2014b;86:2175–2184. doi: 10.1021/ac403845u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhukar NS, Warmoes MO, Locasale JW. Organization of enzyme concentration across the metabolic network in cancer cells. PloS one. 2015;10:e0117131. doi: 10.1371/journal.pone.0117131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulleder M, Calvani E, Alam MT, Wang RK, Eckerstorfer F, Zelezniak A, Ralser M. Functional Metabolomics Describes the Yeast Biosynthetic Regulome. Cell. 2016;167:553–565. e512. doi: 10.1016/j.cell.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németi B, Gregus Z. Reduction of arsenate to arsenite by human erythrocyte lysate and rat liver cytosol–characterization of a glutathione-and NAD-dependent arsenate reduction linked to glycolysis. Toxicological Sciences. 2005;85:847–858. doi: 10.1093/toxsci/kfi157. [DOI] [PubMed] [Google Scholar]
- Noor E, Bar-Even A, Flamholz A, Reznik E, Liebermeister W, Milo R. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput Biol. 2014;10:e1003483. doi: 10.1371/journal.pcbi.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Raez LE, Papadopoulos K, Ricart AD, Chiorean EG, DiPaola RS, Stein MN, Lima CMR, Schlesselman JJ, Tolba K, Langmuir VK. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer chemotherapy and pharmacology. 2013;71:523–530. doi: 10.1007/s00280-012-2045-1. [DOI] [PubMed] [Google Scholar]
- Sakai K, Hasumi K, Endo A. Inactivation of rabbit muscle glyceraldehyde-3-phosphate dehydrogenase by koningic acid. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology. 1988;952:297–303. doi: 10.1016/0167-4838(88)90130-6. [DOI] [PubMed] [Google Scholar]
- Sakai K, Hasumi K, Endo A. Two glyceraldehyde-3-phosphate dehydrogenase isozymes from the koningic acid (heptelidic acid) producer Trichoderma koningii. European journal of biochemistry/FEBS. 1990;193:195–202. doi: 10.1111/j.1432-1033.1990.tb19323.x. [DOI] [PubMed] [Google Scholar]
- Sakai K, Hasumi K, Endo A. Identification of koningic acid (heptelidic acid)-modified site in rabbit muscle glyceraldehyde-3-phosphate dehydrogenase. Biochimica et biophysica acta. 1991;1077:192–196. doi: 10.1016/0167-4838(91)90058-8. [DOI] [PubMed] [Google Scholar]
- Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, Doyle J, Kitano H. Next generation simulation tools: the Systems Biology Workbench and BioSPICE integration. Omics A Journal of Integrative Biology. 2003;7:355–372. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- Shestov AA, Liu X, Ser Z, Cluntun AA, Hung YP, Huang L, Kim D, Le A, Yellen G, Albeck JG. Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. eLife. 2014;3:e03342. doi: 10.7554/eLife.03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nature Reviews Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, Lombardo PS, Van Nostrand JL, Hutchins A, Vera L, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med. 2016;22:1108–1119. doi: 10.1038/nm.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman LA, Holton T, Yuneva M, Louie RJ, Padró M, Daemen A, Hu M, Chan DA, Ethier SP, van’t Veer LJ. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer cell. 2013;24:450–465. doi: 10.1016/j.ccr.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG. Exploiting tumor metabolism: challenges for clinical translation. J Clin Invest. 2013;123:3648–3651. doi: 10.1172/JCI72391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. The Journal of antibiotics. 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Hasumi K, Fukushima Y, Sakai K, Endo A. Cloning of two isozymes of Trichoderma koningii glyceraldehyde-3-phosphate dehydrogenase with different sensitivity to koningic acid. Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression. 1993;1172:43–48. doi: 10.1016/0167-4781(93)90267-h. [DOI] [PubMed] [Google Scholar]
- Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic acids research. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.