Abstract
The adherence of bacteria to epithelial cells and mucosal surfaces is a prerequisite for their colonization in the gut and a key criterion for the selection of probiotics. In this study, the eleven indigenous lactic acid bacterial isolates obtained from traditional fermented foods of Western Himalayas were screened for their adherence potential to intestinal epithelial cell lines. The level of adherence of eleven indigenous isolates to Caco-2 and HT-29 cell lines varied from 2.45 ± 0.5 to 9.55 ± 0.76% and 4.11 ± 0.68 to 12.88 ± 0.63%, respectively. Percent adhesion of indigenous isolates to Caco-2 cells was relatively lower as compared to HT-29 cells. Indigenous isolate AdF10 (L. plantarum) was found to be the most adhesive to HT-29 and Caco-2 with corresponding figures of 12.88 ± 0.63 and 9.55 ± 0.76%, respectively. AdF4 (B. coagulans) was found to be least adhesive to HT-29 and Caco-2 with respective corresponding figures of 4.11 ± 0.68 and 2.45 ± 0.5%. Based on the percent adhesion values, indigenous isolate AdF10 (L. plantarum) was comparable to the reference probiotic strain L. rhamnosus GG-ATCC-53103 with respective adhesion of 13.5 ± 1.19 and 10.33 ± 0.64% to HT-29 and Caco-2 cell lines. It was closely followed by indigenous isolates AdF5 (L. plantarum) and AdF6 (L. plantarum); thus, indicating their potential as a promising probiotic candidates.
Keywords: Adherence, Indigenous, Lactic acid bacteria, Intestinal cell lines
Introduction
Fermented foods have been a well established part of the human diet that not only provide the important source of nutrients but also have a great potential in maintaining health (Chilton et al. 2015). The claimed beneficial effects of these fermented foods is chiefly ascribed to their associated microflora particularly probiotic organisms which upon ingestion in adequate amount exert health benefits beyond inherent basic nutrition (Gobbetti et al. 2010). A large number of evidences are available that prove the ability of probiotic lactic acid bacteria in restoring the composition of gut microbiome during disbalanced conditions (Hemarajata and Versalovic 2013; Vandenplas et al. 2015). Most probiotics fall into the category of organisms known as lactic acid-producing bacteria which are normally consumed in the form of fermented foods and beverages (Parvez et al. 2006). Himachal Pradesh, India, is having a rich repository of indigenous fermented foods which are traditionally prepared and consumed since ages (Kanwar et al. 2007). The increasing applications of probiotics in functional foods, highlight the need to explore the functional attributes of beneficial bacteria obtained from traditional fermented foods so that the potential isolates could further be used in enriching the traditional foods or in the development of food products at the commercial scale. A variety of indigenous fermented food products and beverages of Western Himalayas were already documented by our research group with respect to their microbial diversity (Kanwar et al. 2007; Sourabh et al. 2010), probiotic and protective attributes (Sourabh et al. 2011a, b; Walia et al. 2014, 2015). However, these indigenous microorganisms are yet to be evaluated with respect to their adherence to human epithelial cell lines as it is the prerequisite step for optimal expression of physiological functions of probiotics. The adherence of lactic acid bacteria to epithelial cells and mucosal surfaces has been considered as a potential probiotic marker along with other desirable attributes as it stimulates the host-microbe interactions, promote their persistence time in the gut, and provides protection to the intestinal barrier by various mechanisms (Tuomola et al. 2001; Servin 2004). The Caco-2 and HT-29 cell lines derived from human colonic adenocarcinoma have been extensively used models to assess the adhesion ability of probiotics in vitro as they closely mimic the normal intestinal epithelium (Wang et al. 2008; Moussavi and Adams 2009; Duary et al. 2011). The variability in adherence pattern of probiotic isolates to intestinal cell lines has been reported by various workers (Chauviere et al. 1992; Duary et al. 2011) indicating that this property is highly strain specific. Thus, the present study was undertaken with the objective to assess the adhesion potential of indigenous bacterial isolates obtained from traditional fermented foods of Western Himalayas to epithelial cell lines under in vitro conditions.
Materials and methods
Indigenous bacterial isolates and growth conditions
The study was performed with eleven indigenous lactic acid bacterial (LAB) isolates obtained from traditional fermented foods of Western Himalayas (Table 1). These isolates have already been molecularly characterized and their nucleotide sequences were submitted to Genbank, National Centre for Biotechnology Information (NCBI), USA (Sourabh et al. 2010). Lactobacillus rhamnosus GG (LGG; ATCC 53103) procured from American Type Culture Collection (Manassas, VA, USA) was used as a reference probiotic strain. The lactic acid bacteria were grown anaerobically in de Man, Rogosa, and Sharpe (MRS) broth (Merck) at 37 °C for 24 h in a modular atmosphere controlled system (Don Whitley DG250, Scientific Ltd. United Kingdom). For long-term storage, the isolates were maintained at −20 °C in 30% glycerol as well as in skim milk medium at 4 °C until further use. All isolates were subcultured twice prior to the experiment.
Table 1.
Indigenous lactic acid bacterial isolates
| Indigenous isolates | Isolation sourcea | Identification using BLAST n algorithm | GenBank Accession no.b | NBAIM Accession no.c |
|---|---|---|---|---|
| AdF1 | Bhaturu | Enterococcus faecium | GU396270 | NAIMCC-B-01045 |
| AdF2 | Bhaturu | Enterococcus faecium | GU396271 | NAIMCC-B01046 |
| AdF3 | Chilra | Enterococcus faecium | GU396272 | NAIMCC-B-01094 |
| AdF4 | Fermented milk | Bacillus coagulans | GU396273 | NAIMCC-B-01047 |
| AdF5 | Fermented milk | Lactobacillus plantarum | GU396274 | NAIMCC-B-01048 |
| AdF6 | Seera | Lactobacillus plantarum | GU396275 | NAIMCC-B-01095 |
| AdF7 | Seera | Lactobacillus fermentum | HQ677597 | NAIMCC-B-01096 |
| AdF8 | Jan Chang | Lactobacillus fermentum | GU396276 | NAIMCC-B-01049 |
| AdF9 | Jan Chang | Lactobacillus fermentum | GU396277 | NAIMCC-B-01050 |
| AdF10 | Jan Chang | Lactobacillus plantarum | GU396278 | NAIMCC-B-01051 |
| AdF11 | Jan Chang | Enterococcus faecium | GU396279 | NAIMCC-B-01052 |
aFermented foods and beverages
bGenbank, National Centre for Biotechnology Information (NCBI), USA
cNational Bureau of Agriculturally Important Microorganisms, Maunath Bhanjan, Uttar Pradesh, India
Cell culture
The human colonic cell lines HT-29 (mucus secreting) and Caco-2 (non mucus secreting) were procured from National Center of Cell Sciences, Pune, India. The cells were routinely cultured in Dulbecco’s modified Eagle’s minimal essential medium (DMEM; Sigma) supplemented with 10% (v/v) heat-inactivated (30 min, 56 °C) foetal bovine serum in case of HT-29 cells and 20% (v/v) in case of Caco-2 cells. The cells in a medium were also supplemented with 1% (v/v) penicillin–streptomycin solution to a final concentration of 100 U mL−1 penicillin and 100 μg mL−1 streptomycin (Gibco, Life Technologies). The incubation was carried out at 37 °C in an atmosphere of 5% CO2 and 95% air. The cells were fed with complete DMEM every alternate day until the cells reached 80% confluency.
Preparation of cell lines and lactic acid bacterial cultures for adhesion assay
The concentration of cells in a monolayer was determined by trypsinizing the adhered cells with 3 mL of 0.25% trypsin–EDTA solution for 3–4 min at 37 °C. The final cell count in suspension was measured with the help of haemocytometer. For adhesion assay, Caco-2 and HT-29 cells were seeded separately in each well of standard tissue culture plates at a concentration of 1x105 cells/mL and incubated until a complete monolayer was obtained. Change of medium was performed every 48 h. The spent medium was completely removed 24 h before adhesion assay and cells were fed with DMEM lacking antibiotics. The indigenous lactic acid bacterial isolates for adhesion assay were propagated in MRS broth and cultures obtained after 18 h of growth at 37 °C were centrifuged at 5500×g for 10 min. The pellet was washed once with phosphate-buffered saline (PBS; pH 7.4). The cell density was adjusted approximately to the desired levels by measuring the absorbance at 600 nm. The exact number of viable bacteria used in the assay was determined by plate counting on MRS agar.
In vitro adhesion assay
Adhesion of indigenous lactic acid bacterial isolates was measured as per the method described by Jacobsen et al. (1999). The Caco-2 and HT-29 cells in a monolayer were washed twice with 3 mL of phosphate-buffered saline (PBS; pH 7.4). The two mL of DMEM without serum and antibiotics was added to each well and incubated at 37 °C for 30 min before inoculation of bacteria. Different bacterial cultures with concentration of approximately 1 × 109 CFU suspended in 1 mL DMEM without serum and antibiotics were used to inoculate each well of tissue culture plates. The plates were incubated at 37 °C in an atmosphere of 5% CO2 and 95% air for 2 h. After incubation, the monolayer was washed five times with sterile PBS (pH 7.4) to remove non-adherent bacteria. For microscopic examination of adhered lactic acid bacteria, the cells were fixed with 3 mL of methanol and incubated for 10 min at room temperature. After removal of methanol, the cells were stained by adding 3 mL of giemsa stain solution (1:20 dilution of giemsa in PBS). Plates were incubated at room temperature for 20 min and then rinsed extensively with distilled water. The plates were air dried and examined under inverted microscope (EVOS XL Imaging system, Thermo Fisher Scientific).
Percent adhesion
The monolayer was washed five times with sterile PBS (pH 7.4) to remove non-adherent bacteria. In order to enumerate the viable adhered bacteria, the cells from monolayer were detached by trypsinization. Each well was treated with 1 mL of 0.25% trypsin–EDTA solution and incubated for 15 min at room temperature. The suspension of lysed cells and lactic acid bacteria was serially diluted with saline solution and plated on MRS agar. The enumeration was done after 48 h of incubation at 37 °C in an anaerobic atmosphere. The adhesion was expressed as the percentage of number of adhered bacteria to total bacteria used for the experiment and calculated as: Percent adhesion = (B1/B0) * 100, where B0 and B1 CFU/mL are the initial and final count of bacteria.
Statistical analysis
The experiment was performed in triplicate. Results were statistically analyzed by one-way ANOVA and expressed as mean ± standard deviation calculated at 95% confidence level.
Results and discussion
Adhesion potential of indigenous probiotics to epithelial cell lines under in vitro conditions
Adhesion has been considered as an ideal parameter to determine the colonization capability of a promising probiotic strain (Servin and Coconnier 2003). Attachment of probiotic bacteria to gastrointestinal surface extend their residence time in vivo and thus influence the host health by stimulating the gut immune system and it may also a prerequisite step for competitive exclusion of pathogenic bacteria (Tallon et al. 2007). Several earlier studies have been carried out using human epithelial cell lines like HT-29, HT-29MTX (mucus secreting) and Caco-2 (non-mucus secreting) to screen the adherence properties of probiotic strains (Laparra and Sanz 2009; Moussavi and Adams 2009; Duary et al. 2011). The advantage of these cellular models is that they have morphological and functional characteristics of mature enterocytes and express most receptors, enzymes and transporter proteins present in normal human intestinal epithelium (Lesuffleur et al. 1990; Tor Lea 2015).
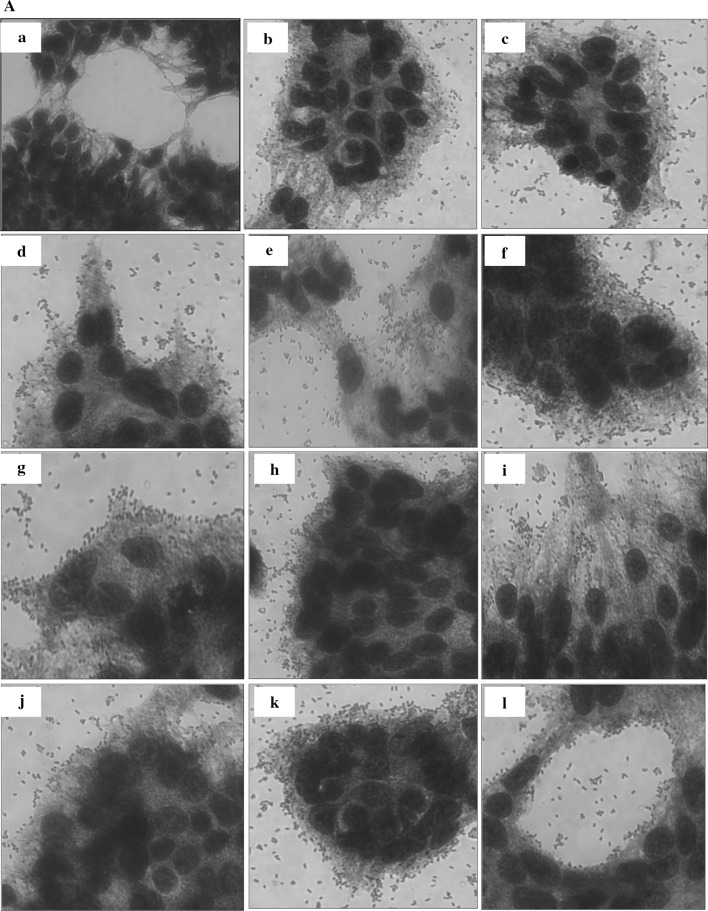
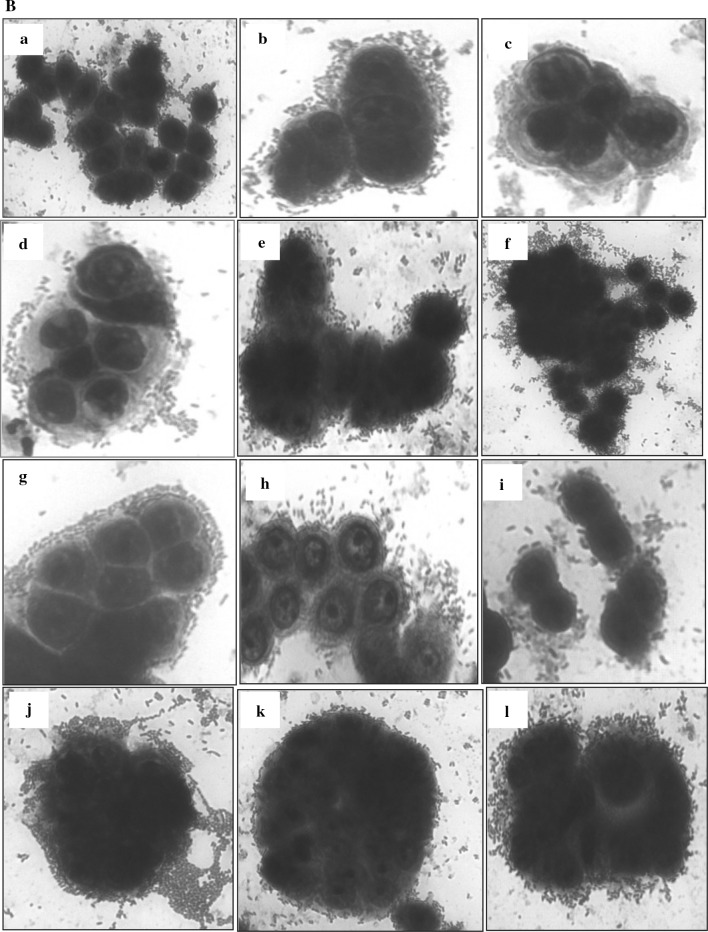
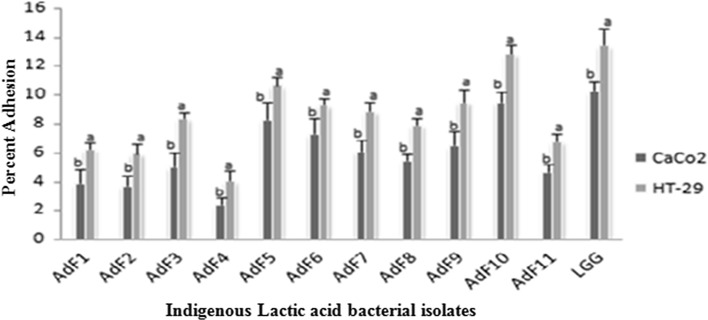
In the present study, the level of adherence of eleven indigenous isolates to Caco-2 and HT-29 cell lines varied from 2.45 ± 0.5 to 9.55 ± 0.76% and 4.11 ± 0.68 to 12.88 ± 0.63%, respectively (Table 2) which is in concurrence with the adherence range reported in previously published studies (Duary et al. 2011; Garcia-Ruiz et al. 2014). Among all indigenous isolates, AdF10 (L. plantarum) showed the highest percent adhesion to HT-29 and Caco-2 with corresponding values of 12.88 ± 0.63 and 9.55 ± 0.76, respectively. It was closely followed by isolate AdF5 (L. plantarum) with respective adhesion of 10.75 ± 0.56 and 8.33 ± 1.21% and AdF6 (L. plantarum) with respective adhesion of 9.36 ± 0.49 and 7.37 ± 1.09% to HT-29 and Caco-2 cell lines. Above findings are supported by the earlier studies where Lactobacillus plantarum strains have demonstrated excellent adhesion to human epithelial cell lines (Anderson et al. 2010; Garcia-Ruiz et al. 2014). Indigenous isolate AdF4 (B. coagulans) was found to be least adhesive with respective corresponding figures of 4.11 ± 0.68 and 2.45 ± 0.5% adhesion to HT-29 and Caco-2 cell lines. The adherence ability of indigenous isolates to intestinal cells was compared with that of the benchmark reference probiotic strain L. rhamnosus GG-ATCC-53103 due to its strong adhesive capacity documented in earlier in vitro and in vivo studies (Goldin et al. 1992; Kankainen et al. 2009; Segers and Lebeer 2014). Based on the percent adhesion values, AdF10 (L. plantarum) was found to be statistically at par with reference probiotic strain with respective percent adhesion of 13.5 ± 1.19 and 10.33 ± 0.64 to HT-29 and Caco-2 cell lines. Hence, this particular isolate AdF10 due to its better adherence capability as compared to others could serve as a promising probiotic candidate and can be targeted for more intensive in vivo studies in future. Adherent strains are preferred, since their establishment in the gut seems to be necessary for the probiotic effects to be exerted (Jacobsen et al. 1999). The variability in adherence pattern of isolates to both cell lines was observed in the present study and these findings are in agreement with the earlier studies where strain and species specific adherence of probiotic isolates was reported including variation depending on the cell culture used (Collado et al. 2006; Tallon et al. 2007; Garcia-Ruiz et al. 2014). The adhered indigenous lactic acid bacterial isolates with both cell lines viz., Caco-2 and HT-29 as examined microscopically after staining with giemsa stain are illustrated in Fig. 1a, b.
Table 2.
Adhesion of eleven indigenous isolates to Caco-2 and HT-29 cell lines
| S. no. | Indigenous isolates | Relative percent adhesion | ||
|---|---|---|---|---|
| Caco-2 | HT-29 | |||
| 1 | AdF1 | E. faecium | 3.96 ± 0.95hij | 6.31 ± 0.43gh |
| 2 | AdF2 | E. faecium | 3.78 ± 0.72hijk | 6.0 ± 0.65gh |
| 3 | AdF3 | E. faecium | 5.08 ± 1.0fgh | 8.43 ± 0.43def |
| 4 | AdF4 | B. coagulans | 2.45 ± 0.5k | 4.11 ± 0.68i |
| 5 | AdF5 | L. plantarum | 8.33 ± 1.21bc | 10.75 ± 0.56b |
| 6 | AdF6 | L. plantarum | 7.37 ± 1.09cd | 9.36 ± 0.49cd |
| 7 | AdF7 | L. fermentum | 6.15 ± 0.82def | 8.95 ± 0.62cde |
| 8 | AdF8 | L. fermentum | 5.51 ± 0.45efg | 8.0 ± 0.39ef |
| 9 | AdF9 | L. fermentum | 6.54 ± 0.97de | 9.57 ± 0.41c |
| 10 | AdF10 | L. plantarum | 9.55 ± 0.76ab | 12.88 ± 0.63a |
| 11 | AdF11 | E. faecium | 4.75 ± 0.55fghi | 6.84 ± 0.56g |
| 12 | Ref. strain | L. rhamnosus GG | 10.33 ± 0.64a | 13.5 ± 1.19a |
| CD (5%) | 1.412 | 1.048 | ||
Results are expressed as mean of triplicate values ± standard deviation. Different superscript letters indicate statistically significant (p < 0.05) differences in a column
Fig. 1.
A Adhesion of eleven indigenous lactic acid bacterial isolates to Caco-2 cell line observed under inverted microscope (40X) after staining with giemsa stain. a Blank Caco-2 cell line; b E. faecium AdF1; c E. faecium AdF2; d E. faecium AdF3; e B. coagulans AdF4; f L. plantarum AdF5; g L. plantarum AdF6; h L. fermentum AdF7; i L. fermentum AdF8; j L. fermentum AdF9; k L. plantarum AdF10; l E. faecium AdF11. B Adhesion of eleven indigenous lactic acid bacterial isolates to HT-29 cell line observed under inverted microscope (40X) after staining with giemsa stain. a E. faecium AdF1; b E. faecium AdF2; c E. faecium AdF3; d B. coagulans AdF4; e L. plantarum AdF5; f L. plantarum AdF6; g L. fermentum AdF7; h L. fermentum AdF8; i L. fermentum AdF9; j L. plantarum AdF10; k E. faecium AdF11; l L. rhamnosus GG ATCC-53103 (reference strain)
Human intestinal epithelial cells are widely used models to study the host-bacterial interactions, with different cell lines exhibiting specific characteristics and functions in the gut. In particular, the presence of mucus may play a significant role in bacterial adhesion (Gagnon et al. 2013). In the present study, percent adhesion of indigenous isolates to Caco-2 cells was relatively lower as compared to HT-29 cells which may be due to the high adhesiveness of probiotic bacteria to low mucus producing HT-29 cells as compared to non-mucus producing Caco-2 cells (Fig. 2). The observation in this regard is in agreement with the earlier studies where the similar adhesion ability of intestinal bacteria to different in vitro intestinal models with or without mucin was reported (Laparra and Sanz 2009; Duary et al. 2011; Gagnon et al. 2013). The epithelial cells of the gastrointestinal tract are covered by a layer of mucus which is the first physical barrier to the host-cell stimulation by bacteria in the gut (Tuomola et al. 1999; Van Tassell and Miller 2011). Therefore, the adhesion to mucus producing matrix is considered to be a prerequisite step required for probiotic organisms to interact with host cells to elicit any particular response (Ouwehand et al. 2001; Tuomola et al. 2000; Swidsinski et al. 2007). Similarly, in our study also, the indigenous isolates on low mucus producing HT29 intestinal cell model exhibited higher adhesion values as compared to non mucus producing Caco-2 cell model.
Fig. 2.
Comparative evaluation of Caco-2 and HT-29 cell lines for adhesion of eleven indigenous lactic acid bacterial isolates. Results are expressed as mean of triplicate values ± standard deviation. Different letters above the bars indicate statistically significant differences (p < 0.05)
Conclusion
The probiotic strains would persist in the gastrointestinal tract for a prolonged period of time if their adhesion is high. In line with this, the indigenous isolates, particularly AdF10 (L. plantarum) followed by AdF5 (L. plantarum) and AdF6 (L. plantarum) exhibit a strong adherence capacity as compared to others and thus could serve as a promising isolates for enriching local foods to harvest probiotic related health benefits. These isolates could further be used for the production of functional foods at the commercial scale.
Acknowledgements
The authors would like to acknowledge the financial assistance provided by the Department of Science and Technology, Govt. of India, New Delhi through an adhoc Project No. SEED/TSP/CODER/007/2012.
Contributor Information
Sakshi Sharma, Email: sakshisharma.pau@gmail.com.
Sarbjit Singh Kanwar, Email: sskanwar1956@gmail.com.
References
- Anderson RC, Cookson AL, McNabb WC, Kelly WJ, Roy NC. Lactobacillus plantarum DSM 2648 is a potential probiotic that enhances intestinal barrier function. FEMS Microbiol Lett. 2010;309:184–192. doi: 10.1111/j.1574-6968.2010.02038.x. [DOI] [PubMed] [Google Scholar]
- Chauviere G, Coconnier MH, Kerneis S, Fourniat J, Servin AL. Adhesion of Lactobacillus acidophilus strain LB to human enterocyte-like Caco-2 cells. J Gen Microbiol. 1992;138:1689–1696. doi: 10.1099/00221287-138-8-1689. [DOI] [PubMed] [Google Scholar]
- Chilton SN, Burton JP, Reid G. Inclusion of fermented foods in food guides around the world. Nutrients. 2015;7:390–404. doi: 10.3390/nu7010390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Jalonen L, Meriluoto J, Salminen S. Protection mechanism of probiotic combination against human pathogens: in vitro adhesion to human intestinal mucus. Asia Pac J Clin Nutr. 2006;15:570–575. [PubMed] [Google Scholar]
- Duary RK, Rajput YS, Batish VK, Grover S. Assessing the adhesion of putative indigenous probiotic Lactobacilli to human colonic epithelial cells. Indian J Med Res. 2011;134:664–671. doi: 10.4103/0971-5916.90992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon M, Zihler Berner A, Chervet N, Chassard C, Lacroix C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J Microbiol Methods. 2013;94:274–279. doi: 10.1016/j.mimet.2013.06.027. [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz A, de Llano DG, Esteban-Fernandez A, Requena T, Bartolome B, Moreno-Arribas MV. Assessment of probiotic properties in lactic acid bacteria isolated from wine. Food Microbiol. 2014;44:220–225. doi: 10.1016/j.fm.2014.06.015. [DOI] [PubMed] [Google Scholar]
- Gobbetti M, Di Cagno R, De Angelis M. Functional microorganisms for functional food quality. Crit Rev Food Sci Nutr. 2010;50:716–727. doi: 10.1080/10408398.2010.499770. [DOI] [PubMed] [Google Scholar]
- Goldin BR, Gorbach SL, Saxelin M, Barakat S, Gualtieri L, Salminen S. Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig Dis Sci. 1992;37:121–128. doi: 10.1007/BF01308354. [DOI] [PubMed] [Google Scholar]
- Hemarajata P, Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol. 2013;6:39–51. doi: 10.1177/1756283X12459294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Moller PL, Michaelsen KF, Paerregaard A, Sandstrom B, Tvede M, Jakobsen M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65:4949–4956. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci USA. 2009;106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar SS, Gupta MK, Katoch C, Kumar R, Kanwar P. Traditional fermented foods of Lahaul and Spiti area of Himachal Pradesh. Indian J Trad Knowl. 2007;6:42–45. [Google Scholar]
- Laparra JM, Sanz Y. Comparison of in vitro models to study bacterial adhesion to the intestinal epithelium. Lett Appl Microbiol. 2009;49:695–701. doi: 10.1111/j.1472-765X.2009.02729.x. [DOI] [PubMed] [Google Scholar]
- Lea T, et al. Caco-2 cell line. In: Verhoeckx K, et al., editors. The impact of food bio-actives on gut health. Cham: Springer International Publishing AG; 2015. pp. 103–111. [Google Scholar]
- Lesuffleur T, Barbat A, Dussaulx E, Zweibaum A. Growth adaptation to methotrexate of HT-29 human colon carcinoma cells is associated with their ability to differentiate into columnar absorptive and mucus-secreting cells. Cancer Res. 1990;50:6334–6343. [PubMed] [Google Scholar]
- Moussavi M, Adams MC. An in vitro study on bacterial growth interactions and intestinal epithelial cell adhesion characteristics of probiotic combinations. Curr Microbiol. 2009;60:327–335. doi: 10.1007/s00284-009-9545-1. [DOI] [PubMed] [Google Scholar]
- Ouwehand AC, Tuomola EM, Tolkko S, Salminen S. Assessment of adhesion properties of novel probiotic strains to human intestinal mucus. Int J Food Microbiol. 2001;64:119–126. doi: 10.1016/S0168-1605(00)00440-2. [DOI] [PubMed] [Google Scholar]
- Parvez S, Malik KA, Ah Kang S, Kim HY. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol. 2006;100:1171–1185. doi: 10.1111/j.1365-2672.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- Segers ME, Lebeer S. Towards a better understanding of Lactobacillus rhamnosus GG–host interactions. Microb Cell Fact. 2014;13:S1–S7. doi: 10.1186/1475-2859-13-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28:405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Servin AL, Coconnier MH. Adhesion of probiotic strains to the intestinal mucosa and interaction with pathogens. Best Pract Res Clin Gastroenterol. 2003;17:741–754. doi: 10.1016/S1521-6918(03)00052-0. [DOI] [PubMed] [Google Scholar]
- Shiby VK, Mishra HN. Fermented milks and milk products as functional foods. Crit Rev Food Sci Nutr. 2013;53:482–496. doi: 10.1080/10408398.2010.547398. [DOI] [PubMed] [Google Scholar]
- Sourabh A, Kanwar SS, Sharma PN. Diversity of bacterial probiotics in traditional fermented foods of Western Himalayas. Int J Probiotics Prebiotics. 2010;5:193–202. [Google Scholar]
- Sourabh A, Kanwar SS, Sharma OP. Antagonistic potential of indigenous bacterial probiotics of Western Himalayas against antibiotic-resistant bacterial pathogens. Curr Sci. 2011;101:1351–1356. [Google Scholar]
- Sourabh A, Walia S, Kanwar SS. Role of probiotics in colorectal cancer. J Pharm Biomed Sci. 2011;5:1–6. [Google Scholar]
- Swidsinski A, Loening-Baucke V, Theissig F, Engelhardt H, Bengmark S, Koch S, Lochs H, Dorffel Y. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007;56:343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallon R, Arias S, Bressollier P, Urdaci MC. Strain-and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J Appl Microbiol. 2007;102:442–451. doi: 10.1111/j.1365-2672.2006.03086.x. [DOI] [PubMed] [Google Scholar]
- Tuomola EM, Ouwehand AC, Salminen SJ. Human ileostomy glycoproteins as a model for small intestinal mucus to investigate adhesion of probiotics. Lett Appl Microbiol. 1999;28:159–163. doi: 10.1046/j.1365-2672.1999.00518.x. [DOI] [PubMed] [Google Scholar]
- Tuomola EM, Ouwehand AC, Salminen SJ. Chemical, physical and enzymatic pre treatments of probiotic lactobacilli alter their adhesion to human intestinal mucus glycoproteins. Int J Food Microbiol. 2000;60:75–81. doi: 10.1016/S0168-1605(00)00319-6. [DOI] [PubMed] [Google Scholar]
- Tuomola E, Crittenden R, Playne M, Isolauri E, Salminen S. Quality assurance criteria for probiotic bacteria. Am J Clin Nutr. 2001;73:393S–398S. doi: 10.1093/ajcn/73.2.393s. [DOI] [PubMed] [Google Scholar]
- Van Tassell ML, Miller MJ. Lactobacillus adhesion to mucus. Nutrients. 2011;3:613–636. doi: 10.3390/nu3050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenplas Y, Huys G, Daube G. Probiotics: an update. J Pediatr (Rio J) 2015;91:6–21. doi: 10.1016/j.jped.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Walia S, Keshani SS, Kanwar SS. Exhibition of DNA-bioprotective activity by microflora of traditional fermented foods of North-Western Himalayas. Food Res Int. 2014;55:176–180. doi: 10.1016/j.foodres.2013.11.001. [DOI] [Google Scholar]
- Walia S, Kamal R, Kanwar SS, Dhawan DK. Cyclooxegenase as a target in chemoprevention by probiotics during 1,2-dimethylhydrazine induced colon carcinogenesis in rats. Nutr Cancer. 2015;67:603–611. doi: 10.1080/01635581.2015.1011788. [DOI] [PubMed] [Google Scholar]
- Wang B, Wei H, Yuan J, Li Q, Li Y, Li N. Identification of a surface protein from Lactobacillus reuteri JCM1081 that adheres to porcine gastric mucin and human enterocyte-like HT-29 cells. Curr Microbiol. 2008;57:33–38. doi: 10.1007/s00284-008-9148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]