Abstract
Fc gamma receptors (FcγR) bind the Fc region of antibodies and therefore play a prominent role in antibody-dependent cell-based immune responses such as ADCC, CDC and ADCP. The immune effector cell activity is directly linked to a productive molecular engagement of FcγRs where both the protein and glycan moiety of antibody and receptor can affect the interaction and in the present study we focus on the role of the FcγR glycans in this interaction. We provide a complete description of the glycan composition of Chinese hamster ovary (CHO) expressed human Fcγ receptors RI (CD64), RIIaArg131/His131 (CD32a), RIIb (CD32b) and RIIIaPhe158/Val158 (CD16a) and analyze the role of the glycans in the binding mechanism with IgG. The interactions of the monoclonal antibody rituximab with each FcγR were characterized and we discuss the CHO-FcγRIIIaPhe158/Val158 and CHO-FcγRI interactions and compare them to the equivalent interactions with human (HEK293) and murine (NS0) produced receptors. Our results reveal clear differences in the binding profiles of rituximab, which we attribute in each case to the differences in host cell-dependent FcγR glycosylation. The glycan profiles of CHO expressed FcγRI and FcγRIIIaPhe158/Val158 were compared with the glycan profiles of the receptors expressed in NS0 and HEK293 cells and we show that the glycan type and abundance differs significantly between the receptors and that these glycan differences lead to the observed differences in the respective FcγR binding patterns with rituximab. Oligomannose structures are prevalent on FcγRI from each source and likely contribute to the high affinity rituximab interaction through a stabilization effect. On FcγRI and FcγRIIIa large and sialylated glycans have a negative impact on rituximab binding, likely through destabilization of the interaction. In conclusion, the data show that the IgG1-FcγR binding kinetics differ depending on the glycosylation of the FcγR and further support a stabilizing role of FcγR glycans in the antibody binding interaction.
Immunoglobulin G1 (IgG1)1 antibodies are glycoproteins that are generated by adaptive immune cells (B cells) as a means to opsonize pathogens such as bacteria, viruses and parasites, to neutralize toxins or to detect transformed cells via recognition of tumor associated antigens. IgG1 opsonized immune complexes induce effector responses including antibody dependent cellular phagocytosis (ADCP), antibody dependent cellular cytotoxicity (ADCC), and complement dependent cytotoxicity (CDC) in innate immune cells such as macrophages, neutrophils, and natural killer cells through engagement with cell surface membrane bound Fc gamma receptors (FcγRs). FcγRs recognize and interact with the Fc region of the antibody and together they play a critical role in the immune response following recognition of invading particles and tumor-associated antigens. FcγRs can therefore be a link between the innate and adaptive immune systems. Depending on the type and abundance of FcγR in the plasma membrane the effector cell can enter either a state of activation through engagement of activating FcγRs (FcγRI/CD64, FcγRIIa/CD32a, and FcγRIIIa/CD16a) or deactivation through the inhibitory receptor FcγRIIb/CD32b following engagement of opsonized immune complexes and receptor clustering in the membrane (1). FcγRs can therefore also be viewed as regulators of the immune system (2). Receptors can be further classified into high affinity FcγRI (KD 10−9 m) and lower affinity FcγRII and FcγRIII (KD 10−6-10−7 m). Except for FcγRIIIb, which is GPI-linked, intracellular signaling cascades are activated following FcγR engagement and activation of immuno-receptor tyrosine-based activation (ITAM) or inhibitory (ITIM) motifs in the cytoplasmic tail of the receptor, which in turn leads to cellular activation/deactivation and effector/inhibitory responses.
Significant heterogeneity exists in FcγRs because of differences in glycosylation and polymorphisms found in the extracellular domain of the receptor. Two polymorphic variants of FcγRIIIa exist with a phenylalanine (Phe) or valine (Val) residue in position 158 (3). These polymorphisms have significant effects on the binding interaction with IgG1 antibodies with the Val 158 variant showing higher affinity (4, 5). This difference in affinity also translates into physiological effects, where patients with the Val 158 form respond better to rituximab treatment (6, 7). Physiological effects are also observed with the Phe 158 variant linked to rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) in patients with this form of FcγRIIIa (8, 9). The observed differences in rituximab binding can be partly explained by greater steric hindrance with the bulkier Phe 158 polymorphism. Polymorphisms of FcγRIIa (Arg 131/His 131) exist, which also result in differences in the binding affinity for IgG with human IgG2 binding efficiently to the His 131 variant and not to the Arg 131 variant (10). Physiological effects also exist with the Arg 131 form causing increased susceptibility to bacterial infections, nephropathy and SLE (11–13). Polymorphic variants have also been reported for FcγRIIb where an isoleucine to threonine switch in position 232 is associated with an increase in susceptibility to SLE and autoimmunity, although no data on the effect of these polymorphisms on IgG1 binding is available (14, 15). A polymorphism in FcγRIIIb in position 233 (cysteine to alanine) is also believed to result in differences in IgG binding and recently increased protection against malaria has been associated with the Ala 233 variant (16, 17).
Glycosylation is an important modification of FcγRs, which further increases their heterogeneity and complexity and the properties of the IgG1-FcγR interaction are influenced by the glycosylation of both binding partners (see Hayes et al., 2014 and Hayes et al., 2016 for reviews of FcγR glycosylation (18, 19)). N-linked glycosylation of IgG1 on Asn 297 in both antibodies heavy chains plays a critical role in the antibodies function and removal of the glycans from the CH2 domain of the Fc region causes a loss of structural integrity, reduced interaction with the FcγR and loss of effector function (20, 21). The IgG N-glycans have been shown to be flexible and dynamic and their compositions have also been shown to differentially affect the binding to each low affinity FcγR (CD16 A/B, CD32 B/C) (22, 23). Several studies have also established the importance of FcγR glycosylation on the binding mechanism with IgG1. N-glycans found on Asn 162 in FcγRIIIa have been shown to be important for antibody binding and mutation of this asparagine residue or elimination of the sugar moiety has been shown to reduce the binding affinity of afucosylated IgG1 by over one order of magnitude (24). This can be explained by carbohydrate-carbohydrate interactions between the glycans present on Asn 162 in FcγRIIIa and the Asn 297 glycans of the IgG1-Fc (25). Glycosylation on an additional asparagine residue in FcγRIIIa (Asn 45) has been shown to negatively regulate IgG1 binding (26). We have recently reported that the glycans of FcγRI and FcγRIIIaPhe158/Val158 produced in HEK293 cells influence the IgG1-FcγR interaction and that there is a decrease in the binding levels and an increase in the dissociation rate of antibody if the receptor glycans are removed (27). Together, these results show that FcγR glycosylation has a direct impact on IgG1 binding and can positively and negatively influence the antibody interaction and likely downstream immune effector cell functions.
There are many studies that have investigated IgG-FcγR binding mechanisms, however, reported affinities of the same IgG1-FcγR interaction may differ up to 10-fold depending on the source of IgG1 and FcγR and the different surface plasmon resonance (SPR) experimental set-ups used (28, 29). Affinities reported are consistently in the micromolar range for FcγRII and FcγRIII and in the nanomolar range for FcγRI. Adding to the complexity of the IgG1-FcγR interaction and increasing the difficulty in analyzing the binding kinetics is the significant heterogeneity found in both antibody (e.g. fucosylated and afucosylated) and the FcγR because of glycosylation. In the present study, we describe the glycosylation of human FcγRs expressed in Chinese hamster ovary cells and compare the CHO-FcγRI and FcγRIIIa forms to those expressed in murine (NS0) and human (HEK293) cells. We identify N-glycans from each of the differently expressed FcγRI and FcγRIIIa preparations and link the observed differences in FcγR glycosylation to differences in antibody binding to identify the glycans that influence the antibody binding interaction. The information adds to our understanding of the IgG1-FcγR interaction, particularly the impact of FcγR glycosylation and our findings suggest that variations in glycan patterns of FcγRs can influence the results obtained from IgG1-FcγR interaction experiments and likely influence the antibody mediated immune response. Fundamentally, results suggest that glycosylation can be used as a mechanism by the immune system to fine-tune antibody mediated immune responses based on how an immune cell can glycosylate FcγRs in a cell type specific manner.
EXPERIMENTAL PROCEDURES
Production of Human FcγRs in CHO Cells
Human FcγRs (FcγRI, FcγRIIaArg131/His131, FcγRIIb, FcγRIIIaPhe158/Val158) were recombinantly produced as C-terminal hexa-histidine tagged proteins through standard cloning procedures. FcγR constructs used for expression purposes have the following predicted N-glycosylation sites: FcγRI (7), FcγRIIa (2), FcγRIIb (3), FcγRIIIa (5), FcγRIII (6), For FcγRIIIa variants Val 158 and Phe 158 single clones were used for production of recombinant receptor and for FcγRI and FcγRIIa/b pools of clones were used. Stably transfected CHO-DG44 cells expressing FcγRs were selected by dihydrofolate reductase and methotrexate selection. Hexa-histidine tagged FcγRs were purified to homogeneity from CHO cell media by a sequence of Ni-NTA affinity chromatography, IgG Sepharose chromatography and size exclusion chromatography. Proteins were concentrated to ∼2–3 mg/ml by ultrafiltration for glycan and SPR analysis. Pooled glycans from different batches of FcγRs were used for analysis. The recombinant FcγRs used in this study were produced for research purposes only.
N-linked Glycan Release and 2-aminobenzamide (2-AB) Labeling of CHO Expressed Human Fcγ Receptors
Fcγ receptors used for glycosylation analysis and IgG1 binding studies were provided by Boehringer-Ingelheim (Germany). N-linked glycans were released and labeled by the SDS-PAGE immobilization and release method described by Royle et al. (30). Approximately 100 μg of individual receptors were resolved by SDS-PAGE and visualized by Coomassie® blue staining. Bands were excised from SDS-PAGE gels and glycans were released by incubation at 37 °C overnight with 0.1 mU recombinant peptide-N-glycosidase F (Prozyme, Hayward, CA). Released glycans were extracted from gel slices and labeled with 2-AB (Ludger, Oxfordshire, UK) according to the manufacturer's instructions and purified using normal phase resin columns. (Phytips, Phynexus, San Jose, CA).
Hydrophilic Interaction Ultra Performance Liquid Chromatography (HILIC UPLC)
2-AB labeled glycans were prepared at 80% (v/v) acetonitrile solutions for analysis by HILIC UPLC using a Waters BEH-Glycan 1.7 μm (150 mm x 2.1 mm) column and fluorescence detection (excitation at 420 nm and emission at 330 nm) as previously described (31). All separations were performed using a Waters Acquity H-Class UPLC instrument over a 30-min period at 40 °C using 50 mm ammonium formate pH 4.4 as solvent A and 100% (v/v) acetonitrile as solvent B. 2-AB-labeled dextran was used as an internal calibration standard and for the generation of glucose unit (GU) values. Retention times for 2-AB labeled dextran peaks were used to fit a fifth order polynomial distribution curve using Waters Empower 3 software, which provided the means for converting chromatographic retention times into standardized GU values as previously described (30). Glycans were named according to the Oxford notation (32).
Weak Anion Exchange HPLC (WAX-HPLC)
WAX-HPLC was used to analyze the extent of terminal sialylation of receptor N-glycans and for fractionation of charged structures. A Waters Biosuite® DEAE 10 μm AXC (7.5 mm × 75 mm) column was used for separation of charged carbohydrate structures. Solvent A was 100 mm ammonium acetate pH 7 in 20% (v/v) methanol and solvent B was 20% (v/v) acetonitrile. All WAX-HPLC separations were performed on a Waters Alliance 2695 instrument with online Waters 4795 fluorescence detector. Carbohydrate samples were separated over a 30-min gradient. The relative proportions of terminal charged glycan structures were determined by comparison to N-linked glycans released from bovine fetuin, which contains neutral, mono-, di-, tri-, and tetra-sialylated structures. For WAX-fractionation experiments glycan pools were separated by WAX-HPLC and peaks from this analysis were manually collected. Glycans were dried, reconstituted in MilliQ water and analyzed by HILIC UPLC. This 2-dimensional approach was used to deconvolute the complex FcγR N-glycan pools. For UPLC-FLD-QToF analysis, peaks from HILIC UPLC analysis of WAX fractionated glycan pools were manually collected, dried and reconstituted in 10 μl MilliQ water.
Exoglycosidase Panel Digests
Exoglycosidase arrays were used to sequence the N-Glycans of human FcγRs. Using this technique, sequence, composition and linkage specificity information was obtained. Fluorescently labeled glycans were routinely digested in 50 mm sodium acetate, pH 5.5 at 37 °C overnight in a volume of 10 μl using a selected panel of exoglycosidase enzymes. The following day enzymes were removed using 10 kDa spin filters (Pall Corp, NY). Digested 2-AB-labeled glycans were analyzed by HILIC UPLC as described previously. The specificities of the exoglycosidase enzymes used are well-established and specific monosaccharides were removed as follows: terminal sialic acid in all linkages: α(2,6), α(2,3), α(2,8) is removed with 1 mU/μl Arthrobacter ureafaciens sialidase (ABS)(Prozyme); terminal galactose monosaccharides were removed using 0.5 mU/μl bovine testes β-galactosidase (BTG)(Prozyme), which releases both β(1,3)- and β(1,4)-linked galactose; terminal N-acetylglucosamine (GlcNAc) monosaccharides in β(1,4) linkage were released with 40 mU/μl Streptococcus pneumoniae hexosaminidase (GUH)(Prozyme); core α(1,6)-fucose was selectively removed using 1 mU/μl bovine kidney α-fucosidase (BKF)(Prozyme); terminal nonreducing end fucose in α(1–3)- and α(1–4)- linkage was removed using 0.004 mU/μl almond meal α-fucosidase (AMF)(Prozyme), α-linked mannose was removed using 150 mU/μl jack bean α-mannosidase (JBM)(Prozyme) and βGalNAc and βGlcNAc were removed using 50 mU/μl jack bean β-N-acetylhexosaminidase (JBH)(Prozyme).
UPLC-FLR-MS of N-Glycans
Labeled N-glycans were separated using a Waters ACQUITY UPLC BEH Amide column (1.0 × 150 mm, 1.7 μm particle) connected to an ACQUITY UPLC equipped with online fluorescence detection (Waters Corporation, Milford, MA) under the control of MassLynx 4.1 Software. The fluorescence excitation and emission wavelength parameters were 350 and 425 nm, respectively. The flow rate was 0.150 ml/min and column temperature was maintained at 60 °C. Solvent A was 50 mm ammonium formate pH 4.4 and solvent B was acetonitrile. A 40-min linear gradient was used and was as follows: 28% A for 1 min, 28–43% A for 30 min, 43–70% A for 1 min, 70% A for 3 min, 70–28% solvent A for 1 min, and finally 28% A for 4 min. Samples were diluted in 75% acetonitrile prior to analysis; the injection volume was 8 μl. The outlet of the chromatographic system was coupled directly to a Waters Xevo G2 QToF mass spectrometer (Milford, MA) equipped with an electrospray ionization interface. The instrument was operated in negative ion mode with a capillary voltage of 1.80 kV. The ion source and nitrogen desolvation gas temperatures were set at 120 °C and 400 °C, respectively. The desolvation gas flow rate was 600 L/h. The cone voltage was maintained at 50 V. Full-scan MS data was acquired over the range of 450 to 2500 m/z. Data collection and processing was controlled using MassLynx 4.1 (Waters Corporation, Milford, MA). To avoid contamination of the instrument, the column flow was diverted to waste for the first 1.2 min and after 32 min of the chromatographic run.
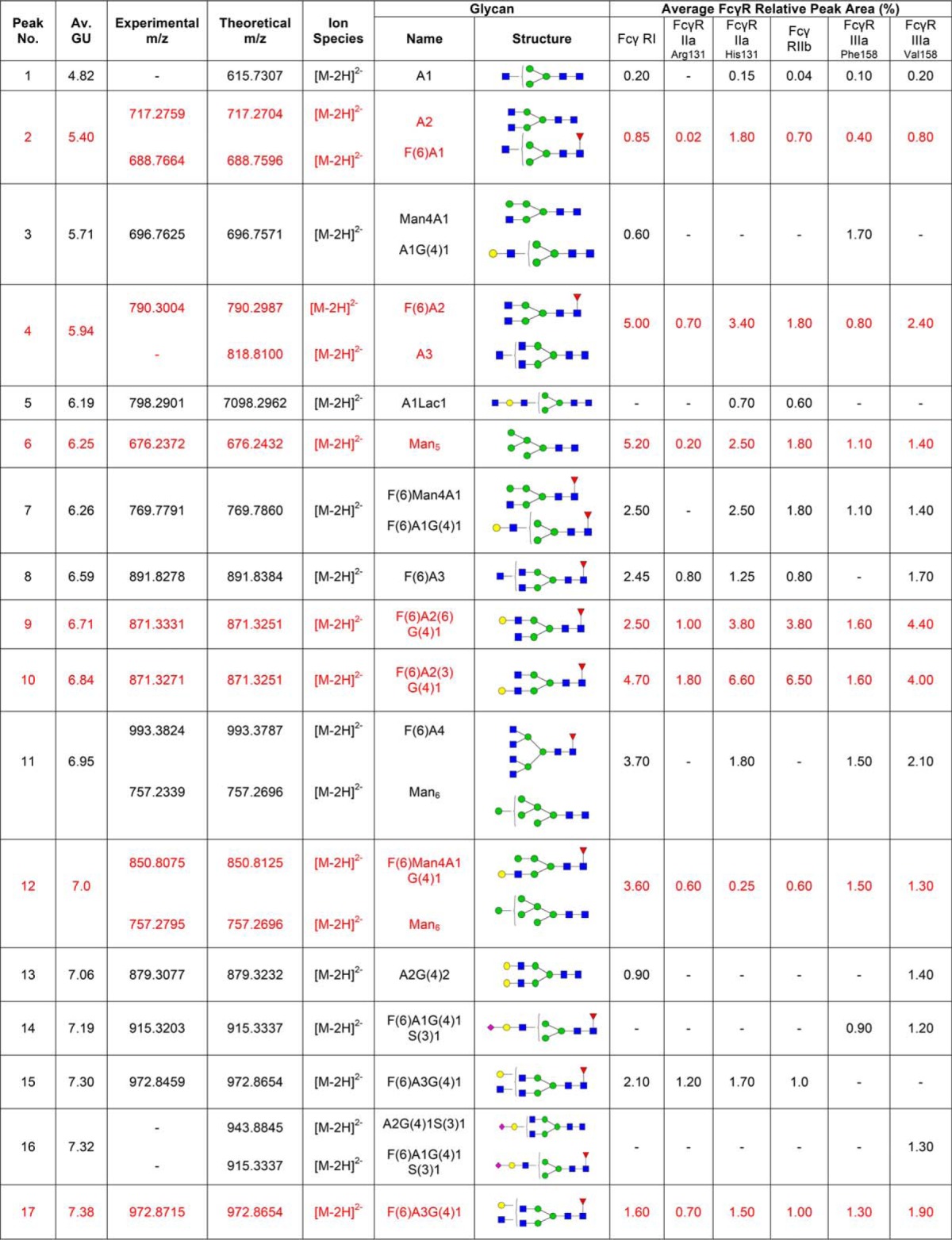
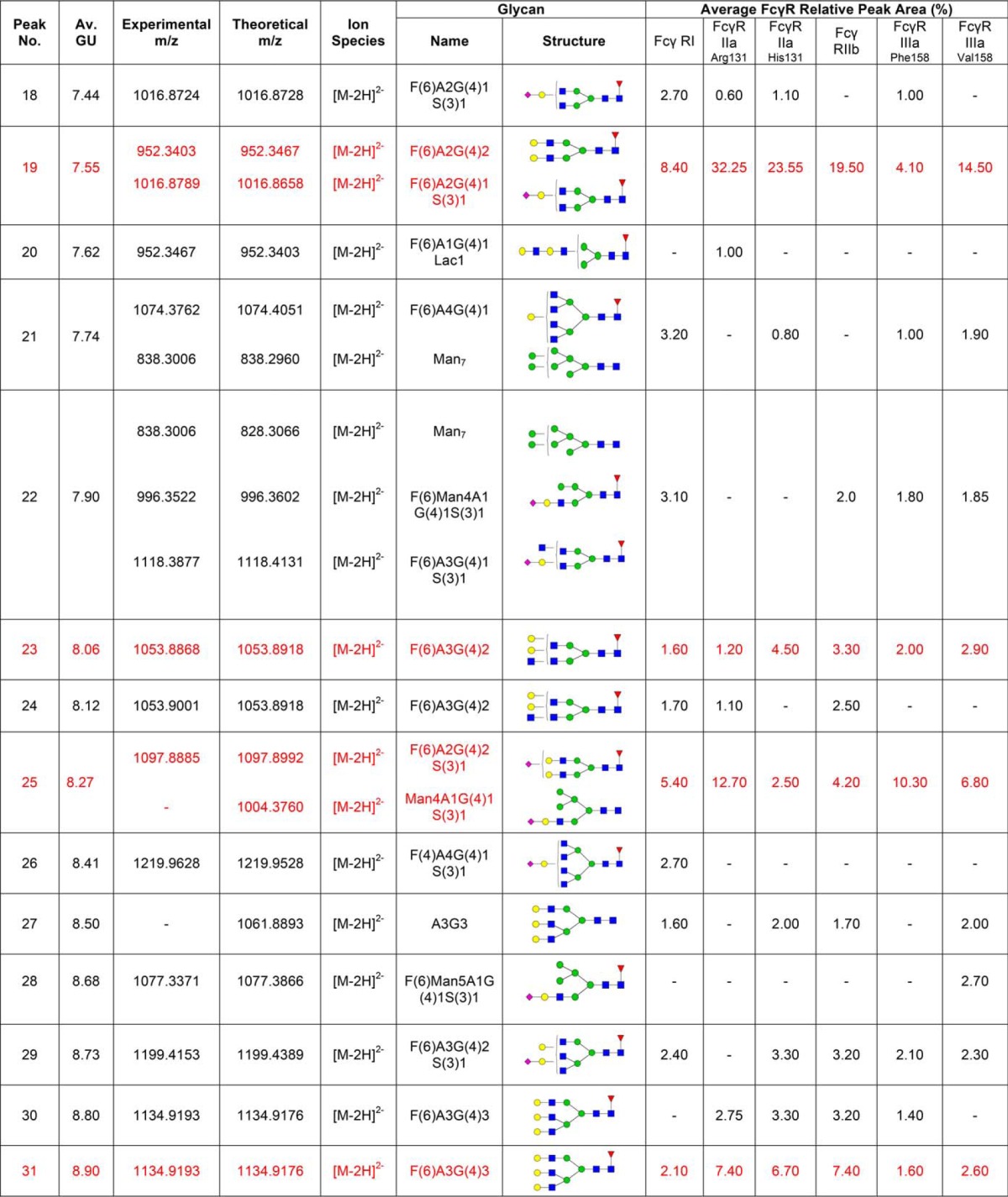
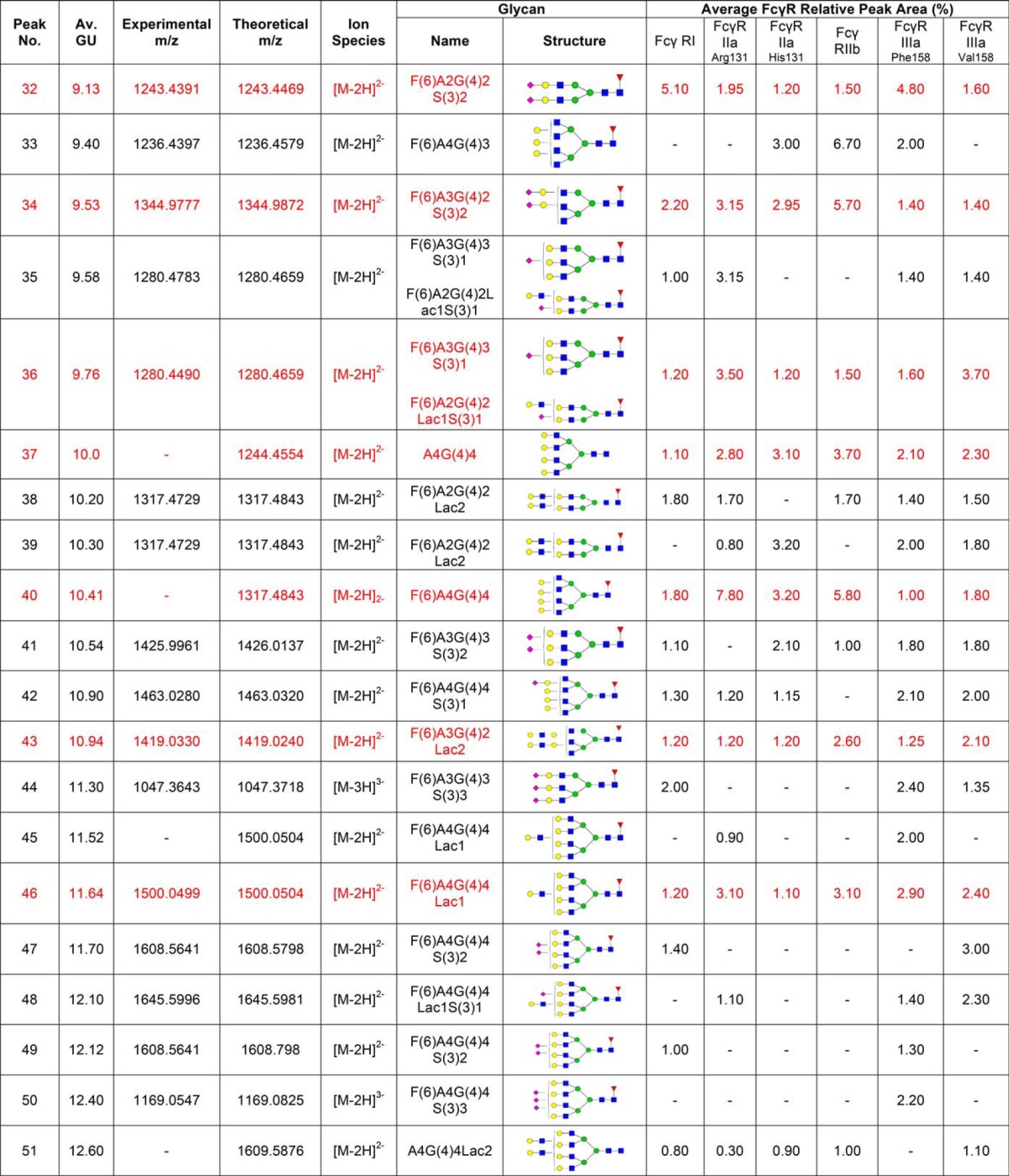
Following acquisition, annotation of mass spectra of glycans was performed manually using GlycoWorkbench 2.0 (33). GlycoWorkbench is a suite of software tools designed for the rapid drawing of glycan structures and for assisting the process of structure determination from mass spectrometry data. Experimental masses observed above a 1500 counts intensity threshold were considered as putative glycan structures, which also showed a peak in the fluorescence trace (Table I). Searching parameters included 2-AB as the reducing end and 20 parts-per-million (ppm) accuracy. Glycan annotation was supported by the following databases: CFG, Carbbank, GlycomeDB, and Glycoscience, as part of the GlycoWorkbench tool.
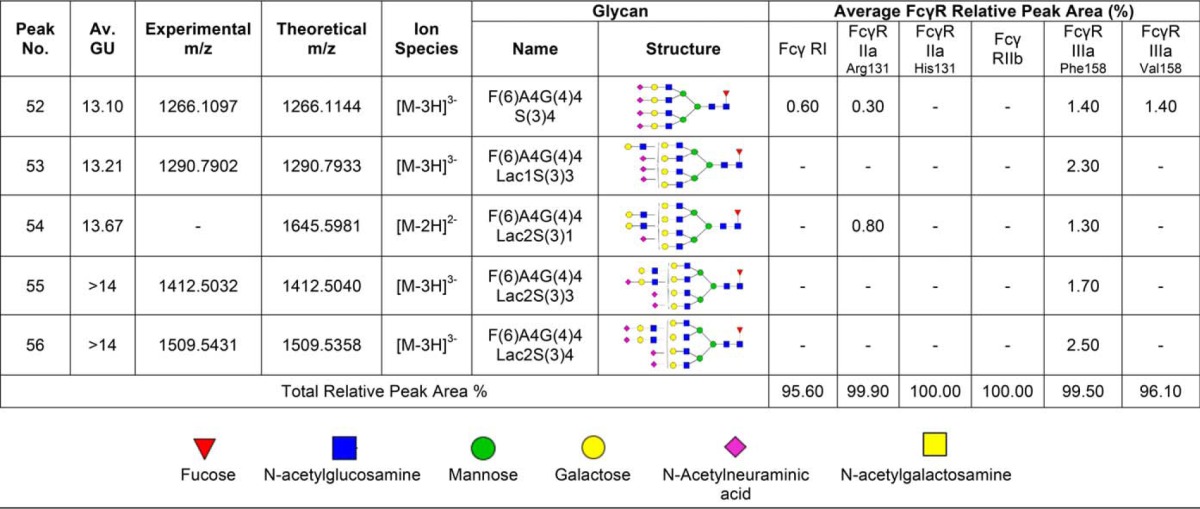
Table I. N-glycans of human FcγRs expressed in CHO cells. Glycan structures shown in red are common to each FcγR and correspond to the peaks indicated by dashed lines shown in Fig. 1A and 1B. Glycans were identified and quantified using a combination of HILIC UPLC, exoglycosidase digestions, WAX HPLC, WAX fractionation and mass spectrometry. Average GU (Glucose Unit) values and % areas are the mean of three individual glycan releases. Glycans shown in red correspond to the common glycans shown in Fig. 1. MS spectral annotation was performed using GlycoWork Bench 2.0 (33). Glycans are named according to the Oxford notation. Glycan symbols used are shown in the legend below Table I.
FcγR-Rituximab Analyses by Surface Plasmon Resonance (SPR)
FcγR-rituximab binding analysis was performed using a Biacore T200 system (GE Healthcare, Uppsala, Sweden) with analysis temperature set to 25 °C and sample compartment temperature to 15 °C. Series S Sensor Chip CM5 and PBS-p + buffer, pH 7.4 were from GE Healthcare. Recombinant C-terminal hexa-histidine tagged extracellular domains of FcγRs expressed in HEK, NS0, and CHO cell lines were from Sino Biological Inc., Beijing, China; R&D Systems, MN; and Boehringer-Ingelheim, Biberach/Riss, Germany, respectively. Rituximab (lot number: B6001B02) was pharmacy obtained and stored according to the manufacturer's instructions. A His Capture Kit (GE Healthcare) was used for capture of His tagged FcγRs. The anti-His antibody was amine coupled to the sensor chip, according to the manufacturer′s instructions. Immobilization levels in the range 6000–8000 RU were used, with similar levels in the active and reference flow cells. His-tagged FcγRs (0.5–1 μg/ml) were injected for 60 s in the active flow cell only, obtaining capture levels in the range 90–250 RU for kinetic data. A capture stabilization time of 1–3 min was typically applied. Rituximab was injected in order of increasing concentration over reference and active flow cell applying a single cycle kinetics procedure (34), except for the injection time experiment. Rituximab concentrations in the range 1.2–300 nm were used for FcγRI and 24.7–2000 nm for FcγRIIIa, using the manufacturer's specifications. The same concentration series of rituximab from the same preparation was used for each comparison results series, to ensure that the receptor would be the only reagent that was varied in the experiment. Following each experiment both flow cells were regenerated. Blank cycles (FcγR capture + buffer injections + regeneration) were performed first and last for each FcγR. For the saturation experiments ∼100 RU FcγR was captured followed by injections of rituximab at a saturating concentration (6 μm) ensuring that saturating responses were obtained for the different injection times, 10, 20, and 60 s. Sensorgrams were then aligned at the end of injection and normalized between 0 and 100. All data were double referenced firstly by subtraction of the reference flow cell data and then subtraction of blank cycles. Data analysis was performed using Biacore T200 evaluation software 3.0.
RESULTS
Glycan Analysis of CHO Expressed FcγRs
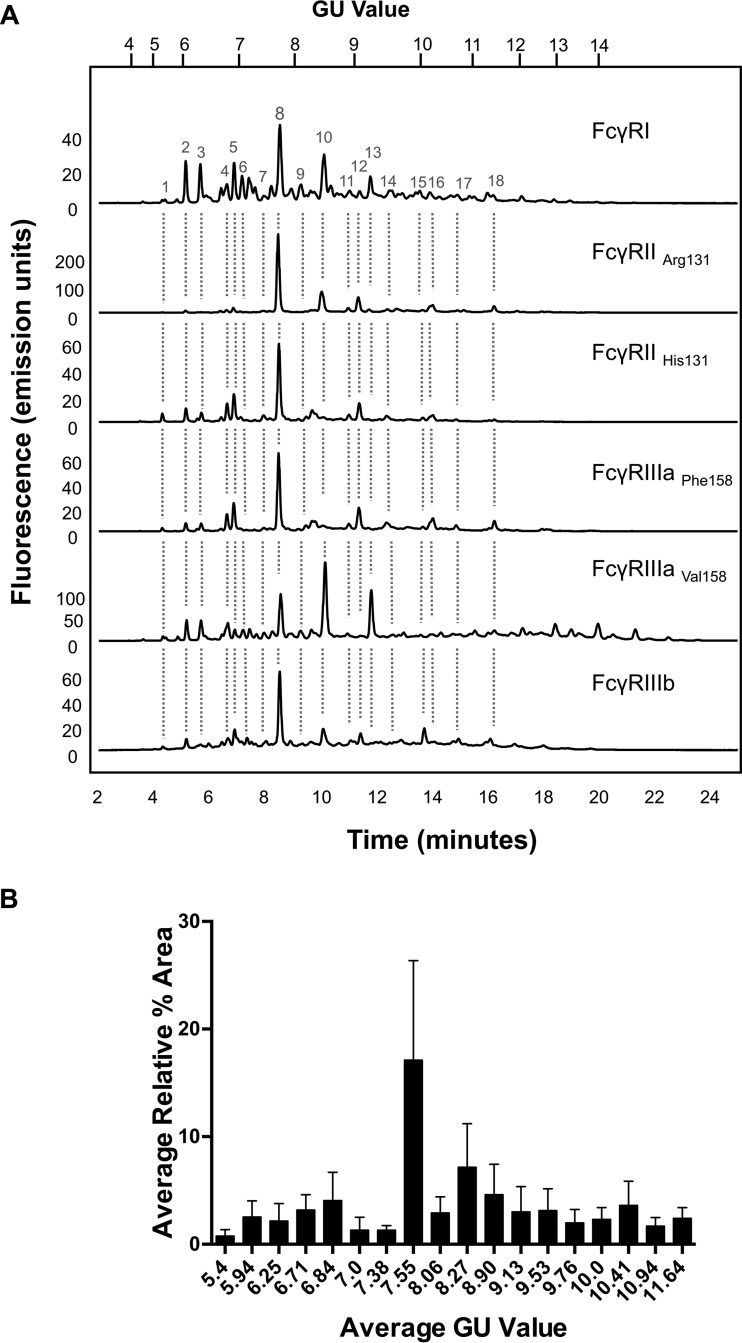
Routine glycan analysis of CHO derived FcγRs (FcγRI, FcγRIIaArg131/His131, FcγRIIb, FcγRIIIaPhe158/Val158) was performed on pooled samples in triplicate by immobilization of proteins in SDS-PAGE gels and in-gel PNGase F release of N-glycans followed by fluorescent labeling with 2-AB and HILIC UPLC analysis. Triplicate glycan analysis of the labeled FcγR N-glycans showed that the results were reproducible with minor differences in relative glycan abundance. When different FcγRs were analyzed large differences in the glycan profiles were observed (see Fig. 1A) with the main difference being in the relative abundance of the individual glycans (see Fig. 1B).
Fig. 1.
CHO expressed human FcγRs display complex and differential glycosylation. A, Glycan analysis of FcγRs expressed in CHO cells. Glycosylation of receptors was complex and differential with 30–40 unique glycan structures for each receptor. Following enzymatic release and HILIC UPLC analysis common structures (18) were identified and are indicated by dashed vertical lines. Numbered peaks represent the common glycan structures shown in Table I in red. GU values were assigned using internal 2-AB labeled dextran standards and integration using Waters Empower 3 software. Glycan structures and relative abundance for each receptor are shown in Table I. Glycan release and analysis experiments were performed in triplicate. B, FcγRs contain common N-glycans with differences in relative abundance. Average peak area percentages were calculated for peaks from individual receptors (FcγRI, FcγRIIaArg131, FcγRIIaHis131, FcγRIIb, FcγRIIIaPhe158, FcγRIIIaVal158). Eighteen common peaks were identified (see Fig. 1A) and values were plotted as the average GU value and average % area from three different releases of individual receptors. This value was then used to calculate the average value across the range of receptors for each peak to show the distribution in relative abundance between different receptors. Error bars represent the standard deviation for the relative % areas. Large error bars as seen for certain peaks e.g. GU 7.55 (FA2G2) signifying large differences in abundance for certain glycans between receptors. N-glycan structures corresponding to the GU values shown can be seen in red in Table I.
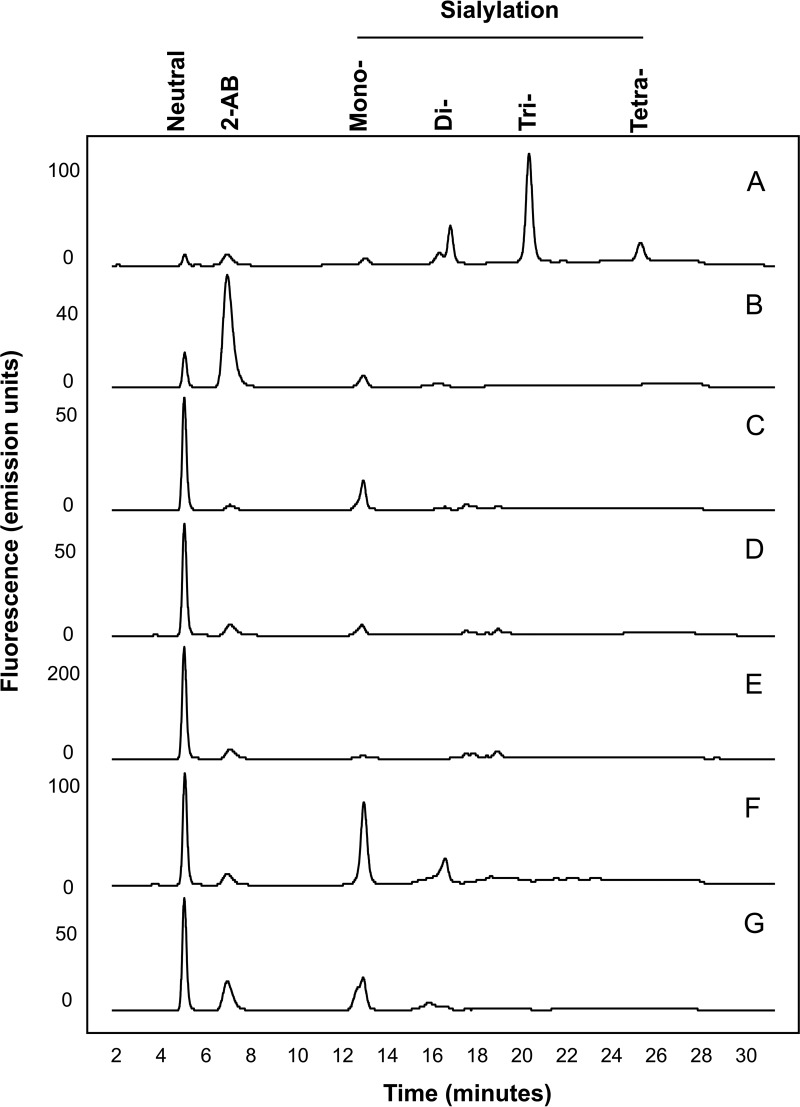
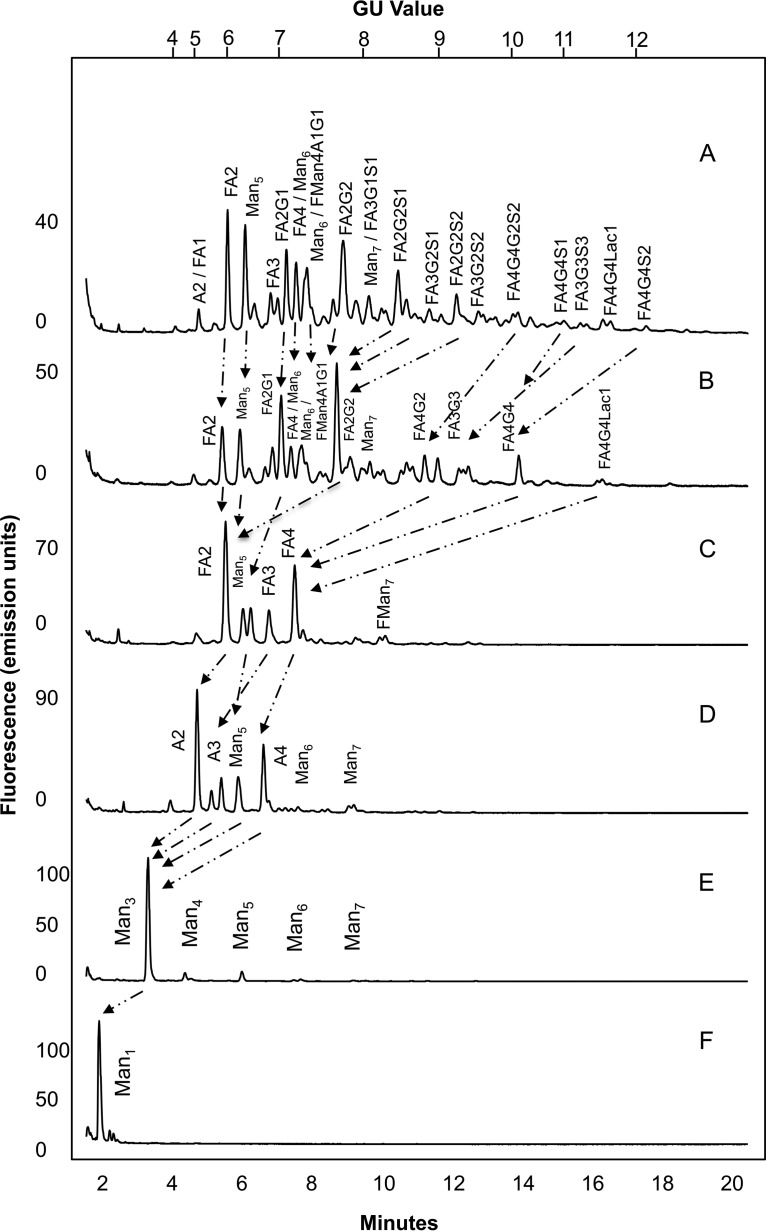
Weak anion exchange (WAX) HPLC was used to analyze the extent of charged glycan structures present on each receptor, typically sialic acids. The majority of N-glycans were found to be composed of mainly neutral and mono-sialylated species with smaller amounts of higher sialylated structures (see Fig. 2). Sialidase treatment followed by WAX analysis was further used to confirm the presence of the mainly neutral and mono-sialylated glycans as the sialylated peaks migrated to the neutral fraction following treatment. WAX HPLC followed by HILIC UPLC was further used to de-convolute the complex pools of glycans in a two-dimensional approach. To sequence the N-glycans of FcγRs and determine their monosaccharide compositions and linkage information a series of exoglycosidase digestions were performed and analyzed by HILIC UPLC (see Fig. 3 and supplementary Figs. 1–5). Panel digests including α-sialidase (ABS), β-galactosidase (BTG), α-fucosidase (BKF), β-hexosaminidase (GUH), and α-mannosidase (JBM) typically reduce N-glycans to common core Man1 structures and this approach allowed the tentative identification of N-glycans for each CHO produced FcγR (see Table I). In summary, typical CHO glycosylation was observed for all FcγRs with complex mulrtiantennary structures and smaller amounts of oligomannose and hybrid structures without bisecting GlcNAc (see Table II). The large majority of glycans were bi-, tri-, and tetra-antennary structures that were core-fucosylated with one, two, three or four galactose residues. Glycans were mainly neutral or mono-sialylated with limited amounts of higher forms of sialylation and minor amounts of tri- and tetra-sialylated structures (see Table II). Also present were minor amounts of poly-N-lactosamine (LacNAc) residues of varying complexity. In addition, comparison of digests following ABS (α-2,3 and α-2,6) and Nan1 (α-2,3) sialidase treatment revealed that all sialic acids were in α-2,3 linkage and comparison of digests with BTG (β-1,3 and β-1,6) and SPG (β-1,4) galactosidases revealed that all galactose residues were in β-1,4 linkage. Mass spectrometry (LC/FLD-MS) was used to verify the glycan compositions (see supplementary Figs. 6–52).
Fig. 2.
WAX HPLC analysis of FcγRs reveals charged glycan species and shows that glycans are mainly neutral and mono-sialylated. N-glycans of FcγRs are mainly neutral and mono-sialylated. Sialidase treatment followed by WAX analysis revealed that peaks are predominantly sialic acids. Bovine fetuin, which contains mono-, di-, tri-, and tetra-sialylated glycans was used as a standard to identify the sialylated species of FcγRs. FcγRIIIaPhe158 has the highest proportion of mono- and di-sialylated glycans, particularly in comparison to FcγRIIIaVal158. A, Bovine Fetuin; B, FcγRI; C, FcγRIIaArg131; D, FcγRIIaHis131; E, FcγRIIb; F, FcγRIIIaPhe158; G, FcγRIIIaVal158.
Fig. 3.
Exoglycosidase sequencing of the FcγR N-glycans was used to identify the monosaccharide compositions and linkage information. Arrows indicate the migrations of peaks following exoglycosidase digestion and HILIC UPLC analysis A. Undigested FcγRI glycan profile B. Arthrobacter ureafaciens sialidase (ABS) C. bovine testes β-galactosidase (BTG) D. bovine kidney α-fucosidase (BKF), E. jack bean β-N-acetylhexosaminidase (JBH), F. jack bean α-mannosidase (JBM). Peaks migrate to Man1 structures following digestion. Exoglycosidase sequencing was performed for each FcγR. The exoglycosidase panel digest for FcγRI is shown. See Supplementary Figs. for the Exoglycosidase panel digests for FcγRI, FcγRIIaArg131, FcγRIIaHis 131, FcγRIIIaPhe 158, FcγRIIIaVal158.
Table II. Summary of CHO expressed human FcγRs. Numbers are expressed as percentage values of the total glycan abundances.
| FcγRI | FcγRIIa Arg131 | FcγRIIa His131 | FcγRIIb | FcγRIIIa Phe158 | FcγRIIIa Val158 | |
|---|---|---|---|---|---|---|
| Complex | 76.00 | 98.10 | 92.90 | 93.10 | 90.55 | 81.75 |
| Hybrids | 7.10 | 1.30 | 2.10 | 3.80 | 4.95 | 9.30 |
| High Mannose | 12.50 | 0.50 | 5.00 | 3.10 | 4.00 | 3.95 |
| Total | 95.60 | 99.90 | 100.00 | 100.00 | 99.50 | 96.10 |
| Antennarity | ||||||
| mono (A1) | 3.40 | 0.80 | 4.00 | 2.20 | 4.20 | 2.90 |
| bi (A2) | 32.00 | 54.50 | 50.55 | 41.90 | 31.90 | 43.45 |
| tri (A3) | 27.20 | 25.05 | 23.70 | 30.35 | 25.75 | 25.50 |
| tetra (A4) | 13.45 | 18.70 | 12.75 | 18.65 | 29.20 | 22.40 |
| Galactosylation | ||||||
| No Galactose (G0) | 25.15 | 4.80 | 10.5 | 8.30 | 7.15 | 11.40 |
| mono (G1) | 23.80 | 5.60 | 15.70 | 19.60 | 10.05 | 18.30 |
| di (G2) | 26.89 | 53.44 | 47.00 | 35.70 | 35.85 | 31.85 |
| tri (G3) | 7.40 | 17.55 | 18.3 | 21.50 | 17.60 | 12.85 |
| tetra (G4) | 12.40 | 14.50 | 9.50 | 14.90 | 28.85 | 21.70 |
| Sialylation | ||||||
| neutral (S0) | 55.60 | 67.90 | 85.65 | 81.70 | 54.40 | 57.65 |
| mono (S1) | 26.60 | 25.80 | 8.10 | 10.1 | 23.30 | 24.90 |
| di (S2) | 10.80 | 5.90 | 6.25 | 8.20 | 9.30 | 7.80 |
| tri (S3) | 2.00 | 0.00 | 0.00 | 0.00 | 8.60 | 1.35 |
| tetra (S4) | 0.60 | 0.30 | 0.00 | 0.00 | 3.90 | 1.40 |
| Core-fucosylation | 74.50 | 95.00 | 89.35 | 76.15 | 73.90 | 78.75 |
Comparison of CHO, NS0 and HEK293 FcγRI Glycosylation
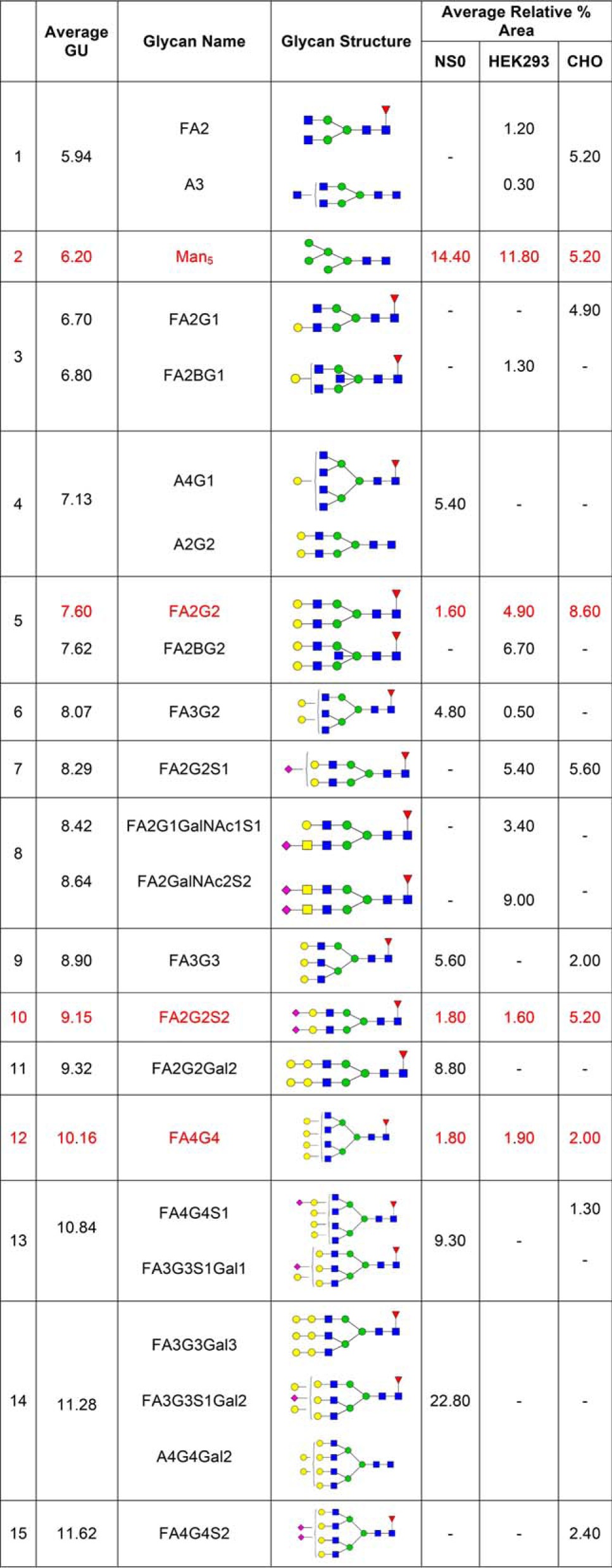
Previous glycan analysis of recombinant human FcγRs expressed in murine (NS0) and human (HEK293) cells (27, 35) was extended to describe the glycan compositions of CHO expressed FcγRs and compare the differently expressed receptors to identify common and mechanistically important glycan structures. Because of the unusual and complex rituximab binding kinetics exhibited by FcγRI and FcγRIIIaPhe158/Val158, which we attribute in part to heterogeneity because of glycosylation of the receptors, our analysis focused on these receptors and the role of receptor N-glycans in the antibody binding mechanism. Common glycans and their relative abundances for FcγRI from the NS0, HEK293, and CHO expression systems are shown in Table III. The most abundant glycan found on FcγRI from each system is the oligomannose structure Man5, which comprises 14.4% in NS0 cells, 11.8% in HEK293 cells, and 5.2% in CHO cells (Table III). The presence of this high-mannose glycan is higher in FcγRI than the other FcγRs and suggests that it is present on a site that is protected from further processing by mannosidases and glycosyltransferases in the Golgi. Another prominent glycan found on FcγRI is the core fucosylated bi-antennary structure FA2G2, comprising 1.6% in NS0 cells (another 4.8% contributed by the tri- antennary galactosylated structures FA3G2 and 5.6% FA3G3), 4.9% in HEK293 (another 6.7% contributed by the related bisecting structure FA2BG2) cells and 8.6% in CHO cells (Table III). These oligomannose and complex bi- and tri-antennary glycans are the most abundant on FcγRI and are therefore likely to be structures that most influence the antibody binding interaction. It should also be noted that glycosylation is cell-type specific and many cell type specific glycan epitopes contribute strongly to the overall glycan composition of FcγRs from these sources. With NS0 cells we observe abundant murine specific glycan epitopes on FcγRs such as gal-α (1, 3)-gal that are not observed in HEK293 or CHO cells (27). Similarly, we observe LacdiNAc structures in HEK293 cells that are not present in the other cell types and we do not observe any bi-secting GlcNAc structures in NS0 or CHO cells because of lack of N-acetylglucosaminyltransferase 3 (GNT3) expression. In addition, sialylation differs between the cell types with N-glycolylneuraminic acid found in NSO cells and predominantly α (2, 3) sialylation in CHO cells.
Table III. The most abundant and common N-glycans found on NS0, HEK293, and CHO FcγRI. Shown in red are the common glycans present on each FcγRI.
Comparison of CHO, NS0, and HEK293 FcγRIIIa Glycosylation
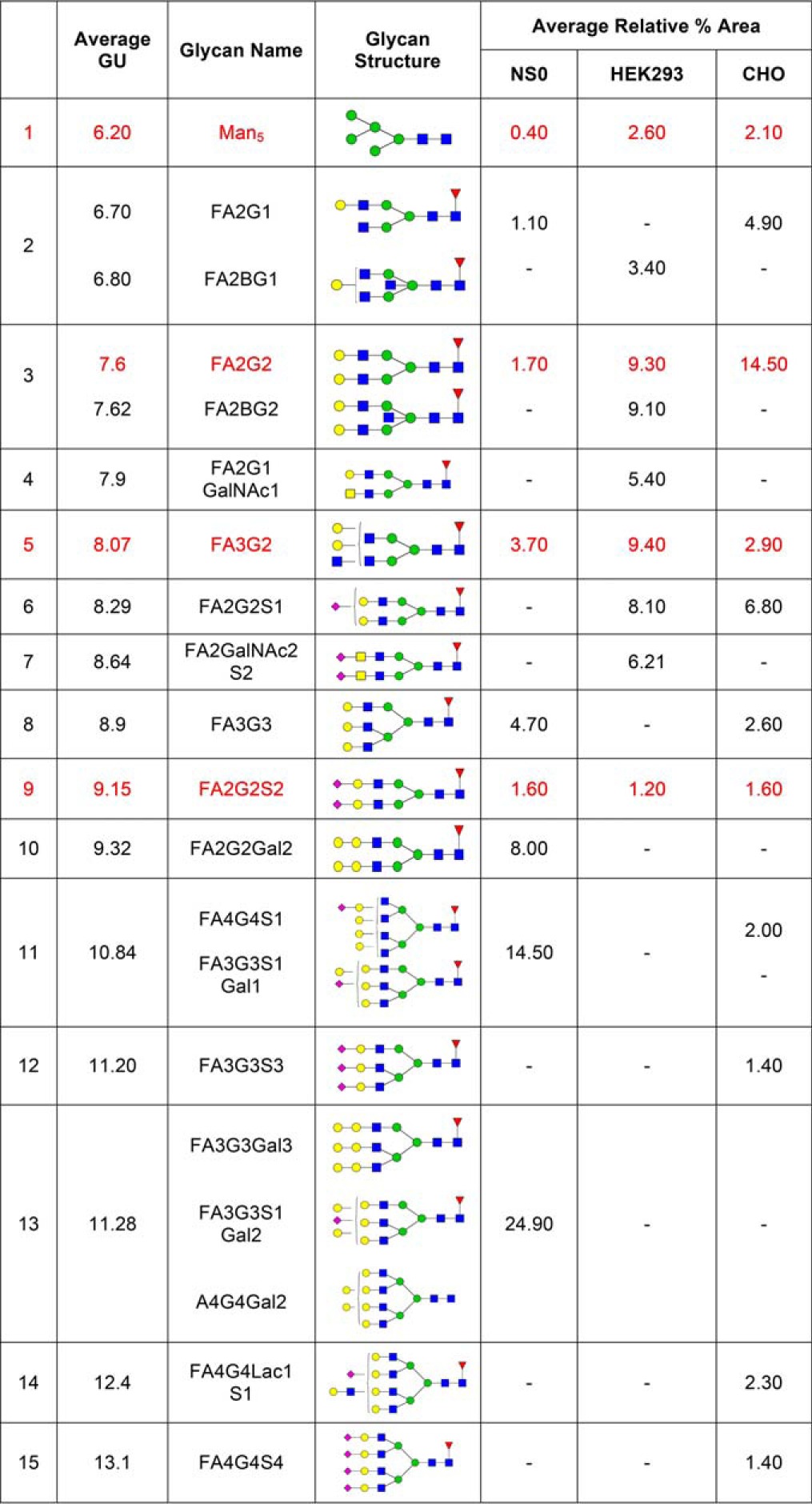
Analysis and comparison of the glycans of FcγRIIIa was performed for the Val 158 variant from NS0, HEK293, and CHO cells and for the Phe 158 variant from HEK293 and CHO cells only as this variant from was not available from NS0 cells. Glycosylation of the Val 158 and Phe 158 variants of FcγRIIIa from HEK293 cells was almost identical but was significantly different from CHO cells with the major differences in the relative abundance of glycan structures. In addition, there are significantly more tetra-antennary and sialylated glycans present on the Phe 158 variant (Tables I and II). The most abundant FcγRIIIaVal158 glycan is a core fucosylated bi-antennary structure (FA2G2), comprising 9.3% in HEK293 cells (another 9.1% contributed by the bisecting form FA2BG2), 14.5% in CHO cells and 1.7% in NS0 cells (another 8% contributed by FA2G2 with 2 α-gal extensions) (Table IV). A further core-fucosylated bi-antennary structure (FA2G1) is also abundant in FcγRIIIaVal158 from NS0, HEK293, and CHO cells accounting for 1.1%, 3.4%, and 4.9% respectively, although the HEK293 form was also a bi-secting structure (FA2BG1) (Table IV). Additional core-fucosylated, galactosylated structures, such as FA3G2 (3.7% in NS0, 9.4% in HEK293, 2.9% in CHO) and FA3G3 (4.7% in NS0 and 2.6% in CHO cells) were also present. Higher antennary galactosylated structures such as FA4G4 are also present in each cell type in lower amounts, which can also have α-gal extensions in NS0 cells but which have limited sialic acid capping. A fully sialylated form of FA2G2 (FA2G2S2) is present on each receptor in minor amounts (1.6% in NS0, 1.2% in HEK293, 1.6% in CHO) (Table IV). However, it is worth noting that a sialylated form of FA2G2 (FA2G2S1) with the 3 arm sialylated is present in substantial amounts in both HEK293 (8.1%) and CHO (6.8%) cells (Tables I and IV). Further higher tri- and tetra-sialylated structures are also present on FcγRIIIa from CHO cells in lower amounts, particularly on the Phe 158 variant (Tables I and II). In summary, the most obvious feature of FcγRIIIa glycosylation is the presence of core-fucosylated, bi-antennary, galactosylated glycans of varying complexity with varying degrees of sialylation (Tables II and IV).
Table IV. The most abundant and common N-glycans found on NS0, HEK293 and CHO FcγRIIIaVal158. Shown in red are the common glycans present on each FcγRIIIaVal158.
Comparison of Rituximab Binding to CHO, NS0, and HEK293 Derived FcγRI
Rituximab was used for FcγR interaction studies, as it is a well-characterized IgG1 therapeutic monoclonal antibody. Low levels of histidine-tagged FcγRI from the three cell types were captured in separate analysis cycles to the anti-His antibody chip. A concentration series of rituximab (1.2–300 nm) was injected and a single cycle kinetics procedure was applied, where all concentrations in a series are injected in the same cycle, followed by dissociation. The binding of human IgG1 to FcγRI and FcγRIIIa is known to be heterogeneous, with heterogeneity observed in both receptors and antibodies, particularly in glycosylation (27). Glycosylation of rituximab used in the present study was analyzed and was consistent with IgG1 type glycosylation with large amounts of fucosylated agalactosylated glycans (G0 45%), fucosylated mono-galactosylated glycans (G1 45%) and fucosylated di-galactosylated glycans (G2 1.6%). Minor amounts of oligomannose and sialylated structures were also present. Approximately 92% of the glycan structures were core-fucosylated leaving the remaining 8% afucosylated. Because of the complex binding kinetics we used the sensorgram comparison functionality in Biacore T200 evaluation software 3.0 to compare binding curves (36). The sensorgram comparison feature compares binding curves directly to upper and lower limit sensorgrams obtained with one or several standard samples. The upper and lower limit sensorgrams are typically generated from an average sensorgram to which n times the standard deviation sensorgram are added or subtracted. A sensorgram that falls between upper and lower limit sensorgrams receives a similarity score of 100%. Sensorgrams that appear partly or completely outside the area defined by limit sensorgrams receive lower similarity scores and the more distant the new sensorgram is the lower the similarity score. The sensorgram comparison feature thus compares sensorgrams directly without the need to assume a binding mechanism.
When analyzing and comparing the FcγRI interaction the CHO-FcγRI-rituximab interaction was selected as the standard sensorgram to which the NS0 and HEK293 FcγRI-rituximab interactions were compared (any of the three receptor types could have been selected as the standard). To obtain an experimental variation of the standard interaction, capture levels of CHO FcγRI receptors were varied (see Fig. 4A). The binding levels for all the five cycles shown in Fig. 4A were then normalized between 0 and 100, focusing on the curve shapes of the sensorgrams (see Fig. 4B). Binding of rituximab to HEK293 derived FcγRI was then compared with the standard CHO-FcγRI sensorgram and showed that there was slower dissociation from the HEK293 and NS0 derived receptors (red curves shown in Fig. 4C and 4D) than from the CHO cell derived receptor. The rate of rituximab dissociation from FcγRI was slowest from NS0, then HEK293, and then CHO receptors with similarity scores of 14, 48, and 100% respectively, when a comparison window of 5 standard deviations (S.D.) for the dissociation phase was applied. With high S.D. settings, the comparison window becomes very wide and eventually all samples will fall inside the window as the number of SDs is increased. A five S.D. window of comparison was chosen here to highlight that the binding of rituximab to the different FcγRs was very different. A slower association rate was also seen for rituximab binding to NS0-FcγRI compared with the CHO-FcγRI but not for the HEK293 derived receptor where there was no observable difference.
Fig. 4.
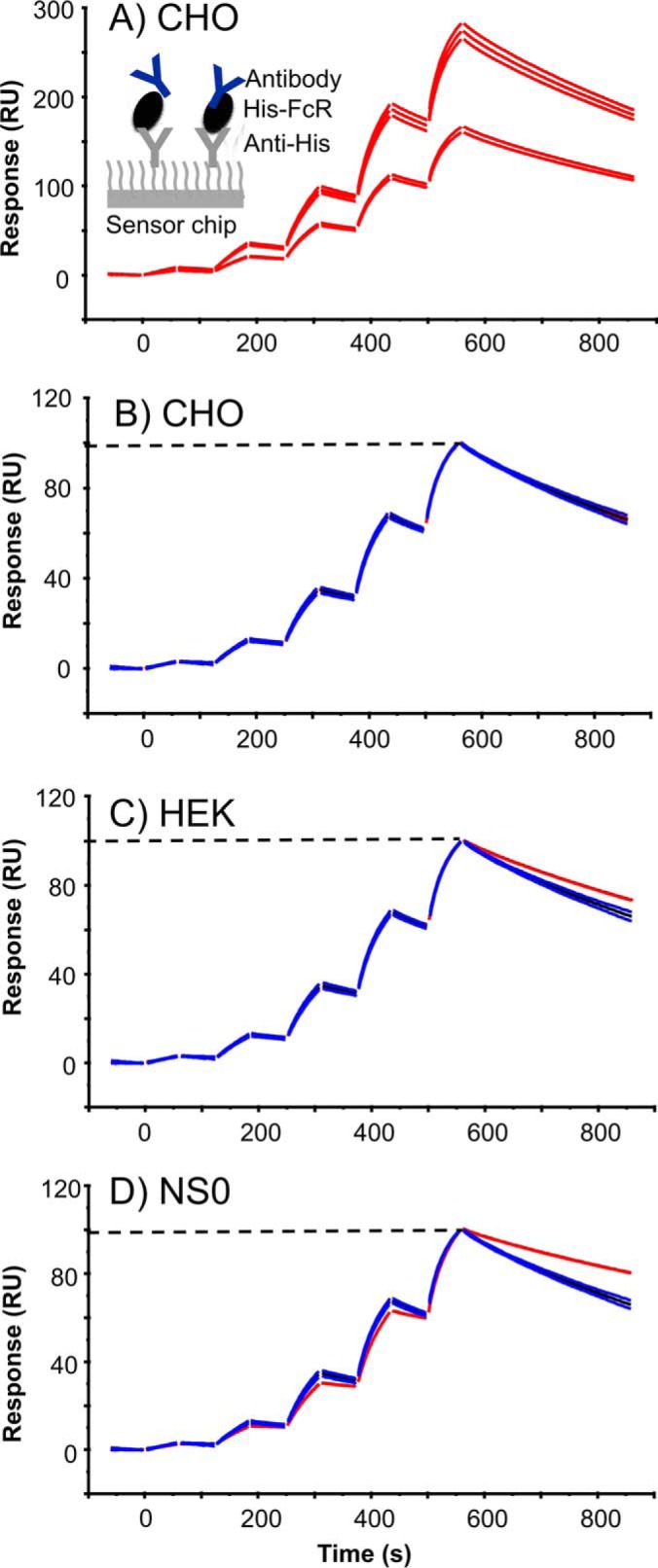
Rituximab binding kinetics depends on the source and glycosylation of FcγRI. Rituximab binding to FcγRI from CHO, HEK293 and NS0 cells is compared; using CHO expressed receptor as the standard. A, Experimental variation (n = 5) of rituximab (1.2, 4.7, 18.8, 75, and 300 nm) binding to two different levels of captured CHO-FcγRI (110 and 210 RU). B, Rituximab-CHO-FcγRI normalized data, used as standard. C, Rituximab-HEK-FcγRI binding (red) compared with the CHO standard. D, Rituximab-NS0-FcγRI binding (red) compared with the CHO standard. The dashed line indicates that the binding data is normalized between 0 and 100. Experimental data is shown in red, the average curve of the CHO-FcγRI standard is shown in black and the average curve standard deviation limits are shown in blue.
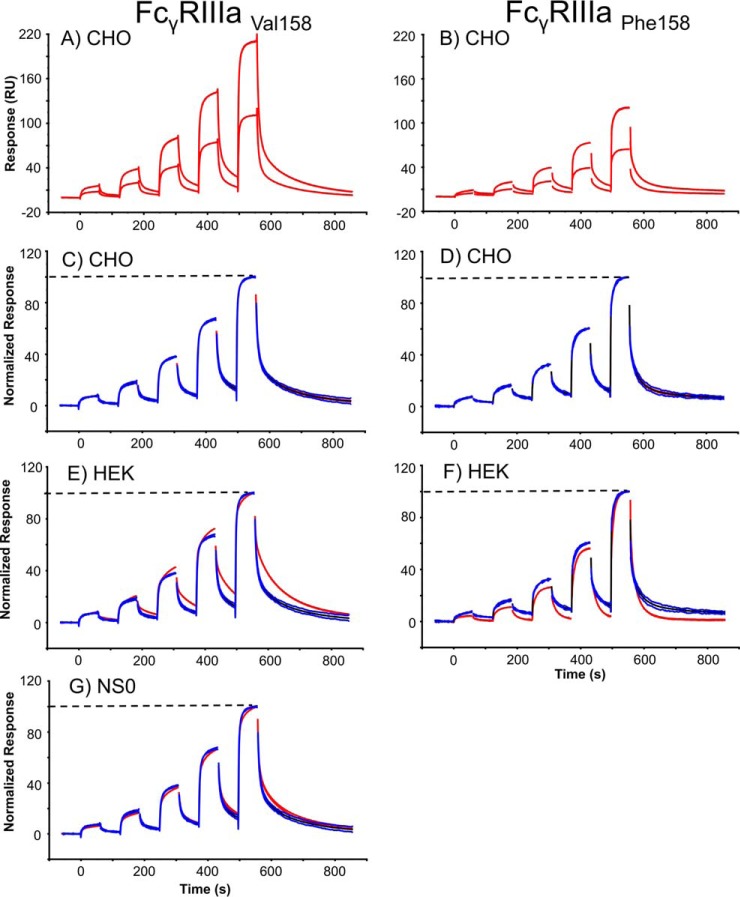
Comparison of Rituximab Binding to CHO, NS0, and HEK293 Derived FcγRIIIa
A similar assay set up to FcγRI was applied for FcγRIIIa comparisons, but with a different rituximab concentration series (25–2000 nm). Comparison of rituximab binding to FcγRIIIa is shown in Fig. 5 where the upper panels show the sensorgrams obtained for rituximab binding to two different capture levels for both receptor variants (Phe 158 and Val 158). Comparison of rituximab binding to FcγRIIIaVal158 (left hand panels in Fig. 5) shows that during the association phase, equilibrium (plateau level) is reached faster when rituximab binds to CHO-FcγRIIIaVal158 (Fig. 5C) when compared with rituximab binding to HEK293 or NS0-FcγRIIIaVal158 (see Fig. 5E and 5G). There are also clear differences in the rituximab dissociation phases with slowest dissociation from HEK293-FcγRI (Fig. 5E), then NS0-FcγRI (Fig. 5G) and then CHO-FcγRIIIaVal158 (Fig. 5C). Similarity scores applied to both binding and dissociation phases were 11, 44, and 100% respectively when a comparison window of 5 S.D. was used. Again, a wide S.D. window was chosen to illustrate how significant these binding differences are. The rituximab dissociation was not analyzed for NS0-FcγRIIIaPhe158, as this receptor was not available, however, rituximab was found to dissociate slower from CHO-FcγRIIIaPhe158 (Fig. 5D) when compared with HEK293-FcγRIIIaPhe158 (Fig. 5F).
Fig. 5.
Rituximab binding kinetics depends on the source and glycosylation of FcγRIIIa and the polymorphic receptor variant. Comparison of rituximab binding to FcγRIIIaVal 158/Phe158, produced in CHO, HEK293 and NS0 cells, using the CHO receptors as the standard. A and B, Experimental variation (n = 5) of rituximab (24.7, 74, 222, 667, and 2000 nm) binding to two different levels (90 and 170 RU) of captured FcγRIIIa for the Phe 158 and Val 158 polymorphic variants. C and D, Rituximab-CHO-FcγRIIIa normalized data, used as the standard. E and F, Rituximab-HEK293-FcγRIIIa binding (red) compared with the CHO FcγRIIIa standards. G, Rituximab-NS0-FcγRIIIa binding (red) compared with the CHO FcγRIIIa standards. The dashed line indicates that the binding data is normalized between 0 and 100. Experimental data is shown in red, the average curve of the CHO FcγRIIIa standard is shown in black and the average curve standard deviation limits are shown in blue.
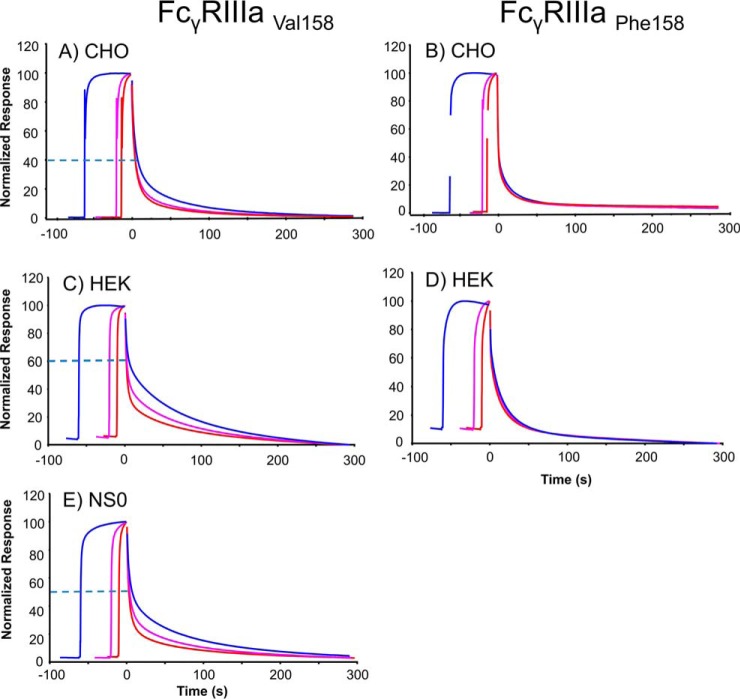
Previous studies have shown that when HEK293 derived FcγRIIIaVal158 was saturated with rituximab the dissociation rate from the receptor was dependent on injection time, where longer injection times lead to slower off-rates (see Fig. 6C) (27). This could be explained by accumulation of afucosylated antibody over time. The binding of rituximab to CHO and NS0-FcγRIIIaVal158 receptors (Figs. 6A and 6E) showed similar but less pronounced time dependent binding, results indicating the least pronounced effect for CHO receptors. In contrast to FcγRIIIaVal158, time dependent binding of rituximab to CHO and HEK293 FcγRIIIaPhe158 was almost undetectable (see Figs. 6B and 6D). It is also evident that rituximab dissociates more completely from HEK293 FcγRIIIaPhe158 if compared with CHO FcγRIIIaPhe158 receptors (see Figs. 6D and 6B).
Fig. 6.
Rituximab FcγRIIIa dissociation rates depend on the interaction time and FcγR variant at saturating conditions. Saturating concentration (6 μm) of rituximab was injected over a sensor chip with ∼100 RU captured FcγRs of different types: A and B, CHO-FcγRIIIa, C and D, HEK293-FcγRIIIa. E, NS0-FcγRIIIa. Injections were performed for 10 s (red curve), 20 s (magenta curve), and 60 s (blue curve) in separate cycles ensuring that saturating levels (∼200 RU for Val 158 and 140 RU for Phe 158) were obtained for all injection times. Curves were overlaid and responses normalized between 0 and 100 and aligned at the end of the injections (time = 0). Dissociation rates were visually compared. Dashed lines indicate the level where the slower dissociation of the 60 s injection visually starts, the higher the dashed line then the larger the time effect and slower dissociation.
DISCUSSION
FcγRs are heavily glycosylated membrane bound receptors for IgG antibodies, which have complex and heterogenous interactions and carbohydrate components that are an important factor in the binding mechanism (25, 27). In the present study, we performed complete glycan analysis of the human FcγRs: FcγRI, FcγRIIaArg131/His131, FcγRIIb, FcγRIIIaPhe158/Val158produced in CHO cells, which has previously only been reported for FcγRIIIa (37) and demonstrate that glycosylation of the receptors is differential and heterogenous with ∼30–40 unique glycan structures per receptor with mainly complex type multiantennary glycans and limited sialic acid capping. Previous reports of the glycan compositions of recombinant FcγRs also show multiantennary structures and characteristics of the expression host (27, 35, 38, 39). It is worth noting that the glycan profiles of the polymorphic variants of FcγRIIaArg131/His131 and FcγRIIIaPhe158/Val158 respectively differed significantly and as these receptors were produced in CHO DG44 cells, the reason for the observed differences in the glycan profiles are likely caused by the different expression clones (FcγRIIIa) or pools (FcγRIIa) that were used to produce the individual receptors and may be also caused by variations in the protein production process. It is also worth noting that FcγRs used in the present study were pooled from several rounds of production and that batch-to-batch variation in glycosylation can occur with recombinantly produced proteins. It is widely known and reported that the bioprocessing conditions of recombinant proteins, including growth conditions and nutrient availability can influence the final protein glycoforms (40–42). Variation in the glycan profiles of recombinant FcγRs can occur because of the cell culture and biological production of the receptors, however, it is also likely that specific sites on the receptors will have conserved glycans, which are important for the antibody interaction, similar to the conserved nature of the IgG Fc glycans produced in biological systems. This highlights the need for site-specific glycan analysis of FcγRs to identify glycan sites and glycans present on these sites that are required for productive antibody interactions. Currently this only exists for FcγRIIIa and shows that glycans on specific sites such as Asn 162 can influence the antibody interaction but does exist for other immune proteins and immunoglobulins (37, 43–45). Glycan analysis was followed by rituximab binding analysis and it was decided to focus on FcγRI and FcγRIIIaPhe158/Val158 for binding studies as the IgG1 binding kinetics for these receptors are complex and glycosylation of these receptors has been reported to influence the IgG1 interaction (24–27). Rituximab binding analysis was also performed for FcγRIIa and FcγRIIb; however, they have not been discussed in this study as the data are characterized by rapid approach to equilibrium followed by rapid dissociation (“square-wave”-like kinetic profiles). The determined affinities are in the 2–12 μm range as reported widely in the literature (27, 28, 46).
Glycosylation of CHO FcγRs is complex, however, there are a number of characteristics worth noting. Bi-antennary galactosylated glycans are the most abundant type of glycan structure across the range of CHO FcγRs, comprising 30–50% of the entire pool (Table II). Oligomannose glycans (Man5, Man6, Man7) are also a prominent feature of CHO FcγR glycosylation but differ dramatically in their diversity and relative abundance with large amounts detected for FcγRI, FcγRIIaHis131, FcγRIIb and FcγRIIIa but lower amounts for FcγRIIaArg131 (Table II). Oligomannose structures were detected on FcγRIIIa (4% and 3.5% on the Phe 158 and Val 158 variants respectively), however, a previous site specific analysis of CHO and HEK293 FcγRIIIa by Zeck et al., 2011 did not report the presence of oligomannose structures on the Asn 45 or Asn 162 amino acids of FcγRIIIaVal158 (37). In contrast to this analysis we have observed the high mannose glycans Man5, Man6, and Man7 on the Val 158 and Phe 158 variants of FcγRIIIa (Table II). A further noticeable feature of CHO FcγR glycosylation, except for FcγRIIIaPhe158, which contains 8.6% and 3.9% tri- and tetra-sialylated glycans respectively, is a lack of higher order sialylated (S3 and S4) glycan structures, even though substrates for sialylation (tri- and tetra-galactosylated glycans) are present. This lack of higher order sialylation is likely because of the FcγR amino acid sequence and secondary structure and has potential physiological relevance, particularly in modulating the antibody-mediated immune response. Interestingly, a larger amount of higher antennary galactosylated (tri- and tetra-galactosylated) glycans were also found on the Phe 158 variant of FcγRIIIa than the Val 158, which has a higher affinity for IgG (see Table II). It is worth noting that the glycan analysis performed on each FcγR was from total denatured and reduced protein immobilized in SDS-PAGE gels and site-specific glycan analysis of each receptor would provide more detailed information on the glycans that are present or conserved on specific sites, such as oligomannose structures that are highly abundant on FcγRI and are likely to be conserved. Significant heterogeneity exists in both glycosylation and glycosylation site number of FcγRs and interestingly, microheterogeneity that exists on glycan sites of FcγRs has the potential to modulate the antibody interaction and this can be explored further with site-specific analysis and more detailed knowledge of natural forms of these immune receptors.
Glycan analysis of FcγRs has provided detailed descriptions of the glycan composition and abundance for receptors from different sources and expression systems; however, it remains a challenge to fully characterize the influence of this glycosylation on the binding interaction with IgG. Our previous deglycosylation studies of HEK293 FcγRI and FcγRIIIaPhe158/Val158 showed that the binding affinities and kinetics of rituximab were altered, with faster dissociation if the receptor glycans were removed, suggesting a modulatory effect of receptor glycosylation (27). Previous studies have also shown that glycosylation of FcγRIIIa influences IgG1 binding (24–26). To investigate and identify FcγR glycans that are important for the IgG1 interaction we compared the glycosylation of receptors from different expression sources, which are described here and are available elsewhere in the literature (27, 35, 37) and identified the most abundant and common glycan structures for NS0, HEK293, and CHO FcγRI and FcγRIIIaVal158 (Table III and IV). The most obvious feature of FcγRI glycosylation is the presence of the oligomannose structure Man5, which we consistently observe in large amounts on this receptor from different sources and it is likely that this high mannose glycan influences the binding of rituximab to FcγRI because of its large abundance on each FcγRI form. Supporting this is the observation that the relative abundance of Man5 is highest for NS0 FcγRI (14.4%), then HEK293 FcγRI (11.8%) and lowest for CHO FcγRI (5.2%), which directly correlates with the rituximab binding patterns that we observe, where dissociation is fastest from CHO FcγRI and slowest from NS0 FcγRI (Tables III and V). This suggests that Man5 has a stabilizing effect on IgG1 binding. It is also worth noting that Man5 is an afucosylated structure and although there is no evidence in the literature to suggest that afucosylated glycan structures on FcγRs lead to increased antibody affinity it is possible that afucosylation of FcγR glycans have a similar effect to IgG1 afucosylation in the IgG-FcγR interaction, possibly because of steric hindrance effects. It has previously been reported that oligomannose structures on FcγRIIIa have a stabilizing effect on antibody binding (25). Furthermore, high-mannose structures have been reported on FcγRIIIa from human NK cells but not monocytes and it has been suggested that this glycan structure increases the binding of IgG1 to these cells (47, 48).
Table V. Correlation of FcγR-rituximab binding patterns with glycan profiles.
For FcγRI Man5 is found in the highest abundance on the NS0 receptor and lowest on the CHO receptor, matching the binding order with fastest dissociation from CHO-FcγRI. Man5 therefore appears to have a stabilizing effect on rituximab binding. The glycan FA2G2 is in the highest abundance on CHO FcγRI and together with other higher antennary glycan structures such as FA3G2 have a possible destabilizing effect on rituximab binding. For FcγRIIIa bi-antennary galactosylated glycans such as FA2G2 and FA2BG2 are the most abundant glycans and are also present in the highest amounts on HEK-FcγRIIIa, which has the slowest dissociation, which indicates that these glycans have a possible stabilizing effect on IgG1 binding to this receptor. Sialylation also appears to influence binding of rituximab to FcγRIIIa with CHO-FcγRIIIa having the highest amount of sialylation and the fastest dissociation. Higher amounts of sialylation and higher order sialylated structures are associated with less stable rituximab binding and increased rates of dissociation. Larger glycans such as FA4G4 also appear to have a destabilizing effect on IgG1 binding to FcγRIIIa.
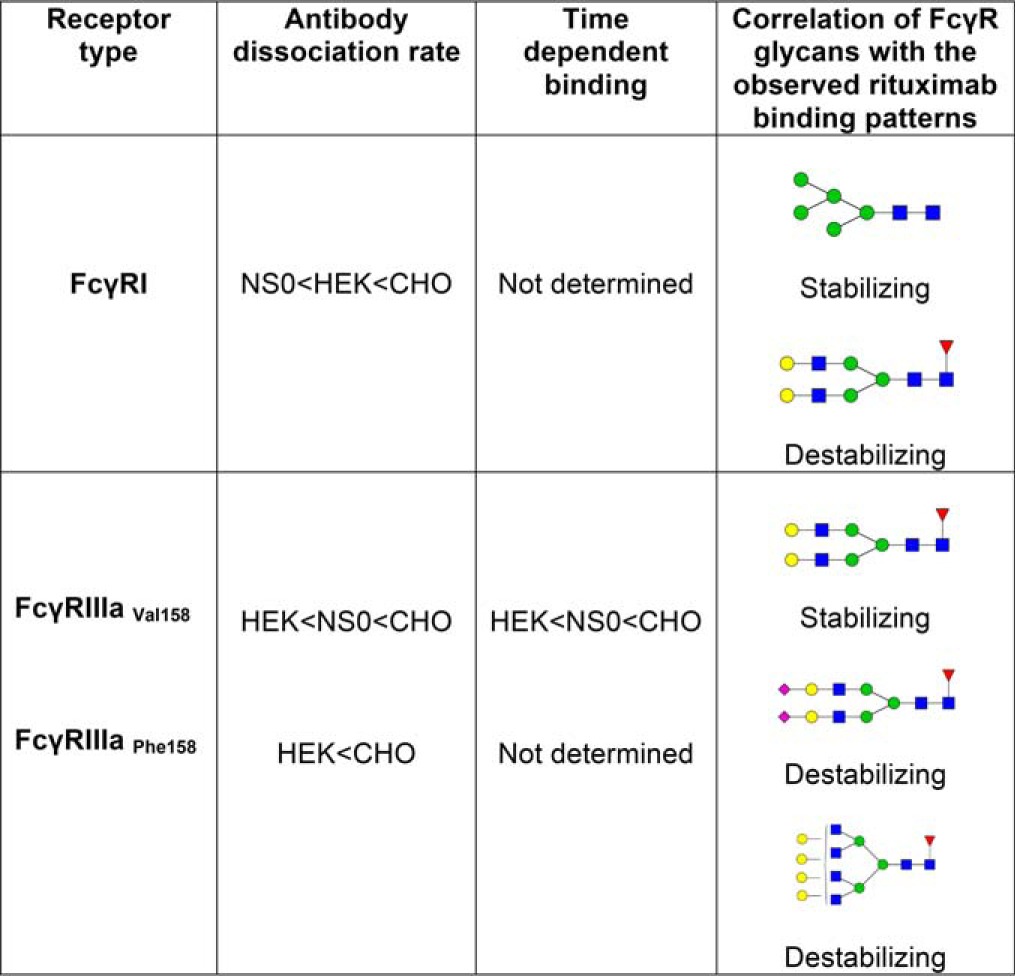
Bi-antennary glycans and similar multiantennary structures that are core-fucosylated are also abundant on FcγRI and are likely to influence the binding of IgG1. The bi-antennary glycan FA2G2 is consistently present on this receptor and is highest on CHO FcγRI (8.6%), then HEK293 FcγRI (4.9%) and lowest on NS0 FcγRI (1.6%), which correlates with the binding patterns that we observe, where dissociation is fastest from CHO FcγRI and slowest from NS0 FcγRI and suggests that this glycan and similar structures have a possible destabilizing effect on IgG1 binding (Tables III and V). Other large multiantennary structures are also present on FcγRI and may be present on specific glycan sites that can influence and destabilize the IgG interaction, further highlighting the need for site-specific glycan analysis of FcγRI. Crystal structures exist for FcγRI and recently FcγRI in complex with IgG1 was described, which show the importance of the IgG1 glycan for high affinity binding but not of the receptor glycosylation (49–51). It is also worth noting that several of the N-glycan sites of FcγRI are located in the high affinity D3 domain making them more likely to be involved in high affinity IgG1 binding.
Glycan analysis of FcγRIIIaPhe158/Val158 from NS0, HEK293 and CHO cells consistently reveals large amounts of bi-antennary, galactosylated glycans with smaller amounts of higher antennary structures, which are not fully sialylated, except for FcγRIIIa Phe158, which does contain a significant amount of larger sialylated structures. For both polymorphic variants of FcγRIIIa it is evident that there is a direct correlation between receptor sialylation and glycan size and antibody dissociation rate. This property is particularly evident for FcγRIIIaVal158 as data is available for this receptor from the three expression systems. We consistently observe that a higher proportion of larger sialylated glycans on FcγRIIIaVal158 results in faster dissociation of rituximab. CHO FcγRIIIa has the highest amount of larger sialylated glycans and the fastest dissociation of rituximab, whereas HEK293 FcγRIIIa has the lowest amount of sialylated glycans and the slowest dissociation of rituximab (Tables IV and V). Larger multiantennary glycans that are not sialylated, such as FA4G4 are also more abundant on CHO FcγRIIIa than the NS0 and HEK293 FcγRIIIa forms and also appear to have a destabilizing effect. Sialylation and glycan size therefore appear to have a destabilizing effect on IgG1 binding and the sialylation status of an FcγR, which can depend on where the receptor is expressed and on what cell-type therefore has the potential to modulate the binding interaction with IgG and control the activation/inhibition of effector responses by cells of the innate immune system. Zeck et al., 2011 also report different binding patterns for CHO and HEK293 FcγRIIIa with slightly faster dissociation rates from CHO-FcγRIIIaVal158 (37). Crystal structures exist for FcγRIIIa and FcγRIIIa-IgG1 complexes, however, they do not provide much information on the nature of the glycans involved in the interaction as fully processed glycans are underrepresented in the electron density maps and are largely absent from the three-dimensional structure as carbohydrates are usually refractory to protein crystallization because of the flexibility and heterogeneity of the glycans (52, 53).
The binding experiments of rituximab to FcγRI and FcγRIIIaPhe158/Val158 from the NS0, HEK293 and CHO production systems described in the present study (Figs. 4 and 5) used the sensorgram comparison and evaluation method described by Karlsson et al. 2016 because of the complexity in the binding kinetics and the inability to fit the data to standard models (36). Where available, time dependent dissociation experiments from different receptors agree with the order of dissociation observed from the sensorgram comparison method. The time dependent data indicate that binding of afucosylated antibody could be accumulating on the receptor and show that these types of experiments could potentially be used to quantitate the amount of afucosylated antibody present, as previously suggested (27). Time dependent binding to FcγRIIIaPhe158 was minimal but with a similar tendency as for FcγRIIIaVal158, agreeing with Cardarelli et al. where low binding of afucosylated antibody to FcγRIIIaPhe158 was shown (54). The observed binding patterns together with the most abundant glycan structures associated with these binding patterns are summarized in Table V. The rate of rituximab dissociation from FcγRI was slowest from NS0 and fastest from CHO receptors and the rate of dissociation from FcγRIIIaVal158 was slowest from HEK293 and fastest from CHO receptors. We attribute these differences in dissociation to FcγR glycosylation with larger more complex mulrtiantennary structures and increased sialylation causing faster dissociation of antibody from CHO expressed receptors. Fastest dissociation of rituximab from CHO FcγRI is consistent with the faster dissociation of rituximab described for CHO FcγRIIIa. The results presented here also suggest that there are a range of dissociation rates because of heterogeneity in glycosylation of the FcγR and the number of glycosylated variants of the receptor, which would have implications for IgG1 and therapeutic drug interactions in vivo.
Evidence presented here that glycosylation influences the kinetics of IgG-FcγR interactions suggests that glycosylation can play an important role in cellular activation/inhibition and the immune response following opsonization. It is important to note that cell-type specific glycoforms of FcγRs exist and have been described here for recombinant forms such as abundant α-gal glycans and N-glycolylneuraminic acid in NSO cells, abundant LacdiNAc and bisecting glycans in HEK293 cells and α (2, 3) sialylation in CHO cells, which can influence the antibody interaction. Many of these glycoforms do not exist on human proteins, particularly proteins of the immune system, which are described here suggesting limitations in the recombinant expression systems and highlighting the need for detailed analysis of FcγRs as they are expressed by cells of the immune system, although limited information on these natural forms exists. Cell-type specific glycoforms also exist for natural forms of the FcγRs, which are highly likely to have profound physiological effects. Detailed descriptions of natural FcγR glycosylation by immune cells are currently lacking, however, Kimberly et al. and Edberg et al. noted differences in glycosylation when FcγRIIIa was expressed by isolated monocytes and NK cells and differences in the binding of IgG1 to these differently glycosylated receptor (47, 48, 55). This finding shows that different cells of the innate immune system interact with IgG differently depending on how the FcγR is glycosylated in a cell-type specific manner with implications for activation and control of the antibody-mediated immune response. It is likely, based on the demonstration that glycosylation of FcγRs can influence and modulate the IgG interaction that the immune system can use this as a mechanism to fine-tune the antibody response, to decrease immune complex activated inflammation in times of over activation or promote antibody effector responses when needed. The data described here provide further evidence for the role of FcγR glycosylation in the binding interaction with IgG, however, it remains a challenge to describe the glycosylation of these receptors in their natural forms, in both healthy and disease states and the full impact of FcγR glycosylation on the immune response.
Supplementary Material
Acknowledgments
We thank Dieter Nachtigal, Thomas Waizenegger, Barbara Enenkel, Volker Mahlbacher (Boehringer Ingelheim) for general support and review of the manuscript and Vera Grathwohl (Boehringer Ingelheim) for cell culture support.
Footnotes
Author contributions: J.M.H., A.F., R.K., S. Müller, E.C., C.A., G.P.D., and P.M.R. designed research; J.M.H., A.F., R.K., S. Millan-Martin, M.P., and F.R. performed research; S. Müller contributed new reagents or analytic tools; J.M.H., A.F., and R.K. analyzed data; J.M.H., A.F., R.K., and S. Müller wrote the paper.
* This work was part supported by an EU Initial Training Network, Project No. 608381 and Science Foundation Ireland Grant No. SFI-13/SP SSPC/I2893. This work was supported by the National Institute for Bioprocessing, Research and Training (NIBRT), GE Healthcare and the Industrial Development Authority (IDA) Ireland. A.F., R.K., and C.A. are employees of GE Healthcare. S.M., M.P., and F.R. are employees of Boehringer Ingelheim.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- IgG
- immunoglobulin
- ABS
- α-sialidase
- ADCC
- antibody-dependent cell mediated cytotoxicity
- ADCP
- antibody-dependent cell mediated cytotoxicity
- BKF
- α-fucosidase
- BTG
- β-galactosidase
- CDC
- complement dependent cytotoxicity
- CHO
- Chinese hamster ovary
- FcγR
- Fc gamma receptor
- GalNAc
- N,-acetylgalactosamine
- GlcNAc
- N-acetylglucosamine
- GNT
- N-acetylglucosaminyltransferase
- GUH
- β-hexosaminidase
- HEK
- human embryonic kidney
- HILIC
- Hydrophilic interaction liquid chromatography
- JBM
- α-mannosidase
- LacdiNAc
- N-acetyllactosamine
- RA
- rheumatoid arthritis
- SLE
- systemic lupus erythematosus
- SPR
- surface plasmon resonance
- WAX
- weak anion exchange.
REFERENCES
- 1. Daeron M. (1997) Fc receptor biology. Annu. Rev. Immunol. 15, 203–234 [DOI] [PubMed] [Google Scholar]
- 2. Nimmerjahn F., and Ravetch J. V. (2008) Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 8, 34–47 [DOI] [PubMed] [Google Scholar]
- 3. Ravetch J. V., and Perussia B. (1989) Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J. Exp. Med. 170, 481–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koene H. R., Kleijer M., Algra J., Roos D., von dem Borne A. E., and de Haas M. (1997) Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood 90, 1109–1114 [PubMed] [Google Scholar]
- 5. de Haas M., Koene H. R., Kleijer M., de Vries E., Simsek S., van Tol M. J., Roos D., and von dem Borne A. E. (1996) A triallelic Fc gamma receptor type IIIA polymorphism influences the binding of human IgG by NK cell Fc gamma RIIIa. J. Immunol. 156, 2948–2955 [PubMed] [Google Scholar]
- 6. Weng W. K., and Levy R. (2003) Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J. Clin. Oncol. 21, 3940–3947 [DOI] [PubMed] [Google Scholar]
- 7. Cartron G., Dacheux L., Salles G., Solal-Celigny P., Bardos P., Colombat P., and Watier H. (2002) Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 99, 754–758 [DOI] [PubMed] [Google Scholar]
- 8. Wu J., Edberg J. C., Redecha P. B., Bansal V., Guyre P. M., Coleman K., Salmon J. E., and Kimberly R. P. (1997) A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J. Clin. Invest. 100, 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nieto A., Caliz R., Pascual M., Mataran L., Garcia S., and Martin J. (2000) Involvement of Fcgamma receptor IIIA genotypes in susceptibility to rheumatoid arthritis. Arthritis Rheum. 43, 735–739 [DOI] [PubMed] [Google Scholar]
- 10. Sanders L. A., Feldman R. G., Voorhorst-Ogink M. M., de Haas M., Rijkers G. T., Capel P. J., Zegers B. J., and van de Winkel J. G. (1995) Human immunoglobulin G (IgG) Fc receptor IIA (CD32) polymorphism and IgG2-mediated bacterial phagocytosis by neutrophils. Infect. Immun. 63, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duits A. J., Bootsma H., Derksen R. H., Spronk P. E., Kater L., Kallenberg C. G., Capel P. J., Westerdaal N. A., Spierenburg G. T., Gmelig-Meyling F. H., and Van de Winkel J. G. (1995) Skewed distribution of IgG Fc receptor IIa (CD32) polymorphism is associated with renal disease in systemic lupus erythematosus patients. Arthritis Rheum. 38, 1832–1836 [DOI] [PubMed] [Google Scholar]
- 12. Haseley L. A., Wisnieski J. J., Denburg M. R., Michael-Grossman A. R., Ginzler E. M., Gourley M. F., Hoffman J. H., Kimberly R. P., and Salmon J. E. (1997) Antibodies to C1q in systemic lupus erythematosus: characteristics and relation to Fc gamma RIIA alleles. Kidney Int. 52, 1375–1380 [DOI] [PubMed] [Google Scholar]
- 13. Sanders L. A., van de Winkel J. G., Rijkers G. T., Voorhorst-Ogink M. M., de Haas M., Capel P. J., and Zegers B. J. (1994) Fc gamma receptor IIa (CD32) heterogeneity in patients with recurrent bacterial respiratory tract infections. J. Infect. Dis. 170, 854–861 [DOI] [PubMed] [Google Scholar]
- 14. Kono H., Kyogoku C., Suzuki T., Tsuchiya N., Honda H., Yamamoto K., Tokunaga K., and Honda Z. (2005) FcgammaRIIB Ile232Thr transmembrane polymorphism associated with human systemic lupus erythematosus decreases affinity to lipid rafts and attenuates inhibitory effects on B cell receptor signaling. Hum. Mol. Genet. 14, 2881–2892 [DOI] [PubMed] [Google Scholar]
- 15. Chu Z. T., Tsuchiya N., Kyogoku C., Ohashi J., Qian Y. P., Xu S. B., Mao C. Z., Chu J. Y., and Tokunaga K. (2004) Association of Fcgamma receptor IIb polymorphism with susceptibility to systemic lupus erythematosus in Chinese: a common susceptibility gene in the Asian populations. Tissue Antigens 63, 21–27 [DOI] [PubMed] [Google Scholar]
- 16. Chen J. Y., Wang C. M., Ma C. C., Luo S. F., Edberg J. C., Kimberly R. P., and Wu J. (2006) Association of a transmembrane polymorphism of Fcgamma receptor IIb (FCGR2B) with systemic lupus erythematosus in Taiwanese patients. Arthritis Rheum. 54, 3908–3917 [DOI] [PubMed] [Google Scholar]
- 17. Adu B., Jepsen M. P., Gerds T. A., Kyei-Baafour E., Christiansen M., Dodoo D., and Theisen M. (2014) Fc gamma receptor 3B (FCGR3B-c.233C>A-rs5030738) polymorphism modifies the protective effect of malaria specific antibodies in Ghanaian children. J. Infect. Dis. 209, 285–289 [DOI] [PubMed] [Google Scholar]
- 18. Hayes J. M., Cosgrave E. F., Struwe W. B., Wormald M., Davey G. P., Jefferis R., and Rudd P. M. (2014) Glycosylation and Fc receptors. Current Topics Microbiol. Immunol. 382, 165–199 [DOI] [PubMed] [Google Scholar]
- 19. Hayes J. M., Wormald M. R., Rudd P. M., and Davey G. P. (2016) Fc gamma receptors: glycobiology and therapeutic prospects. J. Inflamm. Res. 9, 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jefferis R., Lund J., and Pound J. D. (1998) IgG-Fc-mediated effector functions: molecular definition of interaction sites for effector ligands and the role of glycosylation. Immunol. Rev. 163, 59–76 [DOI] [PubMed] [Google Scholar]
- 21. Allhorn M., Olin A. I., Nimmerjahn F., and Collin M. (2008) Human IgG/Fc gamma R interactions are modulated by streptococcal IgG glycan hydrolysis. PLoS ONE 3, e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subedi G. P., and Barb A. W. (2016) The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc gamma receptor. MAbs 8, 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barb A. W., and Prestegard J. H. (2011) NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat. Chem. Biol. 7, 147–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferrara C., Stuart F., Sondermann P., Brunker P., and Umana P. (2006) The carbohydrate at FcgammaRIIIa Asn-162. An element required for high affinity binding to non-fucosylated IgG glycoforms. J. Biol. Chem. 281, 5032–5036 [DOI] [PubMed] [Google Scholar]
- 25. Ferrara C., Grau S., Jager C., Sondermann P., Brunker P., Waldhauer I., Hennig M., Ruf A., Rufer A. C., Stihle M., Umana P., and Benz J. (2011) Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. U.S.A. 108, 12669–12674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shibata-Koyama M., Iida S., Okazaki A., Mori K., Kitajima-Miyama K., Saitou S., Kakita S., Kanda Y., Shitara K., Kato K., and Satoh M. (2009) The N-linked oligosaccharide at Fc gamma RIIIa Asn-45: an inhibitory element for high Fc gamma RIIIa binding affinity to IgG glycoforms lacking core fucosylation. Glycobiology 19, 126–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hayes J. M., Frostell A., Cosgrave E. F., Struwe W. B., Potter O., Davey G. P., Karlsson R., Anneren C., and Rudd P. M. (2014) Fc Gamma Receptor Glycosylation Modulates the Binding of IgG Glycoforms: A Requirement for Stable Antibody Interactions. J. Proteome Res. 13, 5741–5485 [DOI] [PubMed] [Google Scholar]
- 28. Bruhns P., Iannascoli B., England P., Mancardi D. A., Fernandez N., Jorieux S., and Daeron M. (2009) Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 [DOI] [PubMed] [Google Scholar]
- 29. Visser J., Feuerstein I., Stangler T., Schmiederer T., Fritsch C., and Schiestl M. (2013) Physicochemical and functional comparability between the proposed biosimilar rituximab GP2013 and originator rituximab. BioDrugs 27, 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Royle L., Radcliffe C. M., Dwek R. A., and Rudd P. M. (2006) Detailed structural analysis of N-glycans released from glycoproteins in SDS-PAGE gel bands using HPLC combined with exoglycosidase array digestions. Methods Mol. Biol. 347, 125–143 [DOI] [PubMed] [Google Scholar]
- 31. Ahn J., Bones J., Yu Y. Q., Rudd P. M., and Gilar M. (2010) Separation of 2-aminobenzamide labeled glycans using hydrophilic interaction chromatography columns packed with 1.7 microm sorbent. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878, 403–408 [DOI] [PubMed] [Google Scholar]
- 32. Harvey D. J., Merry A. H., Royle L., Campbell M. P., Dwek R. A., and Rudd P. M. (2009) Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics 9, 3796–3801 [DOI] [PubMed] [Google Scholar]
- 33. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., and Haslam S. M. (2008) GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 34. Karlsson R., Katsamba P. S., Nordin H., Pol E., and Myszka D. G. (2006) Analyzing a kinetic titration series using affinity biosensors. Analytical Biochem. 349, 136–147 [DOI] [PubMed] [Google Scholar]
- 35. Cosgrave E. F., Struwe W. B., Hayes J. M., Harvey D. J., Wormald M. R., and Rudd P. M. (2013) N-Linked Glycan Structures of the Human Fcgamma Receptors Produced in NS0 Cells. J. Proteome Res. 12, 3721–3737 [DOI] [PubMed] [Google Scholar]
- 36. Karlsson R., Pol E., and Frostell A. (2016) Comparison of surface plasmon resonance binding curves for characterization of protein interactions and analysis of screening data. Anal. Biochem. 502, 53–63 [DOI] [PubMed] [Google Scholar]
- 37. Zeck A., Pohlentz G., Schlothauer T., Peter-Katalinic J., and Regula J. T. (2011) Cell type-specific and site directed N-glycosylation pattern of FcgammaRIIIa. J. Proteome Res. 10, 3031–3039 [DOI] [PubMed] [Google Scholar]
- 38. Takahashi N., Cohen-Solal J., Galinha A., Fridman W. H., Sautes-Fridman C., and Kato K. (2002) N-glycosylation profile of recombinant human soluble Fcgamma receptor III. Glycobiology 12, 507–515 [DOI] [PubMed] [Google Scholar]
- 39. Takahashi N., Yamada W., Masuda K., Araki H., Tsukamoto Y., Galinha A., Sautes C., Kato K., and Shimada I. (1998) N-glycan structures of a recombinant mouse soluble Fcgamma receptor II. Glycoconj. J. 15, 905–914 [DOI] [PubMed] [Google Scholar]
- 40. Nyberg G. B., Balcarcel R. R., Follstad B. D., Stephanopoulos G., and Wang D. I. (1999) Metabolic effects on recombinant interferon-gamma glycosylation in continuous culture of Chinese hamster ovary cells. Biotechnol. Bioengineer. 62, 336–347 [DOI] [PubMed] [Google Scholar]
- 41. Butler M. (2006) Optimisation of the cellular metabolism of glycosylation for recombinant proteins produced by Mammalian cell systems. Cytotechnology 50, 57–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burleigh S. C., van de Laar T., Stroop C. J., van Grunsven W. M., O'Donoghue N., Rudd P. M., and Davey G. P. (2011) Synergizing metabolic flux analysis and nucleotide sugar metabolism to understand the control of glycosylation of recombinant protein in CHO cells. BMC Biotechnol. 1, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Plomp R., Bondt A., de Haan N., Rombouts Y., and Wuhrer M. (2016) Recent advances in clinical glycoproteomics of immunoglobulins (Igs). Mol. Cell. Proteomics 15, 2217–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Plomp R., Hensbergen P. J., Rombouts Y., Zauner G., Dragan I., Koeleman C. A., Deelder A. M., and Wuhrer M. (2013) Site-specific N-glycosylation analysis of human immunoglobulin E. J. Proteome Res. 13, 536–546 [DOI] [PubMed] [Google Scholar]
- 45. Cao L., Diedrich J. K., Kulp D. W., Pauthner M., He L., Park S. R., Sok D., Su C. Y., Delahunty C. M., Menis S., Andrabi R., Guenaga J., Georgeson E., Kubitz M., Adachi Y., Burton D. R., Schief W. R., Yates Iii J. R., and Paulson J. C. (2017) Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat. Commun. 8, 14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maenaka K., van der Merwe P. A., Stuart D. I., Jones E. Y., and Sondermann P. (2001) The human low affinity Fcgamma receptors IIa, IIb, and III bind IgG with fast kinetics and distinct thermodynamic properties. J. Biol. Chem. 276, 44898–44904 [DOI] [PubMed] [Google Scholar]
- 47. Edberg J. C., Barinsky M., Redecha P. B., Salmon J. E., and Kimberly R. P. (1990) Fc gamma RIII expressed on cultured monocytes is a N-glycosylated transmembrane protein distinct from Fc gamma RIII expressed on natural killer cells. J. Immunol. 144, 4729–4734 [PubMed] [Google Scholar]
- 48. Edberg J. C., and Kimberly R. P. (1997) Cell type-specific glycoforms of Fc gamma RIIIa (CD16): differential ligand binding. J. Immunol. 159, 3849–3857 [PubMed] [Google Scholar]
- 49. Lu J., Ellsworth J. L., Hamacher N., Oak S. W., and Sun P. D. (2011) Crystal structure of Fcgamma receptor I and its implication in high affinity gamma-immunoglobulin binding. J. Biol. Chem. 286, 40608–40613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lu J., Chu J., Zou Z., Hamacher N. B., Rixon M. W., and Sun P. D. (2015) Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding. Proc. Natl. Acad. Sci. U.S.A. 112, 833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kiyoshi M., Caaveiro J. M., Kawai T., Tashiro S., Ide T., Asaoka Y., Hatayama K., and Tsumoto K. (2015) Structural basis for binding of human IgG1 to its high-affinity human receptor FcgammaRI. Nat. Commun. 6, 6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sondermann P., Huber R., Oosthuizen V., and Jacob U. (2000) The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature 406, 267–273 [DOI] [PubMed] [Google Scholar]
- 53. Radaev S., Motyka S., Fridman W. H., Sautes-Fridman C., and Sun P. D. (2001) The structure of a human type III Fcgamma receptor in complex with Fc. J. Biol. Chem. 276, 16469–16477 [DOI] [PubMed] [Google Scholar]
- 54. Cardarelli P. M., Rao-Naik C., Chen S., Huang H., Pham A., Moldovan-Loomis M. C., Pan C., Preston B., Passmore D., Liu J., Kuhne M. R., Witte A., Blanset D., and King D. J. (2010) A nonfucosylated human antibody to CD19 with potent B-cell depletive activity for therapy of B-cell malignancies. Cancer Immunol. Immunotherapy 59, 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kimberly R. P., Tappe N. J., Merriam L. T., Redecha P. B., Edberg J. C., Schwartzman S., and Valinsky J. E. (1989) Carbohydrates on human Fc gamma receptors. Interdependence of the classical IgG and nonclassical lectin-binding sites on human Fc gamma RIII expressed on neutrophils. J. Immunol. 142, 3923–3930 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.