Abstract
The human interferon α2 (IFNα2) was the first highly active IFN subtype to be cloned in the early eighties. It was also the first IFN and the first cytokine to be produced and commercialized by the pharmaceutical industry. Ipso facto it became the favourite IFNα subtype for academic researchers. For this fortunate reason IFNα2 has been at the origin of most discoveries related to the mechanism of action of type I interferons.
Keywords: Cytokine, type I interferon, interferon α2
1- Introduction
Type I interferons (IFNs) were discovered in 1957 by Isaacs and Lindenman who reported that cells infected with an inactivated virus release a soluble factor exerting an antiviral action (Isaacs and Lindenmann, 1957). We now know that IFN is a key cytokine of the innate immune response which is produced upon recognition of many pathogens and damage-associated molecular patterns released by infected cells or dying cells (Tomasello et al., 2014).
Type I IFNs are capable to act on virtually all body cells since they recognize a receptor which is ubiquitously expressed. In addition to their function in establishing an antiviral state, they are also capable to decrease the proliferation rate of dividing cells and to exert immunomodulatory activities. Type I IFN action impacts not only innate immunity, but also on almost every aspect of cellular and humoral adaptive immune responses. In particular, the action of IFN on dendritic cells (DC) is crucial. It induces IL-15 trans-presentation for NK cells activation. More importandly, IFN can modulate all 3 types of signals delivered by DC to T cells: MHC-antigenic peptide complex, costimulation and cytokine production (Tomasello et al., 2014; Rizza et al., 2015). Type I IFNs are thus critical cytokines for the generation of a protective immune response. When given exogenously as a drug to adult mice, IFN exert a potent antitumor effect (Gresser, 2007). In human, IFNα2 has the longest record of clinical use for the treatment of many types of cancer, including some hematological malignancies and solid tumors (Antonelli et al., 2015). In the last 3 years, it has been demonstrated that type I IFNs are essential in processes of immunosurveillance, tumor rejection and regulation of metastasis spread (Diamond et al., 2011; Fuertes et al., 2011; Bidwell et al., 2012). Moreover, IFNs were shown to improve the efficacy of classical antitumor treatments such as radiotherapy, chemotherapy and monoclonal antibodies-based therapy (Burnette et al., 2011; Schiavoni et al., 2011; Stagg et al., 2011).
The type I IFNs represent a family of several closely related subtypes. At least 8 subclasses: α, β, δ, ε, κ, τ, ω and limitin have been described in different mammalian species. They all act through the same plasma membrane receptor made of IFNAR1 and IFNAR2. Humans have 17 subtypes: 13α, 1β, 1ω, 1ε and 1κ (Pestka et al., 2004).
Genes and cDNAs encoding human type I IFNs were first cloned in the early eighties (Derynck et al., 1980; Goeddel et al., 1980; Nagata et al., 1980b; Taniguchi et al., 1980). Among them, the IFNα1 and α2 were the first two α subtypes characterized. Given the low specific activity exhibited by IFNα1, IFNα2 was the first highly active human IFN made available to scientists and physicians. IFNα2 thus became the prototypic type I IFN subtype used in fundamental research and most clinical applications. With some rare exceptions, the basic knowledge generated by these studies is essentially valid for the other α’s, β and ω IFN subtypes in humans and higher mammals. In this review, we will summarize what we know on the structure, mechanism of action and biological activities of IFNα2. When pertinent, we will emphasize its uniqueness with respect to the other family subtypes.
2- IFNα2 gene and expression
Soon after the cloning of the human IFNα2 cDNA (Goeddel et al., 1980; Streuli et al., 1980), the gene was isolated and mapping of the type I IFN gene family began (Nagata et al., 1980a; Lawn et al., 1981). All type I IFN genes are clustered on a region covering 400 kb on the short arm of chromosome 9 (Diaz et al., 1994). All are intronless genes, suggesting that the family has originated from a retroposition event replacing, in higher vertebrates, ancestral intron-containing IFN genes (Qi et al., 2010). If all eutherian mammals have several type I IFN genes, the diversification of the family seems to have arisen independently in each species. Thus, except for very closely related species (eg. human and chimpanzee), there is no orthologues of human IFNα2 in other species (Woelk et al., 2007).
Several IFNα2 alleles have been described. The best known are α2a and α2b, both commercialized for clinical use as RoferonA and IntronA, respectively. They differ by a neutral K/R substitution at position 23 (von Gabain et al., 1990). In human populations, the gene encoding IFNα2 is found to be under constraints which prevent mutations, suggesting an essential role in physiology (Manry et al., 2011).
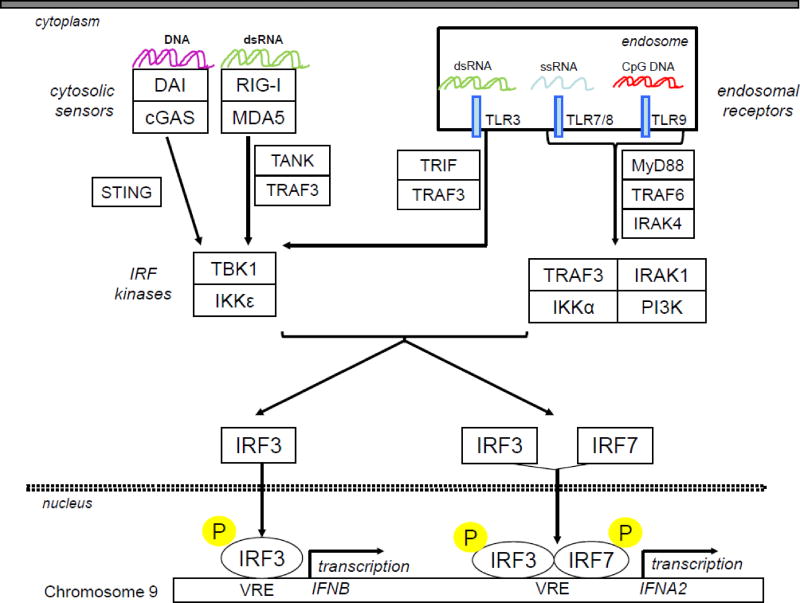
The expression of type I IFN genes is regulated primarily at the transcriptional level. Upstream of the transcription start site and the TATA box, the IFNα2 promoter contains several virus-responsive elements (VREs), also called positive regulatory domain-like elements (PRD-LEs) (Honda et al., 2005; Genin et al., 2009). These elements are found in the promoter of all IFNα genes. Of relevance, the IFNβ promoter contains two additional regulatory domains engaging NF-κB and ATF-2/c-Jun transcription factors. The VREs are activated by two IFN regulatory factors (IRF) primarily responsible for the initiation of IFNα transcription, IRF-3 and IRF-7. The small variations in the VRE sequences of the different IFNα genes affect the affinities of IRF-3 and IRF-7 binding and may account for some temporal and quantitative differences in gene expression (Genin et al., 2009). While IRF-3 is constitutively expressed in almost all cells, IRF-7 is constitutively expressed at high level mainly in plasmacytoïd dendritic cells (pDCs). IRF-7 is however robustly induced in all cells by type I IFNs. In most cell types, a full IFNα gene expression is thus dependent on a positive feedback loop where the de novo synthesis of IRF-7 is critical (Honda et al., 2005).
To act as transcription factors IRF-3 and IRF-7 need to be serine phosphorylated, to homo- or hetero-dimerize and translocate to the nucleus (Fig. 1). Several kinases such as Tank-binding kinase 1 (TBK1) or inducible IκB kinase (IKKε) can mediate phosphorylation. These IRF kinases are themselves activated through signalling cascades initiated by the recognition of many molecular patterns signing virus replication, bacterial infections, viral nucleotide sequences or cellular stresses (Tomasello et al., 2014). If all cells are equipped with the necessary sensors of viral replication, only specific cell types can detect danger patterns upon endocytosis of infected material. Among them pDCs selectively express Toll-like receptor (TLR) 7 and 9 which recognize endosomal ssRNA and DNA with unmethylated CpG motifs, respectively. Since pDCs constitutively express IRF7 and are equipped with a robust protein synthesis and secretion system, they are able to produce very high levels of all IFNα subtypes (Coccia et al., 2004; Ito et al., 2006).
Figure 1. Induction of type I-IFNs.
Damage and pathogen associated molecular patterns are recognized by cytosolic sensors and/or endosomal receptors which activate several pathways leading to the activation of IRF kinases and the subsequent phosporylation of IRF3 and IRF7.Once phosphorylated, these IRF translocate into the nucleus, bind to the promoter of type I IFN genes and activate their transcription. Unlike IFNB gene, IFNA2 gene requires both IRF3 and IRF7 activation to be induced. cGAS: Cyclic GMP-AMP synthase; CpG DNA: DNA with cytidine-phosphate-guanosine motifs; DAI: DNA-dependent activator of IFNregulatory factors; dsRNA: double strand RNA; IKK: inhibitor of kappaB kinase; IRAK: Interleukin-1 receptor-associated kinase; IRF: Interferon regulatory factors; MDA5: Melanoma Differentiation-Associated protein 5; MyD88: myeloid differentiation primary response protein 88; RIG-I: retinoic acid-inducible gene 1; STING: Stimulator of interferon genes; TBK: TANK-binding kinase; TLR: Toll-like receptor; TRIF: Toll/IL-1 receptor (TIR) domain-containing adaptor inducing IFNβ, TRAF: TNF receptor associated factors; VRE: virus responsive element.
3- IFNα2 structure, receptor binding and signalling
The IFNα2 gene encodes a 188 amino acids precursor that contains a N-terminal 23 amino acids leader sequence that is cleaved during the secretion process. The size of the mature IFNα2 is thus 165 amino acids, which is one amino acid shorter than all other human IFNα subtypes. From sequence alignment, it is clear that the aspartic acid at position 44 in all human IFNα subtypes is missing in IFNα2. No functional property was associated with this deletion. Because IFNα2 is not glycosylated, the biopharmaceutics industry has chosen to produce IFNα2 in Escherichia coli fermenter. Up to now, IFNα2 has been classically purified from inclusion bodies and refolded but new methods achieving high soluble protein expression and fast purification are being developed (Bis et al., 2014). The three-dimensional structure of IFNα2 was revealed by nuclear magnetic resonance (NMR) spectroscopy (Klaus et al., 1997) and X-ray crystallography (Radhakrishnan et al., 1996). It contains five α-helices (A to E), the helices A, B, C and E forming a left-handed four helix-bundle characteristic of the helical cytokine family. The long loop between helix A and helix B is perpendicular to the bundle axis; it is linked to helix E by a disulfide bond between cysteine 29 and 138. A second disulfide bond connects the amino-terminal cysteine to position 98 in the helix C.
Type I IFNs exert their biological activity by assembling a ternary IFN-receptor complex with the IFNAR1 and IFNAR2 chains. The structure of the ternary complex IFNAR1-IFNα2-IFNAR2 has been recently solved by X-ray crystallography (Thomas et al., 2011). An excellent recent review analyses in-depth all structural and dynamic aspects of IFNα2 binding (Piehler et al., 2012) and the cartography of the IFNα2 residues interacting with IFNAR1 and IFNAR2 are easily found at Protopedia (http://www.proteopedia.org/wiki/index.php/Journal:Cell:1). Briefly, a surface area of 18 nm2 formed by part of the helices A and E, and the A–B loop interacts with IFNAR2. The domain of interaction with IFNAR1 is located on the opposite side of the IFN molecule and covers a surface area of 22 nm2, containing residues located in the helices B, C and D. It is highly probable that all type I IFN subtypes, α, β and ω, form a similar ternary complex as suggested by the perfect superimposition of two ternary complexes assembled by two different IFNs described by Thomas et al. (Thomas et al., 2011), and by other low and high resolution structural data (Chill et al., 2003; Quadt-Akabayov et al., 2006; Li et al., 2008; Strunk et al., 2008; de Weerd et al., 2013).
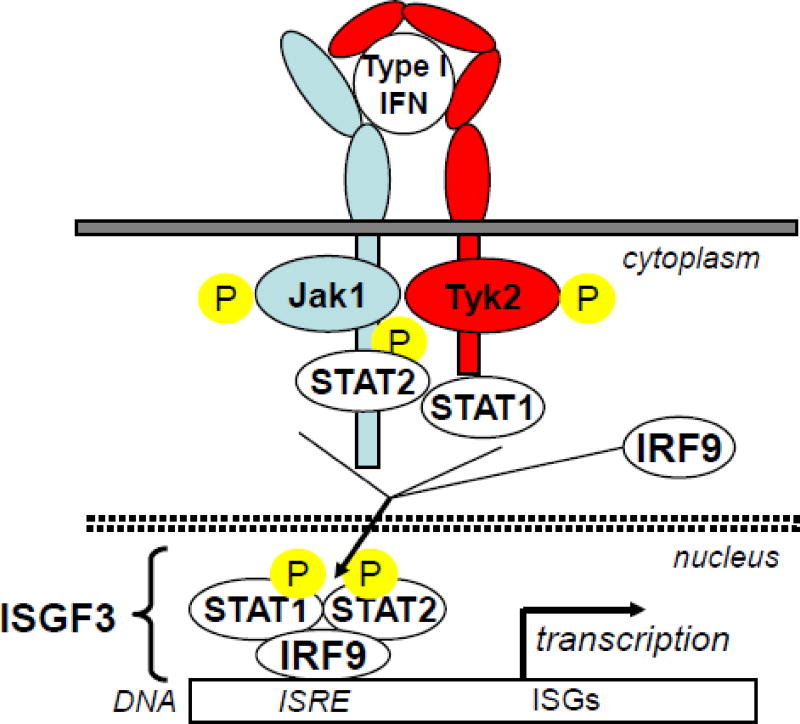
All IFN subtypes show a dissymmetry in their affinities for the individual IFNAR chains. Typically, the constant of dissociation (Kd) of the interaction with IFNAR1 is in the μM range and the Kd for IFNAR2 is much lower, being in the nM range (Piehler et al., 2012). Since IFNAR1 and IFNAR2 are not pre-assembled at the cell surface (Wilmes et al., 2015), it is likely that IFN binds first to IFNAR2 and then to IFNAR1 in a bi-dimensional reaction to form a ternary complex. Only the ternary complex is capable of signal transduction through the activation of the JAK kinases associated with the receptor chains, the ensuing phosphorylation of STAT1 and STAT2 and the formation of the ISGF3 transcription factor which ultimately induces the transcription of the large family of IFN-induced genes (Borden et al., 2007) (Fig. 2). In addition to this main JAK/Stat signalling pathway initiated by IFN in all cell types, other signalling factors can be activated in a cell type-dependent manner. These include other STAT proteins such as STAT3, STAT4 and STAT5, and elements of p38 and ERK MAPK pathways, PI3K signalling and PKC isoforms (Platanias, 2005). In fine, the expression of over 1000 genes can potentially be induced by IFN. It was suggested that such a high number of induced genes (ISG) is necessary to ensure a large spectrum of antiviral action, the replication of each virus being targeted by a unique set of antiviral proteins (Schoggins and Rice, 2011). As a consequence, IFN exhibits a high level of pleiotropic action and can deeply affect the homeostasis of several biological systems. Among these IFN-induced genes, several encode proteins needed for signalling attenuation (Coccia et al., 2006). The most specific and critical negative feed-back regulator of the JAK/STAT pathway activated by IFN is USP18 (Malakhova et al., 2006). This protein binds to the receptor and decreases the stability of the IFN-receptor ternary complex. Moreover, cells where USP18 accumulates become more refractory to IFNα2 than IFNβ, indicating that USP18 is an important determinant of IFN subtype differential activity (Francois-Newton et al., 2011; Wilmes et al., 2015).
Figure 2. Canonical JAK/STAT signalling pathway of type I-IFNs.
Type I-IFN/IFNAR2/IFNAR1 ternary complex induces transphosphorylation of receptor associated JAK kinases which in turn phosphorylate specific tyrosine residues on IFNAR1 and IFNAR2. These phosphorylated tyrosine residues are docking sites for signal transducers and activators of transcription (STAT) which are then recruited to the receptor complex and phosporylated (pSTAT) on specific tyrosine residue. pSTAT2 and pSTAT1 heterodimerize to form together with IRF9 the ISGF3 complex which translocates into the nucleus and binds to IFN stimulated responsive elements (ISRE) in the regulatory region of IFN-stimulated genes (ISG)
The specific activity of a given IFN for a given biological response is function of the stability of the IFN-receptor ternary complex, stability being determined by the individual affinity to IFNAR1 and IFNAR2 (Kalie et al., 2008). The slope of the linear affinity-activity correlation is not necessary the same for all biological activities. For example, on WISH cells, the slope of the antiproliferative specific activity versus affinity is higher than the slope establishing the relationship between the anti-VSV specific activity and the global affinity of the IFN for IFNAR1 and IFNAR2. This explains why the different type I IFN subtypes exhibit differential activities (Kalie et al., 2008; Levin et al., 2011; Piehler et al., 2012). The binding and activity of all human IFNα subtypes has been compared in a recent study (Lavoie et al., 2011). Except IFNα1 that binds weakly, all other IFNα, including IFNα2, have a similar range of affinity and activity, all weaker than those of IFNβ.
4- IFNα2 biological functions
Stricto sensu, nothing is known about functions unique to IFNα2 in human physiology. Probably its function is integrated with that of all other IFNα since, contrary to IFNβ, there is no known context where IFNα2 is specifically produced as an isolated subtype. Indeed, if IFNα2 is the main IFN subtype that has been used for most in vitro experiments to elucidate IFN signalling and mechanism of action, almost all physiological and functional data of type I IFN have been generated in mice, notably through the study of knock-in and knock-out animal models generated in the last decade.
Many aspects of the cellular functions that are modulated by type I IFNs. have been reviewed recently (Trinchieri, 2010; Gough et al., 2012; Tomasello et al., 2014; Crouse et al., 2015; Gajewski and Corrales, 2015; McNab et al., 2015). The important point that emerged is that type I IFN, probably only IFNβ, must be produced at low level under steady-state conditions to ensure the ontogeny and function of several cell types. In the absence of such constitutive production, the homeostasis of the hematopoietic system is perturbed and mice are more susceptible to infection, cancer and have increased bone degradation. Following viral infections or pathogen and stress-associated molecular patterns recognition, IFNα are transiently produced and become detectable in peripheral blood. This IFN not only will prevent viral spread but also will critically orchestrate the set-up of an immune adaptive response. Many pathogenic viruses have developed mechanisms to escape IFN action by inhibiting its production and/or its action (Haller and Weber, 2007; Misasi and Sullivan, 2014). On the other hand, IFN can have detrimental effects in bacterial, parasitic and fungal infections (Trinchieri, 2010; Tomasello et al., 2014; McNab et al., 2015; Stifter and Feng, 2015). These observations explain the well known risk of secondary bacterial infection upon a primary viral attack. Although the mechanisms are complex, and often differ depending of the secondary infection, the general concept is that the IFN produced in response to the primary viral infection inappropriately polarizes the immune responses, compromising the recruitment of IFNγ-dependent anti-microbial innate effector cells.
A balanced IFN production and a balanced IFN response are mandatory to avoid deleterious effects. Subtle dysregulations in the control of these pathways can be disruptive to the equilibrium of the immune system (Tomasello et al., 2014). Systemic lupus erythematous and Sjogren's syndrome are two examples of autoimmune/inflammatory diseases caused and/or worsened by unbridled IFN responses. Compelling evidence of the harmful effect of excess IFNα has come in recent years from the identification of rare monogenic Mendelian disorders which have been grouped as interferonopathies. Patients with the rare Aicardi-Goutières syndrome (AGS) have been extensively studied and causal mutations identified. AGS patients present with severe auto-inflammation affecting mostly the brain and the skin, though a broad spectrum of other clinical phenotypes have been found (Crow, 2015). The genetic basis of AGS has been defined in a number of families. These disorders are genetically different (caused by mutations in different genes), but are all associated with a disturbance in IFNα homeostasis. Rare patients with an impaired response to type I IFN have been reported (Bogunovic et al., 2012; Zhang et al., 2015). In these individuals the complete deficiency of ISG15, a ubiquitin-like modifier, is responsible of high susceptibility to the Bacillus Calmette-Guérin strain used in vaccination against tuberculosis. Moreover, ISG15 deficiency leads to sustained signaling to type I IFNs, autoantibodies production and the development of inflammation targeting the brain. Hence this autoimmune disorder shares similarities with AGS. Importantly, a deregulation of the IFNα system - whether excess production or response - gives rise to a high baseline level of ISG transcripts (a type I IFN signature) in blood cells. Thus, measuring such signature may become a routine screening test in the clinic to identify new interferopathies. Moreover, the recognition of a diverse spectrum of autoimmune phenotypes with excess IFNα calls for strategies of rational intervention with anti-IFN, anti-IFN receptor antibodies or inhibitors of Janus kinases.
5- IFNα2 in the clinic
Except in intra-lesional application, IFNα2 is administered parenterally in order to avoid proteolysis. Among IFNα formulations available in clinical practice, recombinant IFNα2 and pegylated forms (PEG-IFNα2) are the most widely used as antiviral and antitumoral agents. Polyethylene glycol (PEG) covalently linked to IFNα2 improves its pharmacokinetic properties, allowing single weekly administration instead of several administrations per week with standard IFNα2.
Since its first approval for the treatment of hairy cell leukemia in 1986, recombinant IFNα2 has been approved for the treatment of chronic viral hepatitis B (HBV), chronic viral hepatitis C (HCV), chronic myeloid leukemia (CML), Kaposi sarcoma, follicular lymphoma, renal cell carcinoma (RCC), melanoma, T cell lymphoma, multiple myeloma and condylomata acuminate (Antonelli et al., 2015). IFNα2 was also the first available therapy for the treatment of chronic HCV (Hoofnagle et al., 1986). The association of ribavirin and IFNα2 significantly increased the rate of sustained virological response (SVR) (McHutchison et al., 1998). The availability of PEG-IFNα2, more efficient in HCV than IFNα2 (Zeuzem et al., 2000), further enhanced the SVR rate when associated with ribavirin (Manns et al., 2001). PEG-IFNα2/ribavirin was thus the standard of care of chronic HCV.
Antiviral agents against HBV, and more recently against HCV, have challenged the use of IFNα2. Directly acting antiviral agents (DAA), including protease and polymerase inhibitors interfering with HCV replication, are now available. These DAA are well-tolerated drugs that induce a high rate of SVR in a shorter period of treatment. Thus, DAA, initially administered in combination with PEG-IFNα2/ribavirin, are currently investigated in IFN-free regimens (Cortez and Kottilil, 2015). Similarly, targeted therapies are progressively replacing IFN-based therapy of metastatic RCC (VEGF and mTOR inhibitors) and melanoma (BRAF inhibitors). For advanced follicular lymphoma, rituximab is approved in the induction phase (associated with chemotherapy) and in the maintenance setting. Similarly, drugs such as thalidomide, lenalidomide, bortezomib have replaced IFNα2 in the treatment of multiple myeloma (Antonelli et al., 2015).
IFNα2 has re-emerged in the treatment of patients suffering from CML (chronic myelogenous leukemia). IFNα2 was the first drug to significantly improve CML patients’ survival compared to chemotherapy (Talpaz et al., 1987). Since the availability of tyrosine kinase inhibitors (TKI), IFNα2 was no longer considered, until clinical data suggested that IFNα2 pre-treated patients experienced delayed relapses after TKI discontinuation (Rousselot et al., 2007). It was also reported that the association of IFNα2 to TKI significantly improves the major molecular response rate (Preudhomme et al., 2010). Several mechanisms could explain the potential benefit of combining IFNα2 to TKI (Talpaz et al., 2013). The combo therapy is thus currently investigated in clinical trials since TKI do not appear to be curative in CML. Yet, the major limitation of IFNα2 use in CML is its adverse effects which limit its full clinical benefit (Guilhot et al., 2009).
In general, the major limiting factor in the clinical use of IFNα2 is the occurrence of induced side effects that alter the patients’ quality of life and often require dose reduction or premature interruption (Sleijfer et al., 2005). When monitoring patients receiving IFNα2, the main concern is neurological toxicity, with depression and suicidal ideation, symptoms that should be detected and carefully managed. Flu-like syndrome, characterized by fever, chills, myalgia, and headache, almost always occurs but is rarely dose-limiting contrary to the other adverse effects. Among these latter, hematotoxicity results from different mechanisms including inhibition of lymphocyte egress from lymph nodes (Shiow et al., 2006) and inhibition of platelet production (Yamane et al., 2008). Chronic side effects include asthenia, weight loss and autoimmune complications such as thyroid dysfunction, diabetes and dermatological manifestations. Other side effects (hepatic, renal, pulmonary, cardiovascular, gastro-intestinal) are less frequent but can be life-threatening.
A correlation has been found between clinical efficacy of IFNα2 in patients treated for melanoma and the appearance of autoimmune manifestations (Gogas et al., 2006). Thus, side effects can greatly limit treatment efficacy and are likely related to the pleiotropic activity of IFN and its ability to act on all cells in the body. In this regard, methodologies to increase the cellular specificity of IFN action are being developed (Garcin et al., 2014; Tomasello et al., 2014; Uze and Tavernier, 2015). These targeting strategies may provide new IFN formulations, more efficient, better tolerated, and could lead to a revival of this old drug with untapped potential. However, the cell types on which IFN has to act in order to establish the wanted physiological effects are far from being identified. Their characterization is undoubtedly a challenge to the ongoing academic research.
Highlights.
IFNA2 was one of the first type I interferon gene cloned in the 1980's
IFNα2 is one of the most potent interferon α
IFNα2 is the interferon α subtype used in the clinic
Its gene structure, expression, signalling and biological functions are reviewed
Acknowledgments
This review and the corresponding Gene Wiki article are written as part of the Gene Wiki Review series - a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM083924). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. The authors wish to acknowledge grant funding from Canceropôle - INCa, the LabEx MabImprove and the Institut Carnot CALYM and institutional funding from Institut Pasteur. FP was supported by the Fondation ARC and the SIRIC Montpellier Cancer (Grant INCa-DGOS-Inserm 6045).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonelli G, Scagnolari C, Moschella F, Proietti E. Twenty-five years of type I interferon-based treatment: A critical analysis of its therapeutic use. Cytokine Growth Factor Rev. 2015 doi: 10.1016/j.cytogfr.2014.12.006. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwell BN, Slaney CY, Withana NP, Forster S, Cao Y, Loi S, Andrews D, Mikeska T, Mangan NE, Samarajiwa SA, de Weerd NA, Gould J, Argani P, Moller A, Smyth MJ, Anderson RL, Hertzog PJ, Parker BS. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med. 2012;18:1224–31. doi: 10.1038/nm.2830. [DOI] [PubMed] [Google Scholar]
- Bis RL, Stauffer TM, Singh SM, Lavoie TB, Mallela KM. High yield soluble bacterial expression and streamlined purification of recombinant human interferon alpha-2a. Protein Expr Purif. 2014;99:138–46. doi: 10.1016/j.pep.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, Mansouri N, Okada S, Bryant VL, Kong XF, Kreins A, Velez MM, Boisson B, Khalilzadeh S, Ozcelik U, Darazam IA, Schoggins JW, Rice CM, Al-Muhsen S, Behr M, Vogt G, Puel A, Bustamante J, Gros P, Huibregtse JM, Abel L, Boisson-Dupuis S, Casanova JL. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–8. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–90. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette BC, Liang H, Lee Y, Chlewicki L, Khodarev NN, Weichselbaum RR, Fu YX, Auh SL. The efficacy of radiotherapy relies upon induction of type i interferon-dependent innate and adaptive immunity. Cancer Res. 2011;71:2488–96. doi: 10.1158/0008-5472.CAN-10-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chill JH, Quadt SR, Levy R, Schreiber G, Anglister J. The human type I interferon receptor: NMR structure reveals the molecular basis of ligand binding. Structure. 2003;11:791–802. doi: 10.1016/s0969-2126(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, Uze G. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. 2004;34:796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- Coccia EM, Uze G, Pellegrini S. Negative regulation of type I interferon signaling: facts and mechanisms. Cell Mol Biol (Noisy-le-grand) 2006;52:77–87. [PubMed] [Google Scholar]
- Cortez KJ, Kottilil S. Beyond interferon: rationale and prospects for newer treatment paradigms for chronic hepatitis C. Ther Adv Chronic Dis. 2015;6:4–14. doi: 10.1177/2040622314551934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 2015;15:231–42. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- Crow YJ. Type I interferonopathies: Mendelian type I interferon up-regulation. Curr Opin Immunol. 2015;32C:7–12. doi: 10.1016/j.coi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- de Weerd NA, Vivian JP, Nguyen TK, Mangan NE, Gould JA, Braniff SJ, Zaker-Tabrizi L, Fung KY, Forster SC, Beddoe T, Reid HH, Rossjohn J, Hertzog PJ. Structural basis of a unique interferon-beta signaling axis mediated via the receptor IFNAR1. Nat Immunol. 2013;14:901–7. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- Derynck R, Remaut E, Saman E, Stanssens P, De Clercq E, Content J, Fiers W. Expression of human fibroblast interferon gene in Escherichia coli. Nature. 1980;287:193–7. doi: 10.1038/287193a0. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, Murphy KM, Schreiber RD. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz MO, Pomykala HM, Bohlander SK, Maltepe E, Malik K, Brownstein B, Olopade OI. Structure of the human type-I interferon gene cluster determined from a YAC clone contig. Genomics. 1994;22:540–52. doi: 10.1006/geno.1994.1427. [DOI] [PubMed] [Google Scholar]
- Francois-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-Garcia L, Piehler J, Pellegrini S, Uze G. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS One. 2011;6:e22200. doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, Gajewski TF. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208:2005–16. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski TF, Corrales L. New perspectives on type I IFNs in cancer. Cytokine Growth Factor Rev. 2015 doi: 10.1016/j.cytogfr.2015.01.001. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin G, Paul F, Staufenbiel M, Bordat Y, Van der Heyden J, Wilmes S, Cartron G, Apparailly F, De Koker S, Piehler J, Tavernier J, Uze G. High efficiency cell-specific targeting of cytokine activity. Nat Commun. 2014;5:3016. doi: 10.1038/ncomms4016. [DOI] [PubMed] [Google Scholar]
- Genin P, Vaccaro A, Civas A. The role of differential expression of human interferon--a genes in antiviral immunity. Cytokine Growth Factor Rev. 2009;20:283–95. doi: 10.1016/j.cytogfr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Goeddel DV, Yelverton E, Ullrich A, Heyneker HL, Miozzari G, Holmes W, Seeburg PH, Dull T, May L, Stebbing N, Crea R, Maeda S, McCandliss R, Sloma A, Tabor JM, Gross M, Familletti PC, Pestka S. Human leukocyte interferon produced by E. coli is biologically active. Nature. 1980;287:411–6. doi: 10.1038/287411a0. [DOI] [PubMed] [Google Scholar]
- Gogas H, Ioannovich J, Dafni U, Stavropoulou-Giokas C, Frangia K, Tsoutsos D, Panagiotou P, Polyzos A, Papadopoulos O, Stratigos A, Markopoulos C, Bafaloukos D, Pectasides D, Fountzilas G, Kirkwood JM. Prognostic significance of autoimmunity during treatment of melanoma with interferon. N Engl J Med. 2006;354:709–18. doi: 10.1056/NEJMoa053007. [DOI] [PubMed] [Google Scholar]
- Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–74. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresser I. The antitumor effects of interferon: a personal history. Biochimie. 2007;89:723–8. doi: 10.1016/j.biochi.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Guilhot F, Roy L, Saulnier PJ, Guilhot J. Interferon in chronic myeloid leukaemia: past and future. Best Pract Res Clin Haematol. 2009;22:315–29. doi: 10.1016/j.beha.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Haller O, Weber F. Pathogenic viruses: smart manipulators of the interferon system. Curr Top Microbiol Immunol. 2007;316:315–34. doi: 10.1007/978-3-540-71329-6_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Takaoka A, Taniguchi T. Regulation of the type I IFN induction: a current view. Int Immunol. 2005;17:1367–78. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- Hoofnagle JH, Mullen KD, Jones DB, Rustgi V, Di Bisceglie A, Peters M, Waggoner JG, Park Y, Jones EA. Treatment of chronic non-A, non-B hepatitis with recombinant human alpha interferon. A preliminary report. N Engl J Med. 1986;315:1575–8. doi: 10.1056/NEJM198612183152503. [DOI] [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–67. [PubMed] [Google Scholar]
- Ito T, Kanzler H, Duramad O, Cao W, Liu YJ. Specialization, kinetics, and repertoire of type 1 interferon responses by human plasmacytoid predendritic cells. Blood. 2006;107:2423–31. doi: 10.1182/blood-2005-07-2709. [DOI] [PubMed] [Google Scholar]
- Kalie E, Jaitin DA, Podoplelova Y, Piehler J, Schreiber G. The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J Biol Chem. 2008;283:32925–36. doi: 10.1074/jbc.M806019200. [DOI] [PubMed] [Google Scholar]
- Klaus W, Gsell B, Labhardt AM, Wipf B, Senn H. The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution. J Mol Biol. 1997;274:661–75. doi: 10.1006/jmbi.1997.1396. [DOI] [PubMed] [Google Scholar]
- Lavoie TB, Kalie E, Crisafulli-Cabatu S, Abramovich R, DiGioia G, Moolchan K, Pestka S, Schreiber G. Binding and activity of all human alpha interferon subtypes. Cytokine. 2011;56:282–9. doi: 10.1016/j.cyto.2011.07.019. [DOI] [PubMed] [Google Scholar]
- Lawn RM, Adelman J, Dull TJ, Gross M, Goeddel D, Ullrich A. DNA sequence of two closely linked human leukocyte interferon genes. Science. 1981;212:1159–62. doi: 10.1126/science.6165082. [DOI] [PubMed] [Google Scholar]
- Levin D, Harari D, Schreiber G. Stochastic receptor expression determines cell fate upon interferon treatment. Mol Cell Biol. 2011;31:3252–66. doi: 10.1128/MCB.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Strunk JJ, Lamken P, Piehler J, Walz T. The EM structure of a type I interferon-receptor complex reveals a novel mechanism for cytokine signaling. J Mol Biol. 2008;377:715–24. doi: 10.1016/j.jmb.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–67. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–65. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- Manry J, Laval G, Patin E, Fornarino S, Itan Y, Fumagalli M, Sironi M, Tichit M, Bouchier C, Casanova JL, Barreiro LB, Quintana-Murci L. Evolutionary genetic dissection of human interferons. J Exp Med. 2011;208:2747–59. doi: 10.1084/jem.20111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485–92. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi J, Sullivan NJ. Camouflage and misdirection: the full-on assault of ebola virus disease. Cell. 2014;159:477–86. doi: 10.1016/j.cell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Mantei N, Weissmann C. The structure of one of the eight or more distinct chromosomal genes for human interferon-alpha. Nature. 1980a;287:401–8. doi: 10.1038/287401a0. [DOI] [PubMed] [Google Scholar]
- Nagata S, Taira H, Hall A, Johnsrud L, Streuli M, Ecsodi J, Boll W, Cantell K, Weissmann C. Synthesis in E. coli of a polypeptide with human leukocyte interferon activity. Nature. 1980b;284:316–20. doi: 10.1038/284316a0. [DOI] [PubMed] [Google Scholar]
- Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Piehler J, Thomas C, Garcia KC, Schreiber G. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol Rev. 2012;250:317–34. doi: 10.1111/imr.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Preudhomme C, Guilhot J, Nicolini FE, Guerci-Bresler A, Rigal-Huguet F, Maloisel F, Coiteux V, Gardembas M, Berthou C, Vekhoff A, Rea D, Jourdan E, Allard C, Delmer A, Rousselot P, Legros L, Berger M, Corm S, Etienne G, Roche-Lestienne C, Eclache V, Mahon FX, Guilhot F. Imatinib plus peginterferon alfa-2a in chronic myeloid leukemia. N Engl J Med. 2010;363:2511–21. doi: 10.1056/NEJMoa1004095. [DOI] [PubMed] [Google Scholar]
- Qi Z, Nie P, Secombes CJ, Zou J. Intron-containing type I and type III IFN coexist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J Immunol. 2010;184:5038–46. doi: 10.4049/jimmunol.0903374. [DOI] [PubMed] [Google Scholar]
- Quadt-Akabayov SR, Chill JH, Levy R, Kessler N, Anglister J. Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex. Protein Sci. 2006;15:2656–68. doi: 10.1110/ps.062283006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan R, Walter LJ, Hruza A, Reichert P, Trotta PP, Nagabhushan TL, Walter MR. Zinc mediated dimer of human interferon-alpha 2b revealed by X-ray crystallography. Structure. 1996;4:1453–63. doi: 10.1016/s0969-2126(96)00152-9. [DOI] [PubMed] [Google Scholar]
- Rizza P, Moretti F, Capone I, Belardelli F. Role of type I interferon in inducing a protective immune response: Perspectives for clinical applications. Cytokine Growth Factor Rev. 2015 doi: 10.1016/j.cytogfr.2014.10.002. In Press. [DOI] [PubMed] [Google Scholar]
- Rousselot P, Huguet F, Rea D, Legros L, Cayuela JM, Maarek O, Blanchet O, Marit G, Gluckman E, Reiffers J, Gardembas M, Mahon FX. Imatinib mesylate discontinuation in patients with chronic myelogenous leukemia in complete molecular remission for more than 2 years. Blood. 2007;109:58–60. doi: 10.1182/blood-2006-03-011239. [DOI] [PubMed] [Google Scholar]
- Schiavoni G, Sistigu A, Valentini M, Mattei F, Sestili P, Spadaro F, Sanchez M, Lorenzi S, D'Urso MT, Belardelli F, Gabriele L, Proietti E, Bracci L. Cyclophosphamide synergizes with type I interferons through systemic dendritic cell reactivation and induction of immunogenic tumor apoptosis. Cancer Res. 2011;71:768–78. doi: 10.1158/0008-5472.CAN-10-2788. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–25. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–4. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- Sleijfer S, Bannink M, Van Gool AR, Kruit WH, Stoter G. Side effects of interferon-alpha therapy. Pharm World Sci. 2005;27:423–31. doi: 10.1007/s11096-005-1319-7. [DOI] [PubMed] [Google Scholar]
- Stagg J, Loi S, Divisekera U, Ngiow SF, Duret H, Yagita H, Teng MW, Smyth MJ. Anti-ErbB-2 mAb therapy requires type I and II interferons and synergizes with anti-PD-1 or anti-CD137 mAb therapy. Proc Natl Acad Sci U S A. 2011;108:7142–7. doi: 10.1073/pnas.1016569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifter SA, Feng CG. Interfering with Immunity: Detrimental Role of Type I IFNs during Infection. J Immunol. 2015;194:2455–2465. doi: 10.4049/jimmunol.1402794. [DOI] [PubMed] [Google Scholar]
- Streuli M, Nagata S, Weissmann C. At least three human type alpha interferons: structure of alpha 2. Science. 1980;209:1343–7. doi: 10.1126/science.6158094. [DOI] [PubMed] [Google Scholar]
- Strunk JJ, Gregor I, Becker Y, Li Z, Gavutis M, Jaks E, Lamken P, Walz T, Enderlein J, Piehler J. Ligand binding induces a conformational change in ifnar1 that is propagated to its membrane-proximal domain. J Mol Biol. 2008;377:725–39. doi: 10.1016/j.jmb.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Talpaz M, Hehlmann R, Quintas-Cardama A, Mercer J, Cortes J. Re-emergence of interferon-alpha in the treatment of chronic myeloid leukemia. Leukemia. 2013;27:803–12. doi: 10.1038/leu.2012.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talpaz M, Kantarjian HM, McCredie KB, Keating MJ, Trujillo J, Gutterman J. Clinical investigation of human alpha interferon in chronic myelogenous leukemia. Blood. 1987;69:1280–8. [PubMed] [Google Scholar]
- Taniguchi T, Guarente L, Roberts TM, Kimelman D, Douhan J, 3rd, Ptashne M. Expression of the human fibroblast interferon gene in Escherichia coli. Proc Natl Acad Sci U S A. 1980;77:5230–3. doi: 10.1073/pnas.77.9.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, Lee C, Yarden G, Vleck SE, Glenn JS, Nolan GP, Piehler J, Schreiber G, Garcia KC. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 2011;146:621–32. doi: 10.1016/j.cell.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasello E, Pollet E, Vu Manh TP, Uze G, Dalod M. Harnessing Mechanistic Knowledge on Beneficial Versus Deleterious IFN-I Effects to Design Innovative Immunotherapies Targeting Cytokine Activity to Specific Cell Types. Front Immunol. 2014;5:526. doi: 10.3389/fimmu.2014.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–63. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uze G, Tavernier J. High efficiency targeting of IFN-alpha activity: Possible applications in fighting tumours and infections. Cytokine Growth Factor Rev. 2015 doi: 10.1016/j.cytogfr.2014.10.006. In Press. [DOI] [PubMed] [Google Scholar]
- von Gabain A, Lundgren E, Ohlsson M, Holmgren E, Josephsson S, Alkan SS. Three human interferon-alpha 2 subvariants disclose structural and functional differences. Eur J Biochem. 1990;190:257–61. doi: 10.1111/j.1432-1033.1990.tb15570.x. [DOI] [PubMed] [Google Scholar]
- Wilmes S, Beutel O, Li Z, Francois-Newton V, Richter CP, Janning D, Kroll C, Hanhart P, Hötte K, You C, Uzé G, Pellegrini S, Piehler J. Receptor dimerization dynamics as regulatory valve for plasticity of type I interferon signaling. J. Cell. Biol. 2015 doi: 10.1083/jcb.201412049. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk CH, Frost SD, Richman DD, Higley PE, Kosakovsky Pond SL. Evolution of the interferon alpha gene family in eutherian mammals. Gene. 2007;397:38–50. doi: 10.1016/j.gene.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Nakamura T, Suzuki H, Ito M, Ohnishi Y, Ikeda Y, Miyakawa Y. Interferon-alpha 2b-induced thrombocytopenia is caused by inhibition of platelet production but not proliferation and endomitosis in human megakaryocytes. Blood. 2008;112:542–50. doi: 10.1182/blood-2007-12-125906. [DOI] [PubMed] [Google Scholar]
- Zeuzem S, Feinman SV, Rasenack J, Heathcote EJ, Lai MY, Gane E, O'Grady J, Reichen J, Diago M, Lin A, Hoffman J, Brunda MJ. Peginterferon alfa-2a in patients with chronic hepatitis C. N Engl J Med. 2000;343:1666–72. doi: 10.1056/NEJM200012073432301. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, Tezcan I, Rice GI, Chen C, Mansouri N, Mahdaviani SA, Itan Y, Boisson B, Okada S, Zeng L, Wang X, Jiang H, Liu W, Han T, Liu D, Ma T, Wang B, Liu M, Liu JY, Wang QK, Yalnizoglu D, Radoshevich L, Uze G, Gros P, Rozenberg F, Zhang SY, Jouanguy E, Bustamante J, Garcia-Sastre A, Abel L, Lebon P, Notarangelo LD, Crow YJ, Boisson-Dupuis S, Casanova JL, Pellegrini S. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]