Abstract
Eukaryotic genomes are rich in transcription units encoding “long noncoding RNAs” (lncRNAs). The purpose of all this transcription is unclear since most lncRNAs are quickly targeted for destruction during synthesis or shortly thereafter. As debates continue over the functional significance of many specific lncRNAs, support grows for the notion that the act of transcription rather than the RNA product itself is functionally important in many cases. Indeed, this alternative mechanism might better explain how low-abundance lncRNAs transcribed from noncoding DNA function in organisms. Here, we highlight some of the recently emerging features that distinguish coding from noncoding transcription and discuss how these differences might have important implications for the functional consequences of noncoding transcription.
Keywords: nascent transcription, noncoding transcription, transcription cycle, transcriptional interference, long noncoding RNA (lncRNA), gene regulation, RNA Polymerase II transcription, chromatin
RNA Polymerase II (RNAPII) pervasively transcribes genomes, generating all protein-coding messenger RNAs (mRNAs) and many noncoding RNAs longer than 200 nucleotides (long noncoding RNAs; lncRNAs) (Jensen et al. 2013). Today, it is accepted that most eukaryotic genomes generate an abundance of lncRNA transcripts but, for technical reasons discussed below, many lncRNAs escaped detection until recently. Consequently, only a relatively small fraction of lncRNAs have been functionally characterized, unlike other classes of well-characterized short noncoding RNAs generated by RNAPII (e.g., micro RNAs, small interfering RNAs, small nuclear RNAs, and small nucleolar RNAs). The fact that lncRNAs are defined by their size rather than any common function is itself evidence that this grouping is arbitrary and, indeed, relatively little is yet known about the roles of this heterogeneous class of noncoding transcripts.
Superficially, mRNAs and lncRNAs share many common features. Both classes of transcripts are 5′-capped, may be multiexonic, alternatively spliced, and 3′ polyadenylated. However, unlike stable mature mRNAs that are exported to the cytoplasm for protein synthesis, lncRNAs are predominantly nuclear and often rapidly degraded. In fact, subclasses of lncRNAs are distinguished by different decay pathways responsible for their degradation (Marquardt et al. 2011), which are often targeted during or shortly after synthesis (Houseley and Tollervey 2009). As a class, lncRNAs exhibit poor sequence conservation between species. However, lack of sequence similarity does not necessarily rule out function as several functionally conserved lncRNAs exhibit little or no detectable primary sequence homology between closely related species, as is the case for the X-inactivation-specific transcript XIST, which is responsible for initiating dosage compensation in female mammals (Pang et al. 2006). Moreover, some lncRNA promoters, splice sites, and positions with respect to flanking genes are preserved through evolution (Ulitsky 2016), indicating that lncRNA transcription at equivalent positions can be conserved, even though this results in RNA products that lack obvious homology.
Putative functions have been assigned to an ever-increasing number of relatively high- and low-abundance lncRNAs in diverse organisms [as reviewed extensively in Mercer et al. (2009), Ponting et al. (2009), Rinn and Chang (2012), Geisler and Coller (2013), Kung et al. (2013), Engreitz et al. (2016), Quinn and Chang (2016)]. However, the mechanisms by which specific lncRNAs achieve their effects on gene regulation are complicated by the fact that lncRNA transcription can itself impinge on the activity of nearby genes (Kornienko et al. 2013). This is problematic since it is experimentally difficult to distinguish the roles of any given lncRNA from the unexpected consequences resulting from the act of its transcription. For example, the imprinted mouse lncRNA Airn was initially proposed to induce targeted repression of the Igf2r gene by mediating chromatin looping, but later analyses revealed that the Airn lncRNA is itself dispensable as the act of transcribing this locus is sufficient to induce Igf2r silencing (Latos et al. 2012). Similarly, new lncRNA studies regularly challenge or even overturn the interpretations of earlier ones (Cech and Steitz 2014). This is partly because generally accepted methods for clearly assigning function to lncRNAs have only recently been set out and adopted (Bassett et al. 2014; Goff and Rinn 2015). Ultimately, the idea that the act of noncoding transcription can itself have important functional outcomes provides an intriguing paradigm for how widespread transcription, even when such acts fail to produce stable lncRNAs (“cryptic transcription”), can serve important biological roles in both dense yeast genomes as well as in much larger genomes of multicellular eukaryotes.
Here, we discuss recently emerging features that distinguish coding from noncoding transcription in yeast and human, and outline how these differences might have important implications for the functional consequences of pervasive noncoding transcription both proximal to and far from protein-coding genes.
Protein-Coding Gene Transcription Cycle
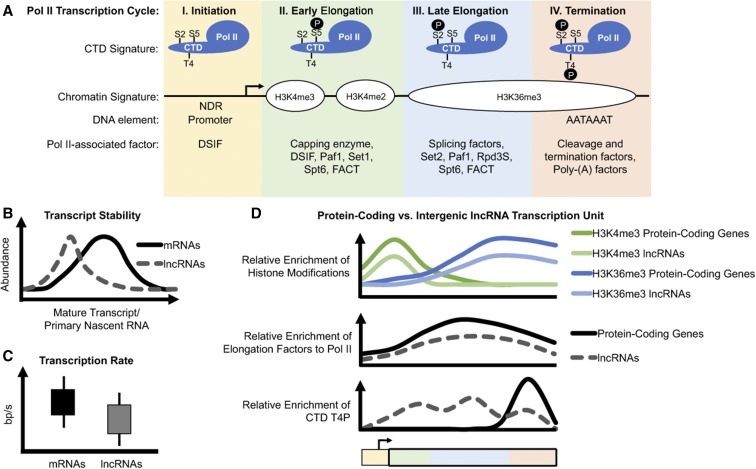
RNAPII transcription is tightly coordinated with chromatin changes and nascent RNA processing. Many of the factors mediating these cotranscriptional activities are targeted by interactions with nascent RNA (Bentley 2014; Battaglia et al. 2017), post-translational histone modifications (Jonkers and Lis 2015), and/or post-translational modifications of the C-terminal domain (CTD) of the largest subunit of RNAPII, which is composed of heptapeptide repeats (Y1S2P3T4S5P6S7) (Corden 2013). Different CTD modifications recruit/repel distinct RNA-processing and chromatin-modifying complexes and therefore accompany different stages in the transcription cycle (Figure 1A). For example, promoter-associated RNAPII acquires Ser5 phosphorylation (Ser5P), which recruits Set1 to trimethylate histone H3 lysine 4 (H3K4me3) on promoter-proximal nucleosomes (Ng et al. 2003). H3K4me3, along with histone acetylation, are defining features of active protein-coding gene promoters. Well-conserved histone chaperones FACT and Spt6 travel with RNAPII and facilitate transcription through chromatin (Kwak and Lis 2013). As transcription proceeds from the site of initiation, Set1 restricts histone acetylation to the promoter by depositing H3K4me2, which creates a platform for the Set3 histone deacetylase complex (HDAC) (Kim and Buratowski 2009). Elongating RNAPII quickly loses Ser5P, acquires Ser2P, and attracts Set2 to deposit H3K36me3, which is required to stabilize the Rpd3S HDAC over active gene bodies (Li et al. 2003; Brodsky et al. 2005; Carrozza et al. 2005; Keogh et al. 2005; Drouin et al. 2010; Govind et al. 2010). Cells lacking these factors tend to accumulate lncRNAs that initiate within or overlap with and interfere with protein-coding gene expression (Kaplan et al. 2003; Li et al. 2007; Cheung et al. 2008). In some cases, alternative intragenic promoters can also give rise to alternative transcript isoforms that generate N-terminally truncated proteins with distinct functional outcomes (Wiesner et al. 2015). Thus, RNAPII transcription-coupled chromatin changes both suppress alternative coding transcript isoforms and prevent noncoding transcription initiation from “cryptic” intragenic promoters. Finally, transcription termination is characterized by Ser2P and Thr4P, which help slow RNAPII and trigger nascent transcript cleavage/3′-end formation (Harlen and Churchman 2017). This last step is particularly important as inefficient termination causes RNAPII to continue unabated and interfere with the expression of genes positioned downstream (Proudfoot 1986), a phenomenon frequently observed in dense yeast genomes (Gullerova and Proudfoot 2010). In addition, unauthorized interfering acts of transcription have also been observed in various human diseases (Proudfoot 2016). Thus, pervasive transcription implies that most acts of transcription are likely to impinge on the activity of other closely positioned or overlapping transcription units (Mellor et al. 2016), even in mammalian genomes that show much greater spacing between genes.
Figure 1.
Emerging differences in RNAPII behaviors at protein-coding and lncRNA transcription units. (A) The RNAPII transcription cycle at protein-coding genes is defined by four distinct stages. These stages are characterized by the association of RNAPII with transcription elongation factors and RNA processing factors, many of which are recruited by different post-translational modifications to the RNAPII CTD and/or histones. Only some of these factors are listed here. Active promoters are NDRs, while terminators are generally enriched for the AATAAAT consensus motif. (B) In general, lncRNAs are much less abundant than mRNAs, as lncRNAs are predominantly localized in chromatin where they are rapidly degraded (Schlackow et al. 2017). (C) Metabolic labeling studies using 4sU or 4tU reveal that RNAPII transcription rates are on average slower for lncRNAs (Barrass et al. 2015; Eser et al. 2016; Mukherjee et al. 2017). (D) Many lncRNAs also exhibit reduced levels of transcription-coupled histone marks such as H3K4me3 and H3K36me3 (Sun et al. 2015), decreased recruitment of many common elongation factors (Battaglia et al. 2017; Fischl et al. 2017), inefficient processing (Eser et al. 2016; Mukherjee et al. 2017; Schlackow et al. 2017), and early termination (Milligan et al. 2016; Schlackow et al. 2017). 4sU, 4-thiouridine; 4tU, 4-thiouracil; CTD, C-terminal domain; H3K, histone H3 lysine; lncRNA, long noncoding RNA; NDR, nucleosome-depleted regions; RNAPII, RNA Polymerase II.
Distinct Behaviors of RNAPII on Coding and Noncoding Transcription Units
In multicellular organisms, tissue-specific lncRNA expression and methods relying on steady-state RNA levels [e.g., total RNA sequencing (RNA-seq)] can result in lower estimates of lncRNA abundance through undersampling of lncRNA in complex mixtures (Deveson et al. 2017). Many lncRNAs escape detection since RNAPII transcription over noncoding DNA frequently fails to produce stable RNAs. Previously, these cryptic lncRNAs had only been observed in cells depleted of RNA quality control/decay factors (Jensen et al. 2013), implying that they are rapidly targeted for degradation and might simply be aberrantly produced. However, mounting evidence suggests that the transcription of both stable and cryptic lncRNAs can be sensitive to environmental cues and hence dynamically regulate the expression of adjacent genes (described below). The development of less intrusive methods to profile nascent transcription in wild-type cells now allows the annotation and characterization of previously undetected cryptic transcription units across entire genomes.
One of these less disruptive methods to profile nascent transcription involves quickly labeling nascent RNA by exposing cells to 4-thiouracil (4tU) or 4-thiouridine (4sU). Once isolated from cells, labeled RNA is then enriched from total RNA, sequenced, and compared with steady-state RNA-seq data to calculate the rate of RNA synthesis and degradation. This approach has revealed recently that coding and noncoding transcription units display distinct patterns of synthesis and degradation. In general, yeast and human lncRNAs are transcribed slower and degraded faster than mRNAs (Barrass et al. 2015; Eser et al. 2016; Mukherjee et al. 2017), consistent with their lower overall levels (Figure 1, B and C). Although there is evidence to suggest that lncRNA splice sites are conserved (Haerty and Ponting 2015; Nitsche et al. 2015), 4tU-/4sU-sequencing (-seq) analyses demonstrate that most human and yeast lncRNAs are only weakly spliced (Eser et al. 2016; Mukherjee et al. 2017). Overall, lncRNA transcription and processing appear, on average, much less efficient than that of mRNAs.
Nascent transcription has also been profiled by sequencing RNAPII-associated RNAs. By capturing native RNAPII–DNA–RNA complexes from chromatin and sequencing, the 3′-most nucleotide of RNAPII-associated RNAs, NET-seq (native elongating transcript sequencing) visualizes active transcription at single-nucleotide resolution (Churchman and Weissman 2011). This method has been used to profile nascent transcription in different yeast species and mutants (Churchman and Weissman 2011; Marquardt et al. 2014; Nguyen et al. 2014; Fischl et al. 2017; Shetty et al. 2017) and has recently been adapted for human cells (Nojima et al. 2015; Fong et al. 2017). Data generated by related methods that rely on UV cross-linking RNAPII to nascent RNA (e.g., cross-linking and cDNA analysis, otherwise known as CRAC) or genome-wide nuclear run-on analyses [e.g., global or precision run-on sequencing (GRO-seq or PRO-seq)] strongly correlate with NET-seq data sets (Mahat et al. 2016; Milligan et al. 2016; Nojima et al. 2016). Therefore, while each method has different limitations and applicability, they all provide relatively accurate depictions of nascent transcription across genomes.
In both Saccharomyces cerevisiae and human cells, distinct patterns of behavior between genic and intergenic lncRNA transcription cycles are also emerging from studies using NET-seq or CRAC to map specific RNAPII CTD modifications. Notably, lncRNA transcription only infrequently transitions from initiation to the elongation phase and, when it does, exhibits a great deal of early termination (Milligan et al. 2016; Schlackow et al. 2017). For example, strong CTD-Thr4P peaks are observed almost exclusively at transcription termination sites for human protein-coding genes, while this termination-specific modification is enriched throughout intergenic lncRNA transcription units (Schlackow et al. 2017) (Figure 1D). Schlackow et al. (2017) also compared RNA-seq profiles for transcripts isolated from distinct subnuclear compartments (e.g., chromatin-enriched and nucleoplasm-enriched) and found that lncRNAs are inefficiently spliced, infrequently polyadenylated, and enriched in chromatin where they are targeted for exosome-mediated degradation. These findings corroborate observed differences in lncRNA processing and stability from kinetic studies utilizing 4tU- or 4sU-seq.
Differential Recruitment of Elongation Factors to RNAPII During Coding and Noncoding Transcription
The positions of RNAPII-associated factors recruited during nascent transcription, as well as the interactions of many of these factors with nascent RNAs, have recently been profiled genome-wide. For example, NET-seq in S. cerevisiae has been adapted to capture elongation factors in complex with RNAPII-associated RNAs to visualize the position of such factors during transcription (Fischl et al. 2017). These analyses demonstrate that the termination-specific factor Pcf11 is enriched on RNAPII during lncRNA transcription, while many common elongation factors (including Paf1, Set2, FACT, and Spt6) on average exhibit decreased recruitment (Figure 1D). Although it is currently unclear what determines these differences, recent findings suggest that many elongation factors in S. cerevisiae are partially recruited to RNAPII through relatively nonspecific direct interactions with nascent RNA and that they show much greater affinity for mRNAs over lncRNAs (Battaglia et al. 2017). These results imply that stable coding transcripts disproportionately recruit elongation factors required for productive RNAPII transcription, since the targeted association of RNA surveillance factors to specific motifs in lncRNAs might physically block elongation factors from binding. Importantly, these findings are consistent with abovementioned analyses showing poor processivity, inefficient processing, and early termination of RNAPII during lncRNA transcription.
lncRNA-Specific Chromatin Features
Differences in the recruitment of core elongation factors to RNAPII during noncoding transcription explain the unique chromatin signatures found at many lncRNA loci (Figure 2). For example, many human lncRNAs exhibit reduced levels of common transcription-coupled histone modifications, such as H3K4me3 and H3K36me3 (Sun et al. 2015). Additionally, active human lncRNA promoters appear to be enriched for H3K9me3 (Méle et al. 2017), a modification typically associated with heterochromatin and gene silencing. In S. cerevisiae, lncRNA transcription antisense to protein-coding genes is characterized by significantly higher levels of H3 acetylation and reduced H3K36me3 (Murray et al. 2015). Within transcribed regions of centromeres in higher eukaryotes, nucleosomes containing canonical H3 are depleted for marks characteristic of active promoters and are instead enriched for H3K4me1/2 and H3K36me2/3 (Sullivan and Karpen 2004; Bergmann et al. 2011). Noncoding transcription at active enhancer elements in metazoans also coincides with an absence of many active promoter marks, instead displaying high levels of H3K4me1 (Li et al. 2016). Collectively, these findings provide compelling evidence distinguishing transcription-associated chromatin signatures for protein-coding genes from that of emerging subclasses of noncoding RNAPII transcription units.
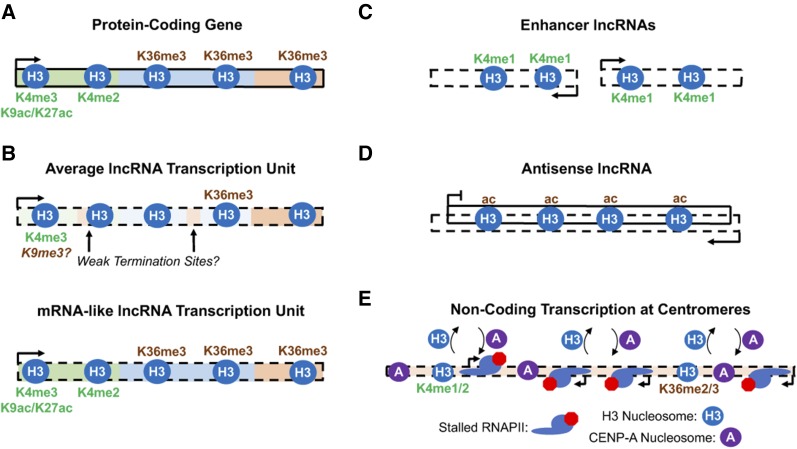
Figure 2.
Chromatin signatures of coding and noncoding transcription. (A) Distribution of transcription-coupled histone modifications across active protein-coding genes. H3K4me3 and H3 acetylation at K9 and K27 are enriched at promoters while H3K4me2 and H3K36me3 accumulate over gene bodies (Corden 2013). (B) Intergenic lncRNAs on average display reduced levels of H3K4me3 and H3K36me3 (Sun et al. 2015) and exhibit premature termination and defective nascent transcript processing (Schlackow et al. 2017). H3K9me3, a mark generally characteristic of repressive heterochromatin, has been detected at active lncRNA promoters in human cells (Méle et al. 2017). Importantly, some lncRNA transcription units also have more mRNA-like patterns of CTD and histone modifications. (C) Chromatin marks typically associated with active promoters (H3K4me3 and H3 acetylation) are depleted from noncoding transcription units at enhancer elements in higher eukaryotes. Instead, enhancers are enriched in H3K4me1 (Li et al. 2016). (D) lncRNA transcription antisense to protein-coding genes has been observed to correspond with reduced H3K36me3 and other marks commonly associated with active transcription elongation and instead enriched for H3 acetylation (Murray et al. 2015). (E) Noncoding transcription in centromeres has been observed to correspond with an absence of H3K4me3 and H3 acetylation. Instead, centromeric nucleosomes containing canonical H3 are enriched in H3K4me1/2 and H3K36me2/3 (Sullivan and Karpen 2004; Bergmann et al. 2011). Frequent transcription stalling in fission yeast centromeres has also been found to stimulate the incorporation of nucleosomes containing the H3-variant CENP-A, which epigenetically specifies centromeres (Catania et al. 2015). CENP-A, centromere protein-A; CTD, C-terminal domain; H3K, histone H3 lysine; lncRNA, long noncoding RNA; RNAPII, RNA Polymerase II.
Divergent Coding/Noncoding Promoters Provide a Unique Opportunity to Compare RNAPII Transcription Cycles Across lncRNA/mRNA Pairs
Profiling nascent transcription has been particularly informative in demonstrating that most nucleosome-depleted regions initiate transcription in both directions (Wei et al. 2011; Scruggs et al. 2015) (Figure 3). Cap analysis gene expression (CAGE) analyses demonstrate that transcription cycles in both directions begin by promoting the same 5′-cap modification to the first transcribed base (Andersson et al. 2014). However, divergent lncRNAs are often targeted for exosome-mediated degradation (Preker et al. 2008), with sense-strand mRNA being noticeably more stable. This is even the case for divergent lncRNA/mRNA pairs that show similar levels of nascent transcription (Sigova et al. 2013). Thus, most promoters appear to preferentially drive transcription in the coding direction. The mechanism(s) by which directionality is achieved are not well-understood. Canonical transcript cleavage, polyadenylation, and termination sequences are enriched in some divergent lncRNAs (Almada et al. 2013). Interestingly, while these cis DNA elements lead to the formation of stable mRNA in the coding direction, divergent lncRNA termination triggers exosome-mediated lncRNA degradation (Ntini et al. 2013). The different effects of cleavage and polyadenylation cis-elements on mRNAs and lncRNAs reconcile differences in the stability of transcript pairs from such promoters. Additionally, the strength of transcription in either direction is selectively regulated at the chromatin level, since divergent lncRNA transcription can be suppressed or activated by a number of chromatin remodelling factors (Churchman and Weissman 2011; Marquardt et al. 2014; Scruggs et al. 2015). RNAPII-associated factors such as PAFI, DSIF, and Ssu72 are also implicated in controlling directionality (Tan-Wong et al. 2012; Fischl et al. 2017; Shetty et al. 2017). Interestingly, there is evidence to suggest that the RNAPII CTD is enriched for phosphorylation of Tyr1 during divergent noncoding transcription (Descostes et al. 2014; Hsin et al. 2014), further revealing the unique properties of lncRNA transcription. Although transcription from coding/noncoding pairs is tightly controlled, our understanding of how RNAPII, RNAPII-associated factors, and chromatin-remodellers discriminate between coding and noncoding orientations is rudimentary. Moreover, the purpose of noncoding transcription from bidirectional promoters remains poorly understood. Considering that widespread divergent noncoding transcription is detected in many organisms, it may prove to be an unavoidable consequence of eukaryotic promoter structure. However, the identification of factors required for specific activation or repression of divergent lncRNA transcription illustrates tight regulation of this process, which is indicative of functional significance although that significance is yet to be fully explained.
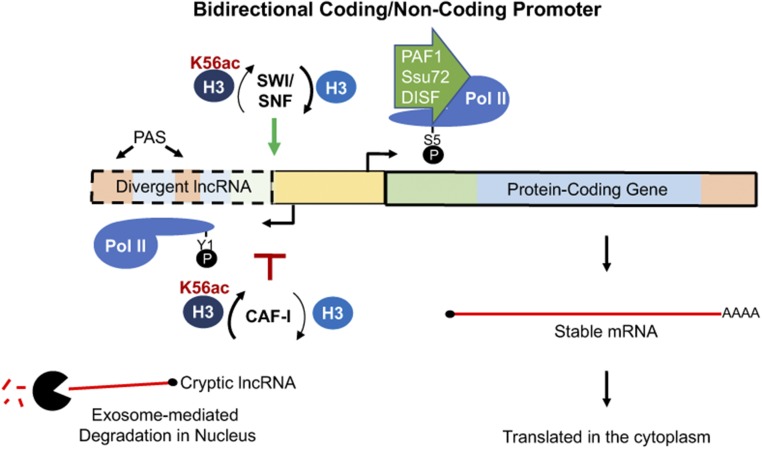
Figure 3.
Bidirectional promoters. Although most promoters can initiate transcription in either direction, the sense orientation is generally favored. Several RNAPII-associated factors and chromatin changes appear to control directionality. For example, PAF1, DSIF, and Ssu72 appear to stimulate RNAPII transcription in the sense direction (Tan-Wong et al. 2012; Fischl et al. 2017; Shetty et al. 2017). Chromatin remodellers also compete to regulate divergent transcription. Notably, CAF-I suppresses divergent transcription by favoring the incorporation of nucleosomes with H3K56ac, while the SWI/SNF complex opposes this activity and thereby promotes divergent transcription (Marquardt et al. 2014). Divergent RNAPII transcription can also be enriched for Tyr1P (Y1P) (Descostes et al. 2014; Hsin et al. 2014). Finally, many divergent lncRNAs are enriched for promoter-proximal polyadenylation sites and targeted for early termination and exosome-mediated degradation in the nucleus (Preker et al. 2008; Almada et al. 2013; Ntini et al. 2013). In contrast, stable mature mRNAs are transported to the cytoplasm for protein synthesis. H3, histone H3; lncRNA, long noncoding RNA; poly-(A) site (PAS), ; RNAPII, RNA Polymerase II; switch/sucrose non-fermentable(SWI/SNF).
Functional lncRNAs Display mRNA-Like Transcriptional Properties
Predictably, there are exceptions to the dominant patterns highlighted above. Nascent transcription analyses have also uncovered lncRNAs that exhibit more mRNA-like transcriptional properties (See Figure 2B). For example, two such transcripts detected in human HeLa cells include the highly abundant transcripts NORAD and TINCR (Schlackow et al. 2017). NORAD localizes to the cytoplasm where it controls gene expression by interacting with and regulating Pumilio RNA-binding proteins (Lee et al. 2016), while TINCR maintains somatic cell differentiation by interacting with and stabilizing differentiation-specific mRNAs (Kretz et al. 2013). Since lncRNAs generally display stronger tissue-specific expression patterns than protein-coding genes (Derrien et al. 2012; Kornienko et al. 2016), nascent transcript analyses must be expanded to other human cell types to identify more mRNA-like lncRNAs since this subclass is highly likely to have specific and discernible functions.
Gene Regulation by the Act of Transcription
The differences in coding and noncoding transcription that have been highlighted above suggest that lncRNA transcription is almost exclusively cryptic. Even some of the more abundant human lncRNAs are predominantly limited to chromatin where they are rapidly degraded (Schlackow et al. 2017). Overall, findings from diverse eukaryotic systems suggest a role for noncoding DNA sequences beyond the relatively short-lived cryptic lncRNAs produced. The idea that the act of noncoding transcription can have functional consequences is supported by a growing number of intergenic lncRNAs whose transcription regulates nearby genes.
The best-characterized example of adjacent gene regulation by noncoding RNAPII transcription involves the S. cerevisiae SER3 gene (encoding a 3-phosphoglycerate dehydrogenase). SER3 is repressed by transcription of the SRG1 lncRNA initiating ∼500 bp upstream of the SER3 promoter when cells are grown in the presence of serine (Martens et al. 2004). Consequently, reduced SRG1 transcription following serine starvation permits SER3 induction (Martens et al. 2005). The SER3 mRNA also accumulates in cells grown in serine-replete conditions following the disruption of numerous transcription elongation factors known to be involved in transcription-coupled chromatin changes (e.g., Spt6, FACT, and Paf1), even though SRG1 is still expressed in these mutants (Hainer et al. 2011; Pruneski et al. 2011). Notably, transcription-mediated SER3 regulation occurs in both a Set1- and Set2-independent manner (Hainer et al. 2011), indicating that H3K4 and H3K36 methylation do not play a significant role in this mechanism. In contrast, key histone H3 and H4 residues are required for upstream lncRNA transcription to allow increased nucleosome density over the SER3 promoter (Hainer and Martens 2011), but the precise role of these histone residues is still unclear. Collectively, these analyses revealed that SER3 is repressed by the act of upstream transcription, which mediates specific transcription-coupled chromatin changes over the SER3 promoter in a process referred to as “transcriptional interference.”
Other acts of transcription-mediated gene regulation have also been reported in S. cerevisiae and in other systems. In S. cerevisiae, upstream lncRNA transcription represses the meiosis-inducing IME1 gene and thereby governs sexual differentiation (van Werven et al. 2012), while cell–cell adhesion is in part controlled by lncRNA transcription upstream of the FLO11 gene (Bumgarner et al. 2009). In the fission yeast Schizosaccharomyces pombe, nutrient homeostasis is controlled by upstream lncRNA transcription-mediated regulation of numerous genes, including fbp1+, pho1+, and tgp1+ (Hirota et al. 2008; Ard et al. 2014; Ard and Allshire 2016). Related examples of gene regulation by adjacent acts of transcription have also been described in fruit flies, mouse, and human cells (Corbin and Maniatis 1989; Bird et al. 2006; Petruk et al. 2006; Abarrategui and Krangel 2007; Martianov et al. 2007). In addition, aberrant transcription-mediated gene repression has been observed in human diseases (De Gobbi et al. 2006; Grosso et al. 2015; Rutkowski et al. 2015). It has also been suggested that some cancers may be initiated by reactivated retrotransposons, the transcription of which suppresses the expression of adjacent genes (Wilkins 2010). Similarly, RNAPII transcription initiating from within an intergenic T-DNA insertion was observed to overlap and repress a downstream gene in Arabidopsis thaliana (Hedtke and Grimm 2009), demonstrating that this mechanism is also likely to be functionally conserved in plants. Together, these examples emphasize that gene regulation by overlapping transcription is widely conserved, not strictly limited to canonical lncRNA transcription, and can have important consequences for human health and disease.
Diverse Mechanisms of Gene Regulation by the Act of Transcription
Noncoding transcription near or overlapping protein-coding genes can have diverse, locus-specific outcomes (Mellor et al. 2016). In most cases, tandem transcription into a downstream promoter results in gene repression by transcriptional interference (Proudfoot 1986). High-resolution analysis of chromatin-isolated RNA shows that transcription factor binding induces strong RNAPII pausing in human cells (Mayer et al. 2015), arguing against a model whereby transcription-mediated gene repression simply involves displacing downstream transcription factors. As seen for SER3 gene regulation by the transcription of the SRG1 lncRNA, transcription-coupled chromatin changes mediated by RNAPII-recruited factors at least partially contribute to the repression of gene promoters by interfering lncRNAs in diverse systems (Petruk et al. 2006; van Werven et al. 2012; Ard and Allshire 2016; Kim et al. 2016). Thus, such chromatin remodelling activities are recruited by RNAPII during these acts of transcription in sufficient amounts to downregulate underlying promoter activity. Additionally, these findings suggest that factors involved in suppressing cryptic intragenic promoters during mRNA synthesis also repress gene promoters by transcriptional interference. However, the specific factors involved differ in many of the cases studied (Figure 4). Some of these differences are likely attributed to the length and/or other properties specific to individual acts of lncRNA transcription. For example, shortening the distance between an interfering transcription unit and its target promoter reduces the requirement of Set2-mediated repression by H3K36me3 (Kim et al. 2016), consistent with the observation that Set2 is recruited during transcription elongation and predominantly suppresses cryptic promoters toward the 3′-end of longer yeast genes (Li et al. 2007). Moreover, Set2 silences gene promoters occluded by longer, but not shorter, interfering lncRNAs (Hainer et al. 2011; van Werven et al. 2012; Ard and Allshire 2016). Conversely, upstream transcription can also positively influence downstream gene expression in certain instances. Notably, cascading transcription of multiple lncRNAs into the S. pombe fbp1+ gene promoter is required to displace promoter-bound transcriptional repressors and activate gene expression (Takemata et al. 2016). Likewise in Drosophila, intergenic transcription has been reported to stimulate nearby gene activation by displacing repressive chromatin modifying complexes (Schmitt et al. 2005). An intriguing possibility is that reduced recruitment of particular factors to RNAPII at specific noncoding transcription units [as proposed by Fischl et al. (2017) and Battaglia et al. (2017)] might prevent otherwise repressive effects on an underlying promoter. Clearly, a comprehensive mechanistic understanding is needed to determine how some acts of lncRNA transcription result in gene activation whereas others repress expression.
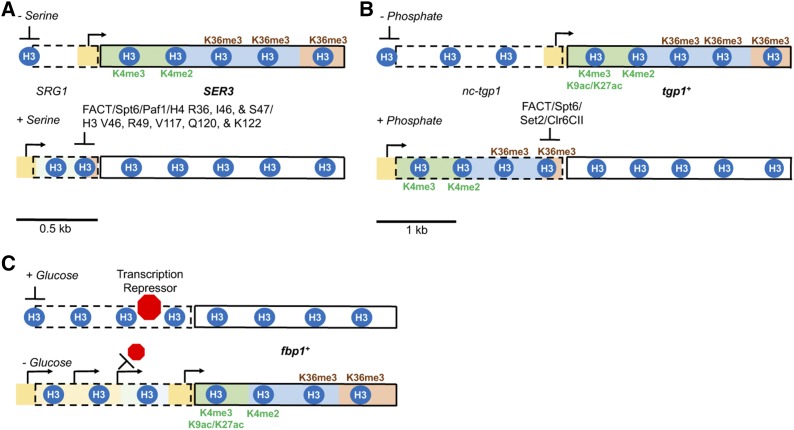
Figure 4.
Distinct mechanisms of lncRNA transcription-mediated gene regulation. (A) Serine-starvation induces SER3 expression in S. cerevisiae. In presence of serine, SER3 is repressed by transcription of the SRG1 lncRNA, which is initiated from a promoter positioned roughly 0.5 kb upstream of the SER3 transcription start site (Martens et al. 2004, 2005). Repression in this instance requires cotranscriptional changes in nucleosome positions over the SER3 promoter mediated by histone chaperones FACT and Spt6 (Hainer et al. 2011). Efficient SRG1-mediated SER3 repression also requires the PAF1 complex (Pruneski et al. 2011). SRG1 is too short for transcription elongation to deposit significant levels of H3K4me2 or H3K36me3, which explains why Set1 and Set2 are not required for regulating SER3 expression (Hainer et al. 2011). In contrast, other less characterized histone tail residues are implicated in SER3 repression by SRG1 (e.g., H3V46, H3R49, H3V117, H3Q120, H3K122, H4R36, H4I46, and H4S47) (Hainer and Martens 2011) (B) In S. pombe, the 2 kb cryptic lncRNA nc-tgp1 represses the tgp1+ gene in response to phosphate availability (Ard et al. 2014). FACT and Spt6, along with Set2 and Clr6Rpd3S, all partially contribute to the suppression of tgp1+ by nc-tgp1 transcription (Ard and Allshire 2016). The role of the PAF1 complex in tgp1+ regulation has yet to be explored. Related mechanisms have been reported for lncRNA-mediated repression of the pho1+ gene in S. pombe (Ard and Allshire 2016) and the IME1 gene in S. cerevisiae (van Werven et al. 2012). (C) The S. pombe fbp1+ gene is repressed when cells are grown in the presence of glucose. Unlike the examples described in (A and B), fbp1+ expression following glucose starvation is stimulated by the transcription of multiple upstream lncRNAs (Hirota et al. 2008). While the act of transcription is thought to help displace transcriptional repressors Tup11 and Tup12 (Takemata et al. 2016), it is unclear why cotranscriptional chromatin changes do not repress the fbp1+ promoter. Whether differences in the recruitment of elongation factors to RNAPII during some acts of noncoding transcription (Battaglia et al. 2017; Fischl et al. 2017) help to explain mechanistic differences is worth exploring in future studies. FACT, facilitates chromatin transcription; H3, histone H3; lncRNA, long noncoding RNA; RNAPII, RNA Polymerase II.
Elucidating the Mechanisms and Scale of Functional Noncoding Transcription
A combination of classical genetic approaches and cutting-edge genomics will be required to better understand the mechanisms and scale of functional noncoding transcription. RNAPII transcription originating within intentional mutagens in S. cerevisiae, such as Ty elements, led to the original characterization of transcription-mediated gene repression and the isolation of suppressor mutants (Winston et al. 1984). Indeed, this is how the histone chaperone Spt6 (suppressor of Ty 6) was initially identified. Given the accumulating number of examples of transcription-mediated gene regulation in higher eukaryotes, forward genetic screens will be particularly useful to better understand the diversity of mechanisms operating in multicellular organisms. Such approaches are proving much less difficult since the advent of efficient clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9-mediated genome-wide screens in human cells (Shalem et al. 2014; Wang et al. 2014). Further advantages can be gained by using a fully haploid cell line (Blomen et al. 2015). Reverse genetic screens informed by studies in yeast will be equally informative to test the conservation of mechanisms described above. In addition, overlaying nascent transcriptome data with the genomic positions of RNAPII-associated factors and histone modifications will provide important clues regarding the chromatin and transcription features that are unique to gene regulation by acts of transcription and should aid in the identification of novel examples of transcription-mediated gene regulation. In particular, TIF-seq (transcript isoform sequencing) sequences transcription start/end sites simultaneously and can therefore distinguish altered transcript isoforms from upstream lncRNAs or readthrough transcripts that overlap downstream promoters (Pelechano et al. 2013). Overlapping transcripts in particular are underrepresented in conventional total RNA-seq and nascent transcriptome data sets as these methods do not accurately predict transcription start/stop sites. TIF-seq is therefore a useful and complementary technique to better characterize the prevalence of potential interfering lncRNAs and identify genes with lncRNA isoforms whose transcription and/or products might be functionally relevant for their regulation. Comparing data generated by these various approaches will be instrumental for understanding the diversity of molecular mechanisms involved in gene regulation by transcription itself as well as the extent to which pervasive transcription contributes to fine-tuning gene expression across genomes.
Functional Consequences of Transcription Beyond Local Gene Regulation
Apart from illuminating how nearby genes are regulated, new genomic approaches will also identify noncoding transcription events further away from protein-coding genes that may still be functionally relevant, such as those involved in larger changes to chromatin/chromosome structure and organization. For example, the act of persistent RNAPII transcription initiation and pausing within S. pombe centromeres is implicated in chromatin changes that facilitate the incorporation of the centromere-specifying H3-variant centromere protein-A (Catania et al. 2015) (See Figure 2E). Such a mechanism might be widely conserved given that centromeric transcription has been detected in organisms ranging from S. cerevisiae to plants and animals (Scott 2013). Additionally, the act of transcription at enhancers has been proposed to help remodel chromatin structure in a manner that mediates contact with their target promoters (Li et al. 2016). In fact, the process of both coding and noncoding transcription can lead to changes in three-dimensional chromatin structure, including higher-order chromatin interactions and the subnuclear localization of specific chromatin regions (Melé and Rinn 2016). Pervasive transcription might therefore play a central role not only in regulating specific genes and epigenetic processes but also in the spatial and structural organization of chromosomes within the nucleus.
Final Thoughts
Recent advances in nascent transcriptome profiling have uncovered many distinct patterns of RNAPII behavior along different coding and noncoding regions of the genome. While these differences are themselves not sufficient to resolve whether any given noncoding transcription unit is functionally relevant or merely inconsequential “transcriptional noise,” important clues are emerging to aid categorization.
Notably, eukaryotic genomes show evidence of widespread, low-quality noncoding transcription by RNAPII. Consequently, stable RNA synthesis is predominantly restricted to protein-coding genes and a subset of lncRNAs exhibiting similar “mRNA-like” properties and/or RNAPII related behavior. Complementary approaches have simultaneously uncovered general differences between coding and noncoding transcription cycles and have also allowed unique RNAPII behaviors of distinct types of noncoding transcription units to be distinguished. It is therefore reasonable to propose that subclasses of lncRNAs can be distinguished based on the properties of their transcription since observed differences in RNAPII behavior and chromatin status are likely to account for contrasting RNA stabilities and functional outcomes. For example, helping to distinguish functional lncRNA products from functional acts of noncoding transcription, albeit that such distinctions might not be absolute.
It is currently unclear what specifies observed differences in RNAPII behavior at any given transcription unit. Transcript length, RNA secondary structure, and specific DNA or RNA sequence motifs can all influence RNAPII transcription and are therefore likely to, at least, partially contribute to the observed differences in RNAPII behavior (Barrass et al. 2015; Eser et al. 2016; Schwalb et al. 2016). Whether these differences are the cause or consequence of reduced recruitment of specific factors to RNAPII during some acts of noncoding transcription remains unclear. Importantly, differential recruitment of RNAPII elongation factors during coding vs. lncRNA transcription have been observed through metagene analyses (Battaglia et al. 2017; Fischl et al. 2017), yet gene repression by transcriptional interference requires many core elongation factors (Hainer et al. 2011; van Werven et al. 2012; Ard and Allshire 2016). It is tempting to speculate that selected lncRNA transcription units with uncharacteristically high RNAPII elongation factor load may pinpoint transcription events affecting nearby expression through lncRNA transcription-mediated gene repression. Likewise, lncRNA transcription units depleted of common RNAPII-associated factors might display other transcription features and/or chromatin signatures that mediate distinct outcomes.
Further investigation is required to determine to what extent histone modifications unique to noncoding regions of the genome direct the observed differences in transcription or are predominantly imposed by RNAPII properties unique to different types of noncoding transcription. Ultimately, post-translational histone modifications could allow for the persistence of chromatin states over transcribed noncoding DNA through epigenetic processes. Such a mechanism would retain not only an epigenetic memory of recent transcriptional activity, but also information about recent transcriptional behavior and therefore have important functional implications for rapid switches in regulated noncoding transcription.
In sum, elongation factors and chromatin remodellers are implicated in favoring productive RNAPII transcription as much as they act to restrict aberrant noncoding transcription that could interfere with gene expression. Although many of these same factors also contribute to the regulation of gene expression through acts of lncRNA transcription, further research is required to determine what underlies the diversity of mechanisms by which noncoding transcription is functionally relevant. Compared to lncRNAs, which have emerged in recent years as important players in cell biology, relatively limited focus has been placed on the functional consequences of transcription itself. Since accumulating evidence supports the notion that most lncRNA transcription does not generate stable products, cryptic transcription likely plays even more diverse and important roles in cells than is currently appreciated. For this reason, the functional consequences of transcription deserve much greater attention. Noncoding transcription units are often sensitive to the cellular environment and tightly regulated changes in their transcriptional activity can profoundly affect local gene expression, the propagation of epigenetically-specified chromatin domains, and likely also contribute to larger changes in chromosome architecture. Future studies focusing on functional roles of transcribed noncoding DNA in genomes informed by the differences in noncoding transcription cycles will determine to what extent the process of pervasive transcription underpins these important cellular processes.
Acknowledgments
This work is supported by a European Molecular Biology Organization Long-Term Postdoctoral Fellowship (ALTF: 463-2016) to R.A., a Copenhagen Plant Science Centre Young Investigator Starting Grant to S.M., and a Novo Nordisk Foundation Hallas-Møller Investigator Award to S.M. R.C.A. is a Wellcome Principal Research Fellow; his research is supported by the Wellcome Trust (200885) and core funding of the Wellcome Centre for Cell Biology (203149).
Footnotes
Communicating editor: A. Wilkins
Literature Cited
- Abarrategui I., Krangel M. S., 2007. Noncoding transcription controls downstream promoters to regulate T-cell receptor alpha recombination. EMBO J. 26: 4380–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almada A. E., Wu X., Kriz A. J., Burge C. B., Sharp P. A., 2013. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R., Andersen P. R., Valen E., Core L. J., Bornholdt J., et al. , 2014. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat. Commun. 5: 5336. [DOI] [PubMed] [Google Scholar]
- Ard R., Allshire R. C., 2016. Transcription-couple changes to chromatin underpin gene silencing by transcriptional interference. Nucleic Acids Res. 44: 10619–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ard R., Tong P., Allshire R. C., 2014. Long non-coding RNA-mediated transcriptional interference of a permease gene confers drug tolerance in fission yeast. Nat. Commun. 5: 5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrass J. D., Reid J. E., Huang Y., Hector R. D., Sanguinetti G., et al. , 2015. Transcriptome-wide RNA processing kinetics revealed using extremely short 4tU labeling. Genome Biol. 16: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Akhtar A., Barlow D. P., Bird A. P., Brockdroff N., et al. , 2014. Considerations when investigating lncRNA function in vivo. Elife 3: e03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia S., Lidschreiber M., Baejen C., Torkler P., Vos S. V., et al. , 2017. RNA-dependent chromatin association of transcription elongation factors and Pol II CTD kinases. Elife 6: 25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., 2014. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann J. H., Rodriguez M. G., Martins N. M., Kimura H., Kelly D. A., et al. , 2011. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 30: 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. J., Gordon M., Eide D. J., Winge D. R., 2006. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 25: 5726–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen V. A., Majek P., Jae L. T., Bigenzahn J. W., Nieuwenhuis J., et al. , 2015. Gene essentiality and synthetic lethality in haploid human cells. Science 350: 1092–1096. [DOI] [PubMed] [Google Scholar]
- Brodsky A. S., Meyer C. A., Swinburne I. A., Hall G., Keenan B. J., et al. , 2005. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 6: R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner S., Dowell R. D., Grisafi P., Gifford D. K., Fink G. R., 2009. Toggle involved cis-interfering noncoding RNAs controls gene expression in yeast. Proc. Natl. Acad. Sci. USA 106: 18321–18326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., et al. , 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Catania S., Pidoux A. L., Allshire R. C., 2015. Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet. 11: e1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Steitz J. A., 2014. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157: 77–94. [DOI] [PubMed] [Google Scholar]
- Cheung V., Chua G., Batada N. N., Landry C. R., Michnick S. W., et al. , 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman L. S., Weissman J. S., 2011. Nascent transcript sequencing visualizes transcription at single nucleotide resolution. Nature 469: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin V., Maniatis T., 1989. Role of transcriptional interference in the Drosophila melanogaster Adh promoter switch. Nature 337: 279–282. [DOI] [PubMed] [Google Scholar]
- Corden J. L., 2013. RNA polymerase II C-terminal domain: tethering transcription to transcript and template. Chem. Rev. 113: 8423–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gobbi M., Viprakasit V., Hughes J. R., Fisher C., Buckle V. J., et al. , 2006. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science 312: 1215–1217. [DOI] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., et al. , 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descostes N., Heidemann M., Spinelli L., Schüller R., Maqbool M. A., et al. , 2014. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalians cells. Elife 3: e02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveson I. W., Hardwick S. A., Mercer T. R., Mattick J. S., 2017. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 33: 464–478. [DOI] [PubMed] [Google Scholar]
- Drouin S., Laramée L., Jacques P. E., Forest A., Bergeron M., et al. , 2010. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 6: e1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J. M., Ollikainen N., Guttman M., 2016. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17: 756–770. [DOI] [PubMed] [Google Scholar]
- Eser P., Wachutka L., Maier K. C., Demel C., Boroni M., et al. , 2016. Determinants of RNA metabolism in the Schizosaccharomyces pombe genome. Mol. Syst. Biol. 12: 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl H., Howe F. S., Furger A., Mellor J., 2017. Paf1 has distinct roles in transcription elongation and differential transcript fate. Mol. Cell 65: 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N., Saldi T., Sheridan R. M., Cortazar M. A., Bentley D. L., 2017. RNA Pol II dynamics modulate co-transcriptional chromatin modification, CTD phosphorylation, and transcriptional direction. Mol. Cell 66: 546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Coller J., 2013. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff L. A., Rinn J. L., 2015. Linking RNA biology to lncRNAs. Genome Res. 25: 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C. K., Qiu H., Ginsburg D. S., Ruan C., Hofmeyer K., et al. , 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell 39: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso A. R., Leite A. P., Carvalho S., Matos M. R., Martins F. B., et al. , 2015. Pervasive transcription read-through promoters aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife 4: e09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M., Proudfoot N. J., 2010. Transcription interference and gene orientation in yeast: noncoding RNA connections. Cold Spring Harb. Symp. Quant. Biol. 75: 299–311. [DOI] [PubMed] [Google Scholar]
- Haerty W., Ponting C. P., 2015. Unexpected selection to retain high GC content and splicing enhancers within exons of multiexonic lncRNA loci. RNA 21: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Martens J. A., 2011. Identification of histone mutants that are defective for transcription-coupled nucleosome occupancy. Mol. Cell. Biol. 31: 3557–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Pruneski J. A., Mitchell R. D., Monteverde R. M., Martens J. A., 2011. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25: 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlen K. M., Churchman L. S., 2017. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18: 236–273. [DOI] [PubMed] [Google Scholar]
- Hedtke B., Grimm B., 2009. Silencing of a plant gene by transcriptional interference. Nucleic Acids Res. 37: 3739–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Miyoshi T., Kugou K., Hoffman C. S., Shibata T., et al. , 2008. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456: 130–134. [DOI] [PubMed] [Google Scholar]
- Houseley J., Tollervey D., 2009. The many pathways of RNA degradation. Cell 136: 763–776. [DOI] [PubMed] [Google Scholar]
- Hsin J. P., Li W., Hoque M., Tian B., Manley J. L., 2014. RNAPII CTD tyrosine 1 performs diverse functions in vertebrate cells. Elife 3: e02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T. H., Jacquier A., Libri D., 2013. Dealing with pervasive transcription. Mol. Cell 52: 473–484. [DOI] [PubMed] [Google Scholar]
- Jonkers I., Lis J. T., 2015. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C. D., Laprade L., Winston F., 2003. Transcription elongation factors repression transcription initiation from cryptic sites. Science 301: 1096–1099. [DOI] [PubMed] [Google Scholar]
- Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., et al. , 2005. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Lee B. B., Oh Y. M., Zhu C., Steinmetz L. M., et al. , 2016. Modulation of mRNA and lncRNA expression dynamics by the Set2-Rpd3S pathway. Nat. Commun. 7: 13534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Buratowski S., 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko A. E., Guenzl P. M., Barlow D. P., Pauler F. M., 2013. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko A. E., Dotter C. P., Guenzl P. M., Gisslinger H., Gisslinger B., et al. , 2016. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. 17: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M., Siprashvili Z., Chu C., Webster D. E., Zehnder A., et al. , 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493: 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J. T., Colognori D., Lee J. T., 2013. Long noncoding RNAs: past, present, and future. Genetics 193: 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H., Lis J. T., 2013. Control of transcriptional elongation. Annu. Rev. Genet. 47: 483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos P. A., Pauler F. M., Koerner M. V., Senergin H. B., Hudson Q. J., et al. , 2012. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338: 1469–1472. [DOI] [PubMed] [Google Scholar]
- Lee S., Kopp F., Chang T. C., Sataluri A., Chen B., et al. , 2016. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Howe L., Anderson S., Yates J. R., Workman J. L., 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278: 8897–8903. [DOI] [PubMed] [Google Scholar]
- Li B., Gogol M., Cary M., Pattenden S. G., Seidel C., et al. , 2007. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 21: 1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Notani D., Rosenfeld M. G., 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 17: 207–223. [DOI] [PubMed] [Google Scholar]
- Mahat D. B., Kwak H., Booth G. T., Jonkers I. H., Danko C. G., et al. , 2016. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat. Protoc. 11: 1455–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S., Hazelbaker D. Z., Buratowski S., 2011. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription 2: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S., Escalante-Chong R., Pho N., Wang J., Churchman L. S., et al. , 2014. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell 157: 1712–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. A., Laprade L., Winston F., 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574. [DOI] [PubMed] [Google Scholar]
- Martens J. A., Wu P. Y., Winston F., 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19: 2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A., 2007. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666–670. [DOI] [PubMed] [Google Scholar]
- Mayer A., di Iulio J., Maleri S., Eser U., Vierstra J., et al. , 2015. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161: 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M., Rinn J. L., 2016. “Cat’s Cradling” the 3D genome by the act of lncRNA transcription. Mol. Cell 62: 657–664. [DOI] [PubMed] [Google Scholar]
- Melé M., Mattioli K., Mallard W., Shechner D. M., Gerhardinger C., et al. , 2017. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 27: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Woloszczuk R., Howe F. S., 2016. The interleaved genome. Trends Genet. 32: 57–71. [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Mattick J. S., 2009. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10: 155–159. [DOI] [PubMed] [Google Scholar]
- Milligan L., Huynh-Thu V. A., Delan-Forino C., Tuck A., Petfalski E., et al. , 2016. Strand-specific, high-resolution mapping of modified RNA polymerase II. Mol. Syst. Biol. 12: 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N., Calviello L., Hirsekorn A., de Pretis S., Pelizzola M., et al. , 2017. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 24: 86–96. [DOI] [PubMed] [Google Scholar]
- Murray S. C., Haenni S., Howe F. S., Fischl H., Chocian K., et al. , 2015. Sense and antisense transcription are associated with distinct chromatin architectures across genes. Nucleic Acids Res. 43: 7823–7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Robert F., Young R. A., Struhl K., 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709–719. [DOI] [PubMed] [Google Scholar]
- Nguyen T., Fischl H., Howe F. S., Woloszczuk R., Serra Barros A., et al. , 2014. Transcription mediated insulation and interference direct gene cluster expression switches. Elife 3: e03635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche A., Rose D., Fasold M., Reiche K., Stadler P. F., 2015. Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved. RNA 21: 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T., Gomes T., Grosso A. R., Kimura H., Dye M. J., et al. , 2015. Mammalian NET-Seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161: 526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T., Gomes T., Carmo-Fonseca M., Proudfoot N. J., 2016. Mammalian NET-seq analysis defines nascent RNA profiles and associated RNA processing genome-wide. Nat. Protoc. 11: 413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntini E., Jarvelin A. I., Bornholdt J., Chen Y., Boyd M., et al. , 2013. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 20: 923–928. [DOI] [PubMed] [Google Scholar]
- Pang K. C., Frith M. C., Mattick J. S., 2006. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 22: 1–5. [DOI] [PubMed] [Google Scholar]
- Pelechano V., Wei W., Steinmetz L. M., 2013. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497: 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S., Sedkov Y., Riley K. M., Hodgson J., Schweisguth F., et al. , 2006. Transcription of bxd noncoding RNAs promoter by trithorax represses Ubx in cis by transcriptional interference. Cell 127: 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L., Reik W., 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641. [DOI] [PubMed] [Google Scholar]
- Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M. S., et al. , 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., 1986. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature 322: 562–565. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., 2016. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneski J. A., Hainer S. J., Petrov K. O., Martens J. A., 2011. The Paf1 complex represses SER3 transcription in Saccharomyces cerevisiae by facilitating intergenic transcription-dependent nucleosome occupancy of the SER3 promoter. Eukaryot. Cell 10: 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. J., Chang H. Y., 2016. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genetics 17: 47–62. [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Chang H. Y., 2012. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski A. J., Erhard F., L’Hernault A., Bonfert T., Schilhabel M., et al. , 2015. Widespread disruption of host transcription termination in HSV-1 infection. Nat. Commun. 6: 7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlackow M., Nojima T., Gomes T., Dhir A., Carmo-Fonseca M., et al. , 2017. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell 65: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S., Prestel M., Paro R., 2005. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 19: 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B., Michel M., Zacher B., Frühauf K., Demel C., et al. , 2016. TT-seq maps the human transient transcriptome. Science 352: 1225–1228. [DOI] [PubMed] [Google Scholar]
- Scott K. C., 2013. Transcription and ncRNAs: at the cent(rome)re of kinetochore assembly and maintenance. Chromosome Res. 21: 643–651. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., et al. , 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A., Kallgren S. P., Demel C., Maier K. C., Spatt D., et al. , 2017. Spt5 plays vital roles in the control of sense and antisense transcription elongation. Mol. Cell 66: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova A. A., Mullen A. C., Molinie B., Gupta S., Orlando D. A., et al. , 2013. Divergent transcription of long noncoding RNA /mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. USA 110: 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B. A., Karpen G. H., 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11: 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Gadad S. S., Kim D. S., Kraus W. L., 2015. Discovery, annotation, and functional analysis of long non-coding RNAs controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol. Cell 59: 698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemata N., Oda A., Yamada T., Galipon J., Miyoshi T., et al. , 2016. Local potentiation of stress-responsive genes by upstream noncoding transcription. Nucleic Acids Res. 44: 5174–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong S. M., Zaugg J. B., Camblong J., Xu Z., Zhang D. W., et al. , 2012. Gene loops enhance transcriptional directionality. Science 338: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I., 2016. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 17: 601–614. [DOI] [PubMed] [Google Scholar]
- van Werven F. J., Neuert G., Hendrick N., Lardenois A., Buratowski S., et al. , 2012. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150: 1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M., Lander E. S., 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Pelechano V., Jarvelin A. I., Steinmetz L. M., 2011. Functional consequences of bidirectional promoters. Trends Genet. 27: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T., Lee W., Obenauf A. C., Ran L., Murali R., et al. , 2015. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature 526: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins A. S., 2010. The enemy within: an epigenetic role of retrotransposons in cancer initiation. BioEssays 32: 856–865. [DOI] [PubMed] [Google Scholar]
- Winston F., Chaleff D. T., Valent B., Fink G. R., 1984. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107: 179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]