Abstract
A fundamental tenet of inheritance in sexually reproducing organisms such as humans and laboratory mice is that gametes combine randomly at fertilization, thereby ensuring a balanced and statistically predictable representation of inherited variants in each generation. This principle is encapsulated in Mendel’s First Law. But exceptions are known. With transmission ratio distortion, particular alleles are preferentially transmitted to offspring. Preferential transmission usually occurs in one sex but not both, and is not known to require interactions between gametes at fertilization. A reanalysis of our published work in mice and of data in other published reports revealed instances where any of 12 mutant genes biases fertilization, with either too many or too few heterozygotes and homozygotes, depending on the mutant gene and on dietary conditions. Although such deviations are usually attributed to embryonic lethality of the underrepresented genotypes, the evidence is more consistent with genetically-determined preferences for specific combinations of egg and sperm at fertilization that result in genotype bias without embryo loss. This unexpected discovery of genetically-biased fertilization could yield insights about the molecular and cellular interactions between sperm and egg at fertilization, with implications for our understanding of inheritance, reproduction, population genetics, and medical genetics.
Keywords: gametes, fertilization, transmission ratio distortion, meiosis, segregation
The Problem
Our understanding of inheritance in sexually reproducing organisms assumes, with good evidence, that the combination of egg and sperm that join at fertilization is largely independent of their genetic content. Equal transmission of alternative alleles through meiosis in heterozygotes ensures a balanced parental genetic contribution to offspring at each generation. Independent segregation and random union of gametes at fertilization are foundations of classical, quantitative, population, evolutionary, and medical genetics (East 1916; Holliday 1984; Crow 1991). Mendel’s First Law captures this principle, which is one of the few that applies generally in biology.
The most prominent exceptions to random segregation are the rare, naturally occurring examples of transmission ratio distortion (TRD) that have been described in fungi (Turner and Perkins 1979), corn (Rhoades and Vilkomerson 1942), flies (Morgan et al. 1925; Gershenson 1928; Sandler and Novitski 1957; Pimpinelli and Dimitri 1989; Larracuente and Presgraves 2012), mice (Dunn 1957; Montagutelli et al. 1996; Lyon 2003; Wu et al. 2005; Didion et al. 2015, 2016), humans (Chen et al. 2012; Huang et al. 2013), and other species (Scofield et al. 1982; Fishman and Willis 2005; Koide et al. 2008; Hoang et al. 2016). Biased sex ratios have also been reported (Hamilton 1967; Jaenike 2001; Tao et al. 2007a,b; Helleu et al. 2014, 2016; Rice 2014). These exceptions arise despite strong selective pressures to maintain normal segregation (Crow 1991) and sex ratios (Fisher 1930; Crow and Kimura 1970). Based on alternative functions in haploid gametes, one allele is preferentially transmitted to offspring at the expense of other alleles. In these cases, TRD drives allelic preference in one sex, regardless of the genetics of the mating partner. Reproductive performance is often not reduced because the normal number of gametes and zygotes is produced. TRD may arise during chromosome segregation in meiosis (meiotic drive), gametogenesis (gamete competition), or embryonic development (preferential lethality). These examples of TRD probe the nature of Mendel’s First Law by illuminating genetic, molecular, and cellular mechanisms that underlie meiosis, recombination, gametogenesis, and early development (East 1916; Crow 1979, 1988, 1991; Holliday 1984; Lyttle 1993; Herrmann et al. 1999; Pardo-Manuel de Villena and Sapienza 2001; Lyon 2003). Many of these “selfish genetic” systems are composed of several closely linked elements that not only lead to preferential transmission of the chromosome on which they are carried, but also confer sterility or lethality on homozygous carriers, thereby preventing fixation at the cost of reduced population fitness (Dunn 1957). Without these counterbalances, TRD systems would quickly replace their wild-type (WT) alleles in the population, their competitive advantage would be lost, and selective sweeps affecting variation at neighboring loci would be their only legacy.
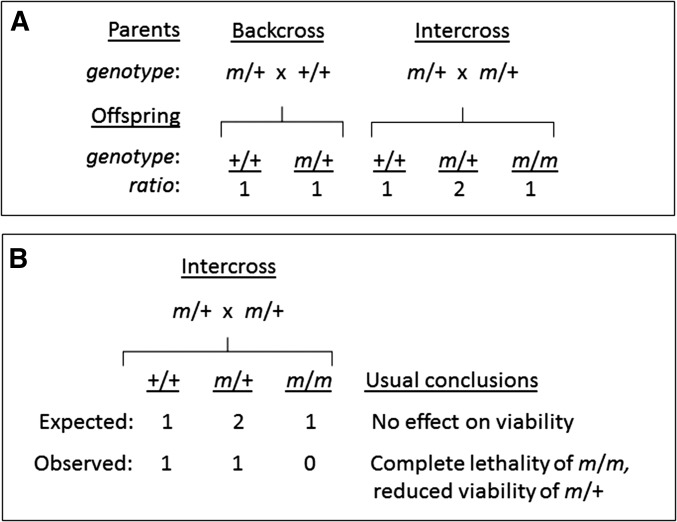
With spontaneous, induced, and engineered single-gene mutations, one of the first tasks is to assess consequences on viability, fertility, and other phenotypes (White et al. 2013; Hrabe de Angelis et al. 2015; Dickinson et al. 2016; Meehan et al. 2017). Absence of mutant homozygotes is usually accepted as evidence for induced lethality and reduced numbers of heterozygotes is taken as evidence for a detrimental dosage effect (Figure 1). But sometimes the fit to Mendelian segregation is not explicitly examined. Litter size, which should be reduced proportionately to the number of missing genotypes, is often not reported. Backcrosses, which can provide information about parent-of-origin effects on gametogenesis and embryogenesis, are sometimes not included in study designs. As a result, whether particular cases of non-Mendelian segregation result from lethality or from other phenomena such as TRD remains ambiguous, despite claims in publications.
Figure 1.
Regular and irregular outcomes of Mendelian segregation. (A) Conventional segregation in backcrosses and intercrosses for a single gene with two alleles, mutant (m) and wild-type (+). (B) Irregular segregation in an intercross with deficiency of all m/m homozygotes and half of +/m heterozygotes as an exemplar.
Consider an early controversy in mammalian genetics. Cuenot (1905), studying the absence of pure yellow segregants in crosses between mice heterozygous for the dominantly-acting agouti yellow (Ay) mutation, argued that gametes carrying the agouti yellow allele never join at fertilization. By contrast, Castle and Little, based on considerations of both segregation ratios and litter size, correctly concluded that homozygous yellow mice fail to complete development, with reduced litter size providing the critical evidence for lethality (Castle and Little 1910; Heaney et al. 2009). As Castle and Little showed, a full analysis is needed to establish with confidence the basis for unusual segregation.
Four steps are needed to transfer genetic and epigenetic information from one generation to the next through the germline. Meiosis converts the chromosome complement from diploid to haploid in the parental generation. Gametogenesis provides a cellular vehicle for the haploid genome. From a pair of haploid gametes, fertilization restores diploidy in the zygote. Finally, the germline is set aside early in embryonic development to renew these steps in the offspring generation. TRD has been reported for three of these steps: meiotic drive, gamete competition, and preferential embryo survival. In each case, dysfunction in one sex is sufficient for TRD that is largely independent of the genetics of mating partners.
The evidence reviewed here provides examples for TRD in the third step, fertilization, where genetic variants, acting in both sexes and in some cases depending on environmental (dietary) conditions, control the combination of gametes that join at fertilization to create zygotes. Our work on epigenetic inheritance in mice and a review of the mouse literature revealed strong evidence that mutations in any of 12 genes (Box 1), where either too many or too few heterozygotes were found among intercross progeny, together with absent or deficient homozygotes, were found without evidence for dead embryos or reduced litter size (Table 1 and Table 2; see also Supplemental Material, Table S1 and Table S2). Of these 12 genes, six involve single spontaneous or engineered mutations on an inbred genetic background. In another case, biased segregation was found only in crosses involving a pair of mutant genes (epistasis). Finally, five cases involved dietary folic acid supplementation of mice carrying a single-gene mutation affecting neural tube development, where segregation was biased on one diet but normal on the alternative diet, with similar litter sizes and rates of prenatal lethality. Normal segregation in backcrosses between heterozygotes and WT homozygotes argues that meiosis and gametogenesis function normally in each sex. These unusual results suggest that fertilization is genetically biased toward particular gametes based on their genetic content.
BOX 1. Genes that bias fertilization. Additional information can be found in the Mouse Genome Database (informatics.jax.org).
A1cf
Apobec1 complementation factor, chromosome (Chr) 19, 26.6 cM. A1cf is expressed primarily in the nucleus where it encodes the RNA-binding subunit for Apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (APOBEC1), a cytidine deaminase that edits specific bases, sometimes in coding sequences but more usually in 3′-UTRs (Blanc and Davidson 2010; Blanc et al. 2014). It must have additional functions since deficiency leads to early embryonic lethality (Blanc et al. 2005), while APOBEC1-deficient mice are fully viable and fertile (Hirano et al. 1996; Nelson et al. 2012). Partial deficiency increases TGCT risk in a parent-of-origin manner (Carouge et al. 2016).
Ago2
Argonaute RNA-induced silencing complex (RISC) catalytic subunit 2, Chr 15, 33.9 cM. AGO2 is required for RNA-mediated gene silencing (RNAi) by RISC (Ender and Meister 2010). Guide RNAs (miRNAs and siRNAs) direct RISC to complementary RNAs that are targets for RISC-mediated gene silencing. AGO2 and serine/threonine protein phosphatase 2 catalytic subunit β isoform (PPP2CB) (see below) promote mitotic chromosome segregation in the Drosophila and Caenorhabditis elegans germline (Claycomb et al. 2009; Pek and Kai 2011a,b). Interestingly, AGO3 piRNA component Aubergine enhances transmission distortion for the segregation distortion (SD) in Drosophila (Gell and Reenan 2013), raising the possibility that AGO2 might have similar effects under appropriate circumstances. Kirsten RAS oncogene homologue (KRAS) signaling controls AGO2 sorting into exosomes (McKenzie et al. 2016) that transport RNAs, including tRNA fragments (Sharma et al. 2016) for intercellular signaling. RNAs transferred to, as well as produced in, sperm could have significant effects on gamete functions (Hosken and Hodgson 2014). Selectivity in exosome targeting could lead to functional differences among haploid gametes. Loss of siRNA but not miRNA AGO2 activity leads to meiotic catastrophe in MI oocytes (Galgano et al. 2008; Leibovich et al. 2010; Stein et al. 2015). In AGO2-deficient mice, miRNA levels are reduced substantially in oocytes (Kaneda et al. 2009) and in AGO2-deficient oocytes siRNA levels are reduced while retrotransposons and selected mRNA levels are increased (Watanabe et al. 2008). Homozygous-deficient mice display embryonic lethality with various defects in embryonic and extraembryonic organs and tissues. Partial deficiency increases TGCT risk in a parent-of-origin manner (Carouge et al. 2016).
Apob
Apolipoprotein B, Chr 12, 3.53 cM. APOB is widely expressed where it transports lipids such as cholesterol. APOB is encoded as a single, long mRNA. The shorter apoB-48 protein is produced after RNA editing of the apoB-100 transcript at residue 2180 (CAA→UAA), resulting in the creation of a stop codon and early translation termination. Homozygous deficiency leads to embryonic lethality, with embryo loss by embryonic day E9. Heterozygotes tend to have incomplete neural tube closure. Partial deficiency severely reduces fertility in males with sperm showing impaired motility and reduced ability to fertilize both in vivo and in vitro, arguing for a diploid rather than a haploid effect (Huang et al. 1995, 1996).
Apobec1
Apobec1, Chr 6, 57.7 cM. Apobec1 encodes a cytidine deaminase that edits C to U (read as T in coding regions) primarily in 3′-UTRs (Blanc and Davidson 2010). A1CF is the RNA-binding protein that targets specific mRNA sites for editing (Blanc et al. 2005). Apobec1 interacts with the Dnd1Ter mutation to increase TGCT risk in a conventional manner in the male germline and in a transgenerational manner in the female germline (Nelson et al. 2012).
Dnd1
Dead-end microRNA-mediated repression inhibitor 1, Chr 18, 19.5 cM. Dnd1 is an A1cf-related RNA-binding protein expressed in many tissues (Youngren et al. 2005). DND1 controls access of particular miRNAs to their mRNA targets in human TGCTs (Kedde et al. 2007). It is essential for germ cell differentiation (Bustamante-Marin et al. 2013) and also acts like A1cf and Ago2 in offspring to process inherited epigenetic changes from parents (Carouge et al. 2016). DND1 and NANOS2 load RNAs onto the CCR4-Not (CNOT) deadenylase complex for germ cell differentiation (Suzuki et al. 2016) and to suppress specific RNAs (Suzuki et al. 2016). The Ter mutation severely reduces fertility and is a potent modifier of TGCT susceptibility (Stevens 1973; Youngren et al. 2005), whereas the targeted deficiency results in biased fertilization (Zechel et al. 2013).
Ddx1
DEAD (Asp-Glu-Ala-Asp) box polypeptide 1, Chr 12, 6.4 cM. Ddx1 is expressed primarily in the nucleus of the fetal and adult testis and ovary, where it functions as an ATP-dependent RNA helicase to unwind RNA–RNA and RNA–DNA secondary structures for translation initiation, nuclear and mitochondrial splicing, ribosome and spliceosome assembly (including tRNAs), and pre-miRNA and polyA processing (Han et al. 2014; Popow et al. 2014). Reduced DDX1 activity promotes ovarian tumor growth (Han et al. 2014). In Drosophila, Ddx1 deficiency results in stress (starvation)-induced sterility in males and autophagy in egg chambers (Germain et al. 2015).
L3P
No information except Marean et al. (2011).
Lrp6
Low density lipoprotein receptor-related protein 6, Chr 6, 65.4 cM. Lrp6 is expressed at E11.5 and after in the female reproductive system and in fetal testes (Kofron et al. 2007). Lrp6 encodes a transmembrane cell surface protein involved in receptor-mediated endocytosis of lipoprotein and protein ligands. It can function alone or as a coreceptor with Frizzed for canonical Wnt/β-catenin signaling. LRP6 and other members of the Hedgehog and WNT pathways are expressed in human embryonic stem cells and testicular cancers (Dormeyer et al. 2008). Partial embryonic lethality, growth retardation, and various vertebral and skeletal abnormalities are found in mutant homozygotes, and subtle skeletal defects in mutant heterozygotes.
Ppp2cb
Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform, Chr 8, 20.6 cM. Ppp2cb encodes the 2A catalytic subunit of the protein phosphatase 2A (PP2A) heterodimer and is expressed in the female and male reproductive systems from E15 through adulthood. In particular, it is highly expressed in mature spermatozoa and MII oocytes where it localizes at centromeres in meiosis and spindle poles in mitosis (Su et al. 2012a). It is a negative regulator of the mitogen-activated protein kinase (MAPK) pathway and plays a role in the DNA damage response, cell cycle control, apoptosis, and mRNA splicing. Inhibition of PPP2CB releases meiotic arrest and enables meiotic progression (Su et al. 2012a,b). Loss of PPP2CB in oocytes causes both failure of meiosis II exit and reduced fertility in females (Su et al. 2012a,b) and males (Pan et al. 2015). Related proteins play similar roles in meiotic control and chromosome segregation (Bizzari and Marston 2011; Chang et al. 2011; Chambon et al. 2013). PPP2CB deficiency affects sperm tails (Soler et al. 2009; Pan et al. 2015) and chromosome segregation in females (Su et al. 2012a). Other reports find viable and fertile Ppp2cb mutant homozygotes without obvious phenotype (Gu et al. 2012).
Pum1
Pumilio1 RNA-binding family member 1, Chr 4, 63.4 cM. Pum1 is a widely expressed cytoplasmic protein found in the ovary and testis throughout fetal development and adulthood. It encodes an RNA-binding protein that targets Pumilio Response Elements (PRE) in 3′-UTRs to recruit both CCR4-NOT deadenylase, other deadenylases, and miRNAs such as miR221 and miR222, which together repress the expression of genes such as p27 that maintain genome integrity (Kedde et al. 2010; Miles et al. 2012; Van Etten et al. 2012). Interestingly, DND1 blocks access of miR221 to its p27 mRNA target (Kedde et al. 2010), suggesting that DND1 acts competitively with PUM1 to control miRNA actions. In the testis, PUM1 acts as a post-transcriptional regulator of spermatogenesis by binding to the 3′-UTR of mRNAs coding for regulators of transformation-related protein 53 (TRP53) (Subasic et al. 2016) and also suppresses caspase- and TRP53-apoptosis in germ cells (Chen et al. 2012; Subasic et al. 2016). It is involved in embryonic stem cell renewal by facilitating the transition from pluripotency to differentiation (Leeb et al. 2014). PUM1-deficient males exhibit reduced weight of testes and seminiferous tubules, reduced number of sperm, and increased germ cell apoptosis and infertility (Chen et al. 2012). PUM1 also controls to the number of primordial ovarian follicles, meiosis, and reproductive competence in females (Mak et al. 2016). Finally, PUM1 contributes to the antiviral response (Narita et al. 2014), suggesting that it might play a more general role in the stress response to environmental and physiological conditions that often lead to transgenerationally-inherited epigenetic changes (Schaefer and Nadeau 2015).
Vangl2
Vang-like planar cell polarity protein 2, Chr 1, 79.5 cM. VANGL2 is found in the plasma membrane and cytoskeleton where it provides directional signals to cilia (Torban et al. 2012). It is expressed in female and male reproductive systems from E15 through adulthood. Both male homozygous mutants (Lp/Lp) and female heterozygous mutants (Lp/+) are sterile (Strong and Hollander 1949).
Zic2
Zinc finger protein of the cerebellum 2, Chr 14, 66.0 cM. ZIC2 is expressed in the female and male reproductive system from E15 through adulthood and represses transcription in the nucleus. ZIC2 promotes self-renewal of liver cancer stem cells by recruiting the nuclear remodeling factor complex to activate the octamer-binding transcription factor 4 (OCT4) pluripotency factor (Zhu et al. 2015). It also enhances transcription to promote differentiation of embryonic stem cells in Drosophila (Luo et al. 2015) and controls naïve vs. primed pluripotency state (Buecker et al. 2014). Deficiency results in neurulation defects and embryonic lethality in mice (Marean et al. 2011) and holoprosencephaly in humans (Brown et al. 1998).
Table 1. Summary of genetic effects.
| Genes | Observed Ratio (m+: ++) | Average Effect Size | Litter Size (No. Litters) | Conclusion for Intercrosses | |||
|---|---|---|---|---|---|---|---|
| Backcross (1.00) | Intercross (2.00) | Backcross | Intercross | Backcross | Intercross | ||
| Ago2 | 0.89 | 1.03 | 0.07 | 0.34 | 4.9 (216) | 3.9 (33) | Too few heterozygotes |
| Dnd1 | 0.89 | 1.35 | 0.04 | 0.20 | 5.7 (48) | 5.2 (19) | |
| A1cf | 1.10 | 5.00 | 0.05 | 0.35 | 5.8 (178) | 5.9 (51) | Too many heterozygotes |
| Ddx1 | 1.15 | 8.21 | 0.07 | 0.48 | 8.1 (81) | 7.3 (52) | |
| Ppp2cb | 0.79 | 3.47 | 0.12 | 0.23 | 7.6 (73) | na | |
| Pum1 | 1.68 | 5.17 | 0.25 | 0.36 | 6.9 (26) | 6.9 (53) | |
Ratio (m/+: +/+) for backcrosses and intercrosses with a Mendelian expectation of 1 (= 1:1) for backcrosses and 2 (= 2:1) for intercrosses (cf. Figure 1). This ratio emphasizes the relative number of heterozygotes, especially in cases where the frequency of the m allele did not deviate markedly among offspring (cf. Table S1 and Table S2). Weighted averages were used for the multiple evidence for A1cf, Ddx1, and Ppp2cb. Strong differences from expectations are highlighted in gray. Effect size assesses the difference between the observed transmission and Mendelian expectations. Cohen’s w for a goodness-of-fit test was used as a standardized measure of effect size, with a difference between observation and expectation of w = 0.10 as the threshold for declaring a small effect, 0.30 for a medium effect, and 0.50 for a strong effect (Cohen 1988), with medium and strong differences highlighted in gray. Additional information is provided in Table S1. m, m allele; +, wild-type allele; No., number; na, not applicable.
Table 2. Summary of gene–folic acid effects.
| Genes | Folic Acid Dose (ppm) | Observed Ratio | Effect Size | Litter Size (No. Litters) | |
|---|---|---|---|---|---|
| mm: ++ (1.00) | m+: ++ (2.00) | ||||
| L3p | 2 | 0.39 | 1.43 | 0.30 | 7.2 (13) |
| 10 | 1.14 | 2.00 | 0.06 | 4.2 (10) | |
| Zic2 | 2 | 0.57 | 1.56 | 0.20 | 6.1 (18) |
| 10 | 0.95 | 2.57 | 0.14 | 5.7 (10) | |
| Apob | 2 | 1.13 | 2.00 | 0.05 | 6.2 (16) |
| 10 | 0.68 | 0.95 | 0.33 | 5.8 (18) | |
| Lrp6 | 2 | 0.54 | 1.97 | 0.22 | 7.9 (20) |
| 10 | 0.29 | 1.56 | 0.36 | 8.5 (22) | |
| Vangl2 | 2 | 1.15 | 3.15 | 0.16 | 4.3 (16) |
| 10 | 0.48 | 1.39 | 0.26 | 4.4 (15) | |
Two diets (2 and 10 ppm,) were tested on a panel of single-gene mutations that cause neural tube defects (Gray et al. 2010; Marean et al. 2011; Nakouzi and Nadeau 2014). Five genes provided evidence for biased fertilization depending on the level of dietary folate supplementation. Mendelian expectations were 1 (= 1:1) for backcrosses and 2 (= 2:1) for intercrosses (cf. Figure 1). Effect size is Cohen’s w for a goodness-of-fit test (Cohen 1988) (see Table 1 for details). Additional information is provided in Table S2. Strong differences from expectations are highlighted in gray. ppm, parts per million; m, m allele; +, wild-type allele; No., number.
Historically, the genetics of fertilization has been largely resistant to molecular studies (Furnes and Schimenti 2007; Wright and Bianchi 2016). Discovery of genes and gene–diet conditions that bias fertilization may be a breakthrough in understanding mechanisms of sperm–egg interactions at fertilization. Here, evidence for TRD resulting from nonrandom union of gametes in mice is reviewed, possible mechanisms proposed, genetic and reproductive considerations discussed, and genetic implications considered.
Evidence: Reanalysis of the Literature
Genetics
The principles and methods are outlined and then evidence summarized for simple and complicated single-gene effects, as well as an unusual two-gene effect.
As expected, most genetic variants segregate normally and show reduced litter size in proportion to genotype bias, both in our hands and in the literature (informatics.jax.org; www.komp.org), suggesting that biased fertilization is exceptional and results from specific rather than general dysfunctions. The evidence reported here involves single-gene mutations on an inbred strain background. Two cases of fertilization bias have been reported previously (Agulnik et al. 1993; Wedekind et al. 1996), although the responsible genetic factor is not known in either case.
Principles and methods:
Central to this evidence are three expected outcomes: (1) a biased genotype distribution and reduced litter size suggesting either selective embryonic lethality or reduced oocyte production; (2) biased genotype distributions in both backcrosses and intercrosses together with normal litter sizes suggesting a bias in meiotic segregation or gamete numbers or function; and (3) biased genotype distributions in intercrosses but not backcrosses together with normal litter sizes suggesting that gametogenesis is normal but fertilization is biased.
Three measures of TRD were used, depending on the particular question: (1) the frequency of m/+ heterozygotes among m/+ and +/+ segregants; (2) the frequency of the m allele among all offspring; (3) the ratio of m/+ heterozygotes to +/+ homozygotes, and (4) the ratio of m/m to +/+ homozygotes. To test for departures from expectations and to summarize evidence for nonrandom fertilization, emphasis was placed first on testing departures from Mendelian expectations (hypothesis H1), namely 1:1 in backcrosses and 1:2:1 in intercrosses (Figure 1), and then on measures of the nature and magnitude of these departures (Table 1 for genetic effects and Table 2 for gene–folic acid interactions, see also Table S1 and Table S2 for complete data, analytical methods, and results).
Effect size is an important but often neglected measure of phenotypic variation and was used here to provide a normalized quantitative measure of departures from expectations for genetic and gene–diet effects (Cohen 1988). According to accepted standards (Cohen 1988), effect sizes > 0.10 are classified as “small,” > 0.30 “medium,” and > 0.50 “strong.” These measures are independent of sample size. This combination of statistical tests and effect size estimates ensured that significant results and their phenotypic effects were highlighted.
Single genes, simple cases:
For Dnd1 and Ago2, segregation was highly unusual with significant deficiencies of m/+ and m/m genotypes among intercross progeny, - examples of “too few heterozygotes” (Table 1, see also Table S1). For backcrosses, genotype transmission ratios were close to 1:1 expectations and effect sizes were small, whereas for intercrosses these ratios were closer to 1 (=1:1) than 2 (=2:1) and effect sizes were medium (Table 1). Heterozygotes that are missing in intercrosses are found in expected numbers in backcrosses, suggesting that they do not carry intrinsic deficits. In addition, embryonic lethality does not account for the observed genotype ratios because litter sizes were similar among intercrosses and backcrosses (Table S1). For Dnd1, neither mutant homozygotes nor dead embryos were found at embryonic day E3.5 (Zechel et al. 2013). For Ago2, a 25% reduction in litter size would be consistent with the reported lethality of m/m mutant homozygotes (Morita et al. 2007), but not with the observed 50% deficiency of m/+ heterozygotes.
For A1cf, Ppp2cb, and Pum1, highly significant excesses of m/+ heterozygotes were found together with an absence of mutant homozygotes among intercross progeny, - examples of “too many heterozygotes” (Table 1, see also Table S1). For backcrosses, the ratios of m/+ heterozygotes to WT were close to 1:1 expectations and effect sizes were small, whereas for intercrosses, these ratios ranged from 3.1 to 9.7, instead of the expected ratio of 2, and effect sizes were large (Table 1). Despite these differences, average litter sizes were remarkably similar for backcrosses and intercrosses (Table S1). For Pum1, m/m embryos were not detected at E3.5 and litter size did not differ between intercrosses and backcrosses (Zhang et al. 2015) (Significant departures from expectations were also found in intercrosses after accounting for the modest departures in backcrosses, χ2 = 12.64, P < 0.001, see Table S1). Pum1 also showed modest departures in backcrosses, For A1cf, loss of homozygous embryos between E3.5 and 4.5 does not account for excess heterozygosity or for normal litter size (Blanc et al. 2005). Although litter size was not reported for Ppp2cb (Sasaki et al. 2007), the highly significant heterozygote excess (> 3:1) in intercrosses vs. backcrosses is striking and consistent with results for A1cf, Ddx1 (see below), and Pum1. Normal segregation in backcrosses with mutant heterozygotes shows that gametes are produced in comparable (1:1) numbers and functionality in each sex.
Ddx1: complicated single-gene effect:
An engineered deficiency of DEAD Box 1 helicase (Ddx1) and an induced epigenetic change in its WT allele provide strong evidence for biased fertilization (Hildebrandt et al. 2015; Hollick 2017). For the engineered mutation, m/m embryos were missing with no homozygotes detected at E3.5 in test crosses. In addition, a substantial deficit of WT segregants relative to m/+ heterozygotes was also observed in some crosses (Table 3 and Table S1), leading the authors to conclude that the engineered mutation induced a modified, perhaps paramutated allele (Hollick 2017) (designated “*”) that leads to lethality in both WT (+/+) and */* embryos resulting from crosses involving a */+ parent. However, review of litter sizes for test and control crosses revealed remarkably little evidence for embryo loss that would account for the putative loss of m/m, */*, and +/+ embryos. Strongly biased transmission without embryo loss argues that preferential fertilization is a more likely explanation.
Table 3. Ddx1 transmission.
| Cross | Offspring Genotypes | Proposed Loss (%) (Lost Genotypes) | Observed Litter Size (No. Pups, No. Litters) | Conclusion About Litter Size |
|---|---|---|---|---|
| +/+ × +/+ | +/+ | 0 | 6.7 (187, 28) | Reference |
| +/+ × +/m | +/+, +/m | 0 | 8.1 (657, 81) | Baseline |
| +/+ × +/* | +/+, +/* | 0 | 5.3 (42, 8) | Reduced 25% |
| +/+ × m/* | +/m, +/* | 0 | 7.7 (370, 48) | Baseline |
| +/m × +/m | +/+, +/m, m/m | 25% (m/m) | 7.3 (378, 52) | Not reduced |
| +/* × +/* | +/+, +/*, */* | 25% (*/*) | 10 (20, 2) | Not reduced |
| m/* × m/* | m/m, m/*, */* | 100% | 7.0 (861, 123) | Not reduced |
| +/m × +/* | +/+, +/*, m/+, m/* | 25% (m/*) | 6.0 (72, 12) | Not reduced |
| */* × m/* | */m, */* | 100% | 7.1 (57, 8) | Not reduced |
| */* × */* | */* | most | 5.5 (44, 8) | Reduced 25% |
Three alleles are shown, wild-type (+), mutant (m) and modified wild-type (*). Various crosses were used to examine the effect of m and * on embryonic viability. Offspring genotypes are shown for each cross. Proposed loss is based on the hypothesis that the genotypes m/m and */* result in embryonic lethality and where m/*, m/+, and */+ are viable (Hildebrandt et al. 2015). Loss is summarized as the deviation (percentage) from Mendelian expectations for these genotypes in each cross. “Conclusion about litter size” is based on comparing the observed litter size with the “reference” for the wild-type cross. Data are from Hildebrandt et al. (2015) and from R. Godbout (personal communication).
Apobec1 and Dnd1: a complicated two-gene effect:
The last genetic example emerged in tests to determine whether Apobec1 and Dnd1 interact to modulate inherited risk for spontaneous testicular germ cell tumors (TGCTs; see below for additional information about TGCT origins, genetics, and biology) (Nelson et al. 2012). These genes independently affect risk in both a conventional and an inherited epigenetic manner (Nelson et al. 2012), but whether they interact in the genetic sense was uncertain. Surveys for TGCTs among intercross offspring revealed unexpected evidence for biased fertilization (Table 4). With maternal heterozygosity for Dnd1 and paternal heterozygosity for Apobec1, offspring occurred in the expected (1:1:1:1) Mendelian ratio for two independently segregating genes, but results for the reciprocal cross revealed a marked deficiency of all single- and double-mutant genotypes. If the number of WT (+/+, n = 64) progeny is accepted as the proper reference for the three other genotypic classes, where 1:1:1:1 = 64:64:64:64, then only 27% (51/192, where 192 = 3 × 64) of the expected single- and double-mutant segregants was found. Again, litter size did not differ between the reciprocal crosses (Nelson et al. 2012). Remarkably, both heterozygous and homozygous mutants for each gene are fully viable in separate crosses (Table S1) (Hirano et al. 1996; Blanc et al. 2005; Youngren et al. 2005; Zechel et al. 2013). Thus, in this but not the reciprocal cross, the majority of mutant segregants are missing, with evidence for full viability in other crosses and with no evidence for reduced litter size.
Table 4. Transmission in Apobec1, Dnd1Ter intercrosses.
| Ter/+ × m/+ | Offspring Genotype | m/+ × Ter/+ | ||
|---|---|---|---|---|
| No. Obs | % Obs | No. Obs | % Obs | |
| 27 | 24 | m/+, Ter/+ | 13 | 10 |
| 30 | 26 | +/+ Ter/+ | 19 | 17 |
| 25 | 22 | m/+, +/+ | 19 | 17 |
| 32 | 28 | +/+, +/+ | 64 | 56 |
| 114 | Total | 115 | ||
Ter is a spontaneous mutation in the Dnd1 gene (Youngren et al. 2005). Reciprocal crosses were made between Apobec1 targeted deficiency mutation (m) and Dnd1Ter. Crosses are presented as “female × male.” “No. Obs” is number observed for each genotype and cross. “% Obs” is the percentage for that genotype among all offspring for each cross. For two segregating genes in these intercrosses, a 1:1:1:1 Mendelian ratio is expected. Results are from Nelson et al. (2012).
Gene–folate diet interactions
A related class of TRD involves dietary effects on single-gene models of neural tube defects (NTDs). The beneficial effect of dietary folate supplementation on a common birth defect is one of the greatest achievements in epidemiology (Smithells et al. 1980). NTDs such as anencephaly and spina bifida are the second most common congenital defect, occurring in ∼1 per 1000 live births and often leading to disability and mortality (Detrait et al. 2005; Copp et al. 2013). Mothers and fetuses frequently show reduced folate and elevated homocysteine levels (Smithells et al. 1980; Carmel and Jacobsen 2001). Dietary folate supplementation before and during pregnancy significantly reduces the occurrence and severity of cases, but many (∼50%) remain resistant to the beneficial effects of folate supplementation (Smithells et al. 1980; Blom et al. 2006; Copp et al. 2013). Reliable prediction of individual response to supplementation is currently impossible.
Several studies examined the effects of dietary supplementation with folic acid on development of the neural tube in mouse models (Juriloff and Harris 2000; Greene and Copp 2005; Zohn 2012). Some models respond favorably to folic acid (Barbera et al. 2002) and several respond to other nutrients such as methionine and inositol (Essien 1992; Greene and Copp 1997), but as in humans many are resistant (Juriloff and Harris 2000; Franke et al. 2003; Greene and Copp 2005; Zohn 2012). In the design for these studies, developmental response to various nutrients was tested among intercross progeny. In some cases, +/m heterozygotes are maintained on a test or control dietary formulation, bred together (+/m × +/m), and developmental consequences evaluated among +/+, +/m, and m/m offspring, where offspring are expected to occur in a 1:2:1 Mendelian ratio. In other cases, pregnant +/m females are treated only during critical developmental windows and then embryos or offspring are examined. The former is the only protocol that is instructive about possible dietary effects on gametogenesis and fertilization. Attributing effects to diet simply involves comparing results for the high- vs. low-dosage treatment groups for the identical genotypes on the same defined genetic background. Efficacy is indicated if the number (proportion) of affected m/m individuals is reduced.
During studies to identify mouse NTD models that are resistant to the benefits of folic acid treatments, two examples of fertilization bias were found (Nakouzi and Nadeau 2014). Reanalysis of published reports then revealed three additional NTD models that show biased fertilization in response to folic acid treatment (Table 2). In several cases (Apob, Lrp6, and Vangl2), significant deviations are found in the high folic acid (10 ppm; parts per million) test, but in other cases (L3P and Zic2) deviations were found in the low (2 ppm) test, with similar litter sizes for each mutant on the two diet protocols (Table S2). On one diet, transmission ratios were consistent with normal segregation and effect sizes were small, whereas on the alternative diet, transmission ratios were more divergent and effect sizes were medium (Table 2, see also Table S2). None showed too many heterozygotes. Departures from Mendelian expectations with dietary supplementation were not as strong as the genetic results, perhaps because optimal supplementation levels were not tested and perhaps because dietary consumption and metabolism differ among the various cohorts and are generally more difficult to control. Interestingly, for most models, the percentage of affected m/m was similar in the test and control protocols, suggesting that supplemental folic acid did not reduce the proportion of affected mutant homozygotes (Gray et al. 2010; Marean et al. 2011).
These results raise a provocative question: does folate correct a developmental defect in the neural tube, or do other explanations apply such as reducing the incidence of cases by biasing fertilization away from at-risk genotypes (Nakouzi and Nadeau 2014)? In humans, where NTD genetics is not as clearly understood as in mouse models, with genetic heterogeneity a significant issue and isolated cases common, distinguishing a protective effect vs. biased fertilization would be difficult.
Gene functions
Six of the seven TGCT genes encode proteins that are directly involved in various aspects of RNA biology: A1cf, RNA editing (Blanc and Davidson 2010; Snyder et al. 2017a,b); Apobec1, RNA editing (Blanc and Davidson 2010); Ago2, RNA interference (RNAi) (Ender and Meister 2010); Dnd1, miRNA control (Kedde et al. 2007); Ddx1, RNA helicase (Hildebrandt et al. 2015); and Pum1, translation repression (Zhang et al. 2015) (Box 1). The seventh gene, Ppp2cb, is a serine/threonine phosphatase (Sasaki et al. 2007) (Box 1). Four show inherited epigenetic effects on TGCT risk (A1cf, Apobec1, Ago2, and Dnd1) and one shows epigenetic effects on embryonic viability (Ddx1). Neither Ppp2cb nor Pum1 have been tested for TGCT or epigenetic effects. These proteins (except Ppp2cb) have specific RNA targets that are in turn effectors of molecular, developmental, and physiological functions. These functions could be shared or distinct in males and females depending on the nature of the targeted RNAs in each sex. Identifying these targets is critical for understanding the ways that these genes control gamete choice at fertilization.
By contrast, the five NTD genes appear to have diverse, seemingly unrelated functions without an obvious theme (Apob, L3P, Lrp6, Vangl2, and Zic2; Box 1). Perhaps RNA genes and Ppp2cb are directly involved in molecular and cellular mechanisms of gametogenesis and fertilization, whereas the various NTD genes sensitize folate and homocysteine metabolism to adverse interactions with pathways that directly affect gamete interactions at fertilization.
Hypotheses
Reversed meiosis
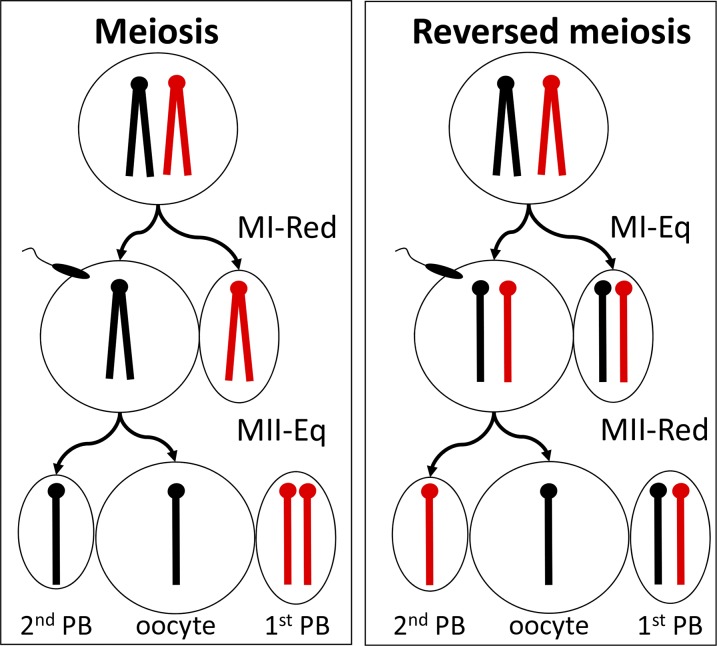
If meiotic divisions in females are ordered the way we have been taught, with the reductional division (MI) preceding the equational division (MII) (Figure 2), biased fertilization without conventional TRD would be difficult. During ovulation, primary oocytes resume meiosis and, over the next few hours, complete MI and arrest at metaphase in MII (Edwards and Gates 1959) Homologous (nonsister) chromatid pairs segregate at MI with one product going to the secondary oocyte and the other to the first polar body, which may divide again at MII (Zanders and Malik 2015; Dean 2016). Fertilization triggers completion of MII with sister chromatids segregating to the second polar body and the oocyte (Zanders and Malik 2015; Dean 2016).
Figure 2.
Regular and reversed meiosis. Chromatids from alternative chromosomes are marked in red and black. MI-reductional (MI-Red) precedes MII-equational (MI-Eq) in regular meiosis, whereas in reversed meiosis, MI-Eq occurs first (Zanders and Malik 2015; Dean 2016). MII arrests at metaphase until sperm entry at fertilization. PB, polar body. Recombination is not included in these scenarios, although normally ∼40% of chromatids at the haploid phase have a crossover (Singer et al. 2004).
Recently, “reversed meiosis” was reported after induced ovulation in humans (Ottolini et al. 2015). Reversing the order of meiotic divisions during oogenesis, with the equational division occurring at MI rather than MII, leaves the secondary oocyte heterozygous for marker genes at fertilization (Figure 2). The genetics of fertilizing sperm could then bias MII segregation with one chromosome preferentially remaining in the oocyte and the other segregating to the second polar body. Although formal tests have yet not been reported in mice, the study by Agulnik et al. (1993) is consistent with reversed meiosis. Interestingly, genes such as Ago2 and Ppp2cb that control chromosome segregation in mitosis are strong candidates for determining the sequence of meiotic divisions (Pek and Kai 2011a,b; Huang et al. 2015), both of which are expressed in germ cells (Su et al. 2012a). Whether meiotic reversal is adaptive or is an anomaly resulting from reduced gene dosage and physiological stresses such as induced ovulation is unclear.
The alternative hypothesis that meiosis is conventional cannot account for fertilization bias in intercrosses but not backcrosses, because completion of MI prior to fertilization makes the reductional division independent of the mating partner and its fertilizing sperm. Bias would therefore be evident in both backcrosses and intercrosses.
Polyamines
The next question concerns the links between folate metabolism and gamete function. Although anomalies in folate metabolism are associated with reduced fertility (Forges et al. 2008; Mora-Esteves and Shin 2013; Aarabi et al. 2015; Ly et al. 2017), there is as yet no evidence for genotype-specific effects on haploid gametes. The challenging task then is to find an explanation connecting folate metabolism with TRD, given that transcription and translation are limited but not absent during spermatogenesis (Dadoune et al. 2004; Shima et al. 2004; Hammoud et al. 2009; Licatalosi 2016; Schuster et al. 2016; Zuo et al. 2016; Zhang et al. 2017).
A strong argument can be made that polyamine metabolism is the missing link. Most of this evidence involves spermatogenesis; the evidence for oogenesis is meager by comparison. Polyamines such as spermine, spermidine, putrescine, and cadaverine are low-molecular weight organic molecules that have at least two amino groups. They are present in all cells and most are associated with nucleic acids (Matthews 1993; Wallace et al. 2003; Persson 2009; Kahana 2016). They are involved in transcription, translation, histone modifications, autophagy, apoptosis, and many other molecular, cellular, and physiological activities (Hesse and Hoefgen 2003; Wallace et al. 2003; Alhonen et al. 2009; Lefevre et al. 2011; Igarashi and Kashiwagi 2015; Miller-Fleming et al. 2015).
Given their interdependent roles in essential molecular, epigenetic, and cellular functions, the polyamine and folate pathways are highly conserved and highly regulated, from yeast to plants and mammals, with sometimes overlooked roles in gametogenesis and fertility (Rubinstein and Breitbart 1991; Wolukau 2004; Roje 2006; Lefevre et al. 2011; Bauer et al. 2013; Aloisi et al. 2016; Chen et al. 2016). The substrates for polyamine synthesis are ornithine, which is derived from arginine and proline, and S-adenosylmethione (SAM), which is part of the homocysteine cycle in folate metabolism (Figure 3). SAM is the methyl donor for all methylation reactions for nucleic acids (DNA and RNAs, including tRNAs), proteins (including histones), lipids, and other molecules (Matthews 1993; Stover 2011; Salbaum and Kappen 2012; Gueant et al. 2013). Moreover, by utilizing acetyl CoA, polyamine catabolism magnifies the effects of folate deficiency (Figure 3). Acetyl CoA is the substrate for synthesizing betaine, which is the alternative methyl donor to synthesize SAM from homocysteine. Despite the essential role for methylation in many molecular and cellular functions, cells preserve polyamine metabolism at the expense of methylation, at least under in vitro and in vivo stress conditions (Kramer et al. 1987, 1988; Sun et al. 2002; Bistulfi et al. 2009, 2010).
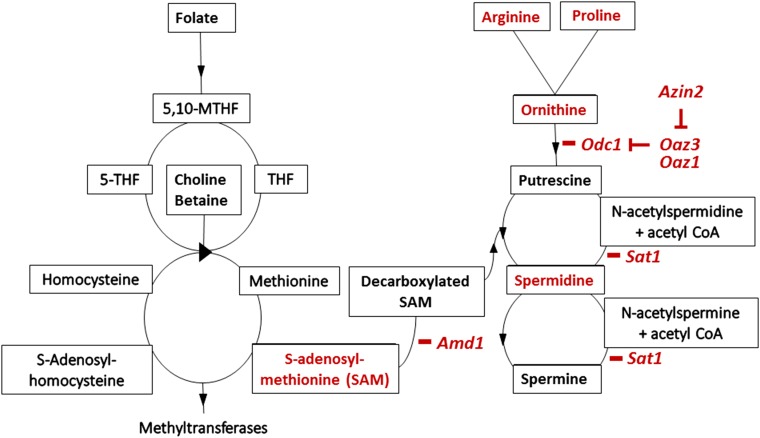
Figure 3.
Folate and polyamine metabolic pathways. [Adapted from references Carmel and Jacobsen (2001) and Wallace et al. (2003)]. Enzymes and metabolites that affect fertility, haploid gamete function, or embryonic stem cell (ESC) pluripotency are highlighted in red (see text for background and references). Genes that encode relevant enzymes are shown in italics with a bar showing their site of action. Blocked bars show proteins with inhibitory effects. Folate is the primary methyl donor; choline and betaine are alternative methyl donors. Amd1 links folate–homocysteine metabolism with polyamine metabolism by converting S-adenosylmethionine to decarboxylated S-adenosylmethionine, which with putrescine produces spermidine. Odc1 converts ornithine to putrescine and is the heavily regulated rate-limiting step in polyamine synthesis. Catabolism of spermine and spermidine consumes acetyl CoA, which also serves as a substrate for choline and betaine synthesis. 5-THF, 5-tetrahydrofolate; 5,10-THF, 5,10-methylenetetrahydrofolate; Amd1, S-adenosylmethione decarboxylase 1; Odc1, ornithine decarboxylase 1; THF, tetrahydrofolate.
Functionality of haploid gametes depends heavily on polyamine metabolism in many species. In Arabidopsis, the MAT3 SAM synthase gene is expressed in pollen, with mutants showing reduced pollen tube growth and seed set as well as changes in polyamine biosynthesis, tRNA levels, and histone methylation (Chen et al. 2016). In humans, polyamine deficiency results in infertility, which can be corrected with SAM or polyamine supplementation (Shohat et al. 1990; Rubinstein et al. 1995; Calandra et al. 1996; Lefevre et al. 2011). In cattle, spermine is essential for acrosomal function (Rubinstein et al. 1995). Overexpression of ornithine decarboxylase 1 (ODC1), the rate-limiting step in polyamine synthesis, also leads to infertility (Qian et al. 1985; Halmekyto et al. 1991; Kilpelainen et al. 2001; Tokuhiro et al. 2009). Several polyamine genes are expressed in haploid gametes (Figure 3) (Lefevre et al. 2011). ODC antizyme 3 (OAZ3) is a testis-specific inhibitor of ODC1 (Qian et al. 1985; Tosaka et al. 2000; Kilpelainen et al. 2001; Ike et al. 2002; Tokuhiro et al. 2009). Deficiency leads to sperm that cannot fertilize (Kilpelainen et al. 2001; Tokuhiro et al. 2009). Antizyme inhibitor 2 blocks the inhibitory effects of OAZ3 in haploid cells (Lopez-Contreras et al. 2009) as well as ODC1 overexpression (Suzuki et al. 1994). Spermidine/spermine N1-acetyltransferase and OAZ1 are differentially expressed with folate supplementation in the LRP6 mouse NTD model (Gray et al. 2010; Nakouzi and Nadeau 2014). OAZ1 is also differentially expressed in sperm from folate-deficient mice (Lambrot et al. 2013). Finally, the primary inputs for polyamine metabolism, namely the enzyme SAM decarboxylase 1 (AMD1), which catalyzes the conversion of SAM to decarboxylated SAM, and the amino acids arginine, ornithine, and proline, are critical for pluripotency control in embryonic stem cells (ESCs) (Figure 3) (Pierce et al. 1990; Nishimura et al. 2002; Casalino et al. 2011; Zhang et al. 2012; Zhao et al. 2012; D’Aniello et al. 2015; Lee et al. 2016). Given the close lineage relationships between germ cells and embryonic stem cells (ESCs) (Zwaka and Thomson 2005; Bustamante-Marin et al. 2013), similar effects in gametes would not be surprising. Polyamine metabolism is therefore a compelling candidate for biased fertilization, given its established impact on haploid gametes and its strong connection to folate metabolism.
Genetic and Reproductive Considerations
Sperm in the epididymis, gametes in the oviduct
The issue here is whether reproductive organs and sperm–egg recognition provide opportunities for gametic selection based on haploid effects (Holt and Fazeli 2015). Semen, as well as oviductal and uterine fluids, contain various proteins, nucleic acids, and small molecules that provide a chemically appropriate environment for sperm maturation and capacitation, for fertilization, and for implantation of genetically nonself embryos (Nieder and Macon 1987; Vinijsanun and Martin 1991; Wolfner 2002; Huang et al. 2004; Waberski et al. 2006; Holt and Fazeli 2010; Lane et al. 2014; Johnson et al. 2015; Jodar et al. 2016). Because mating usually occurs prior to ovulation, sperm can be in the oviduct hours before ovulation, often needing just minutes from ejaculation to arrive in the oviduct (Lewis and Wright 1935). Sperm and eggs induce gene expression changes in the oviduct that alter the biochemistry of oviductal fluids (Fazeli et al. 2004) and that in turn restrict sperm access (Holt and Fazeli 2015, 2016). Millions of sperm are released at ejaculation but usually < 100 reach the oviduct (Braden and Austin 1954). Not only do sperm compete for access to eggs (Clark et al. 1999; Clark 2002), the oviduct can select sperm based in part on their genetic content, including sex chromosomes (X or Y) (Alminana et al. 2014; Holt and Fazeli 2016) and on chromatin stability (Ardon et al. 2008).
Signaling before contact between sperm and egg has obvious advantages for haploid gametes. There are reports that small, transiently expressed peptides attract a minority of sperm that are only briefly responsive, presumably representing the 10% of sperm that are appropriately capacitated (Eisenbach and Ralt 1992; Eisenbach 1999). Thermotaxis (Bahat and Eisenbach 2010), chemotaxis (Holt and Fazeli 2015, 2016), and signaling molecules including olfactory receptors (Flegel et al. 2015), trace-amine-associated receptors (Zucchi et al. 2006; Chiellini et al. 2012), cannabinoid receptors (Agirregoitia et al. 2010), and calcium receptors (Dell’Aquila et al. 2006; De Santis et al. 2009; Mendoza et al. 2012), have been reported in gametes and gonads. These could play a role in sperm–egg attraction, but the evidence is limited both about mechanisms and especially about the impact of genetic variation on molecular interactions.
In Drosophila and other species, success of fertilizing sperm depends on the genetics of both the male and female mating partners, suggesting that ligand–receptor interactions are critical (Braden 1958; Chow et al. 2010). But how genetic variation affects affinity and signaling in these ligand–receptor pairs is largely unknown. Once sperm penetrate the glycoprotein coat surrounding the egg, the sperm and egg must recognize each other and fuse membranes. The zona pellucida hardens irreversibly to prevent polyspermy (Braden et al. 1954). Presumably fertilization bias must occur before membrane fusion and zona pellucida hardening, otherwise the conceptus would either persist as a viable embryo, or be lost with a corresponding reduction in litter size. Recognition between sperm and egg is based on ligand–receptor binding between Izumo1 (sperm) and Juno (egg); absence of either protein leads to infertility (Bianchi et al. 2014; Aydin et al. 2016; Bianchi and Wright 2016; Ohto et al. 2016). Although Juno, which is a member of the folate receptor family, no longer binds folate (Bianchi et al. 2014), residual functions might still depend on folate levels. Unexplained anomalies between other ligand–receptor pairs such as loss of one but not the other member of the pair resulting in infertility suggests that models of sperm–egg recognition remain incomplete (Cho et al. 1998). In at least one instance, union of sperm and egg brings together two distinct proteins that act together as a dimer to suppress mutagenesis in early embryos (Lord and Aitken 2015).
Several approaches have been employed to define mechanisms for sperm–egg recognition. For example, ENU mutagenesis has been used to find genes controlling gametogenesis and fertilization (Hrabe de Angelis et al. 2000; Clark et al. 2004; Sakuraba et al. 2005). Although many of these mutated genes affect germ cell biology and meiosis, genes affecting fertilization were not found (Furnes and Schimenti 2007). Fortuitously, our genetic studies of epigenetic TGCT risk and NTD dietary response discovered several such genes, thereby enabling new genetic approaches for studying mechanisms of gamete function at fertilization.
Haploid effects
Functional effects confined to haploid gametes are critical for fertilization bias. The four products of meiosis produced in males usually have an equal chance of fertilization, in part because syncytia provide small intercellular channels through which developing sperm share molecules, thereby minimizing the impact of haploid effects (Greenbaum et al. 2011). However, gametes carrying a t-haplotype, which are a classic example of TRD in the mouse (Dunn 1957; Lyon 2003), produce two critical elements that control sperm motility in a haploid-specific manner. Gametes that carry a t-haplotype produce molecules that pass through syncytial bridges to hyperactivate Rho signaling in both t- and +-bearing sperm, thereby compromising their motility. However, t-bearing sperm also produce a protective, haploid-specific variant of SMOK1 (sperm motility kinase) that does not pass through bridges and protects t-bearing but not +-bearing sperm, thereby providing a motility advantage to t-bearing sperm (Herrmann et al. 1999; Veron et al. 2009; Bauer et al. 2012). Because such effects are intrinsic to heterozygous males and would be found in both backcrosses and intercrosses, a corresponding effect would need to operate in females to be a sufficient explanation for biased fertilization.
Competition between diploid and haploid phases
Discoveries about biased fertilization are relevant to theories concerning sexual antagonism, namely the contrasting priorities between diploid organisms and their haploid gametes. Diploids strive for reproductive success vs. other diploids, whereas haploid gametes compete with each other for successful fertilization. Because gametic competition could reduce parental fertility, diploid cells should seek to reduce functional differences among gametes by limiting their transcription and translation. As long as gametes are functionally equivalent, diploids have the advantage over their haploid gametes. It is noteworthy then that many of the genes that bias fertilization affect aspects of RNA biology that impact translation (Box 1). Partial loss-of-function in mutant heterozygotes might enable phenotypic differences among gametes, leading to gamete competition and biased fertilization.
The window for bias
The length of the haploid phase in spermatogenesis and oogenesis places conditions on the mechanisms that might contribute to biased fertilization. In mammals, the haploid phase is long in males, with the MI division arrested during embryonic development and recommencing at puberty. Spermatogenesis then continues throughout reproductive life. The haploid phase begins with completion of meiosis in the testis and continues as they mature and capacitate, and while they pass through the epididymis, vas deferens, urethra, and uterus to the site of fertilization in the oviduct, a period that can last several days. By contrast, the haploid phase is remarkably short in females, lasting only from completion of MII, which fertilization triggers, until female and male pronuclei fuse. This brief window raises the likelihood that fertilization drives the bias in oocytes.
Centromeric- or gene-specific effects
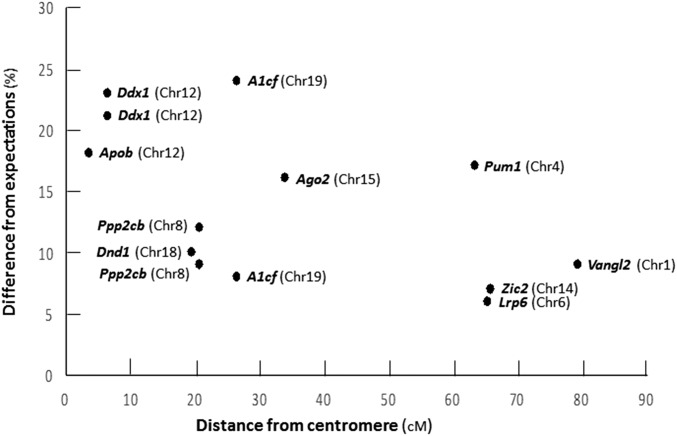
Sometimes, TRD results from preferential segregation of centromeric elements that guide chromosome movement and segregation during karyokinesis (Lyttle 1989; Pimpinelli and Dimitri 1989; Pardo-Manuel de Villena and Sapienza 2001; Axelsson et al. 2010; Chmatal et al. 2014; Wei et al. 2017). Genes that are closely linked to the centromere would also show TRD with the degree of distortion depending on the recombination distance from the centromere and with genes located 50 cM or more showing normal 1:1 segregation. However, several genes that are located far from the centromere show substantial TRD and overall there is little evidence that TRD declines as a strong function of recombination distance from the centromere (Figure 4). However, the propensity of recombined chromatids to segregate preferentially to oocytes during reversed meiosis precludes a firm conclusion (Ottolini et al. 2015). Finally, the dispersed chromosomal locations for these genes (Figure 4, Table 1, and Table 2) argue against a selfish gene complex, as is found frequently with other TRD systems (Lyttle 1993; Lyon 2003). Together, these observations are consistent with gene-specific TRD rather than hitch-hiking resulting from close linkage to centromere-driven elements.
Figure 4.
The difference (%) from expectations was calculated as the absolute value of the deviation between observed and expected frequency of m/+ heterozygotes among heterozygous and wild-type (WT) segregants. Difference provides a direct measure of the departure from expectations. Recombination distances are from the Mouse Genome Database (informatics.jax.org). The correlation between deviation and distance was modest (r = −0.54) and accounted for only 29% of the variation (r2), suggesting that centromeric drivers, if any, had modest effect on transmission ratios. Limiting the analysis to data points < 50 cM yielded r = −0.39 and r2 = 0.15.
Litter size
Although many factors such as the number of fertilized eggs, pre- and postimplantation mortality, and uterine capacity can affect litter size, various evidence shows that the primary determinant is the number of ovulated eggs. Selection for larger litter size increases ovulation rate (Falconer 1960; Joakimsen and Baker 1977; Bakker and Politiek 1978; Durrant et al. 1980), while selection for increased ovulation rate results in larger litters (Bradford 1969). Inbreeding reduces litter size because fewer eggs are ovulated (McCarthy 1967). Unilateral ovariectomy reduces litter size by 50% (Vinijsanun and Martin 1991), arguing against eggs held in reserve to compensate for failed fertilizations and embryo loss. In parallel, eggs mature within ovarian follicles that rupture to release eggs at ovulation, with the number of growing follicles determined by host genetics long before and independent of fertilization. Thus, the number of ovulated eggs available for fertilization appears to be the primary determinant of litter size. Whether any of these 12 mutant genes, together with regular or folate-supplemented diets, affect the number of ovulated eggs remains to be determined, although any such effects should be found in both backcrosses and intercrosses.
Lethality
Three lines of evidence argue against lethality as the explanation for departures from Mendelian expectations (Figure 1). First, litter size is not reduced in intercrosses vs. backcrosses for any genes that bias fertilization (Table 1, Table 2, and Table 3). Again, if mutant homozygotes show embryonic lethality, then litter size should be reduced by 25%, and if half the heterozygotes are also missing, then litter size should be further reduced to 50%. Second, in some cases [Dnd1 (Zechel et al. 2013), Ddx1], surveys at E3.5 failed to find mutant homozygotes or dead embryos (Hildebrandt et al. 2015), and Pum1 (Zhang et al. 2015). Third, loss of particular genotypes is not based on their inherently deleterious nature, given that mutant heterozygotes that are missing among intercross progeny are found in expected numbers among backcross progeny (Table 1), in reciprocal crosses (Table 4), or on alternative folic acid diets (Table 2). Thus, negative selection against embryos having particular genotypes does not seem to be involved.
Perspectives
Biased fertilization may be a previously unrecognized manifestation of genes that control TGCT susceptibility, epigenetic inheritance, and related germline abnormalities. The first evidence for fertilization bias was discovered during work on the control of inherited TGCT risk in the 129 family of inbred strains (Lam et al. 2007; Heaney et al. 2008; Nelson et al. 2012). These TGCTs are models for several classes of TGCTs in humans (Bustamante-Marin et al. 2013; Rijlaarsdam and Looijenga 2014). Like gametes, many originate from the germ cell lineage during fetal development (Kleinsmith and Pierce 1964; Stevens 1967). The germline origin of TGCTs and gametes suggests that they may share similar vulnerabilities to genetic and environmental perturbations. Perhaps no other lineage undergoes such dramatic transitions in developmental potential and with such profound implications for health, fertility, and perpetuation of the germline across generations (Jablonka and Lamb 1999). The germline is unipotent until fertilization, when it transitions to pluripotent embryonic cells that in turn differentiate into specialized somatic and germ cell lineages (Saitou and Yamaji 2012; Hackett and Surani 2014; Reik and Surani 2015). Various mechanisms preserve the genomic integrity and developmental capacity of the “mother of all stem cell lineages” (Donovan 1998) by repairing DNA defects (Khurana et al. 2010) maintaining cellular conditions (Gemble et al. 2015), suppressing transposon activity (Bourc’his and Bestor 2004; Bourc’his and Voinnet 2010; Senti and Brennecke 2010; Barau et al. 2016; Wylie et al. 2016a,b), and programming epigenetic state (Buecker et al. 2014; Oliveros-Etter et al. 2015). Failure of pluripotency control can lead to precocious differentiation of germ cells (Hargan-Calvopina et al. 2016; Sanchez et al. 2016), spontaneous transformation of germ cells during fetal development (Kleinsmith and Pierce 1964; Stevens 1967; Bustamante-Marin et al. 2013; Ferguson and Agoulnik 2013), infertility (Skakkebaek et al. 2001, 2016; Carouge et al. 2016), and other reproductive disorders (Ferguson and Agoulnik 2013). Dysfunction can also lead to inherited epigenetic changes that affect risk for TGCTs and urogenital abnormalities in offspring and later generations in the absence of genes that originally triggered these transgenerational effects (Lam et al. 2007; Heaney et al. 2008; Nelson et al. 2012; Carouge et al. 2016). Genetic anomalies together with dietary, hormonal, and other environmental influences may induce germline dysfunctions that manifest as conventional genetic effects, unconventional epigenetic inheritance, and now perhaps preference for particular gamete combinations at fertilization.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300109/-/DC1.
Acknowledgments
I thank Rosaline Godbout and her group for generously sharing litter size and segregation data from Hildebrandt et al. (2015) and for sharing her thoughts about their study and the interpretation proposed here. I also thank David Crews, Mary Ann Handel, and especially Hamish Spencer and Monika Ward for comments on a draft of this paper. Andy Clark, Aimee Dudley, John Eppig, Fernando Pardo de Villena, Jasper Rine, Jesse Riordan, Carmen Sapienza, and Patrick Stover provided helpful suggestions, insights, and speculations. I thank the reviewers for their many helpful comments. The following National Institutes of Health grants supported this work: Eunice Kennedy Shriver National Institute of Child Health and Human Development Pioneer Award DP1 HD075624, National Cancer Institute CA75056, and National Institute of Neurological Disorders and Stroke NS058979.
Footnotes
Communicating editor: J. C. Schimenti
Literature Cited
- Aarabi M., San Gabriel M. C., Chan D., Behan N. A., Caron M., et al. , 2015. High-dose folic acid supplementation alters the human sperm methylome and is influenced by the MTHFR C677T polymorphism. Hum. Mol. Genet. 24: 6301–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirregoitia E., Carracedo A., Subiran N., Valdivia A., Agirregoitia N., et al. , 2010. The CB(2) cannabinoid receptor regulates human sperm cell motility. Fertil. Steril. 93: 1378–1387. [DOI] [PubMed] [Google Scholar]
- Agulnik S. I., Sabantsev I. D., Ruvinsky A. O., 1993. Effect of sperm genotype on chromatid segregation in female mice heterozygous for aberrant chromosome 1. Genet. Res. 61: 97–100. [DOI] [PubMed] [Google Scholar]
- Alhonen L., Uimari A., Pietila M., Hyvonen M. T., Pirinen E., et al. , 2009. Transgenic animals modelling polyamine metabolism-related diseases. Essays Biochem. 46: 125–144. [DOI] [PubMed] [Google Scholar]
- Alminana C., Caballero I., Heath P. R., Maleki-Dizaji S., Parrilla I., et al. , 2014. The battle of the sexes starts in the oviduct: modulation of oviductal transcriptome by X and Y-bearing spermatozoa. BMC Genomics 15: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloisi I., Cai G., Serafini-Fracassini D., Del Duca S., 2016. Polyamines in pollen: from microsporogenesis to fertilization. Front. Plant Sci. 7: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardon F., Helms D., Sahin E., Bollwein H., Topfer-Petersen E., et al. , 2008. Chromatin-unstable boar spermatozoa have little chance of reaching oocytes in vivo. Reproduction 135: 461–470. [DOI] [PubMed] [Google Scholar]
- Axelsson E., Albrechtsen A., van A. P., Li L., Megens H. J., et al. , 2010. Segregation distortion in chicken and the evolutionary consequences of female meiotic drive in birds. Heredity 105: 290–298. [DOI] [PubMed] [Google Scholar]
- Aydin H., Sultana A., Li S., Thavalingam A., Lee J. E., 2016. Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature 534: 562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahat A., Eisenbach M., 2010. Human sperm thermotaxis is mediated by phospholipase C and inositol trisphosphate receptor Ca2+ channel. Biol. Reprod. 82: 606–616. [DOI] [PubMed] [Google Scholar]
- Bakker H. W. J., Politiek R. D., 1978. Reproduction and body weight of mice after long-term selection for large litter size. J. Anim. Sci. 46: 1572–1580. [Google Scholar]
- Barau J., Teissandier A., Zamudio N., Roy S., Nalesso V., et al. , 2016. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354: 909–912. [DOI] [PubMed] [Google Scholar]
- Barbera J. P., Rodriguez T. A., Greene N. D., Weninger W. J., Simeone A., et al. , 2002. Folic acid prevents exencephaly in Cited2 deficient mice. Hum. Mol. Genet. 11: 283–293. [DOI] [PubMed] [Google Scholar]
- Bauer H., Schindler S., Charron Y., Willert J., Kusecek B., et al. , 2012. The nucleoside diphosphate kinase gene Nme3 acts as quantitative trait locus promoting non-Mendelian inheritance. PLoS Genet. 8: e1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M. A., Carmona-Gutierrez D., Ruckenstuhl C., Reisenbichler A., Megalou E. V., et al. , 2013. Spermidine promotes mating and fertilization efficiency in model organisms. Cell Cycle 12: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E., Wright G. J., 2016. Sperm meets egg: the genetics of mammalian fertilization. Annu. Rev. Genet. 50: 93–111. [DOI] [PubMed] [Google Scholar]
- Bianchi E., Doe B., Goulding D., Wright G. J., 2014. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508: 483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistulfi G., Diegelman P., Foster B. A., Kramer D. L., Porter C. W., et al. , 2009. Polyamine biosynthesis impacts cellular folate requirements necessary to maintain S-adenosylmethionine and nucleotide pools. FASEB J. 23: 2888–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistulfi G., Vandette E., Matsui S., Smiraglia D. J., 2010. Mild folate deficiency induces genetic and epigenetic instability and phenotype changes in prostate cancer cells. BMC Biol. 8: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzari F., Marston A. L., 2011. Cdc55 coordinates spindle assembly and chromosome disjunction during meiosis. J. Cell Biol. 193: 1213–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V., Davidson N. O., 2010. APOBEC-1-mediated RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2: 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V., Henderson J. O., Newberry E. P., Kennedy S., Luo J., et al. , 2005. Targeted deletion of the murine apobec-1 complementation factor (acf) gene results in embryonic lethality. Mol. Cell. Biol. 25: 7260–7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V., Park E., Schaefer S., Miller M., Lin Y., et al. , 2014. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 15: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom H. J., Shaw G. M., den Heijer M., Finnell R. H., 2006. Neural tube defects and folate: case far from closed. Nat. Rev. Neurosci. 7: 724–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D., Bestor T. H., 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99. [DOI] [PubMed] [Google Scholar]
- Bourc’his D., Voinnet O., 2010. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 330: 617–622. [DOI] [PubMed] [Google Scholar]
- Braden A. W., Austin C. R., 1954. The number of sperms about the eggs in mammals and its significance for normal fertilization. Aust. J. Biol. Sci. 7: 543–551. [DOI] [PubMed] [Google Scholar]
- Braden A. W., Austin C. R., David H. A., 1954. The reaction of zona pellucida to sperm penetration. Aust. J. Biol. Sci. 7: 391–409. [DOI] [PubMed] [Google Scholar]
- Braden A. W. H., 1958. Variation between strains of mice in phenomena associated with sperm penetration and fertilization. J. Genet. 56: 37–47. [Google Scholar]
- Bradford G. E., 1969. Genetic control of ovulation rate and embryo survival in mice. I. Response to selection. Genetics 61: 905–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. A., Warburton D., Brown L. Y., Yu C. Y., Roeder E. R., et al. , 1998. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 20: 180–183. [DOI] [PubMed] [Google Scholar]
- Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., et al. , 2014. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14: 838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante-Marin X., Garness J. A., Capel B., 2013. Testicular teratomas: an intersection of pluripotency, differentiation and cancer biology. Int. J. Dev. Biol. 57: 201–210. [DOI] [PubMed] [Google Scholar]
- Calandra R. S., Rulli S. B., Frungieri M. B., Suescun M. O., Gonzalez-Calvar S. I., 1996. Polyamines in the male reproductive system. Acta Physiol. Pharmacol. Ther. Latinoam. 46: 209–222. [PubMed] [Google Scholar]
- Carmel R., Jacobsen D., 2001. Homocysteine in Health and Disease. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Carouge D., Blanc V., Knoblaugh S. E., Hunter R. J., Davidson N. O., et al. , 2016. Parent-of-origin effects of A1CF and AGO2 on testicular germ-cell tumors, testicular abnormalities, and fertilization bias. Proc. Natl. Acad. Sci. USA 113: E5425–E5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino L., Comes S., Lambazzi G., De Stefano B., Filosa S., et al. , 2011. Control of embryonic stem cell metastability by L-proline catabolism. J. Mol. Cell Biol. 3: 108–122. [DOI] [PubMed] [Google Scholar]
- Castle W. E., Little C. C., 1910. On a modifier Mendelian ratio among yellow mice. Science 32: 868–870. [DOI] [PubMed] [Google Scholar]
- Chambon J. P., Touati S. A., Berneau S., Cladiere D., Hebras C., et al. , 2013. The PP2A inhibitor I2PP2A is essential for sister chromatid segregation in oocyte meiosis II. Curr. Biol. 23: 485–490. [DOI] [PubMed] [Google Scholar]
- Chang H. Y., Jennings P. C., Stewart J., Verrills N. M., Jones K. T., 2011. Essential role of protein phosphatase 2A in metaphase II arrest and activation of mouse eggs shown by okadaic acid, dominant negative protein phosphatase 2A, and FTY720. J. Biol. Chem. 286: 14705–14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Zheng W., Lin A., Uyhazi K., Zhao H., et al. , 2012. Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr. Biol. 22: 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zou T., McCormick S., 2016. S-Adenosylmethionine Synthetase 3 is important for pollen tube growth. Plant Physiol. 172: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiellini G., Erba P., Carnicelli V., Manfredi C., Frascarelli S., et al. , 2012. Distribution of exogenous [125I]-3-iodothyronamine in mouse in vivo: relationship with trace amine-associated receptors. J. Endocrinol. 213: 223–230. [DOI] [PubMed] [Google Scholar]
- Chmatal L., Gabriel S. I., Mitsainas G. P., Martinez-Vargas J., Ventura J., et al. , 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 24: 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C., Bunch D. O., Faure J. E., Goulding E. H., Eddy E. M., et al. , 1998. Fertilization defects in sperm from mice lacking fertilin beta. Science 281: 1857–1859. [DOI] [PubMed] [Google Scholar]
- Chow C. Y., Wolfner M. F., Clark A. G., 2010. The genetic basis for male x female interactions underlying variation in reproductive phenotypes of Drosophila. Genetics 186: 1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. G., 2002. Sperm competition and the maintenance of polymorphism. Heredity 88: 148–153. [DOI] [PubMed] [Google Scholar]
- Clark A. G., Begun D. J., Prout T., 1999. Female x male interactions in Drosophila sperm competition. Science 283: 217–220. [DOI] [PubMed] [Google Scholar]
- Clark A. T., Goldowitz D., Takahashi J. S., Vitaterna M. H., Siepka S. M., et al. , 2004. Implementing large-scale ENU mutagenesis screens in North America. Genetica 122: 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., et al. , 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J., 1988. Statistical Power Analysis for the Behavioral Sciences. Lawrence Erlbaum Associates, Hillsdale, NJ. [Google Scholar]
- Copp A. J., Stanier P., Greene N. D., 2013. Neural tube defects: recent advances, unsolved questions, and controversies. Lancet Neurol. 12: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J., Kimura M., 1970. An Introduction to Population Genetic Theory. Burgess Publ., Minneapolis. [Google Scholar]
- Crow J. F., 1979. Genes that violate Mendel’s rules. Sci. Am. 240: 134–143, 146. [DOI] [PubMed] [Google Scholar]
- Crow J. F., 1988. The ultraselfish gene. Genetics 118: 389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., 1991. Why is Mendelian segregation so exact? BioEssays 13: 305–312. [DOI] [PubMed] [Google Scholar]
- Cuenot L., 1905. Les races pures et leurs combinaisons chez les souris. Arch. de Zool. Exper. Generale 3: 123. [Google Scholar]
- Dadoune J. P., Siffroi J. P., Alfonsi M. F., 2004. Transcription in haploid male germ cells. Int. Rev. Cytol. 237: 1–56. [DOI] [PubMed] [Google Scholar]
- D’Aniello C., Fico A., Casalino L., Guardiola O., Di Napoli G., et al. , 2015. A novel autoregulatory loop between the Gcn2-Atf4 pathway and (L)-Proline [corrected] metabolism controls stem cell identity. Cell Death Differ. 22: 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J., 2016. Exacting requirements for development of the egg. N. Engl. J. Med. 374: 279–280. [DOI] [PubMed] [Google Scholar]
- Dell’Aquila M. E., De Santis T., Cho Y. S., Reshkin S. J., Caroli A. M., et al. , 2006. Localization and quantitative expression of the calcium-sensing receptor protein in human oocytes. Fertil. Steril. 85(Suppl. 1): 1240–1247. [DOI] [PubMed] [Google Scholar]
- De Santis T., Casavola V., Reshkin S. J., Guerra L., Ambruosi B., et al. , 2009. The extracellular calcium-sensing receptor is expressed in the cumulus-oocyte complex in mammals and modulates oocyte meiotic maturation. Reproduction 138: 439–452. [DOI] [PubMed] [Google Scholar]
- Detrait E. R., George T. M., Etchevers H. C., Gilbert J. R., Vekemans M., et al. , 2005. Human neural tube defects: developmental biology, epidemiology, and genetics. Neurotoxicol. Teratol. 27: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson M. E., Flenniken A. M., Ji X., Teboul L., Wong M. D., et al. , 2016. High-throughput discovery of novel developmental phenotypes. Nature 537: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion J. P., Morgan A. P., Clayshulte A. M., McMullan R. C., Yadgary L., et al. , 2015. A multi-megabase copy number gain causes maternal transmission ratio distortion on mouse chromosome 2. PLoS Genet. 11: e1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion J. P., Morgan A. P., Yadgary L., Bell T. A., McMullan R. C., et al. , 2016. R2d2 drives selfish sweeps in the house mouse. Mol. Biol. Evol. 33: 1381–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan P. J., 1998. The germ cell–the mother of all stem cells. Int. J. Dev. Biol. 42: 1043–1050. [PubMed] [Google Scholar]
- Dormeyer W., van Hoof D., Braam S. R., Heck A. J., Mummery C. L., et al. , 2008. Plasma membrane proteomics of human embryonic stem cells and human embryonal carcinoma cells. J. Proteome Res. 7: 2936–2951. [DOI] [PubMed] [Google Scholar]
- Dunn L. C., 1957. Evidence of evolutionary forces leading to the spread of lethal genes in wild populations of house mice. Proc. Natl. Acad. Sci. USA 43: 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant B. S., Eisen E. J., Ulberg L. C., 1980. Ovulation rate, embryo survival and ovarian sensitivity to gonadotrophins in mice selected for litter size and body weight. J. Reprod. Fertil. 59: 329–339. [DOI] [PubMed] [Google Scholar]
- East E. M., 1916. Inheritance in crosses between Nicotiana langsdorffii and Nicotiana alata. Genetics 1: 311–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. G., Gates A. H., 1959. Timing of the stages of the maturation divisions, ovulation, fertilization and the first cleavage of eggs of adult mice treated with gonadotrophins. J. Endocrinol. 18: 292–304. [DOI] [PubMed] [Google Scholar]
- Eisenbach M., 1999. Sperm chemotaxis. Rev. Reprod. 4: 56–66. [DOI] [PubMed] [Google Scholar]
- Eisenbach M., Ralt D., 1992. Precontact mammalian sperm-egg communication and role in fertilization. Am. J. Physiol. 262: C1095–C1101. [DOI] [PubMed] [Google Scholar]
- Ender C., Meister G., 2010. Argonaute proteins at a glance. J. Cell Sci. 123: 1819–1823. [DOI] [PubMed] [Google Scholar]
- Essien F. B., 1992. Maternal methionine supplementation promotes the remediation of axial defects in Axd mouse neural tube mutants. Teratology 45: 205–212. [DOI] [PubMed] [Google Scholar]
- Falconer D. S., 1960. The genetics of litter size in mice. J. Cell. Comp. Physiol. 56(Suppl. 1): 153–167. [DOI] [PubMed] [Google Scholar]
- Fazeli A., Affara N. A., Hubank M., Holt W. V., 2004. Sperm-induced modification of the oviductal gene expression profile after natural insemination in mice. Biol. Reprod. 71: 60–65. [DOI] [PubMed] [Google Scholar]
- Ferguson L., Agoulnik A. I., 2013. Testicular cancer and cryptorchidism. Front. Endocrinol. 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford. [Google Scholar]
- Fishman L., Willis J. H., 2005. A novel meiotic drive locus almost completely distorts segregation in mimulus (monkeyflower) hybrids. Genetics 169: 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel C., Vogel F., Hofreuter A., Schreiner B. S., Osthold S., et al. , 2015. Characterization of the Olfactory receptors expressed in human spermatozoa. Front. Mol. Biosci. 2: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forges T., Pellanda H., Diligent C., Monnier P., Guéant J. L., 2008. Do folates have an impact on fertility? Gynecol. Obstet. Fertil. 36: 930–939. [DOI] [PubMed] [Google Scholar]
- Franke B., Klootwijk R., Lemmers B., de Kovel C. G., Steegers-Theunissen R. P., et al. , 2003. Phenotype of the neural tube defect mouse model bent tail is not sensitive to maternal folinic acid, myo-inositol, or zinc supplementation. Birth Defects Res. A Clin. Mol. Teratol. 67: 979–984. [DOI] [PubMed] [Google Scholar]
- Furnes B., Schimenti J., 2007. Fast forward to new genes in mammalian reproduction. J. Physiol. 578: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgano A., Forrer M., Jaskiewicz L., Kanitz A., Zavolan M., et al. , 2008. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One 3: e3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell S. L., Reenan R. A., 2013. Mutations to the piRNA pathway component aubergine enhance meiotic drive of segregation distorter in Drosophila melanogaster. Genetics 193: 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemble S., Ahuja A., Buhagiar-Labarchede G., Onclercq-Delic R., Dairou J., et al. , 2015. Pyrimidine pool disequilibrium induced by a cytidine deaminase deficiency inhibits PARP-1 activity, leading to the under replication of DNA. PLoS Genet. 11: e1005384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain D. R., Li L., Hildebrandt M. R., Simmonds A. J., Hughes S. C., et al. , 2015. Loss of the Drosophila melanogaster DEAD box protein Ddx1 leads to reduced size and aberrant gametogenesis. Dev. Biol. 407: 232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenson S., 1928. A new sex-ratio abnormality in Drosophila obscura. Genetics 13: 488–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. D., Nakouzi G., Slowinska-Castaldo B., Dazard J. E., Rao J. S., et al. , 2010. Functional interactions between the LRP6 WNT co-receptor and folate supplementation. Hum. Mol. Genet. 19: 4560–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum M. P., Iwamori T., Buchold G. M., Matzuk M. M., 2011. Germ cell intercellular bridges. Cold Spring Harb. Perspect. Biol. 3: a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene N. D., Copp A. J., 1997. Inositol prevents folate-resistant neural tube defects in the mouse. Nat. Med. 3: 60–66. [DOI] [PubMed] [Google Scholar]
- Greene N. D., Copp A. J., 2005. Mouse models of neural tube defects: investigating preventive mechanisms. Am. J. Med. Genet. C. Semin. Med. Genet. 135C: 31–41. [DOI] [PubMed] [Google Scholar]
- Gu P., Qi X., Zhou Y., Wang Y., Gao X., 2012. Generation of Ppp2Ca and Ppp2Cb conditional null alleles in mouse. Genesis 50: 429–436. [DOI] [PubMed] [Google Scholar]
- Gueant J. L., Namour F., Gueant-Rodriguez R. M., Daval J. L., 2013. Folate and fetal programming: a play in epigenomics? Trends Endocrinol. Metab. 24: 279–289. [DOI] [PubMed] [Google Scholar]
- Hackett J. A., Surani M. A., 2014. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell 15: 416–430. [DOI] [PubMed] [Google Scholar]
- Halmekyto M., Hyttinen J. M., Sinervirta R., Utriainen M., Myohanen S., et al. , 1991. Transgenic mice aberrantly expressing human ornithine decarboxylase gene. J. Biol. Chem. 266: 19746–19751. [PubMed] [Google Scholar]
- Hamilton W. D., 1967. Extraordinary sex ratios. A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science 156: 477–488. [DOI] [PubMed] [Google Scholar]
- Hammoud S. S., Nix D. A., Zhang H., Purwar J., Carrell D. T., et al. , 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460: 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Liu Y., Wan G., Choi H. J., Zhao L., et al. , 2014. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep. 8: 1447–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargan-Calvopina J., Taylor S., Cook H., Hu Z., Lee S. A., et al. , 2016. Stage-specific demethylation in primordial germ cells safeguards against precocious differentiation. Dev. Cell 39: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney J. D., Lam M. Y., Michelson M. V., Nadeau J. H., 2008. Loss of the transmembrane but not the soluble kit ligand isoform increases testicular germ cell tumor susceptibility in mice. Cancer Res. 68: 5193–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney J. D., Michelson M. V., Youngren K. K., Lam M. Y., Nadeau J. H., 2009. Deletion of eIF2beta suppresses testicular cancer incidence and causes recessive lethality in agouti-yellow mice. Hum. Mol. Genet. 18: 1395–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleu Q., Gerard P. R., Montchamp-Moreau C., 2014. Sex chromosome drive. Cold Spring Harb. Perspect. Biol. 7: a017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleu Q., Gerard P. R., Dubruille R., Ogereau D., Prud’homme B., et al. , 2016. Rapid evolution of a Y-chromosome heterochromatin protein underlies sex chromosome meiotic drive. Proc. Natl. Acad. Sci. USA 113: 4110–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann B. G., Koschorz B., Wertz K., McLaughlin K. J., Kispert A., 1999. A protein kinase encoded by the t complex responder gene causes non-Mendelian inheritance. Nature 402: 141–146. [DOI] [PubMed] [Google Scholar]
- Hesse H., Hoefgen R., 2003. Molecular aspects of methionine biosynthesis. Trends Plant Sci. 8: 259–262. [DOI] [PubMed] [Google Scholar]
- Hildebrandt M. R., Germain D. R., Monckton E. A., Brun M., Godbout R., 2015. Ddx1 knockout results in transgenerational wild-type lethality in mice. Sci. Rep. 5: 9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Young S. G., Farese R. V., Jr, Ng J., Sande E., et al. , 1996. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. J. Biol. Chem. 271: 9887–9890. [DOI] [PubMed] [Google Scholar]
- Hoang K. P., Teo T. M., Ho T. X., Le V. S., 2016. Mechanisms of sex determination and transmission ratio distortion in Aedes aegypti. Parasit. Vectors 9: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J. B., 2017. Paramutation and related phenomena in diverse species. Nat. Rev. Genet. 18: 5–23. [DOI] [PubMed] [Google Scholar]
- Holliday R., 1984. The biological significance of meiosis. Symp. Soc. Exp. Biol. 38: 381–394. [PubMed] [Google Scholar]
- Holt W. V., Fazeli A., 2010. The oviduct as a complex mediator of mammalian sperm function and selection. Mol. Reprod. Dev. 77: 934–943. [DOI] [PubMed] [Google Scholar]
- Holt W. V., Fazeli A., 2015. Do sperm possess a molecular passport? Mechanistic insights into sperm selection in the female reproductive tract. Mol. Hum. Reprod. 21: 491–501. [DOI] [PubMed] [Google Scholar]
- Holt W. V., Fazeli A., 2016. Sperm selection in the female mammalian reproductive tract. Focus on the oviduct: hypotheses, mechanisms, and new opportunities. Theriogenology 85: 105–112. [DOI] [PubMed] [Google Scholar]
- Hosken D. J., Hodgson D. J., 2014. Why do sperm carry RNA? Relatedness, conflict, and control. Trends Ecol. Evol. 29: 451–455. [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M. H., Flaswinkel H., Fuchs H., Rathkolb B., Soewarto D., et al. , 2000. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 25: 444–447. [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M. H., Nicholson G., Selloum M., White J. K., Morgan H., et al. , 2015. Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat. Genet. 47: 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang X., Liu X., Cao S., Shan G., 2015. RNAi pathway participates in chromosome segregation in mammalian cells. Cell Discov. 1: 15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. C., Goldsby J. S., Arbab F., Melhem Z., Aleksic N., et al. , 2004. Oviduct prostacyclin functions as a paracrine factor to augment the development of embryos. Hum. Reprod. 19: 2907–2912. [DOI] [PubMed] [Google Scholar]
- Huang L. O., Labbe A., Infante-Rivard C., 2013. Transmission ratio distortion: review of concept and implications for genetic association studies. Hum. Genet. 132: 245–263. [DOI] [PubMed] [Google Scholar]
- Huang L. S., Voyiaziakis E., Markenson D. F., Sokol K. A., Hayek T., et al. , 1995. apo B gene knockout in mice results in embryonic lethality in homozygotes and neural tube defects, male infertility, and reduced HDL cholesterol ester and apo A-I transport rates in heterozygotes. J. Clin. Invest. 96: 2152–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. S., Voyiaziakis E., Chen H. L., Rubin E. M., Gordon J. W., 1996. A novel functional role for apolipoprotein B in male infertility in heterozygous apolipoprotein B knockout mice. Proc. Natl. Acad. Sci. USA 93: 10903–10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Kashiwagi K., 2015. Modulation of protein synthesis by polyamines. IUBMB Life 67: 160–169. [DOI] [PubMed] [Google Scholar]
- Ike A., Yamada S., Tanaka H., Nishimune Y., Nozaki M., 2002. Structure and promoter activity of the gene encoding ornithine decarboxylase antizyme expressed exclusively in haploid germ cells in testis (OAZt/Oaz3). Gene 298: 183–193. [DOI] [PubMed] [Google Scholar]
- Jablonka E., Lamb M. J., 1999. Epigenetic Inheritance and Evolution. Oxford University Press, Oxford. [Google Scholar]
- Jaenike J., 2001. Sex chromosome meiotic drive. Annu. Rev. Ecol. Syst. 32: 25–49. [Google Scholar]
- Joakimsen O., Baker R. L., 1977. Selection for litter size in mice. Acta Agric. Scand. 27: 301–318. [Google Scholar]
- Jodar M., Sendler E., Krawetz S. A., 2016. The protein and transcript profiles of human semen. Cell Tissue Res. 363: 85–96. [DOI] [PubMed] [Google Scholar]
- Johnson G. D., Mackie P., Jodar M., Moskovtsev S., Krawetz S. A., 2015. Chromatin and extracellular vesicle associated sperm RNAs. Nucleic Acids Res. 43: 6847–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juriloff D. M., Harris M. J., 2000. Mouse models for neural tube closure defects. Hum. Mol. Genet. 9: 993–1000. [DOI] [PubMed] [Google Scholar]
- Kahana C., 2016. Protein degradation, the main hub in the regulation of cellular polyamines. Biochem. J. 473: 4551–4558. [DOI] [PubMed] [Google Scholar]
- Kaneda M., Tang F., O’Carroll D., Lao K., Surani M. A., 2009. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A., Slanchev K., et al. , 2007. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131: 1273–1286. [DOI] [PubMed] [Google Scholar]
- Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J. A., Elkon R., et al. , 2010. A Pumilio-induced RNA structure switch in p27–3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 12: 1014–1020. [DOI] [PubMed] [Google Scholar]
- Khurana J. S., Xu J., Weng Z., Theurkauf W. E., 2010. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 6: e1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpelainen P. T., Saarimies J., Kontusaari S. I., Jarvinen M. J., Soler A. P., et al. , 2001. Abnormal ornithine decarboxylase activity in transgenic mice increases tumor formation and infertility. Int. J. Biochem. Cell Biol. 33: 507–520. [DOI] [PubMed] [Google Scholar]
- Kleinsmith L. J., Pierce G. B., Jr, 1964. Multipotentiality of single embryonal carcinoma cells. Cancer Res. 24: 1544–1551. [PubMed] [Google Scholar]
- Kofron M., Birsoy B., Houston D., Tao Q., Wylie C., et al. , 2007. Wnt11/beta-catenin signaling in both oocytes and early embryos acts through LRP6-mediated regulation of axin. Development 134: 503–513. [DOI] [PubMed] [Google Scholar]
- Koide Y., Ikenaga M., Sawamura N., Nishimoto D., Matsubara K., et al. , 2008. The evolution of sex-independent transmission ratio distortion involving multiple allelic interactions at a single locus in rice. Genetics 180: 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer D. L., Sufrin J. R., Porter C. W., 1987. Relative effects of S-adenosylmethionine depletion on nucleic acid methylation and polyamine biosynthesis. Biochem. J. 247: 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer D. L., Sufrin J. R., Porter C. W., 1988. Modulation of polyamine-biosynthetic activity by S-adenosylmethionine depletion. Biochem. J. 249: 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. Y., Heaney J. D., Youngren K. K., Kawasoe J. H., Nadeau J. H., 2007. Trans-generational epistasis between Dnd1Ter and other modifier genes controls susceptibility to testicular germ cell tumors. Hum. Mol. Genet. 16: 2233–2240. [DOI] [PubMed] [Google Scholar]
- Lambrot R., Xu C., Saint-Phar S., Chountalos G., Cohen T., et al. , 2013. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat. Commun. 4: 2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M., Robker R. L., Robertson S. A., 2014. Parenting from before conception. Science 345: 756–760. [DOI] [PubMed] [Google Scholar]
- Larracuente A. M., Presgraves D. C., 2012. The selfish segregation distorter gene complex of Drosophila melanogaster. Genetics 192: 33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Chen T. Y., Dhar S. S., Gu B., Chen K., et al. , 2016. A feedback loop comprising PRMT7 and miR-24–2 interplays with Oct4, Nanog, Klf4 and c-Myc to regulate stemness. Nucleic Acids Res. 44: 10603–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M., Dietmann S., Paramor M., Niwa H., Smith A., 2014. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell 14: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre P. L., Palin M. F., Murphy B. D., 2011. Polyamines on the reproductive landscape. Endocr. Rev. 32: 694–712. [DOI] [PubMed] [Google Scholar]
- Leibovich L., Mandel-Gutfreund Y., Yakhini Z., 2010. A structural-based statistical approach suggests a cooperative activity of PUM1 and miR-410 in human 3′-untranslated regions. Silence 1: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis W. H., Wright E. S., 1935. On the early development of the mouse egg. Carnegie Inst. Washington Publ. 459: 113–144. [Google Scholar]
- Licatalosi D. D., 2016. Roles of RNA-binding proteins and post-transcriptional regulation in driving male germ cell development in the mouse. Adv. Exp. Med. Biol. 907: 123–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Contreras A. J., Ramos-Molina B., Martinez-de-la-Torre M., Penafiel-Verdu C., Puelles L., et al. , 2009. Expression of antizyme inhibitor 2 in male haploid germinal cells suggests a role in spermiogenesis. Int. J. Biochem. Cell Biol. 41: 1070–1078. [DOI] [PubMed] [Google Scholar]
- Lord T., Aitken R. J., 2015. Fertilization stimulates 8-hydroxy-2′-deoxyguanosine repair and antioxidant activity to prevent mutagenesis in the embryo. Dev. Biol. 406: 1–13. [DOI] [PubMed] [Google Scholar]
- Luo Z., Gao X., Lin C., Smith E. R., Marshall S. A., et al. , 2015. Zic2 is an enhancer-binding factor required for embryonic stem cell specification. Mol. Cell 57: 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly L., Chan D., Aarabi M., Landry M., Behan N. A., et al. , 2017. Intergenerational impact of paternal lifetime exposures to both folic acid deficiency and supplementation on reproductive outcomes and imprinted gene methylation. Mol. Hum. Reprod. 23: 461–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F., 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37: 393–408. [DOI] [PubMed] [Google Scholar]
- Lyttle T. W., 1989. The effect of novel chromosome position and variable dose on the genetic behavior of the responder (Rsp) element of the segregation distorter (SD) system of Drosophila melanogaster. Genetics 121: 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyttle T. W., 1993. Cheaters sometimes prosper: distortion of Mendelian segregation by meiotic drive. Trends Genet. 9: 205–210. [DOI] [PubMed] [Google Scholar]
- Mak W., Fang C., Holden T., Dratver M. B., Lin H., 2016. An important role of Pumilio 1 in regulating the development of the mammalian female germline. Biol. Reprod. 94: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marean A., Graf A., Zhang Y., Niswander L., 2011. Folic acid supplementation can adversely affect murine neural tube closure and embryonic survival. Hum. Mol. Genet. 20: 3678–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews H. R., 1993. Polyamines, chromatin structure and transcription. BioEssays 15: 561–566. [DOI] [PubMed] [Google Scholar]
- McCarthy J. C., 1967. The effects of inbreeding on the components of litter size in mice. Genet. Res. 10: 73–80. [DOI] [PubMed] [Google Scholar]
- McKenzie A. J., Hoshino D., Hong N. H., Cha D. J., Franklin J. L., et al. , 2016. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 15: 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan T. F., Conte N., West D. B., Jacobsen J. O., Mason J., et al. , 2017. Disease model discovery from 3,328 gene knockouts by The International Mouse Phenotyping Consortium. Nat. Genet. 49: 1231–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza F. J., Perez-Marin C. C., Garcia-Marin L., Madueno J. A., Henley C., et al. , 2012. Localization, distribution, and function of the calcium-sensing receptor in sperm. J. Androl. 33: 96–104. [DOI] [PubMed] [Google Scholar]
- Miles W. O., Tschop K., Herr A., Ji J. Y., Dyson N. J., 2012. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 26: 356–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Fleming L., Olin-Sandoval V., Campbell K., Ralser M., 2015. Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 427: 3389–3406. [DOI] [PubMed] [Google Scholar]
- Montagutelli X., Turner R., Nadeau J. H., 1996. Epistatic control of non-Mendelian inheritance in mouse interspecific crosses. Genetics 143: 1739–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Esteves C., Shin D., 2013. Nutrient supplementation: improving male fertility fourfold. Semin. Reprod. Med. 31: 293–300. [DOI] [PubMed] [Google Scholar]
- Morgan T. H., Bridges C. B., Sturtevant A. H., 1925. The genetics of Drosophila. Bibliogr. Genet. 2: 1–262. [Google Scholar]
- Morita S., Horii T., Kimura M., Goto Y., Ochiya T., et al. , 2007. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics 89: 687–696. [DOI] [PubMed] [Google Scholar]
- Nakouzi G. A., Nadeau J. H., 2014. Does dietary folic acid supplementation in mouse NTD models affect neural tube development or gamete preference at fertilization? BMC Genet. 15: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita R., Takahasi K., Murakami E., Hirano E., Yamamoto S. P., et al. , 2014. A novel function of human Pumilio proteins in cytoplasmic sensing of viral infection. PLoS Pathog. 10: e1004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson V. R., Heaney J. D., Tesar P. J., Davidson N. O., Nadeau J. H., 2012. Transgenerational epigenetic effects of the Apobec1 cytidine deaminase deficiency on testicular germ cell tumor susceptibility and embryonic viability. Proc. Natl. Acad. Sci. USA 109: E2766–E2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieder G. L., Macon G. R., 1987. Uterine and oviducal protein secretion during early pregnancy in the mouse. J. Reprod. Fertil. 81: 287–294. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Nakatsu F., Kashiwagi K., Ohno H., Saito T., et al. , 2002. Essential role of S-adenosylmethionine decarboxylase in mouse embryonic development. Genes Cells 7: 41–47. [DOI] [PubMed] [Google Scholar]
- Ohto U., Ishida H., Krayukhina E., Uchiyama S., Inoue N., et al. , 2016. Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization. Nature 534: 566–569. [DOI] [PubMed] [Google Scholar]
- Oliveros-Etter M., Li Z., Nee K., Hosohama L., Hargan-Calvopina J., et al. , 2015. PGC reversion to pluripotency involves erasure of DNA methylation from imprinting control centers followed by locus-specific re-methylation. Stem Cell Rep. 5: 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolini C. S., Newnham L. J., Capalbo A., Natesan S. A., Joshi H. A., et al. , 2015. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat. Genet. 47: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Chen X., Tong X., Tang C., Li J., 2015. Ppp2ca knockout in mice spermatogenesis. Reproduction 149: 385–391. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F., Sapienza C., 2001. Nonrandom segregation during meiosis: the unfairness of females. Mamm. Genome 12: 331–339. [DOI] [PubMed] [Google Scholar]
- Pek J. W., Kai T., 2011a DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc. Natl. Acad. Sci. USA 108: 12007–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek J. W., Kai T., 2011b A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr. Biol. 21: 39–44. [DOI] [PubMed] [Google Scholar]
- Persson L., 2009. Polyamine homoeostasis. Essays Biochem. 46: 11–24. [DOI] [PubMed] [Google Scholar]
- Pierce G. B., Gramzinski R. A., Parchment R. E., 1990. Amine oxidases, programmed cell death, and tissue renewal. Philos. Trans. R. Soc. Lond. B Biol. Sci. 327: 67–74. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S., Dimitri P., 1989. Cytogenetic analysis of segregation distortion in Drosophila melanogaster: the cytological organization of the Responder (Rsp) locus. Genetics 121: 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow J., Jurkin J., Schleiffer A., Martinez J., 2014. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature 511: 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z. U., Tsai Y. H., Steinberger A., Lu M., Greenfield A. R., et al. , 1985. Localization of ornithine decarboxylase in rat testicular cells and epididymal spermatozoa. Biol. Reprod. 33: 1189–1195. [DOI] [PubMed] [Google Scholar]
- Reik W., Surani M. A., 2015. Germline and pluripotent stem cells. Cold Spring Harb. Perspect. Biol. 7: a019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. M., Vilkomerson H., 1942. On the anaphase movement of chromosomes. Proc. Natl. Acad. Sci. USA 28: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. R., 2014. An X-linked sex ratio distorter in Drosophila simulans that kills or incapacitates both noncarrier sperm and sons. G3 (Bethesda) 4: 1837–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijlaarsdam M. A., Looijenga L. H., 2014. An oncofetal and developmental perspective on testicular germ cell cancer. Semin. Cancer Biol. 29: 59–74. [DOI] [PubMed] [Google Scholar]
- Roje S., 2006. S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67: 1686–1698. [DOI] [PubMed] [Google Scholar]
- Rubinstein S., Breitbart H., 1991. Role of spermine in mammalian sperm capacitation and acrosome reaction. Biochem. J. 278: 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein S., Lax Y., Shalev Y., Breitbart H., 1995. Dual effect of spermine on acrosomal exocytosis in capacitated bovine spermatozoa. Biochim. Biophys. Acta 1266: 196–200. [DOI] [PubMed] [Google Scholar]
- Saitou M., Yamaji M., 2012. Primordial germ cells in mice. Cold Spring Harb. Perspect. Biol. 4: a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Sezutsu H., Takahasi K. R., Tsuchihashi K., Ichikawa R., et al. , 2005. Molecular characterization of ENU mouse mutagenesis and archives. Biochem. Biophys. Res. Commun. 336: 609–616. [DOI] [PubMed] [Google Scholar]
- Salbaum J. M., Kappen C., 2012. Genetic and epigenomic footprints of folate. Prog. Mol. Biol. Transl. Sci. 108: 129–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C. G., Teixeira F. K., Czech B., Preall J. B., Zamparini A. L., et al. , 2016. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell 18: 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L., Novitski E., 1957. Meiotic drive as an evolutionary force. Am. Nat. 91: 105–110. [Google Scholar]
- Sasaki M., Ohnishi M., Tashiro F., Niwa H., Suzuki A., et al. , 2007. Disruption of the mouse protein Ser/Thr phosphatase 2Cbeta gene leads to early pre-implantation lethality. Mech. Dev. 124: 489–499. [DOI] [PubMed] [Google Scholar]
- Schaefer S., Nadeau J. H., 2015. The genetics of epigenetic inheritance: modes, molecules and mechanisms. Q. Rev. Biol. 90: 381–415. [DOI] [PubMed] [Google Scholar]
- Schuster A., Tang C., Xie Y., Ortogero N., Yuan S., et al. , 2016. SpermBase: a database for Sperm-Borne RNA contents. Biol. Reprod. 95: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield V. L., Schlumpberger J. M., West L. A., Weissman I. L., 1982. Protochordate allorecognition is controlled by a MHC-like gene system. Nature 295: 499–502. [DOI] [PubMed] [Google Scholar]
- Senti K. A., Brennecke J., 2010. The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet. 26: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U., Conine C. C., Shea J. M., Boskovic A., Derr A. G., et al. , 2016. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima J. E., McLean D. J., McCarrey J. R., Griswold M. D., 2004. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 71: 319–330. [DOI] [PubMed] [Google Scholar]
- Shohat B., Maayan R., Singer R., Sagiv M., Kaufman H., et al. , 1990. Immunosuppressive activity and polyamine levels of seminal plasma in azo-ospermic, oligospermic, and normospermic men. Arch. Androl. 24: 41–50. [DOI] [PubMed] [Google Scholar]
- Singer J. B., Hill A. E., Burrage L. C., Olszens K. R., Song J., et al. , 2004. Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304: 445–448. [DOI] [PubMed] [Google Scholar]
- Skakkebaek N. E., Rajpert-De Meyts E., Main K. M., 2001. Testicular dysgenesis syndrome: an increasingly common developmental disorder with environmental aspects. Hum. Reprod. 16: 972–978. [DOI] [PubMed] [Google Scholar]
- Skakkebaek N. E., Rajpert-De Meyts E., Buck Louis G. M., Toppari J., Andersson A. M., et al. , 2016. Male reproductive disorders and fertility trends: influences of environment and genetic susceptibility. Physiol. Rev. 96: 55–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithells R. W., Sheppard S., Schorah C. J., Seller M. J., Nevin N. C., et al. , 1980. Possible prevention of neural-tube defects by periconceptional vitamin supplementation. Lancet 1: 339–340. [DOI] [PubMed] [Google Scholar]
- Snyder E. M., Licht K., Braun R. E., 2017a Testicular adenosine to inosine RNA editing in the mouse is mediated by ADARB1. Biol. Reprod. 96: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder E. M., McCarty C., Mehalow A., Svenson K. L., Murray S. A., et al. , 2017b APOBEC1 complementation factor (A1CF) is dispensable for C-to-U RNA editing in vivo. RNA 23: 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler D. C., Kadunganattil S., Ramdas S., Myers K., Roca J., et al. , 2009. Expression of transgenic PPP1CC2 in the testis of Ppp1cc-null mice rescues spermatid viability and spermiation but does not restore normal sperm tail ultrastructure, sperm motility, or fertility. Biol. Reprod. 81: 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein P., Rozhkov N. V., Li F., Cardenas F. L., Davydenko O., et al. , 2015. Essential role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet. 11: e1005013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L. C., 1967. Origin of testicular teratomas from primordial germ cells in mice. J. Natl. Cancer Inst. 38: 549–552. [PubMed] [Google Scholar]
- Stevens L. C., 1973. A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J. Natl. Cancer Inst. 50: 235–242. [DOI] [PubMed] [Google Scholar]
- Stover P. J., 2011. Polymorphisms in 1-carbon metabolism, epigenetics and folate-related pathologies. J. Nutrigenet. Nutrigenomics 4: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong L. C., Hollander W. F., 1949. Hereditary Looptail in the house mouse. J. Hered. 40: 329–334. [Google Scholar]
- Su Y. Q., Sugiura K., Sun F., Pendola J. K., Cox G. A., et al. , 2012a MARF1 regulates essential oogenic processes in mice. Science 335: 1496–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. Q., Sun F., Handel M. A., Schimenti J. C., Eppig J. J., 2012b Meiosis arrest female 1 (MARF1) has nuage-like function in mammalian oocytes. Proc. Natl. Acad. Sci. USA 109: 18653–18660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subasic D., Stoeger T., Eisenring S., Matia-Gonzalez A. M., Imig J., et al. , 2016. Post-transcriptional control of executioner caspases by RNA-binding proteins. Genes Dev. 30: 2213–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Wollin A., Stephen A. M., 2002. Moderate folate deficiency influences polyamine synthesis in rats. J. Nutr. 132: 2632–2637. [DOI] [PubMed] [Google Scholar]
- Suzuki A., Niimi Y., Shinmyozu K., Zhou Z., Kiso M., et al. , 2016. Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development. EMBO Rep. 17: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., He Y., Kashiwagi K., Murakami Y., Hayashi S., et al. , 1994. Antizyme protects against abnormal accumulation and toxicity of polyamines in ornithine decarboxylase-overproducing cells. Proc. Natl. Acad. Sci. USA 91: 8930–8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Araripe L., Kingan S. B., Ke Y., Xiao H., et al. , 2007a A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol. 5: e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Masly J. P., Araripe L., Ke Y., Hartl D. L., 2007b A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biol. 5: e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuhiro K., Isotani A., Yokota S., Yano Y., Oshio S., et al. , 2009. OAZ-t/OAZ3 is essential for rigid connection of sperm tails to heads in mouse. PLoS Genet. 5: e1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torban E., Iliescu A., Gros P., 2012. An expanding role of Vangl proteins in embryonic development. Curr. Top. Dev. Biol. 101: 237–261. [DOI] [PubMed] [Google Scholar]
- Tosaka Y., Tanaka H., Yano Y., Masai K., Nozaki M., et al. , 2000. Identification and characterization of testis specific ornithine decarboxylase antizyme (OAZ-t) gene: expression in haploid germ cells and polyamine-induced frameshifting. Genes Cells 5: 265–276. [DOI] [PubMed] [Google Scholar]
- Turner B. C., Perkins D. D., 1979. Spore killer, a chromosomal factor in Neurospora that kills meiotic products not containing it. Genetics 93: 587–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J., Schagat T. L., Hrit J., Weidmann C. A., Brumbaugh J., et al. , 2012. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 287: 36370–36383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veron N., Bauer H., Weisse A. Y., Luder G., Werber M., et al. , 2009. Retention of gene products in syncytial spermatids promotes non-Mendelian inheritance as revealed by the t complex responder. Genes Dev. 23: 2705–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinijsanun A., Martin L., 1991. Effect of early ovariectomy and steroid hormone replacement of embryo transport, development and implantation in mice. Reprod. Fertil. Dev. 3: 35–50. [DOI] [PubMed] [Google Scholar]
- Waberski D., Dohring A., Ardon F., Ritter N., Zerbe H., et al. , 2006. Physiological routes from intra-uterine seminal contents to advancement of ovulation. Acta Vet. Scand. 48: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace H. M., Fraser A. V., Hughes A., 2003. A perspective of polyamine metabolism. Biochem. J. 376: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., et al. , 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543. [DOI] [PubMed] [Google Scholar]
- Wedekind C., Chapuisat M., Macas E., Rulicke T., 1996. Non-random fertilization in mice correlates with the MHC and something else. Heredity (Edinb) 77: 400–409. [DOI] [PubMed] [Google Scholar]
- Wei K. H., Reddy H. M., Rathnam C., Lee J., Lin D., et al. , 2017. A pooled sequencing approach identifies a candidate meiotic driver in Drosophila. Genetics 206: 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. K., Gerdin A. K., Karp N. A., Ryder E., Buljan M., et al. , 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154: 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner M. F., 2002. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity (Edinb) 88: 85–93. [DOI] [PubMed] [Google Scholar]
- Wolukau J. N., Zhang S. L., Xu G. H., Chen D., 2004. The effect of temperature, polyamines and polyamine synthesis inhibitor on in vitro pollen germination and pollen tube growth of Prunus mume. Sci. Hortic. 99: 289–299. [Google Scholar]
- Wright G. J., Bianchi E., 2016. The challenges involved in elucidating the molecular basis of sperm-egg recognition in mammals and approaches to overcome them. Cell Tissue Res. 363: 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Hao L., Han Z., Gao S., Latham K. E., et al. , 2005. Maternal transmission ratio distortion at the mouse Om locus results from meiotic drive at the second meiotic division. Genetics 170: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie A., Jones A. E., Abrams J. M., 2016a p53 in the game of transposons. BioEssays 38: 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie A., Jones A. E., D’Brot A., Lu W. J., Kurtz P., et al. , 2016b p53 genes function to restrain mobile elements. Genes Dev. 30: 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren K. K., Coveney D., Peng X., Bhattacharya C., Schmidt L. S., et al. , 2005. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435: 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S. E., Malik H. S., 2015. Chromosome segregation: human female meiosis breaks all the rules. Curr. Biol. 25: R654–R656. [DOI] [PubMed] [Google Scholar]
- Zechel J. L., Doerner S. K., Lager A., Tesar P. J., Heaney J. D., et al. , 2013. Contrasting effects of Deadend1 (Dnd1) gain and loss of function mutations on allelic inheritance, testicular cancer, and intestinal polyposis. BMC Genet. 14: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhu T., Chen Y., Xu E. Y., 2015. Loss of preimplantation embryo resulting from a Pum1 gene trap mutation. Biochem. Biophys. Res. Commun. 462: 8–13. [DOI] [PubMed] [Google Scholar]
- Zhang D., Zhao T., Ang H. S., Chong P., Saiki R., et al. , 2012. AMD1 is essential for ESC self-renewal and is translationally down-regulated on differentiation to neural precursor cells. Genes Dev. 26: 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gao F., Fu J., Zhang P., Wang Y., et al. , 2017. Systematic identification and characterization of long non-coding RNAs in mouse mature sperm. PLoS One 12: e0173402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Goh K. J., Ng H. H., Vardy L. A., 2012. A role for polyamine regulators in ESC self-renewal. Cell Cycle 11: 4517–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Wang Y., He L., Huang G., Du Y., et al. , 2015. ZIC2-dependent OCT4 activation drives self-renewal of human liver cancer stem cells. J. Clin. Invest. 125: 3795–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohn I. E., 2012. Mouse as a model for multifactorial inheritance of neural tube defects. Birth Defects Res. C Embryo Today 96: 193–205. [DOI] [PubMed] [Google Scholar]
- Zucchi R., Chiellini G., Scanlan T. S., Grandy D. K., 2006. Trace amine-associated receptors and their ligands. Br. J. Pharmacol. 149: 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo H., Zhang J., Zhang L., Ren X., Chen X., et al. , 2016. Transcriptomic variation during spermiogenesis in mouse germ cells. PLoS One 11: e0164874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka T. P., Thomson J. A., 2005. A germ cell origin of embryonic stem cells? Development 132: 227–233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.