Abstract
Mutants remain a powerful means for dissecting gene function in model organisms such as Caenorhabditis elegans. Massively parallel sequencing has simplified the detection of variants after mutagenesis but determining precisely which change is responsible for phenotypic perturbation remains a key step. Genetic mapping paradigms in C. elegans rely on bulk segregant populations produced by crosses with the problematic Hawaiian wild isolate and an excess of redundant information from whole-genome sequencing (WGS). To increase the repertoire of available mutants and to simplify identification of the causal change, we performed WGS on 173 temperature-sensitive (TS) lethal mutants and devised a novel mapping method. The mapping method uses molecular inversion probes (MIP-MAP) in a targeted sequencing approach to genetic mapping, and replaces the Hawaiian strain with a Million Mutation Project strain with high genomic and phenotypic similarity to the laboratory wild-type strain N2. We validated MIP-MAP on a subset of the TS mutants using a competitive selection approach to produce TS candidate mapping intervals with a mean size < 3 Mb. MIP-MAP successfully uses a non-Hawaiian mapping strain and multiplexed libraries are sequenced at a fraction of the cost of WGS mapping approaches. Our mapping results suggest that the collection of TS mutants contains a diverse library of TS alleles for genes essential to development and reproduction. MIP-MAP is a robust method to genetically map mutations in both viable and essential genes and should be adaptable to other organisms. It may also simplify tracking of individual genotypes within population mixtures.
Keywords: genetic mapping, Caenorhabditis elegans, temperature-sensitive mutations, molecular inversion probes, massively multiplex sequencing
The Million Mutation Project, a collection of genomic data for thousands of mutagenized Caenorhabditis elegans strains (Thompson et al. 2013), has provided a dense library of mutant alleles with which to study gene function and has been widely used since its release. However, given that such strains generally contain only homozygous viable mutations, they largely exclude strongly deleterious alleles of essential genes. Such genes encompass a range of classes that include roles in cell division, development, and fertility; key components to all multicellular organisms. Temperature-sensitive (TS) lethal alleles can facilitate the genetic analysis of essential genes through the conditional modulation of their function without the complications that balancer chromosomes can introduce when present in nonconditional lethal strains (Golden et al. 2000; O’Rourke et al. 2011a; Lowry et al. 2015). There are several reported screens for TS lethal alleles in C. elegans, but to date there are only a small portion of genes with TS alleles identified (Zonies et al. 2010; Ehmke et al. 2014; Lowry et al. 2015). Generating a comprehensive library of mutant strains with conditional lethal phenotypes has the potential to expand our knowledge of essential genes, their required levels of expression, the timing of their function(s), and the details of their protein and domain structure. Furthermore, such a catalog could have broad implications for elucidating the life cycle of C. elegans and other organisms. However, such an endeavor requires the mapping and characterization of mutant alleles in a systematic and high-throughput manner.
Chemical mutagenesis methods to generate TS alleles typically result in up to several hundred mutations per strain (Sarin et al. 2008; Flibotte et al. 2010; Thompson et al. 2013). Determining which of these molecular alterations is responsible for the TS mutant phenotype is especially challenging when dealing with partially penetrant or leaky alleles. Mutations that alter protein-coding sequence can be initially prioritized as candidates, but even after such filtering, > 1/5 of the collection’s single-nucleotide variants (SNVs) remain. In contrast to identifying constitutive loss-of-function alleles—such as deletions, frameshifts, and nonsense mutations—there is, as yet, no universal predictive method for recognizing which coding or perhaps noncoding molecular change is most likely to give rise to a TS phenotype (Perry et al. 1994; Rogalski et al. 1995; Harfe et al. 1998; Poultney et al. 2011). When feasible, mutants are initially outcrossed or backcrossed to remove extraneous mutations originating from the mutagenesis process. Otherwise, these additional mutations may, to some degree, affect development or other essential pathways, thus obfuscating the process of allele characterization.
Genetic mapping is a general method for identifying the causative mutation in such strains. Mapping with classic visible genetic markers in C. elegans can be a laborious, iterative process. Single-nucleotide polymorphisms (SNPs) between strains, such as the N2 laboratory reference strain and the Hawaiian strain CB4856, act as molecular genetic markers that allow the parallel mapping of multiple sites across the genome in a single cross. The Snip-SNP assay is a popular method exploiting a subset of Hawaiian genome SNVs that alter recognition sites of the restriction enzyme DraI (Davis et al. 2005). However, the Hawaiian genome is proven to harbor various alleles (De Bono and Bargmann 1998; Seidel et al. 2008, 2011; Andersen et al. 2014) that can negatively alter the representation of segregant populations; CB4856 also has phenotypes of its own that may interfere with the scoring of some behavioral phenotypes (Wicks et al. 2001). Despite these issues, the research community continues to leverage CB4856 as a mapping strain to develop new methods of mapping complex mutations (Doitsidou et al. 2010; O’Rourke et al. 2011b; Minevich et al. 2012; Smith et al. 2016).
More recently, approaches and tools have been developed that allow the simultaneous measurement of multiple SNV frequencies in a cross-population. Included among these are a combined-step whole-genome sequencing (WGS) and SNP analysis complemented by analysis via the CloudMap system as well as restriction site-associated DNA polymorphism mapping (Doitsidou et al. 2010; O’Rourke et al. 2011b; Minevich et al. 2012). At their core, these mapping methods still rely on the basic principles of bulk segregant analysis but now benefit from using massively parallel sequencing to examine molecular markers across the genome in a single sequencing library. As currently implemented, these mapping methods use WGS to a depth of 18–40× or more on populations generated from 20 to 50 F2 mutant phenotype animals to capture data on both the recombinant genome landscape and the associated mutant allele (Doitsidou et al. 2010; Wang et al. 2014; Jaramillo-Lambert et al. 2015; Lowry et al. 2015). On a large scale, mapping mutant strains is a labor-intensive exercise. The large amount of sequence generation, the use of the Hawaiian strain (with its limitations), and the issues with scaling to high-throughput suggest there is room for improvement.
In particular, molecular inversion probes (Turner et al. 2009) (MIPs) allow high-throughput, target-based genome amplification and sequencing. A single MIP targets a region via a pair of complementary annealing arms. Once annealed, the genomic sequence between arms, also known as the gap-fill sequence, is copied by a DNA polymerase and the entire single-stranded probe is circularized by ligase. Each circularized probe represents a unique strand of DNA that can now be linearized and sequenced. MIPs have proven very useful in targeted exome sequencing libraries (Mamanova et al. 2010; Kiezun et al. 2012) and in screening samples for subpopulations of variants. Recently, Hiatt et al. (2013) introduced single-molecule molecular inversion probes (smMIPs) containing the addition of a unique molecular identifier (UMI) for use on genomically diverse samples to differentiate between low-frequency variants and those contributed by sequencing or amplification artifacts (Hiatt et al. 2013).
In complement to the Million Mutation Project collection, we have sequenced a group of 173 previously uncharacterized mutants obtained from multiple screens for TS lethality, which reveals a rich catalog of genomic variants. To identify the specific mutation(s) underlying these TS phenotypes, we developed an alternative strategy to WGS genetic mapping that exploits MIPs to specifically assay only regions of the genome containing SNVs of interest. To avoid the pitfalls associated with the Hawaiian strain, we identified a polymorphic mapping strain from the Million Mutation Project that resembles N2 in movement and growth but has < 300 SNVS across the genome. In pilot experiments, we demonstrate the utility of our MIP-based method (MIP-MAP) in generating high-resolution genomic interval maps for C. elegans mutant alleles. We further developed a competitive fitness approach to map the associated genomic intervals for a subset of our TS lethal mutants. In crosses with our mapping strain we use nonpermissive temperatures to select against TS lethal homozygotes over several generations to identify candidate intervals and query our sequencing data to produce a list of candidate mutations. Our studies identify TS alleles for a variety of novel genes not previously classified as essential to development or reproduction, suggesting our collection can be a useful resource for studying essential genes.
Materials and Methods
TS strain isolation and sequencing
Set 1 strains originated from the Bowerman laboratory and were isolated from an ENU mutagenesis protocol (Kemphues et al. 1988b) in EU1700, a lin-2(e1309) background with an integrated pie-1-driven series of GFP fusions to β-tubulin, histone 2B, and a PH domain. Mutagenized animals were screened for conditional TS embryonic lethality by shifting F2 animals as L4s from 15 to 26° for 24–36 hr and identifying those with an accumulation of dead embryos. These F2 animals were then shifted back to 15° with the intention of isolating any strains that managed to recover and produce some progeny. Recovered strains were then grown at 15° for a few generations before rescreening at 26° to confirm for TS lethality.
Set 2 strains originated from the Schnabel laboratory and were isolated from EMS mutagenesis (Brenner 1974; Ehmke et al. 2014) in a wild-type (N2) background. After 8 days of growth at 15°, animals were singled to 96-well plates and visually screened for TS lethality at 25°. Positive wells were regrouped to new 96-well plates twice before testing a second time at 25°. Second-round positive strains were plated on individual NGM plates before confirming TS lethality a third time at the nonpermissive temperature followed by detailed phenotype analysis.
Set 3 strains originated from the Seydoux laboratory and were isolated from EMS mutagenesis (Kemphues et al. 1988b; Golden et al. 2000) in a JH50 background (him-3(e1147)IV; lin-2(e1309) axIs36). F2 animals were screened by bleach synchronization of F1 gravid adults followed by upshift at L4 from 15 to 25° for 20 hr and then down to 15° for an additional 20 hr. F2 animals accumulating dead embryos were singled to NGM plates at 15° and examined 3 days later for F3 progeny, indicating the presence of a maternal effect embryonic TS lethal mutation.
All 173 strains were sequenced and analyzed using the same methods and custom pipelines described previously for the Million Mutation Project (Thompson et al. 2013). Reads were aligned to build WS230 of the C. elegans genome and SNV calls for each strain can be found in Supplemental Material, File S2.
MIP-MAP design
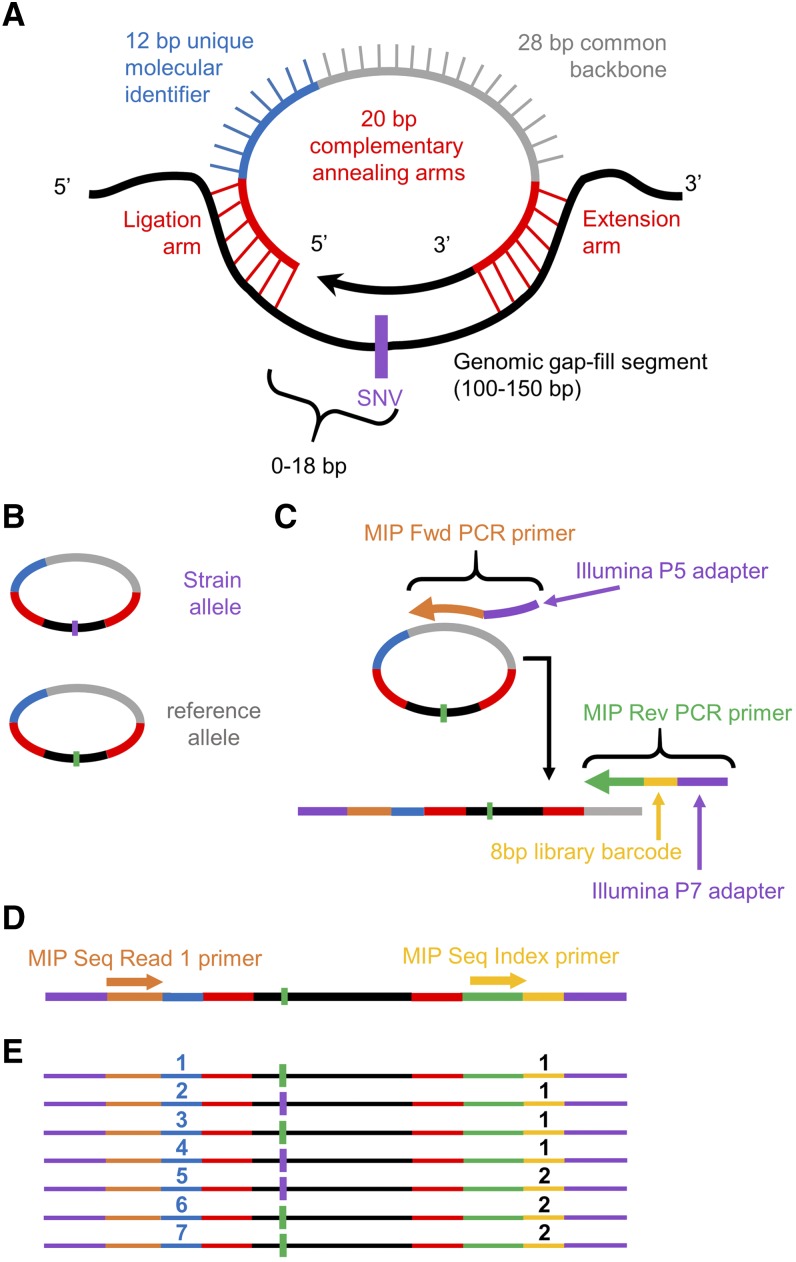
The smMIP design for these experiments was altered from those presented in Hiatt et al. (2013), by placing the 12-bp UMI directly on the 3′-end of the first-read primer sequence. This alteration allows MIP sequencing to proceed in a single-end 50-bp read that includes the UMI, ligation arm sequence, and 18 bp of gap-fill sequence. MIP annealing arms were designed using a custom R script that generated multiple combinations of lengths and locations for the ligation and extension arms. Ligation arm locations were limited to a maximum distance of 18 bp either up- or downstream from the SNV of interest. From these possible variations, the optimal MIP sequences were then chosen based on criteria as described elsewhere (Turner et al. 2009; O’Roak et al. 2012). MIP sequence information for VC20019 probes can be found in File S3.
MIP capture protocol
MIP pools were prepared as described (Turner et al. 2009). Briefly, equimolar amounts of MIPs from 100 µM concentrations were pooled and 85 µl of this pool was treated with 50 units of polynucleotide kinase (from New England Biolabs, Beverly, MA) for 45 min at 37° and then 20 min at 80° in a 100-µl reaction. The 5′-phosphorylated probes were diluted to 330 nM for use in later steps. MIP libraries were based on Hiatt et al. (2013). Annealing reactions containing 500 ng of target genomic DNA, 330 fM of MIP pool, and 1× Ampligase buffer (Epicentre) in 10 µl were treated for 3 min at 98°, 30 min at 85°, 60 min at 60°, and 120 min at 56°. To gap-fill the product, 300 pM dNTPs, 7.5 µM Betaine (Sigma [Sigma Chemical], St. Louis, MO), 20 nM NAD+, 1× Ampligase buffer (Epicentre), 5 units Ampligase, and 2 units Phusion DNA polymerase (New England Biolabs) were added to the 10 µl anneal reaction and incubated for 120 min at 56°, and 20 min at 72°. To degrade genomic template and any remaining linear MIPs, 20 units Exonuclease I (New England Biolabs) and 50 units Exonuclease III (New England Biolabs) were added and incubated for 45 min at 37°, and 20 min at 80°. Then, 10 µl of this capture reaction was amplified by 18 rounds of PCR (15 sec at 98°, 15 sec at 65°, and 45 sec at 72°) with 1 unit Kapa Hifi Hotstart TAQ, 10 nM dNTPs, and 25 pM each of forward and reverse primers in a 50 µl reaction. Libraries were then size-selected between 250 and 450 bp and purified with Agencourt AMPure XP beads before sequencing with Illumina sequencing technology.
MIP sequencing analysis
Sequencing data for each library was analyzed using custom scripts written in R. Briefly, each sequenced library was analyzed for exact matches to the expected MIP ligation arm sequences at their predicted positions within the read. This subset of matched ligation arm reads was then analyzed for specific matches to the expected reference or mutant gap-fill sequence. This set was then trimmed by filtering the quality score at the specific SNV positions to include only those with Phred score ≥ Q30. In addition, any duplicated UMIs from this set were combined to produce a single consensus SNV read. If a duplicated UMI could not find a majority agreement across gap-fill reads, it was removed from further analysis. This uniquely identified final set was used to count reference and SNV representation to produce a percentage calculated using SNV / (reference + SNV) counts.
Normalization of MIP data
Each MIP was analyzed at multiple dilutions to generate a series of curves to account for the difference between any single raw SNV-MIP fraction estimate and the average of the combined SNV-MIP calls at each site. A Bayesian statistical model approximating the observed offset curve for each MIP was inferred using a Markov Chain Monte Carlo technique. Height (h), variance (σ), mean (µ), and offset (o) represent the parameters for the best fitting Gaussian curve. The mode of these parameters (File S4) was then used to readjust raw SNV values to within the expected values using the formula
Mapping strain determination
Based on Million Mutation Project sequencing data, a group of candidate strains was chosen to identify an appropriate mapping strain. We chose candidates with a minimum SNV density of 500 ± 500 kbp while also minimizing for the occurrence of deleterious alleles (nonsense alleles, high Grantham scores, and frameshift mutations etc.). This analysis yielded 28 candidate strains, which were then phenotyped against our reference N2 strain (VC2010) for growth, fecundity, and developmental defects. The Million Mutation Project strain VC20019 exhibited growth and development similar to N2 and was chosen as a mapping strain.
sma-9 bulk segregant mapping
sma-9(tm572) mutants were outcrossed twice to N2 to remove extraneous mutations before crossing with VC20019 males. Outcross (phenotypically wild-type) F1 hermaphrodites were picked to new plates and allowed to self-fertilize. F2 progeny with the Sma body size phenotype were then identified and pooled in groups of varying sizes to grow on 100 mm NGM plates until starved. Pools were isolated for genomic DNA and subsequently sequenced using MIP-MAP.
TS competitive fitness mapping
hlh-1 and TS mutant mapping was conducted using a competitive fitness assay followed by bulk segregant analysis on surviving populations. Briefly, mutant animals were crossed with either VC20019 or DM7448 (VC20019;Ex[pmyo-3::YFP]) males. These crosses were then grown at 15° for 24–48 hr before shifting to 23 or 26°. Plates were allowed to starve and an agar chunk was transferred to fresh 100 mm OP50-seeded plates. Agar chunk size was varied with 4-cm2 pieces having the best mapping results.
Alternatively, nonstarved surviving F1s were individually chosen and plated to fresh 100 mm plates. When a population starved, progeny were subsampled by chunk-transfer to fresh media. This process continued for at least six rounds at the nonpermissive temperature, with each transfer generally producing a new generation of animals. The remaining animals on each plate after a transfer were harvested for genomic samples used in subsequent MIP-MAP analyses.
TS mutant selective bulk segregant mapping
TS mutants were crossed with DM7448 males and allowed to grow at 15°. F1 cross-progeny were chosen based on positive yellow fluorescent protein (YFP) expression. F2 animals were transferred singly to 96-well plates (liquid culture) at 15° and grown to starvation. Each well was subsampled and tested for growth at the nonpermissive temperature in liquid culture. Populations that failed to thrive at the nonpermissive condition were identified and pooled together from the original 15° populations. This pool was expanded and then harvested for genomic DNA used in subsequent MIP-MAP analyses.
MIP-MAP candidate intervals from competitive fitness mapping were determined by identifying the correct TS-associated chromosome, choosing the probe with the highest mapping allele percentage, and using the adjacent probes on either side as the outer boundaries for the interval. In cases where multiple probes were within 1–2% difference between peak and boundaries, intervals were extended outwards to the next nearest probe for variant analysis.
Data availability
Strains are available upon request. File S1 contains strain information including lab origin, allele name, and any available phenotype information. File S2 contains all variant call information from the set of 173 strains described in this manuscript. Raw WGS files of strains are available from the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP111734. File S3 contains all of the 89 MIP sequences and genomic targets used in the mapping analysis presented in the manuscript as well as all primer sequences required for library generation and usage on an Illumina-based sequencer. Sample MIP pools for capture can be made available upon request as well. File S4 contains all of the required parameters for MIP normalization during the processing and analysis of sequence data. File S5 contains a description of all columns and sheets from File S1, File S2, File S3, and File S4. Custom scripts used to analyze sequencing data are available on Github at https://github.com/camok/MIP-MAP-analysis-scripts.
Results
Sequencing a collection of embryonic lethal TS mutant strains
We collected a total of 173 embryonic lethal TS mutant strains from three different sources that were generated either in an N2 or lin-2(e1309) background using different mutagenesis protocols (EMS or ENU) (Kemphues et al. 1988a; Ehmke et al. 2014) (see also Materials and Methods). After mutagenesis, candidate strains were identified through screens for embryonic lethality phenotypes at temperatures of 25 or 26° (Table 1 and File S1). More specifically, the 72 strains in the first set are comprised predominantly of mutant strains with early embryo defects not primarily affecting cell division or strains with lethality occurring after the four-cell stage. A small portion of strains have some form of sterility, and the remainder of this set is comprised of strains with low-penetrance lethality or unconfirmed TS phenotypes. Sixty-seven strains make up the second set and encompass a wide range of TS phenotypes across seven broad classes from general lethality to tissue-specific developmental defects (Table 1). The third set consists of 34 strains identified from a mutagenesis screen for maternal effect lethality. Combined, these sets cover a diverse range of phenotypes that could reveal information on essential genes in a variety of pathways.
Table 1. Temperature-sensitive collection phenotype summary.
| Set | General Phenotype | Total Strains |
|---|---|---|
| 1 | Failure of fertilization | 8 |
| 1 | P1 delay | 11 |
| 1 | Unclassified sterility | 5 |
| 1 | Early divisions normal, cytoplasmic clearing | 3 |
| 1 | Early division normal, late arrest | 8 |
| 1 | Early division normal, terminal phenotype unknown | 22 |
| 1 | Low penetrance embryonic lethality or unlikely TS strain | 15 |
| 2 | General larval lethality and sterility | 39 |
| 2 | Cytoplasm morphology abnormal | 9 |
| 2 | Cell division abnormalities | 17 |
| 2 | Cell death aberrations | 10 |
| 2 | Early gastrulation defects | 1 |
| 2 | Tissue-specific developmental defects | 15 |
| 2 | Morphology abnormalities | 38 |
| 2 | Undocumented phenotype | 3 |
| 3 | Maternal effect lethality | 34 |
Set 2 encompasses overlapping phenotypes from the seven categories, with undocumented phenotype data for three strains. TS, temperature-sensitive.
Prior to mapping, we performed WGS on each strain to a target minimum of 12-fold coverage (with a mean of ∼14.5-fold coverage). Using the bioinformatic pipeline developed for the Million Mutation Project, we find that, on average, each strain carries 328 SNVs including 70 missense changes, nearly three nonsense mutations, and ∼1 splice site mutation per strain (Table 2), or < 1 change in protein coding potential per megabase of sequence within the C. elegans genome. Among these changes, missense mutations are the most likely to be associated with TS phenotypes (O’Rourke et al. 2011a; Lowry et al. 2015), although in rare instances other alterations may be responsible (Harfe et al. 1998). We have included a copy of the final SNV call data as part of our supplemental data (File S2).
Table 2. SNV summary from 173 strains.
| Set (Unique Genes) | Total SNVs | Coding Exons | Noncoding | Introns | |||||
|---|---|---|---|---|---|---|---|---|---|
| Missense | Nonsense | Synonymous | Exons (UTRs) | Splicing | Other | ncRNAs | Intergenic | ||
| Set 1 (4,579) | 9,285 | 1,971 | 72 | 678 | 316 | 42 | 2,770 | 131 | 3,302 |
| Set 2 (11,943) | 38,965 | 8,291 | 356 | 3,396 | 1,087 | 142 | 11,078 | 595 | 14,020 |
| Set 3 (4,269) | 8,612 | 1,896 | 78 | 723 | 251 | 35 | 2,334 | 124 | 3,171 |
| All strains | 56,803 | 12,144 | 505 | 4,793 | 938 | 217 | 16,164 | 850 | 20,478 |
| Unique genes | 14,327 | 7,972 | 493 | 3,937 | 1,497 | 215 | 7,623 | 779 | |
SNV, single-nucleotide variant.
Given the challenges in de novo approaches to identifying the causative alleles from this collection, we turned to genetic mapping to narrow the list of candidates. We sought to develop a method that: (1) produced a linkage interval at a resolution that was sufficient to limit the list to a handful of candidates and was comparable in resolution to current methods; (2) replaced the Hawaiian isolate as a mapping strain with one more genomically similar to N2; and (3) allowed for a cost-efficient, high-throughput workflow. We postulated that mapping with smMIPs in a targeted sequence capture strategy (Hiatt et al. 2013) would provide similar mapping resolution to current WGS methods, allow the use of a strain minimally divergent from N2, and decrease the overall sequencing burden per sample.
Targeted sequencing of select polymorphisms by MIP-MAP
We modified the smMIP structure, leveraging its single-molecule targeting capabilities to analyze SNV fractions at specific sites across the genomes of bulk segregant mapping populations (hereby referred to as MIP-MAP) (Figure 1). Each MIP is composed of an 80-bp oligonucleotide with a pair of annealing arms at the 3′- (extension arm) and 5′- (ligation arm) ends totalling 40 bp. Connecting the annealing arms of each individual oligonucleotide probe is a 12-bp UMI, as well as a common MIP backbone sequence used in library preparation and amplification steps (Figure 1A). Each MIP is designed to target a locus where a known variant is located within the first 18 bp of the 100–150-bp stretch of gap-fill sequence (Figure 1, A and B). The gap-fill sequence also serves to unambiguously map the recovered sequence. The library is then amplified, at which time an experimental index sequence is incorporated (Figure 1C). This design places all the vital information within a single-end 50-bp Illumina sequencing read (Figure 1D). After sequencing, each sample library is demultiplexed by its experimental index and then UMIs and gap-fill sequence data are used to eliminate PCR duplicates (Figure 1E, Materials and Methods). The remaining unique sequencing information is used to determine a MIP target’s SNV representation vs. the total captures matching that locus as calculated by
With a targeted sequencing strategy in hand, we next examined the parameters that would influence our selection of probes across the C. elegans genome: mapping resolution, mapping strain choice, and MIP accuracy. Mapping resolution is generally limited by the number of F2 animals pooled rather than SNV density. With just 100 F2s picked, average resolution is ≤ 1 cM or ∼1–3 Mbp depending on the region of the genome (Barnes et al. 1995; Rockman and Kruglyak 2009; Doitsidou et al. 2010). Accordingly, sampling ∼100 SNV markers across the genome would match the expected genetic resolution. At this density, 200,000 unique sequencing reads would provide deep coverage of each SNV, allowing the accurate estimation of even relatively rare linked SNVs, a vast improvement in sequencing burden vs. current paradigms.
Figure 1.
MIPs design and workflow. The design of the smMIPs (Hiatt et al. 2013) was modified to relocate the unique molecular identifier. Overall, the MIP design incorporates two 20-bp annealing arms, a 12-bp molecular tag, and a 28-bp common backbone. After annealing to a target segment ranging in size from 100–150 bp (A), the MIP is gap-filled with high-fidelity polymerase capturing the sequences of interest and circularized via ligation (B). The SNV of interest is located within an 18-bp gap-fill window upstream of the ligation arm. Uncircularized DNA is then degraded via exonucleases. The remaining MIPs are linearized by combining with sequencing adapters and library barcodes during PCR amplification (C). A single-end read captures both the molecular tag, 5′-annealing sequence, and enough genomic sequence to confirm correct target capture (D). This information is then demultiplexed (E) and used as a means to compare sequence variant ratios. Fwd, forward; MIPs, molecular inversion probes; Rev, reverse; Seq, sequencing; smMIP, single-molecule MIP; SNV, single-nucleotide variant.
Decreasing the density requirement of our mapping SNVs also permitted us to survey the Million Mutation Project strains for potential alternatives to the Hawaiian strain. The Million Mutation Project strains average ∼400 mutations each, yet many are roughly wild-type in appearance. We screened for strains that carried minimal numbers of predicted deleterious coding variants and a relatively even distribution of SNVs across the genome at a frequency of at least 1 per 500 kbp. From 28 candidates, we identified a strain, VC20019, with developmental timing and fecundity similar to that of our N2 laboratory strain (VC2010). VC20019, also referred to as the mapping strain, has a total of 269 mutations, three nonsense, and 51 missense alleles, thus having enough genomic diversity compared to N2 to meet our molecular marker density requirements while having a reduced likelihood of potentially negative interactions.
We designed MIPs targeting 96 SNV sites across the VC20019 genome, generating intervals spaced an average of 0.98 ± 0.36 Mbp apart (see File S3). To identify the efficiency of annealing and overall fidelity of these probes, we generated a series of VC20019 and N2 genomic DNA mixtures for capture reaction and sequencing. From these libraries, we identified sequence-specific biases for each MIP and generated normalization curves for subsequent capture/sequencing reactions (Figure S1 and File S4, Materials and Methods). We removed seven VC20019 MIPs that consistently produced low sequencing read counts, high ratios of nonspecificity, or unpredictable allelic biases, bringing our total probe count to 89 probes (1.06 ± 0.43 Mbp intervals).
We also considered that the frequency for linked, and therefore rare, SNVs could be adversely impacted by false-positive MIP-MAP reads. To gauge the impact of false-positives on the accuracy of our calls, we designed an additional 176 MIP-MAP targets based on SNVs in 44 strains from the Million Mutation Project collection. We analyzed the final SNV representations of each MIP capture library from a genomic DNA mixture that excluded the selected Million Mutation Project target strains. We sequenced this set of MIPs to an average depth of ∼35,000 reads. We removed data from poorly amplifying MIPs, leaving 174 MIP target loci that were expected to have 0% mutant SNV representation. MIP reads with positive mutant calls must originate from gap-fill, PCR, or sequencing errors, and the overall percent mutant SNV representation was calculated as the false-positive rate. Furthermore, we filtered out low-quality sequencing reads for each probes’ SNV site and calculated that the average false-positive rate was 0.0122% ± 0.0129%. (Figure S2A, Materials and Methods).
Lastly, we determined the sequencing efficiency of the MIP-MAP method by comparing the population of UMIs for each MIP vs. the total reads identified for that MIP. For each MIP in our false-positive set of 174 MIP targets, the mean number of sequencing reads per UMI was 1.05 with a mean depth of 35,000 reads per MIP. We analyzed an additional 22 libraries from another sequencing run, consisting of the 89 mapping strain MIPs in each library. The mean reads per UMI across 1958 data points was 1.01 with a mean depth of 7000 reads per MIP (Figure S2B). These results suggest that we are capturing a nearly 1:1 representation of the genomic sample, with few redundant reads from nonunique capture sequences.
In summary, we reengineered the smMIP format to produce MIP-MAP, which targets the genome with high fidelity and high sequencing efficiency at a mapping resolution comparable to current methods. We identified a polymorphic strain similar to N2 with no observed adverse phenotypes and a reduced likelihood of negative genetic interactions. We next turned to examining the MIP-MAP protocol in mapping known mutant alleles and then uncharacterized strains of varying difficulty, such as those from the TS collection.
Mapping sma-9 and hlh-1 mutations using MIP-MAP
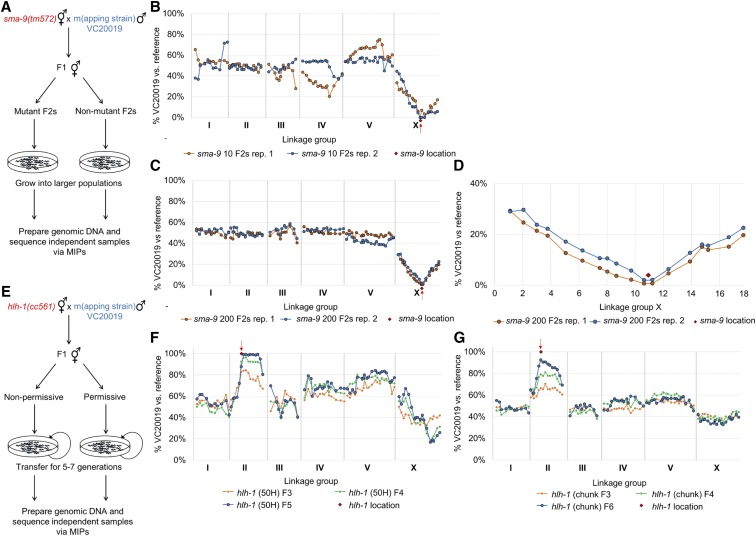
To investigate the mapping capabilities of the MIP-MAP method, we first focused on mapping with two distinct phenotypes; a clearly identifiable sma-9 small body mutant and the well-characterized hlh-1 TS lethal mutant. We proceeded by using the strain LW478 (sma-9(tm572)X) in a standard bulk segregant strategy similar to WGS (Figure 2A). Briefly, we crossed sma-9(tm752)X hermaphrodites with mapping strain males and from F1 cross-progeny chosen groups of F2 Sma phenotype animals (10, 25, 50, 75, 100, and 200) to grow until starvation on OP50-seeded 100 mm NGM plates before isolating genomic DNA for MIP-MAP capture reaction and sequencing (Figure 2 and Figure S3). Our expectation with such a strategy was to observe a region where VC20019 SNVs were nearly depleted due to linkage disequilibrium in selecting for the sma-9 allele from the mutant strain. We generated MIP-MAP data by growing populations seeded with 10 Sma phenotype F2s. These replicate samples had more background signal across the genome than subsequent experiments and the intervals of interest were 1.5 and 4.2 Mbp in these samples (Figure 2B). Seeding populations with 75 Sma phenotype F2s yielded similar mapping profiles that narrowed the likely interval to 2.6 Mbp (Figure S3C). Likewise, two mapping sets seeded with 200 F2 animals also consistently identified a 2.6 Mbp interval containing sma-9 (Figure 2C). Upon closer inspection of our sma-9 mapping intervals, we observed two sites ∼590 kbp apart that typically share nearly identical SNV representation, suggesting linkage disequilibrium. Therefore, it is not surprising to find that sma-9 is located equidistantly between these points, making it less likely to generate a sharp single-point peak even with a high number of F2s as input (Figure 2D). The MIP targets in our 75-F2 sets had a mean of 3778 unique reads, suggesting that each F2 haplotype was sampled an average of 25 times. Therefore, the MIP-MAP technique can sufficiently capture an accurate representation of the recombinant landscape across the population.
Figure 2.
Mapping of sma-9 and hlh-1 via VC20019 and MIP-MAP sequencing. Bulk segregant mapping scheme for sma-9 (A) using MIPs that target a number of VC20019-specific SNVs across the genome. Two replicate MIP-MAP samples of populations grown from 10 Sma phenotype F2 animals sufficiently identified a sma-9-associated interval on X within a 1.5–4.2 Mbp window (B) and replicates using 200 Sma F2 recombinants produced a 2.6 Mbp window (C). Closer examination of the MIP target sites on LGX show sma-9 is located equidistantly between two probes only 590 kbp apart (D). The hlh-1(cc561) temperature-sensitive embryonic lethal allele was mapped using a competitive fitness mapping method (E). PD4605 hermaphrodites were mated with VC20019 males and nonpermissive temperatures were used to select against the subsequent hlh-1(cc561) homozygous progeny. Starting with F1 cross-progeny and transferring mixed-stage subpopulations of animals (F and G) for up to seven generations, a region corresponding to the hlh-1 locus on LGII was successfully identified by the fixation of VC20019-specific SNVs. A mapping interval of 2.8–7.8 Mbp was identified by the F5 generation when transferring populations of 50 L1s (F). A 2.7-Mb interval was observed by the F6 generation when transferring 4-cm2 chunks of mixed-stage animals (G). Each line present in (F and G) represents mapping data from a different generation for the same biological replicate. MIP, molecular inversion probe; MIP-MAP, MIP mapping; SNV, single-nucleotide variant.
We next explored if MIP-MAP was applicable to more challenging mutant phenotypes. For instance, isolating homozygotes of nonconditional lethal/sterile alleles or identifying the correct homozygous F2 populations of low-penetrance alleles may not always be feasible. We approached this scenario with a competitive fitness mapping assay (Figure 2E). We hypothesized that we could exploit the fitness defect of an allele with the gradual fixation of a more “fit” version from the mapping strain. Using the PD4605 (hlh-1(cc561)II) TS embryonic lethal strain, we attempted to map its TS locus by using nonpermissive temperatures to select against this allele over several generations. In contrast to the sma-9 mapping, we expected to identify a region of interest by the fixation of VC20019 SNVs toward 100% representation within the mapping profile. We refined the parameters of the competitive fitness MIP-MAP method by varying the number of progeny passaged at each generation. Briefly, we crossed PD4605 animals to VC20019 males at 15° on 50 mm NGM plates for ∼24 hr before shifting these plates to between 23 and 26° for the remainder of the experiment. We expected that only cross-progeny F1s would thrive at this temperature. F1s were picked and self-fertilized before randomly picking L1- or L2-staged F2 larvae. With each subsequent generation, a subpopulation (50, 100, 200, or 400 animals) was passaged to a new plate for expansion (Figure 2F and Figure S4, A–C). We continued in this manner for 4–7 generations, collecting samples for MIP-MAP library preparation and sequencing at each passage.
Based on our observations from mapping sma-9, we hypothesized that at least 75 F2 animals (upwards of 150 recombinant chromosomes) homozygous for our region of interest were required to consistently generate the smallest mapping interval compatible with the density of our markers. The mapping profile from a competitive fitness assay might therefore be influenced by the number of unique recombinant chromosomes that carry our region of interest in each generation. We reasoned that subsampling in small numbers could limit recombinant representation and increase the incidence of bottlenecking within the population. A chance reduction in recombinant chromosome diversity could lead to quick fixation of a region of interest but with reduced resolution. Conversely, subsampling in larger numbers could give a greater range of recombinant genomes, reduce bottlenecking, and generate better resolution, while likely requiring more time for the fixation of mapping strain alleles.
With this in mind, we varied our passaging size and observed an overrepresented region of VC20019-associated SNVs on LGII with varying degrees of resolution (Figure 2, F and G). This candidate interval encompassed hlh-1 and suggested that we had correctly mapped the cc561 allele. The smallest possible mapping intervals containing cc561 are 1.7 and 1.9 Mbp. When passaging smaller populations (i.e., 50 animals per generation), fixation occurred quickly but in some cases resulted in a larger mapping interval across LGII (Figure 2F and Figure S4A). However, the resolution of MIP-MAP improved in the populations seeded with ≥ 200 randomly chosen F2s (Figure S4, B and C). Thus, sampling more animals generated the optimal minimal mapping interval of 1.71 Mbp with an increase of one to two generations for fixation.
To exclude the possibility of a sampling bias for healthier animals when hand-picking our hlh-1 mapping recombinants, we replicated these experiments with an alternate procedure by transferring completely randomized populations via agar chunks of starved animals for 5–9 generations (Figure 2G and Figure S4, D–F, Materials and Methods). We again observed that larger transfer sizes associated with better resolution but this approach also generated a more pronounced delay of 2–8 additional generations before fixation.
We further validated the competitive fitness assay by investigating the general fitness of the mapping strain in a cross to the reference strain VC2010 and growing cross-progeny at a range of temperatures. We observed only partial fixation of LGV at high temperature over eight generations but other loci preferences within the genome appeared negligible (Figure S5, A–C), thereby confirming that our hlh-1 results were not an artifact of the mapping strain used. Therefore, we reliably identified a strong TS lethal allele through a simplified competitive fitness selection process. In addition, we noted the weak beginnings of fixation on LGII when growing an hlh-1 mapping population at 15° (Figure S6), suggesting that a small population growth defect was conferred by the cc561 allele at the reported permissive growth temperature (Harfe et al. 1998). We next turned to see if the MIP-MAP competitive fitness assay could be applied to genetically mapping uncharacterized TS lethal alleles.
Using MIP-MAP to identify TS mutations that confer lethal phenotypes
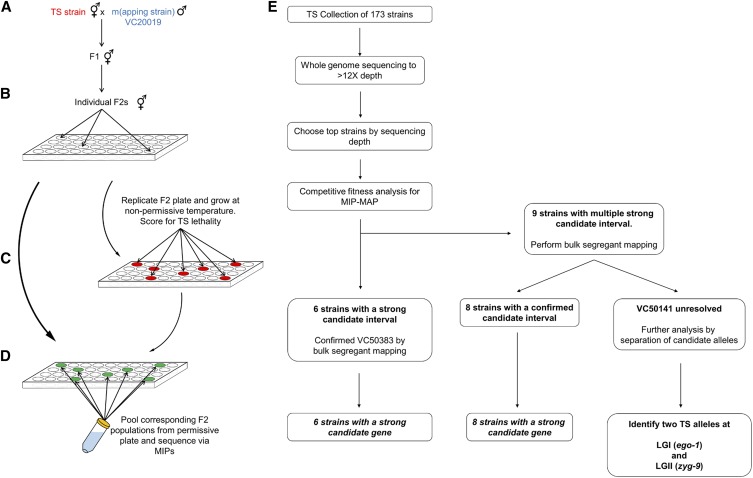
Our success with hlh-1 led us to hypothesize that we could, with good resolution, map mutant alleles from our large collection of sequenced TS lethal strains. Unlike our previous trials, these mutants carry hundreds of variants, any one of which may influence the outcomes of a competitive fitness assay. Furthermore, TS alleles of low or weak penetrance could also prove troublesome given a reduced selection coefficient during the propagation process. Regardless of these factors, from the collection of TS mutants we chose 15 strains (Table 3) of high sequencing depth but varying phenotypes to attempt genetically mapping with MIP-MAP.
Table 3. Table of temperature-sensitive mutants mapped.
| Strain | Set | WGS Coverage | Phenotype Penetrance | Timing of Lethality and Phenotype Observations at 26° |
|---|---|---|---|---|
| VC50022 | 3 | 16.6 | 100% | Arrest in early embryogenesis. |
| VC50028 | 3 | 40.7 | 100% | Sterility of P0 when plated at L4. Any F1s produced arrest at early larval stages. Some sterility also noted at permissive temperatures. |
| VC50031 | 3 | 16.4 | 100% | Sterility of P0 when plated at L4. Any F1s produced arrest at early embryogenesis. |
| VC50141 | 3 | 16.9 | 100% | Sterility of P0 when plated at L4. Any F1s produced arrest at early embryogenesis. |
| VC50174 | 1 | 21.9 | < 80% | Sterility of P0 when plated at L4, but also have bag phenotype so F1 are still produced. Failure of fertilization with vacuolated spermatozoa observed. |
| VC50178 | 1 | 20.7 | 100% | P0 when plated at L4 still produce progeny but F1 population is observed to be sterile. Severe morphogenesis defect, possible twofold arrest. |
| VC50182 | 1 | 19.8 | 100% | Early divisions normal, late arrest. Mostly twofold arrest with movement. |
| VC50255 | 1 | 20.0 | 100% | Sterility of P0 when plated at L4 with bag phenotype of dead eggs likely due to embryonic arrest. Any F1 produced are sterile. P1 delay is observed in embryos. |
| VC50260 | 1 | 22.9 | 99% | Sterility of P0 when plated at L4 with bag phenotype of dead eggs likely due to embryonic arrest. Any F1 produced are sterile. P1 delay is observed in embryos. |
| VC50352 | 2 | 23.4 | < 100% | Slightly leaky as P0 can give rise to F1, and then F2 but these are usually sickly and sterile. In some cases, they can make F3. Pretzel inviable, asynchronous cell cycle, later stage uncoordinated. |
| VC50360 | 2 | 22.2 | 100% | P0 have a mild roller phenotype, sluggish in appearance. F1 progeny appear to arrest in early embryogenesis. Pretzel inviable, pretzel deformed, cell adhesion/migration defect. |
| VC50374 | 2 | 20.9 | 99% | P0 when plated at L4 produce unhatched eggs of various forms and appears embryonic lethal. Any F1s produced are sterile. Arrest of development at premorphogenetic stage. |
| VC50375 | 1 | 19.0 | 100% | F1 sterility with P1 delay observed. |
| VC50380 | 2 | 19.6 | 100% | P0 when plated at L4 appear active and produce eggs. F1 progeny arrest in late embryogenesis and L1. Pretzel inviable, abnormal cytoplasm morphology, spindle defects, aberrant pharyngeal cluster and pretzel deformed. |
| VC50383 | 2 | 21.7 | 100% | P0 plated at L4 appear active but produce eggs that fail to hatch. Pretzel inviable, larval lethal, and muscle defects. |
WGS, whole-genome sequencing.
Since these highly mutated strains could harbor mutations that reduce brood size or mating efficiency, we modified our process to ensure the picking of cross-progeny by crossing our TS mutants with DM7448 (a VC20019 strain carrying an extrachromosomal pmyo-3::YFP marker) males and picking YFP-positive F1 hermaphrodites. These F1s were subjected to the previously-described competitive fitness assay via chunk- or wash-transfer and, in some instances, we supplemented our analysis with a multi-well liquid format of bulk segregant mapping for clarification (Figure 3, A–D, Materials and Methods). Our mapping results (summarized in Figure 3E and Table 4) can be categorized into increasing levels of analysis difficulty: clear single TS loci (Figure 4, A and B and Figure S7), single TS loci with additional partial fitness-defective loci (Figure 4, C and D and Figure S8), and multi-locus profiles resolved by additional investigations (Figure 4, E–G, Figure 5, A–C, and Figure S9).
Figure 3.
Workflows for liquid bulk segregant mapping and TS mutant analysis. (A) VC20019 males were mated to a TS mutant strain with cross-progeny F1 animals used to generate F2 recombinants. Single F2s were used to seed wells (B), growing these populations at permissive temperatures before replicating them to grow in nonpermissive conditions (C) and identifying populations that failed to thrive. Dead populations (red) were then chosen from the original wells (green) and pooled together (D) to prepare a single genomic sample for MIP-MAP sequencing. Samples from multiple plates were combined in the mapping of a single mutant depending on allele penetrance or the number of positive F2 wells identified. The workflow (E) of mapping TS mutants shows six strains could be mapped by competitive fitness MIP-MAP alone, eight strains benefitted from additional mapping via the liquid bulk segregant protocol, and one strain (VC50141) required additional analyses. MIP-MAP, molecular inversion probe mapping; TS temperature-sensitive.
Table 4. Summary of temperature-sensitive mutant candidate mapping intervals.
| Strain | Set | Mapping Methods | Mapping Interval | Candidate Gene(s) |
|---|---|---|---|---|
| VC50022 | 3 | CF | LGIII:3579637-5328496 | tbb-2, F56D2.5 |
| VC50174 | 1 | CF | LGI:3494925-6676830 | C09D4.3 |
| VC50178 | 1 | CF | LGIII:0-3579637 | M01G5.1, let-805 |
| VC50182 | 1 | CF | LGIII:7617321-10853887 | cts-1, enu-3 |
| VC50260 | 1 | CF | LGIV:1125542-3797513 | npp-8 |
| VC50380 | 2 | CF | LGX:15150772-17676467 | lron-3, csb-1, let-2, K09E3.5 |
| VC50028 | 3 | CF and 96-well | LGII:121799849-15279345 | cox-10, jmjd-2, K10H10.4, mog-4 |
| VC50031 | 3 | CF and 96-well | LGV:18400066-20183925 | Y69H2.3, asp-18, emb-4 |
| VC50255 | 1 | CF and 96-well | LGIII:7617321-10853887 | C02F5.10, pri-1 |
| VC50352 | 2 | CF and 96-well | LGIV:5057999-7478926 | vit-2, pqn-62, Y34B4A.6, ifta-1, C09B8.8, pak-1, col-171 |
| VC50360 | 2 | CF and 96-well | LGV:11035658-14223805 | pak-2, F35B12.9, rad-50, sqt-3, F28C1.1, egl-10 |
| VC50374 | 2 | CF and 96-well | LGIV:1125542-3797513 | F42A6.6, hrp-1, Y37E11AL.6, map-1, taf-6.2 |
| VC50375 | 1 | CF and 96-well | LGIV:9147769-12028068 | dct-15, K08F4.5, par-5 |
| VC50383 | 2 | CF and 96-well | LGX:15180772-17676467 | rab-14, let-2, nas-39, acs-9, F52G3.1, gcy-11, crb-1 |
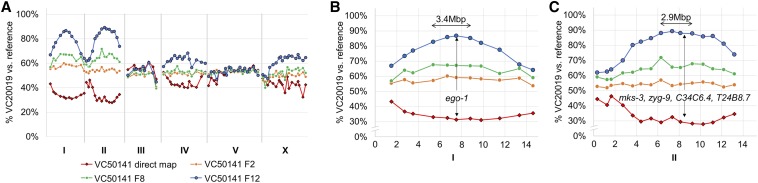
| VC50141 | 3 | CF, 96-well, segregation testing | LGI:6676830-8797510 | ego-1 |
| LGII:6305987-9232191 | mks-3, zyg-9, C34C6.4, T24B8.7 |
CF, competitive fitness mapping; 96-well, liquid bulk segregant mapping.
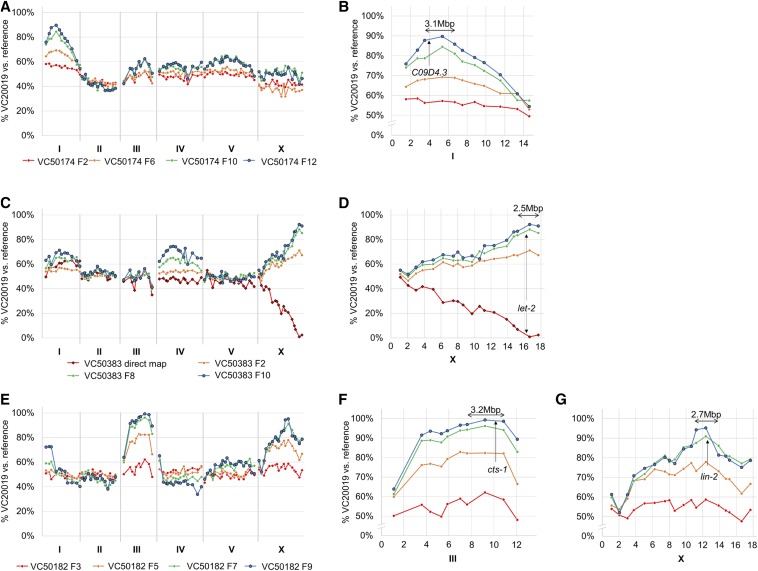
Figure 4.
Successful mapping of TS lethal mutant strains via competitive fitness and liquid-format MIP-MAP techniques. Fifteen TS lethal mutants with genomic sequencing data were mapped by the competitive fitness approach used with hlh-1. Mapping results yielded a diverse group of mapping profiles including clean single-locus mappings as seen in VC50174 (A and B), profiles such as VC50383 with small additional loci or peaks appearing at later generations but resolved by additional liquid-format bulk segregant mapping of phenotypes (C and D), and multiple-locus profiles as observed in VC50182 (E) resolved to a single locus (F) by identifying shared background mutations at competing loci (G). Each line present in a graph represents mapping data from a different generation for the same experimental replicate or the results of a liquid-format bulk segregant mapping experiment (indicated as direct map with diamond points). MIP-MAP, molecular inversion probe mapping; TS temperature-sensitive.
Figure 5.
The TS strain VC50141 has two independent TS alleles of ego-1 and zyg-9. Both competitive fitness mapping and liquid-format bulk segregant mapping confirmed that the TS lethality in VC50141 was related to two loci (A). The first interval located on LGI contains only a single relevant coding mutation in ego-1 (B). The second locus, appearing to be of equal penetrance on LGII, contains four candidates, of which zyg-9 is the most likely causative allele (C). Each line present in the graph represents mapping data from a different generation for the same experimental replicate or the results of a liquid-format bulk segregant mapping experiment (indicated as direct map with diamond points). TS temperature-sensitive.
Three strains yielded clean, single-locus profiles with only one or two TS candidate alleles in each interval. Among these strains, VC50174 mapped to an adenine to thymine transversion resulting in an E196D mutation in the fifth exon of C09D4.3 (Figure 4, A and B), a gene with no prior phenotype data to suggest an essential role in development. Available RNA sequencing (RNAseq) data reports C09D4.3 transcript abundance peaking at the L4 larval stage, most especially in males (Hutter et al. 2009; Boeck et al. 2016), and is suggested to have preferential expression in the germline (Grün et al. 2014). Furthermore, we observed that VC50174 animals exhibited a failure of fertilization and vacuolated spermatozoa. VC50022 mapped to an interval with a coding mutation in tbb-2; one of two nearly functionally redundant C. elegans β-tubulin genes. Although not entirely essential, multiple groups have identified semidominant TS-lethal alleles for this gene (Wright and Hunter 2003; Ellis et al. 2004). In particular, the tbb-2 mutation in VC50174 is located within the same exon as a semidominant embryonic lethal allele t1623 (Gönczy et al. 1999). The other possible candidate, F56D2.5, is at the edge of this interval, shows low expression throughout embryogenesis (Boeck et al. 2016), and has no useful RNA interference (RNAi) phenotype data. VC50260 maps to an interval encompassing a single coding mutation for npp-8, which has also been reported as an essential gene with a documented TS allele, ax146 (Asencio et al. 2012).
In three instances, our TS mapping profiles suggested the presence of additional weak low-fitness alleles at secondary loci (Figure 4, C and D and Figure S8). In these cases, we combined mapping data from multiple time points with the expectation that the TS-associated locus was the least competitive at nonpermissive temperatures and would therefore fixate at a faster rate. When we could not unambiguously resolve the correct chromosome, we further mapped the strain with a liquid bulk-segregant assay (Figure 3A). By growing singled F2 populations in 96-well liquid format we could subsample each and phenotype for TS lethality, going back to the original populations later to pool for MIP-MAP analysis. This method would generally produce a low number of TS F2s (Table S1) but could yield a single mapping signal across the C. elegans genome. Of particular interest, the strain VC50383 yielded a number of candidates within the mapping interval. However, the most likely candidate was a let-2 exon 16 guanine to adenine transition resulting in a G1385E change, which was confirmed through complementation testing. Coincidentally, another set 3 strain, VC50380, also mapped to an interval that included a let-2 guanine to adenine transition (G1110E) in exon 14. From our collection of 173 strains, there are nine separate let-2 mutations that could potentially elucidate the complex nature of this gene (Meneely and Herman 1981).
Our final group of nine TS strains represent multi-locus mapping profiles that fixed to similar proportions within a short period of time (Figure 4, E–G and Figure S9). These strains may carry additional mutations that confer disadvantages during the competitive mapping process. Again, a combination of comparing fixation rates at each locus and the liquid segregant mapping assay were used to resolve the TS-associated locus. In three of these strains, our mappings included a clearly marked interval on chromosome X (Figure 4, E–G) that encompasses the lin-2 gene. Strains from both set 1 and set 3 harbor the lin-2(e1309) mutation that causes egg retention and reduced brood size, which have negative influences in a competitive fitness assay. From these strains in particular, VC50182 yielded two candidates within the interval on LGIII of which cts-1 (a citrate synthase) is the likely prime candidate. VC50182 animals exhibited late arrest at the twofold stage, while reported cts-1 RNAi phenotypes include embryonic lethality and sterility (Kamath et al. 2003; Melo and Ruvkun 2012).
Also of note, in mapping the strain VC50141, we observed the concomitant but incomplete fixation of two loci, one each on LGI and LGII, when using our competitive fitness assay (Figure 5A). Similarly, the liquid-format bulk segregant assay identified the same two LGs at ∼30% mapping allele representation vs. an expected 0% generated from 75 TS-positive wells in 387 randomly chosen F2s. This phenomenon suggested the presence of two TS alleles. The LGI interval contained only a single coding change, which corresponded to a guanine to adenine transition (E994K) in the RNA polymerase homolog ego-1 (Qiao et al. 1995; Smardon et al. 2000) (Figure 5B). There are no previously reported conditional lethal alleles for ego-1. The LGII locus contained four possible candidates including a cytosine to thymine (P632S) zyg-9 mutation (Figure 5C). There are a large number of reported TS alleles located across zyg-9 (Bellanger et al. 2007), suggesting that it is a good candidate for VC50141. To confirm whether these two loci were independent TS alleles or if they perhaps interacted to produce a synthetic TS lethal effect, we proceeded to isolate these two alleles into separate strains followed by scoring for lethal phenotypes at 26°. From recombinants, we observed two independent TS-lethal phenotypes, sterility and embryonic lethality, which were linked to the ego-1 and zyg-9 alleles, respectively. Thus, our mapping methods were able to identify both ego-1(ax458)I and zyg-9(ax3101)II as the primary candidates for two independent TS-lethal mutations in this strain.
Overall, for our set of TS strains, this method yielded relatively small mapping intervals that contained greatly reduced numbers of candidate alleles (Table 4), simplifying the process of finding the causative allele. Of 15 candidate strains, six were mapped with only the competitive fitness approach while nine required additional bulk segregant mapping, with the special case of VC50141 requiring additional intervention (Figure 3E). In three strains, our mapping intervals contained only a single relevant coding sequence candidate, while in the remaining intervals we narrowed candidates to a primary variant based on a combination of gene description, expression pattern, RNAseq, and RNAi phenotype data from WormBase and strain phenotype observations (Table 5). Within the VC50352 mapping interval, pak-1 was the only gene with reported embryonic lethality and an embryonic expression profile that overlapped temporally with the observed pretzel inviable phenotype. VC50352 had the only primary candidate mutation with a predicted splice variant. Of the 16 primary candidate TS genes, seven have no reported TS alleles listed in WormBase and one (C09D4.3) has no previously reported embryonic lethal phenotypes. Of the other candidates with no reported TS alleles, ego-1, let-805, mog-4, and par-5—genes with known mutant phenotypes—are only available from the Caenorhabditis Genetics Center as viable strains with genetic balancers. The gene cts-1 has a single deletion strain available that is listed as sterile/lethal, and pak-1 has two deletion alleles available, one of which is listed as sterile/lethal.
Table 5. TS Candidate mutation information.
| Strain | Primary Candidate | Allele | Candidate Gene Phenotype Information | Examples of TS Allele(s) Reported Prior to This Sequenced Collection |
|---|---|---|---|---|
| VC50022 | tbb-2 | ax123 | Early embryonic lethala | or362, or600 |
| VC50028 | mog-4 | ax87 | Sterilityb | q233 |
| VC50031 | emb-4 | ax968 | Larval lethalc | hc60 |
| VC50141 | ego-1 | ax458 | Embryonic arrest, and small oocytes with unusual chromosome morphology by RNAid. | None reported, in same exon as hypormorphic alleles om54 and om71 |
| zyg-9 | ax3101 | Embryonic lethal by RNAic,e,f. Failure to hatch in TS allele or623g. | or623, or628, or593, or635, or634 | |
| VC50174 | C09D4.3 | or976 | No embryonic phenotypes reported. Peak expression in L4 and male samples. Expression present in sperm transcriptomeh | None reported |
| VC50178 | let-805 | or896 | Embryonic lethale,f, maternal sterilec,e, larval lethalc,e,i. | None reported |
| VC50182 | cts-1 | or1005 | Embryonic lethalc,e,i. | None reported |
| VC50255 | pri-1 | or1187 | Delayed early cell division with prominent 3-cell stagej, and embryonic lethalitye,i. | None reported |
| VC50260 | npp-8 | or1044 | Embryonic lethalc,e. | ax146 |
| VC50352 | pak-1 | t3254 | Low range embryonic lethal, larval lethal phenotypes and larval lethal with muscle detachmentk. | None reported |
| VC50360 | sqt-3 | t3352 | Embryonic lethale, dumpye,f, roller phenotypesl by RNAi and various alleles. | e24, sc8, sc43, sc63, sc80 |
| VC50374 | taf-6.2 | t3399 | Embryonic lethalf | ax701, ax514 |
| VC50375 | par-5 | or996 | Embryonic lethalc,e, and cell cycle delayedm. | None reported |
| VC50380 | let-2 | t3320 | Early larval lethal and embryonic lethaln. | mn114, e1470, b246, mn101, mn109, mn111, g30 |
| VC50383 | t3381 |
TS, temperature-sensitive; RNAi, RNA interference.
O’Rourke et al. (2011).
Discussion
Our collection of sequenced C. elegans conditional mutant strains offers a rare opportunity to study essential genes on a larger scale. However, high-throughput mapping of the causative alleles in this collection is not a trivial task. We began our analysis of these strains with the goal of generating a robust mapping strategy that was affordable, high-throughput, comparable in resolution to current mapping paradigms, and flexible enough for the community’s needs. Using smMIPs we developed MIP-MAP, which relies on targeted loci sequencing of the C. elegans genome at great depth. MIP-MAP is differentiated from other mapping methods by (1) its adaptability to different mapping and segregant selection schema, (2) its independence from the usage of the Hawaiian strain CB4856, (3) by the reduction in sequencing and data generation while maintaining 1 Mbp resolution, and (4) its unique advantage in mapping the more recent collections of sequenced strains.
MIP-MAP successfully identified a phenotype-associated locus in the direct selection of sma-9 F2 segregants, a common mapping paradigm. However, with the hlh-1(cc561) TS lethal allele, we successfully used a competitive selection scheme to identify the hlh-1 locus vs. an alternative approach such as picking dead larvae for WGS (Smith et al. 2016). This indirect approach to identifying lethal loci may also prove useful in gauging the rate of fixation across multiple conditions to elucidate mutation penetrance or gene function. In fact, our hlh-1 data suggested that even at the permissive temperature of 16°, hlh-1(cc561) animals have some level of decreased fitness, despite a lack of reported lethality in this condition (Harfe et al. 1998).
Our results demonstrated the flexibility of this method to work with nonstandard mapping strains. We leveraged a Million Mutation Project strain with a mere 269 SNVs and targeted strain-specific markers at 89 loci, using them to successfully map sma-9 and hlh-1 mutant alleles to within ∼2 Mbp intervals. Furthermore, we have shown with VC20019 that the Million Mutation Project is also a library that contains polymorphic mapping strains with fewer potential issues than more divergent genomes like the Hawaiian strain. From these results, it is also plausible to identify Million Mutation Project strains compatible for mapping synthetic phenotypes. This strategy would reduce the additional steps required in generating homozygous background mutations used in forward genetic screens, simplifying the cloning process.
In terms of cost, the MIPs themselves are a one-time purchase where a single order at 25 nM scale provides > 2 million capture reactions per probe. MIP-MAP also offers comparable resolution to WGS with far less extraneous sequencing data. For example, each MIP-MAP library can be sequenced to an average 200,000 reads to obtain mapping data. In contrast, WGS mapping generally requires 1× coverage to catalog each recombinant haplotype. Recent standards of 18–22X coverage with 50-bp single-end reads (Wang et al. 2014; Jaramillo-Lambert et al. 2015; Smith et al. 2016) require ∼36–48 million reads , an increase of two additional orders of magnitude, with most of the sequence being reference genome data. Mapping 15 candidate strains, we generated a mean candidate interval size of 2.8 Mbp while WGS mapping at a minimum 18× coverage was reported to generate a mean candidate interval of 5.2 Mbp (Wang et al. 2014). MIP-MAP thus permits high multiplexing of samples during the sequencing process while producing comparable mapping resolution. The overall cost savings per strain and capabilities of MIP-MAP are therefore suitable to a high-throughput analysis of collections such as our TS mutations or the Million Mutation Project. More recent large-scale genome-wide association studies may also benefit from this method when verifying hits from among multiple candidate associations (Cook et al. 2016).
Although the MIP-MAP method was demonstrated on sequenced mutant collections, it can also benefit the mapping of de novo strains. The throughput scalability of MIP-MAP provides a significant cost saving but is limited by requiring an additional layer of investigation to identify the specific causal variant. Million Mutation Project findings suggest WGS to a 12-fold depth (Thompson et al. 2013) is sufficient to correctly identify SNVs and other mutations in a heavily mutagenized, homozygous strain. As discussed previously, this level of coverage is insufficient for the current single-step WGS mapping paradigm. Therefore, WGS of a mutant strain can be multiplexed with its own MIP-MAP library at a 30% reduction in overall sequencing burden. Alternatively, MIP-MAP can be applied as an initial step to identify a candidate mapping interval. Afterward, genomic interval pulldown sequencing (O’Rourke et al. 2011b), which uses C. elegans genomic fosmids to target specific regions, can be used to identify the variant of interest. Classical methods, including rescue by fosmid injection and amplicon sequencing, could also be applied after MIP-MAP. Thus, MIP-MAP offers an alternative means of genetic mapping that may better suit the needs of some research groups.
Intriguingly, a number of our competitive fitness mappings identified the presence of additional non-TS loci that conferred fitness defects on the TS strains. This observation is suggestive of two things. The first, that the competitive fitness mapping method may be overly sensitive to secondary mutations of reduced fitness. This issue of sensitivity in the assay was resolved through the addition of bulk segregant analysis of select F2s, but could also be remedied by outcrossing of the mutant strains to the wild-type N2 strain prior to MIP-MAP or careful comparison of parallel MIP-MAP libraries from samples grown at permissive conditions. Second, the observation of strains with multiple mapped loci suggests that our TS collection holds a diverse library of additional mutations related to organismal fitness. In a similar fashion, MIP-MAP could be useful in high-throughput mining of the Million Mutation Project for specific phenotypes in a form of reverse-genetics screen. For instance, rather than testing for TS lethality, MMP strains could be queried for altered fitness phenotypes in normal growth conditions or by investigating the effects of food source, growth media content, or RNAi knockdown. More broadly, MIP-MAP demonstrates, on a focused level, a method of barcoding strains within the context of a mixed population. This low-cost, high-throughput form of analysis could be applied, for example, to the analysis of mixed pools of multiple strains to track variant fitness across a diverse set of conditions. Targeting strains in this manner might reveal the functions of many novel genes. Given that the Million Mutation Project strains have already been sequenced, using MIP-MAP to identify the cause of these phenotypes would require far less sequencing data than WGS methods.
Reviewing our TS-mapping workflow, five of nine strains were bulk segregant mapped with > 60 F2s chosen as TS-positive but none of these generated a concise interval; rather, they complemented the sharp intervals obtained by the competitive fitness approach. Curiously, in three of these five sets, the chosen F2 population was far less than the expected 25% ratio for a homozygous recessive mutation (Table S1). Indeed, in the remaining four strains mapped with F2 counts below 60, these animals were identified at low frequency from very large populations of potential F2s, further suggesting issues in either identifying the TS phenotype or reduced phenotype penetrance in these populations. In toto, using bulk segregant analysis we observed only three strains (VC50028, VC50374, and VC50380) that generated mapping intervals comparable in resolution to data from the competitive fitness approach. In that respect, a MIP-MAP competitive fitness assay over many generations offers the ability to identify mutant loci that are of incomplete penetrance and may prove problematic in standard bulk segregant mapping. From this, we conclude that for the remainder of our TS strains, especially those of apparently lower penetrance, it may be more efficient to complete both mapping approaches in parallel with the benefit of reducing repetition of mapping crosses in downstream workflows.
The collection of C. elegans TS strains we present here is, to our knowledge, the largest, fully-sequenced collection to date. Although we mapped strains based on those with the highest amounts of sequencing coverage, we expect a similar level of mapping success from the remainder of the collection. Based on our mapping results, we suspect that many new and interesting alleles can be mined from these strains. For example, our mapping of the strain VC50174 identified a sperm-specific essential gene C09D4.3. Recent studies suggested that sperm-targeted RNAi screens may be inefficient due to competition between endogenous and exogenous RNAi pathways (Reinke 2003; Ma et al. 2014). This hypothesis supports the literature from multiple RNAi screens that report no embryonic lethal phenotypes for C09D4.3 (Fraser 2000; Maeda et al. 2001; Sönnichsen et al. 2005). Finding this TS allele within our small set suggests that our overall collection of strains has the potential to not only yield the first TS alleles for a number of essential genes, but also the potential to identify new essential genes not discovered by standard RNAi screens. Further analysis of our collection as a whole may identify groups of strains with shared mapping intervals. Candidate genes within these intervals can then be prioritized by their frequency of mutation within these sets. The mapping of C09D4.3 also suggests that ∼6% of the strains in this collection may carry TS alleles for genes not previously characterized as essential. In summary, we present a large collection of sequenced TS mutants housing a variety of embryonic lethal and sterility phenotypes. In conjunction with this collection, we demonstrate the MIP-MAP methodology as a robust, cost-efficient, and high-throughput genetic mapping format. It allows for a wide range of flexible sample generation schema that can be implemented with relative ease. We believe that MIP-MAP can even identify alleles with weak penetrance phenotypes given the proper selection conditions. Taken together, these two tools present a unique opportunity for the nematode research community to investigate a diverse library of mutant strains and to map these and other mutant alleles in an accessible and high-throughput manner. We believe that both of these resources will be a beneficial addition to the nematode and scientific community at large.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300179/-/DC1.
Acknowledgments
We thank Emily Turner, Joseph Hiatt, and Choli Li for advice on molecular inversion probe library preparation and sequencing. We thank Cathrin Struck for expert technical assistance. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD-010440). C.A.M. is supported by the Canadian Institutes for Health Research MFE-135408. N.M. is supported by the Internationales Graduierten Kolleg Niedersachsen. Work in the B.B. laboratory was supported by an R01 grant (GM-114053) from the NIH. Work in the G.S. laboratory was supported by an R01 grant (HD-37047) from the NIH. G.S. is an Investigator of the Howard Hughes Medical Institute. Work in the D.G.M. laboratory was supported by the Canadian Institutes for Health Research and the National Human Genome Research Institute (NHGRI) (through R.H.W.). D.G.M. is a Senior Fellow of the Canadian Institute for Advanced Research. Work in the R.H.W. laboratory was supported by an American Recovery and Reinvestment Act of 2009 Grand Opportunities grant (HG-005921) from the NHGRI, an R21 grant (HG-007201-02) from the NIH, and by the William H. Gates Chair of Biomedical Sciences.
Footnotes
Communicating editor: H. Buelow
Literature Cited
- Andersen E. C., Bloom J. S., Gerke J. P., Kruglyak L., 2014. A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology. PLoS Genet. 10: e1004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asencio C., Davidson I. F., Santarella-Mellwig R., Ly-Hartig T. B. N., Mall M., et al. , 2012. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell 150: 122–135. [DOI] [PubMed] [Google Scholar]
- Barnes T. M., Kohara Y., Coulson A., Hekimi S., 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger J.-M., Carter J. C., Phillips J. B., Canard C., Bowerman B., et al. , 2007. ZYG-9, TAC-1 and ZYG-8 together ensure correct microtubule function throughout the cell cycle of C. elegans embryos. J. Cell Sci. 120: 2963–2973. [DOI] [PubMed] [Google Scholar]
- Boeck M. E., Huynh C., Gevirtzman L., Thompson O. A., Wang G., et al. , 2016. The time-resolved transcriptome of C. Elegans. Genome Res. 26: 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. Caenorhabditis elegans. Methods 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., et al. , 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. E., Zdraljevic S., Roberts J. P., Andersen E. C., 2016. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res. 45: D650–D657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. N., Laufer J. S., Kusch M., Edgar R. S., 1980. Genetic and phenotypic characterization of roller mutants of Caenorhabditis elegans. Genetics 95: 317–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Hammarlund M., Harrach T., Hullett P., Olsen S., et al. , 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bono M., Bargmann C. I., 1998. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94: 679–689. [DOI] [PubMed] [Google Scholar]
- Doitsidou M., Poole R. J., Sarin S., Bigelow H., Hobert O., 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One 5: e15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmke M., Luthe K., Schnabel R., Döring F., 2014. S-adenosyl methionine synthetase 1 limits fat storage in Caenorhabditis elegans. Genes Nutr. 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis G. C., Phillips J. B., O’Rourke S., Lyczak R., Bowerman B., 2004. Maternally expressed and partially redundant beta-tubulins in Caenorhabditis elegans are autoregulated. J. Cell Sci. 117: 457–464. [DOI] [PubMed] [Google Scholar]
- Encalada S. E., Martin P. R., Phillips J. B., Lyczak R., Hamill D. R., et al. , 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228: 225–238. [DOI] [PubMed] [Google Scholar]
- Flibotte S., Edgley M. L., Chaudhry I., Taylor J., Neil S. E., et al. , 2010. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics 185: 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A. G., 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330. [DOI] [PubMed] [Google Scholar]
- Golden A., Sadler P. L., Wallenfang M. R., Schumacher J. M., Hamill D. R., et al. , 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Schnabel H., Kaletta T., Amores A. D., Hyman T., et al. , 1999. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 144: 927–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham P. L., Schedl T., Kimble J., 1993. More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev. Genet. 14: 471–484. [DOI] [PubMed] [Google Scholar]
- Grün D., Kirchner M., Thierfelder N., Stoeckius M., Selbach M., et al. , 2014. Conservation of mRNA and protein expression during development of C. elegans. Cell Rep. 6: 565–577. [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Branda C. S., Krause M., Stern M. J., Fire A., 1998. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development 125: 2479–2488. [DOI] [PubMed] [Google Scholar]
- Hiatt J. B., Pritchard C. C., Salipante S. J., O’Roak B. J., Shendure J., 2013. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 23: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter H., Ng M.-P., Chen N., 2009. GExplore: a web server for integrated queries of protein domains, gene expression and mutant phenotypes. BMC Genomics 10: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Fuchsman A. S., Fabritius A. S., Smith H. E., Golden A., 2015. Rapid and efficient identification of Caenorhabditis elegans legacy mutations using Hawaiian SNP-based mapping and whole genome sequencing. G3 (Bethesda) 5: 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kemphues K. J., Kusch M., Wolf N., 1988a Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. Genetics 120: 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G., Cheng N. S., 1988b Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52: 311–320. [DOI] [PubMed] [Google Scholar]
- Kiezun A., Garimella K., Do R., Stitziel N. O., Neale B. M., et al. , 2012. Exome sequencing and the genetic basis of complex traits. Nat. Genet. 44: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry J., Yochem J., Chuang C.-H., Sugioka K., Connolly A. A., et al. , 2015. High-throughput cloning of temperature-sensitive Caenorhabditis elegans mutants with adult syncytial germline membrane architecture defects. G3 (Bethesda) 5: 2241–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhu Y., Li C., Xue P., Zhao Y., et al. , 2014. Characterisation of Caenorhabditis elegans sperm transcriptome and proteome. BMC Genomics 15: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda I., Kohara Y., Yamamoto M., Sugimoto A., 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11: 171–176. [DOI] [PubMed] [Google Scholar]
- Mamanova L., Coffey A. J., Scott C. E., Kozarewa I., Turner E. H., et al. , 2010. Target-enrichment strategies for next-generation sequencing. Nat. Methods 7: 111–118. [DOI] [PubMed] [Google Scholar]
- Melo J. A., Ruvkun G., 2012. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149: 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely P. M., Herman R. K., 1979. Lethals, steriles and deficiencies in a region of the X chromosome of Caenorhabditis elegans. Genetics 92: 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely P. M., Herman R. K., 1981. Suppression and function of X-linked lethal and sterile mutations in Caenorhabditis elegans. Genetics 97: 65–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minevich G., Park D. S., Blankenberg D., Poole R. J., Hobert O., 2012. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192: 1249–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton D. G., Shakes D. C., Nugent S., Dichoso D., Wang W., et al. , 2002. The Caenorhabditis elegans par-5 gene encodes a 14–3-3 protein required for cellular asymmetry in the early embryo. Dev. Biol. 241: 47–58. [DOI] [PubMed] [Google Scholar]
- O’Roak B. J., Vives L., Fu W., Egertson J. D., Stanaway I. B., et al. , 2012. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S. M., Carter C., Carter L., Christensen S. N., Jones M. P., et al. , 2011a A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS One 6: e16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S. M., Yochem J., Connolly A. A., Price M. H., Carter L., et al. , 2011b Rapid mapping and identification of mutations in Caenorhabditis elegans by restriction site-associated DNA mapping and genomic interval pull-down sequencing. Genetics 189: 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. D., Trent C., Robertson B., Chamblin C., Wood W. B., 1994. Sequenced alleles of the Caenorhabditis elegans sex-determining gene her- 1 include a novel class of conditional promoter mutations. Genetics 138: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poultney C. S., Butterfoss G. L., Gutwein M. R., Drew K., Gresham D., et al. , 2011. Rational design of temperature-sensitive alleles using computational structure prediction. PLoS One 6: e23947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L., Lissemore J. L., Shu P., Smardon A., Gelber M. B., et al. , 1995. Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics 141: 551–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., 2003. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. [DOI] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L., 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski T. M., Gilchrist E. J., Mullen G. P., Moerman D. G., 1995. Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics 139: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S., et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14: 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Prabhu S., O’Meara M. M., Pe’er I., Hobert O., 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5: 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Rockman M. V., Kruglyak L., 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Ailion M., Li J., van Oudenaarden A., Rockman M. V., et al. , 2011. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 9: e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Moorman C., van der Linden A. M., Kuijk E., van den Berghe P. V. E., et al. , 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A., Spoerke J. M., Stacey S. C., Klein M. E., MacKin N., et al. , 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10: 169–178. [DOI] [PubMed] [Google Scholar]
- Smith H. E., Fabritius A. S., Jaramillo-Lambert A., Golden A., 2016. Mapping challenging mutations by whole-genome sequencing. G3 (Bethesda) 6: 1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., et al. , 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. [DOI] [PubMed] [Google Scholar]
- Thompson O., Edgley M., Strasbourger P., Flibotte S., Ewing B., et al. , 2013. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner E. H., Lee C., Ng S. B., Nickerson D. A., Shendure J., 2009. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat. Methods 6: 315–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang J. T., Rasoloson D., Stitzel M. L., O’ Connell K. F., et al. , 2014. Identification of suppressors of mbk-2/DYRK by whole-genome sequencing. G3 (Bethesda) 4: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H., 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Wright, A. J., and C. P. Hunter, 2003 Mutations in a β-Tubulin disrupt spindle orientation and microtubule dynamics in the early Caenorhabditis elegans embryo. Mol. Biol. Cell 14: 4512–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahreddine H., Zhang H., Diogon M., Nagamatsu Y., Labouesse M., 2010. CRT-1/Calreticulin and the E3 ligase EEL-1/HUWE1 control Hemidesmosome maturation in C. elegans development. Curr. Biol. 20: 322–327. [DOI] [PubMed] [Google Scholar]
- Zonies S., Motegi F., Hao Y., Seydoux G., 2010. Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR-2. Development 137: 1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. File S1 contains strain information including lab origin, allele name, and any available phenotype information. File S2 contains all variant call information from the set of 173 strains described in this manuscript. Raw WGS files of strains are available from the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP111734. File S3 contains all of the 89 MIP sequences and genomic targets used in the mapping analysis presented in the manuscript as well as all primer sequences required for library generation and usage on an Illumina-based sequencer. Sample MIP pools for capture can be made available upon request as well. File S4 contains all of the required parameters for MIP normalization during the processing and analysis of sequence data. File S5 contains a description of all columns and sheets from File S1, File S2, File S3, and File S4. Custom scripts used to analyze sequencing data are available on Github at https://github.com/camok/MIP-MAP-analysis-scripts.