Abstract
One difficulty when identifying alternative splicing (AS) events in plants is distinguishing functional AS from splicing noise. One way to add confidence to the validity of a splice isoform is to observe that it is conserved across evolutionarily related species. We use a high throughput method to identify junction-based conserved AS events from RNA-Seq data across nine plant species, including five grass monocots (maize, sorghum, rice, Brachpodium, and foxtail millet), plus two nongrass monocots (banana and African oil palm), the eudicot Arabidopsis, and the basal angiosperm Amborella. In total, 9804 AS events were found to be conserved between two or more species studied. In grasses containing large regions of conserved synteny, the frequency of conserved AS events is twice that observed for genes outside of conserved synteny blocks. In plant-specific RS and RS2Z subfamilies of the serine/arginine (SR) splice-factor proteins, we observe both conservation and divergence of AS events after the whole genome duplication in maize. In addition, plant-specific RS and RS2Z splice-factor subfamilies are highly connected with R2R3-MYB in STRING functional protein association networks built using genes exhibiting conserved AS. Furthermore, we discovered that functional protein association networks constructed around genes harboring conserved AS events are enriched for phosphatases, kinases, and ubiquitylation genes, which suggests that AS may participate in regulating signaling pathways. These data lay the foundation for identifying and studying conserved AS events in the monocots, particularly across grass species, and this conserved AS resource identifies an additional layer between genotype to phenotype that may impact future crop improvement efforts.
Keywords: conserved alternative splicing events, monocot, grass family, RS and RS2Z subfamilies, R2R3-MYB
ALTERNATIVE splicing (AS) is an important post-transcriptional process that can produce two or more transcript isoforms from a pre-mRNA. AS occurs in the spliceosome by removing introns and joining exons through the selective use of splice sites (Lee and Rio 2015), and is governed by cis-regulatory elements such as splicing enhancers/silencers, and trans-regulatory elements including the Serine-Arginine-rich (SR) and Heterogeneous nuclear riboprotein (hnRNP) protein families (Wang and Burge 2008; Busch and Hertel 2012; Reddy et al. 2012, 2013). AS isoforms that are translated can influence proteome diversity, while others that are purposefully nonfunctional can act to post-transcriptionally modulate protein levels (Fu et al. 2009; Hammond et al. 2009). AS participates in many important processes during the lifecycle of plants (Staiger and Brown 2013), and AS occurs in response to many abiotic stresses (Mastrangelo et al. 2012), for example red/blue light (Shikata et al. 2014; Wu et al. 2014), salt stress (Feng et al. 2015), drought (Thatcher et al. 2016), flooding (Syed et al. 2015), and temperature (James et al. 2012; Streitner et al. 2013).
RNA-Seq has become a standard tool to investigate transcriptomes and AS isoforms. While many studies have identified AS in individual species (Filichkin et al. 2010; Li et al. 2014; Shen et al. 2014; Thatcher et al. 2014; Mandadi and Scholthof 2015), there are few reports of conserved AS across multiple plant genomes. Most published studies have focused on identifying conserved AS events between only two species. For example, Severing et al. (2009) identified 56 protein-coding conserved AS events between Arabidopsis and rice orthologous gene sets, and 30 conserved AS events leading to nonfunctional isoforms subjected to nonsense-mediated decay. Similarly, another study detected 537 AS events conserved between Arabidopsis and Brassica (Darracq and Adams 2013), and 71 conserved AS events were identified between Populus and Eucalyptus (Xu et al. 2014). With respect to comparing more than two species, one study found 16 AS transcripts conserved between Brachypodium, rice, and Arabidopsis by performing all-vs-all BLAST between EST sequences (Walters et al. 2013). Recently, discovery and identification of AS events conserved broadly across eudicots has been reported (Chamala et al. 2015). Chuang et al. (2015) used available Sanger EST sets to characterize AS in grass species, but did not focus on conserved AS events. There are no studies that leverage the available deep next-generation sequencing resources to characterize and identify conserved AS events across species within monocots and the grass family.
Many monocot species are economically important and grass species are a particularly important source of calories. Within the grass (Poaceae) there are two major clades. The BEP clade is made up of the Bambusoideae, Ehrhartoideae, and Pooideae, including rice and Brachypodium (Zhao et al. 2013); the PACMAD clade represents the Panicoideae, Arundinoideae, Chloridoideae, Micrairoideae, Aristidoideae, and Danthonioideae subfamilies (Cotton et al. 2015). Maize, sorghum, and foxtail millet are representatives of Panicoideae. Published whole genome sequences exist for many monocot species, including the grass species maize (Schnable et al. 2009), sorghum (Paterson et al. 2009), rice (Goff et al. 2002), Brachypodium (Vogel et al. 2010), and foxtail millet (Bennetzen et al. 2012), and the nongrass species banana (D’Hont et al. 2012) and African oil palm (Singh et al. 2013).
A computational method that identifies high confidence isoforms in any species with RNA-Seq data and a reference genome sequence was used to identify and characterize AS events, and a comparative splice junction-based approach identified conserved AS events across multiple species. This analysis focused on seven monocot species that have available reference genome sequences and deep RNA-Seq data sets. Five of these are grasses (maize, rice, sorghum, foxtail millet, and Brachypodium) with substantial gene collinearity, which eases the identification of synteny and aids ortholog calls (Schnable et al. 2012). The remaining two are banana and African palm oil, which extend the discovery of conserved events into nongrass monocot species. Although there are other nongrass monocot species that have available reference genomes, such as duckweed and orchid, insufficient RNA-Seq data prevented their inclusion in this analysis. Likewise, species representing basal grass lineages were excluded due to lack of reference genome sequences. Finally, the eudicot Arabidopsis thaliana and the basal angiosperm Amborella were included to provide outgroups for comparison. Therefore, the species studied allow identification of conserved AS events at specific reference points across the grasses and extends into nongrass monocots, eudicots, and the sister species to all angiosperms.
Detecting AS events that are broadly conserved between species is likely to identify functionally important AS isoforms. Members of the SR proteins and the hnRNP protein families are splicing regulatory proteins that contain RNA binding domains (reviewed in Barbazuk et al. 2008). AS has been observed in the hnRNP and SR protein splicing factor gene families (Kalyna et al. 2006; Rauch et al. 2014; Chamala et al. 2015), and has been associated with plant stress (Staiger and Brown 2013). We specifically examined the SR and hnRNP genes for conserved AS events to identify isoforms that may be important for the proper function and regulation of these important splice factors, and to provide a platform to examine differences in AS between species. Similarly, we examined transcription factors (TFs) known to be responsive to five major stresses (Naika et al. 2013) for evidence of conserved AS, and applied a STRING functional protein association network analysis (Szklarczyk et al. 2015) to identify interaction networks involving the TFs responsive to the five stresses (above) that also have conserved AS events.
The collection of conserved AS events identified also enables an investigation of the extent of positive selection and functional constraints on AS based on the Ka/Ks, and Ks values of pairwise comparisons between splice anchor sequence tags (SASTs) that flank conserved AS events, as well as a test of the functional sharing hypothesis (Su et al. 2006) between single-copy vs. nonsingle-copy genes. This hypothesis predicts more instances of conserved AS events will be found in single copy genes than in nonsingle-copy genes. Finally, we also present a STRING functional protein association network analysis based on the Arabidopsis protein–protein interaction data to provide evidence of a close association between splicing factors and stress responsive TFs that undergo conserved AS, and between splicing factors and R2R3-MYB TFs.
Materials and Methods
Genome assemblies and annotations
All analyses were performed on publicly available sequence and most reference genome assemblies and annotations were retrieved from Phytozome v10 (Goodstein et al. 2012) (Supplemental Material, Table S1) including.: foxtail millet 164_v2 (Setaria italica), rice 204_v7 (Oryza sativa L. ssp. Japonica), Brachypodium 283_v2 (Brachypodium distachyon), Arabidopsis thaliana 167_TAIR10, and the basal angiosperm Amborella 291_v1 (Amborella trichopoda), which is a sister taxon to all angiosperms. The Maize RefGen_v2 genome sequence and working gene set annotation were retrieved from MaizeGDB (Lawrence et al. 2004). Reference genome sequence and gene annotation for African oil palm v2 (Elaeis guineensis) were retrieved from the Malaysian Palm Oil Board (genomsawit.mpob.gov.my). Reference genome and annotation for banana (Musa acuminata) were retrieved from the banana genome hub (Droc et al. 2013).
Transcriptome data collection
AS in maize (Zea mays) and sorghum (Sorghum bicolor) was previously identified based on public RNA-Seq data (Mei et al. 2017), and available on figshare (https://doi.org/10.6084/m9.figshare.4205079.v3). Transcript sequences and annotations of Arabidopsis thaliana were retrieved from Araport11 PreRelease 20151202 (Cheng et al. 2017), which presents the updated TAIR10 annotation by inclusion of 113 RNA-Seq data sets. For the remaining six species, Illumina RNA-Seq, 454 and Sanger EST transcript sequences were downloaded from NCBI GenBank. The sequence collections are described in Table S2.
Detection of AS
The process and software parameters used to assemble transcripts from Illumina short reads were described previously (Mei et al. 2017). Briefly, three approaches were taken to assemble transcripts from Illumina short reads to maximize isoform detection. The assembly software used was annotation-guided Cufflinks 2.2.1 (Trapnell et al. 2012), genome-guided Trinity 2.0.4 (Grabherr et al. 2011) and annotation-guided StringTie 1.0.0 (Pertea et al. 2015). Isoforms built from Cufflinks were required to have minimal expression FPKM value of 0.1, and required full reads support across the entire isoform.
We masked vector and contaminant sequence in the Sanger ESTs using SeqClean (http://seqclean.sourceforge.net/). A total of 454 transcripts was constructed using Newbler v2.8 2010 (Roche) with parameters “-cdna -urt.” Cufflinks, Trinity, and StringTie assembled isoforms were merged with the Newbler 454 assemblies and the cleaned ESTs. Each specific species sequence collection was aligned back to the appropriate reference genome sequence followed by additional assembly and clustering to remove redundancy using the PASA 2.0 package (Haas et al. 2003). Isoform quality control steps described in Mei et al. (2017) were used on the PASA-generated transcript set to remove isoforms that were poorly supported or potentially poor quality assembled. Briefly, each splice junction in a final retained isoform was required to have a minimum entropy score of two (Sturgill et al. 2013). Retained introns that define potential intron-retention isoforms were expected over a minimum of 90% of their length with a minimal median coverage depth of 10, and a minimum intron retention isoform ratio of 10%. The intron retention rate was calculated as described in Marquez et al. (2012), where the median coverage of the retained intron was divided by the number of reads supporting the splice junction. Lastly, we required each final isoform to have a minimum FPKM of one and compose at least 5% of the total isoform abundance for that gene in at least one tissue examined.
We performed the above methods to identify and define five AS event-types in rice, foxtail millet, Brachypodium, banana, African oil palm, and Amborella. The five AS event types considered are Intron retention, Alternative acceptor, Alternative donor, Exon skipping, and Alternate terminal exon; herein designated as IntronR, AltA, AltD, ExonS, AltTE, respectively. The transcripts from Araport11 PreRelease 20151202 for Arabidopsis were realigned to the Arabidopsis reference genome using PASA to identify and characterize the set of Arabidopsis AS events. AS isoforms defined previously within maize and sorghum Mei et al. (2017) were used to complete the AS isoforms sets analyzed in this study.
Orthofinder clustering across nine species
Orthologous sequences were defined and grouped in orthogroups across the nine species by OrthoFinder with default setting (Emms and Kelly 2015). OrthoFinder was designed for plant genome orthogroup identification and accounts for gene length bias.
Identify sets of syntenic genes across five grass species
Syntenic orthologs across five grass species (Zea mays, Sorghum bicolor, Setaria italica, Oryza sativa, and Brachypodium distachyon) were defined using the methodology employed by Schnable et al. (2012). Gene models were updated according to new annotations from Phytozome 10 (Goodstein et al. 2012); 11,995 syntenic gene lists across five species (sorghum, rice, foxtail millet, Brachypodium, and either maize1/maize2 or both) were used for downstream analysis.
Identification of conserved AS
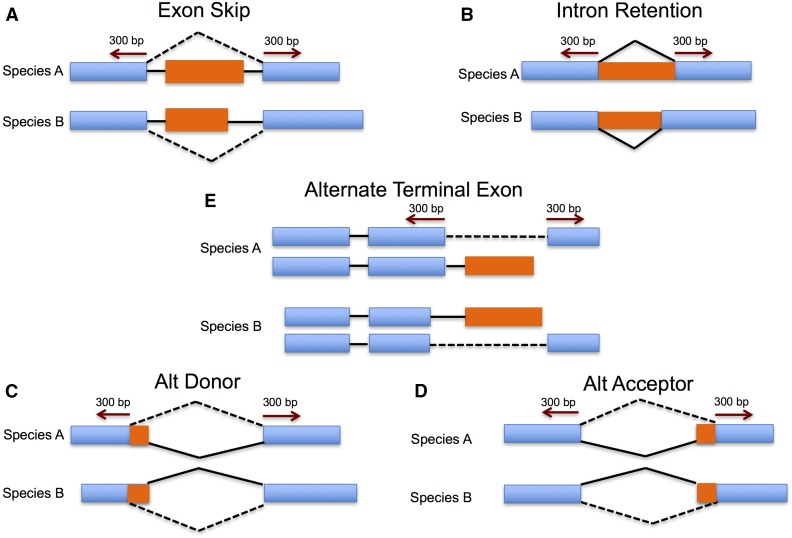
AS was independently identified in each species (see above). The computational methodology to detect conserved AS relies on the creation of SAST for each AS event and then comparing the SASTs for a given AS event type with the SASTs defining the same AS event type between species. SASTs were generated for each AS event by separately extracting up to 300 bp of sequence from both sides of the splice junction that defines the AS event types described in Figure 1. The tags from both sides of splice junction for a particular AS event (i.e., exon skipping SASTs) from each species were compared to one another using tBLASTx from WU-BLAST (Gish, W. (1996–2003) http://blast.wustl.edu) with an e-value cutoff of 1e−5. If the length of two matching tags were both <30 bp, this matching was not considered. In the cases tags being compared were less that 100 bp, the default tBLASTx E-value cutoff value was used. To classify conserved AS, Both SASTs flanking a AS event splice junction are required to be conserved separately between genes across species, and those genes must belong to the same orthogroup. Conserved AS events identified by this process were clustered and each cluster may contain conserved AS events identified between orthologous genes across species (1:1) as well as between paralogs. In an ideal setting, one conserved AS cluster represents one ancient conserved AS event across multiple species. However, if a gene within sorghum is present in two copies (homeologs) in maize, and all three genes harbor a conserved AS event, all three events will be assigned to the same event cluster. Therefore, with respect to the gene in sorghum and the two homeologous copies in maize, this cluster identifies one conserved event defined by two homeologous events in maize and one event in sorghum (three genes total). Therefore, the total number of events making up a cluster may be larger than the number of species harboring the conserved AS event. Because we identify these individual events within each species as instances of a conserved event, we generalize each AS event cluster as a “conserved AS event.” An example illustrating isoform structure, and multiple sequence alignments of conserved events, flanking SASTs and whole transcripts for four randomly picked conserved AS events (two ExonS and two IntronR) are presented in Figure S1 in File S1.
Figure 1.
(A)–(E) Pipeline to identify conserved AS events. Up to 300 bp on either side of a splice junction were extracted. These extracted sequences were binned together with all other similarly extracted, and binned sequences flanking the same type of event for each species (e.g., one bin with sequences flanking junctions involved in intron retention, one bin with sequences flanking junctions involved in exon skipping, etc.). Sequences within each bin were compared with tBLASTx to bins of the same event from other species to identify similar tags. Cases where the sequence tags were similar and the parent genes are from different species but represent orthologs as evidenced by being within the same orthogroup (see Materials and Methods) defined conserved AS events.
Measure of selection pressure on AS
We looked for evidence of selection pressure on AS events that were conserved by examining the difference between pairwise nonsynonymous substitution rates (Ka) vs. synonymous substitution rates (Ks) for residues defined by exon sequences flanking the conserved AS event in grass species. These are the same regions that were used to construct SASTs (Figure 1). SASTs > 150 bp (50 codons) were used in KaKs analysis, and KaKs ratios of those SASTs alignments with Ka < 0.5, Ks < 5, and Ka/Ks < 2 were retained for further analysis (Quint et al. 2012). Since these filtering processes resulted in < 100 SAST alignments associated with AltTE events, we focused only IntronR, ExonS, AltA, and AltD events for this analysis. Selection pressure on AS events at the amino acid level was measured by determining the Ka/Ks ratios using KaKs_Calculator2.0 through model averaging (Wang et al. 2010), and functional constraints were measured with Ks value. To minimize errors introduced by alignments, or due to paralogs that may result when comparing loci across large phylogenetic distances, the Ka/Ks evaluation of conserved AS events was limited to those events originating within orthologous grass genome loci that also exhibit conserved synteny (defined above).
Testing the functional sharing hypothesis: whether single copy genes have more conserved AS events
The functional sharing hypothesis predicts that conserved AS events would have a higher incidence in single-copy genes compared to nonsingle-copy genes. We used a set composed of 2717 multi-exon genes identified as “strictly” and “mostly” single copy identified across 20 flowering plant genomes by De Smet et al. (2013). We compared the number of single-copy genes exhibiting conserved AS events to the number of nonsingle-copy genes with conserved AS events.
Conserved AS in SR and hnRNP proteins
Maize SR protein genes were retrieved from Rauch et al. (2014); Sorghum SR protein genes were retrieved from Richardson et al. (2011). Maize hnRNP protein encoding genes were retrieved from PlantGDB (Duvick et al. 2008). The primary protein sequences of each SR protein gene in maize and sorghum were used for phylogenetic analysis. Protein sequences were aligned with MUSCLE (version 3.8.31) using default parameters (Edgar 2004). The alignments were used to construct a maximum likelihood phylogenetic tree with RAxML (version 8.1.12) software using a gamma distribution and LG4X model (Le et al. 2012; Stamatakis 2014). Conserved AS events in SR families between maize and sorghum exon–intron gene structure were visualized by Fancygene version 1.4 (Rambaldi and Ciccarelli 2009).
Network analysis in stress-responsive TFs and genes with conserved AS
A total of 3150 Arabidopsis TFs responsive to 14 diverse stresses were retrieved from STIFDB V2.0 (Naika et al. 2013). Specific attention was given to the subset of TFs responsive to ABA, drought, cold, salt (NaCl), and light because a substantial number of these were shown to undergo AS, and many were determined to harbor conserved AS events. Protein–protein interaction networks were built around the TFs with conserved events using STRING v10 (Szklarczyk et al. 2015). Additionally, STRING v10 was also used to construct protein–protein interaction networks based around Arabidopsis genes demonstrating conserved AS in Amborella, Arabidopsis, and at least one monocot species. The cluster of constructed networks was identified and analyzed in Cytoscape v3.3.0 (Shannon et al. 2003).
Gene ontology (GO) term annotation
Genes harboring conserved AS events between Amborella, Arabidopsis, and at least one monocot species were further evaluated for functional annotation and GO term enrichment based on the Arabidopsis gene name using the agriGO analysis toolkit (Du et al. 2010). We use the Fisher’s exact test in singular enrichment analysis, followed by multiple test corrections with the Hochberg FDR test. Only associations with a minimum corrected P-value of 0.05 were considered.
Data availability
AS events table and GFF files for the AS isoforms in seven species (Setaria italica, Oryza sativa L. ssp. Japonica, Brachypodium distachyon, Musa acuminata, and Elaeis guineensis, the eudicot Arabidopsis thaliana and the basal angiosperm Amborella) are made available to the community via figshare (https://doi.org/10.6084/m9.figshare.4789153.v2) and github (https://github.com/wenbinmei/Monocot_conserved_splicing), CoGe, the Comparative Genomics Platform. (https://www.genomevolution.org/). Scripts used in the analysis are available at the same github repository above. The Zea mays and Sorghum bicolor data are already available to the community at figshare (https://doi.org/10.6084/m9.figshare.4205079.v3).
Results
Up to 54.6% of expressed multi-exon genes exhibit evidence of AS among nine species
Five types of AS events were considered during this study: IntronR, AltA, AltD, ExonS, and AltTE (Figure 1); 28.7–54.6% of expressed multi-exon genes exhibit AS across these nine species (Table S3). Although the number of Amborella genes that exhibit AS is similar to that of other species examined during this analysis, Amborella has a lower proportion of multi-exon genes in the genome relative to other species. Therefore, despite the similar number of multi-exon genes that undergo AS, Amborella exhibits the highest fraction (54.6%) of multi-exon genes that produce AS transcripts compared to the other species considered here. Intriguingly, Amborella genes that undergo AS produce the highest average number of isoforms/gene compared to other species (Table S3). Whether this feature reflects the evolutionary position of Amborella in angiosperm, or is the result of the absence of lineage specific whole genome duplication events—whereas species with additional lineage specific WGD events have lost or partitioned AS isoforms among paralogues through subfunctionalization (Jiang et al. 2013)—remains to be investigated. Maize has the largest collection of RNA-Seq data among the nine species examined, and also has the largest number of AS events, but the percentage of multi-exon genes that undergo AS is similar to other species. The proportions of genes that undergo AS and the total number of AS events are higher than previously reported for many plant species (Campbell et al. 2006; Amborella Genome Project 2013; Walters et al. 2013; Panahi et al. 2014; Abdel-Ghany et al. 2016). Similar to previous genome-wide AS studies (Li et al. 2014; Thatcher et al. 2014; Chamala et al. 2015; Mandadi and Scholthof 2015), IntronR was the most commonly observed AS event type in monocots. Sorghum and Amborella were found to have the lowest percentage of IntronR events, 35.6 and 37.2%, respectively, while African oil palm and Brachypodium have the highest proportion of intron retention events, 61.4 and 57.6%, respectively (Table S3). While the data available was successful in identifying several AS events, the sequence data used was collected from public sources, and the tissue representation and sequence depth was not consistent between species studied. Therefore, in many cases, the numbers of AS events identified may under-represent the true extent of AS in these species.
AS events conserved between monocots, Arabidopsis, and Amborella
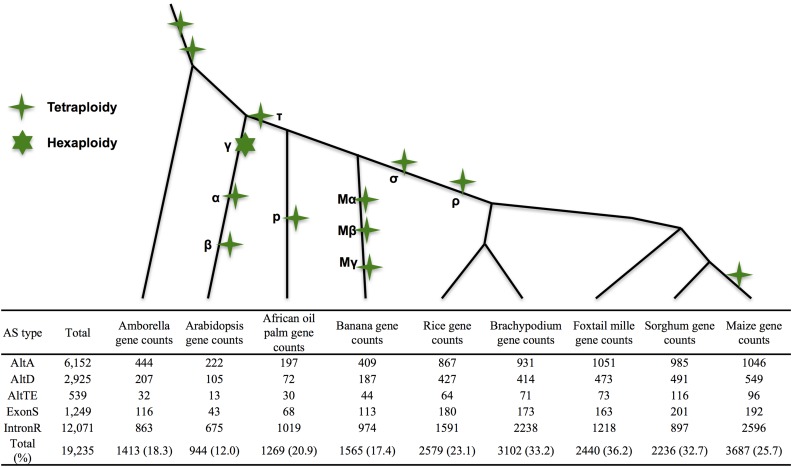
Using a junction-based approach (Figure 1 and File S1; Materials and Methods) to identify conserved AS within nine species revealed 9804 conserved AS events. Each event is present in two or more orthologous genes, and thus distributes across 19,235 genes (Table 1; Materials and Methods). The greatest number of conserved AS events are found in orthologous genes between grasses (Figure 2). Maize had the most (3687) genes harboring conserved AS events, and Arabidopsis the least (944), which may reflect its status as the only eudicot under consideration. Of the conserved AS events, 59% are IntronR; ∼68.7% of conserved AS events are conserved across two species, 18.6% across three species, 7.3% across four species, and 5.4% across five or more species (Table 1). A total of 1816 conserved AS event clusters was found to include Amborella. Of these, 1015 involved conserved IntronR, 450 AltA, 205 AltD, 114 ExonS, and 32 AltTE events (Table 2). In addition, we identified 80 AS events conserved only between Amborella and Arabidopsis and absent from the seven monocot species examined. These 80 AS events likely represent AS events lost after the divergence of eudicots and monocots and include 34 IntronR, 31 AltA, 11 AltD, and four ExonS events (Table 2). As previously mentioned, differences in tissue representation and sequence depth between species make it difficult to ensure that all potential events are discovered. While this does not affect the identification of a conserved event, some will be missed and appear “absent” due to sequence collection coverage and bias.
Table 1. Conserved AS across nine species at the gene family level.
| Number of Species | AltA | AltD | ATE | ExonS | IntronR | Total/% |
|---|---|---|---|---|---|---|
| 2 | 1384 | 684 | 201 | 335 | 4128 | 6732/68.7 |
| 3 | 447 | 181 | 32 | 69 | 1099 | 1828/18.6 |
| 4 | 218 | 112 | 6 | 26 | 351 | 713/7.3 |
| 5 | 117 | 36 | 2 | 21 | 109 | 285/2.9 |
| 6 | 58 | 29 | 0 | 10 | 52 | 149/1.5 |
| 7 | 22 | 12 | 0 | 3 | 32 | 69/0.7 |
| 8 | 7 | 3 | 0 | 4 | 7 | 21/0.2 |
| 9 | 2 | 2 | 0 | 1 | 2 | 7/0.1 |
| Total/% | 2255/23.0 | 1059/10.8 | 241/2.5 | 469/4.8 | 5780/59.0 | 9804 |
Percentage is based on total number of conserved AS clusters.
Figure 2.
Genes with conserved AS events across nine species. We calculate the number of genes with conserved AS events in each species. Tetraploidy and Hexaploidy are labeled on the phylogenetic trees. The length of the phylogenetic tree is not proportional to the phylogenetic distance.
Table 2. Conserved AS clusters among different species and clades.
| Total | IntronR | AltA | AltD | ExonS | ATE | |
|---|---|---|---|---|---|---|
| Conserved between Amborella and other species | 1816 | 1015 | 450 | 205 | 114 | 32 |
| Conserved in Amborella, Arabidopsis, and at least one species of monocots | 129 | 64 | 43 | 11 | 10 | 1 |
| Conserved between Amborella and Arabidopsis not in monocots | 80 | 34 | 31 | 11 | 4 | 0 |
| Conserved across Amborella and seven monocot not in Arabidopsis | 9 | 2 | 3 | 2 | 2 | 0 |
| Conserved in banana, African oil palm and at least one grass species | 239 | 162 | 44 | 20 | 11 | 2 |
| Conserved across five species in grass | 224 | 60 | 106 | 37 | 21 | 0 |
| Conserved across seven species in monocots | 34 | 13 | 12 | 5 | 4 | 0 |
| Conserved between PACMAD and BEP cladesa | 4333 | 2548 | 1052 | 480 | 177 | 76 |
Conserved at least one species in PCAMAD clade and one species in BEP clade.
GO term enrichment analysis was performed on Arabidopsis genes with AS events that are conserved within Amborella, Arabidopsis, and monocots, to determine whether these genes participated in common biological processes. Analysis of 129 AS events representing 64 IntronR, 43 AltA, 11 AltD, 10 ExonS, and one AltTE (Table 2) suggest an over-representation of kinase and phosphorylation activity (Figure S2 in File S1), which is consistent with the proposal that AS impacts protein kinase mediated signaling pathways (Reddy et al. 2013) that may modulate the transfer of developmental or environmental cues.
TFIIIA possesses an ExonS event conserved across land plants (Fu et al. 2009; Barbazuk 2010). This event was also identified in all species examined in our study. Shikata et al. (2014) previously characterized IntronR and AltD events within SPA3—a protein that plays a pivotal role in light signaling as a suppressor of photomorphogenesis (Laubinger and Hoecker 2003). Both splice isoforms harbor premature termination codons resulting in truncated proteins that retain interaction with CONSTITUTIVE PHOTOMORPHOGENIC 1 (COPI) but lose the ability to bind to DAMAGED DNA-BINDING PROTEIN 1 (DDB1). Evidence suggests that phytochrome could mediate the production of AS transcripts of SPA3 in response to some light conditions, thus promoting plant photomorphogenesis (Shikata et al. 2014). Both IntronR and AltD events were identified in our analysis: the AltD event is conserved across all nine species examined and the IntronR event is conserved in four species: maize, foxtail millet, African oil palm, and Arabidopsis. The absence of the IntronR event some species examined could be due to limited data depth or tissue sampling. Further investigation is required to determine whether this intron retention is conserved broadly across the angiosperms.
AS events conserved between Brassica and Arabidopsis are enriched for AltD and AltA events (Darracq and Adams 2013). We found significant enrichment of AltD and AltA events conserved across at least three species relative to events conserved in only two species (P < 0.0001, Fisher’s exact test, Table 1); similar enrichment patterns were observed for ExonS events conserved across at least four species relative to clusters conserved in only two species (P < 0.05, Fisher’s exact test, Table 1). As expected, there are fewer events conserved across multiple species (>3) than between a pair. Additionally, as the number of species that harbor the conserved AS increase, the proportion of conserved AS events that are intron retention decrease relative to the other event types. This suggests that identifying conserved AS events selects for biologically relevant and important events, and that intron retention suffers a higher proportion of “noisy” or nonrelevant splicing relative to other AS types.
The difference in lengths of conserved retained introns vs. nonconserved retained introns (i.e., unique to only one species) varies across the nine species examined. In Arabidopsis, Brachypodium, and foxtail millet, the length of conserved retained introns is significantly shorter than that of nonconserved retained introns (Table 3), but the trend is reversed for Amborella and African oil palm. There is no significant difference in retained intron length between conserved and nonconserved retained introns in banana, maize, sorghum, and rice (Table 3). A previous report suggested that the lengths of retained introns within Arabidopsis that are conserved in Brassica are shorter than nonconserved retained introns (Darracq and Adams 2013). We observed the same length trend in Arabidopsis and Brachypodium, but this is not observed in the other species considered.
Table 3. A comparison of intron length in the conserved intron retention events vs. nonconserved intron retention events across the species examined.
| Amborella | Arabidopsis | Banana | Brachypodium | Foxtail Millet | Maize | Oil Palm | Rice | Sorghum | |
|---|---|---|---|---|---|---|---|---|---|
| Conserved AS | |||||||||
| Mean | 670 | 165 | 329 | 411 | 402 | 440 | 315 | 417 | 409 |
| Median | 390 | 96 | 143 | 233 | 237 | 207 | 131 | 269 | 256 |
| No evidence for conserved AS | |||||||||
| Mean | 651 | 186 | 327 | 474 | 410 | 471 | 287 | 486 | 437 |
| Median | 316 | 113 | 131 | 245 | 201 | 205 | 121 | 247 | 242 |
| Wilcoxon test | |||||||||
| P value | *** | *** | NS | *** | * | NS | * | NS | NS |
(NS, Not significant; * P < 0.05, *** P < 0.001).
Arabidopsis TFs responsive to stress signals were retrieved from STIFDB V2.0 (Naika et al. 2013). Some TFs associated with each stress exhibit AS, and those responsive to ABA, NaCl, drought, light, and cold have the highest proportion with conserved events (Table S4). Two TF genes are particularly interesting: AT4G27410 (NAC TF RD26) and AT1G78070 (Transducin/WD40 repeat-like superfamily protein). These two genes have conserved AS events and are responsive to all five stresses listed above (Figure S3 in File S1). RD26 has an IntronR event conserved across seven of the nine species examined (absent in sorghum and Brachypodium); AT1G78070 has an AltTE event conserved between sorghum, banana, and Arabidopsis. This indicates that there are AS events in stress-response TFs conserved across large phylogenetic distances, and suggests that AS may play an important role in the activity of some TFs during stress response.
AS conserved between monocot grass and nongrass species
There are 239 AS events conserved between banana, African oil palm, and at least one of the five grass species examined; 34 of these are conserved across all seven monocot species examined (Table 2). A total of 204 conserved AS events exist within the banana and African oil palm lineages but are absent from the grass species studied, while 224 conserved AS events are conserved across the five grass species examined (Table 2). Within the PACMAD clade, 1544, 1488, and 1323 AS events are conserved between maize vs. sorghum, maize vs. foxtail millet, and sorghum vs. foxtail millet, respectively. In the BEP clade, 1658 conserved AS events are conserved between rice and Brachypodium; 4333 conserved AS events are conserved in at least one species in the BEP clade and one species in the PACMAD clade (Table 2). Maize clock genes (i.e., circadian rhythm) GRMZM2G033962 (pseudoresponse regulator protein 37, PRR37) and GRMZM2G095727 (pseudoresponse regulator protein 73, PRR73) have an IntronR event conserved across rice, Brachypodium, and foxtail millet. Our data suggests that this intron retention event is likely specific to members of the grass family; the role of intron retention in PRR37 and PRR73 controlling photoperiodic flowering is unknown.
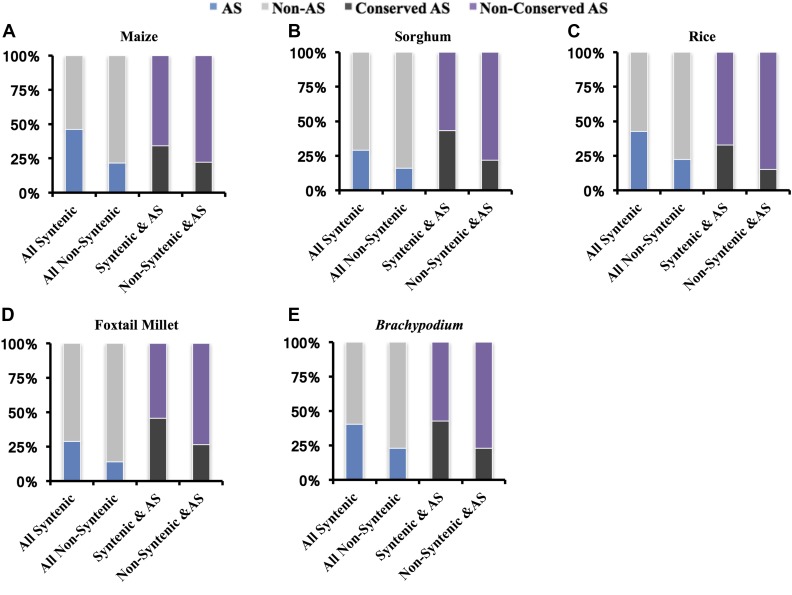
Genes in genomic regions that exhibit synteny between species were more likely to be transcribed and translated than those in nonsyntenic regions (Walley et al. 2016). We tested the hypothesis that AS and conserved AS is enriched in syntenic vs. nonsyntenic regions across the grass species. There are 11,996 syntenic gene clusters across maize, sorghum, foxtail millet, rice, and Brachypodium. In each of five grass species, we compared the proportion of genes within syntenic regions that undergo AS vs. nonsyntenic genes that undergo AS, and the proportion of genes within syntenic regions that have conserved AS events vs. the proportion of nonsyntenic genes that have conserved AS events. The proportion of genes within syntenic regions that exhibit AS is approximately twice that of genes in nonsyntenic regions (Figure 3). This trend is consistent across the grass species analyzed. Similarly, the proportion of genes that have conserved AS events that reside in syntenic regions is approximately twice that of genes with conserved AS events that do not reside within syntenic regions (Figure 3). This suggests that genes residing within conserved synteny blocks in the grass species studied are enriched in both AS, and conserved AS events.
Figure 3.
AS and conserved AS enriched across grass syntenic genes in (A) maize, (B) sorghum, (C) rice, (D) foxtail millet, and (E) Brachypodium. We calculated the percentage of AS in syntenic and nonsyntenic genes (the percentage of AS genes are labeled in blue). In addition, we calculated the percentage of conserved AS in syntenic AS genes and nonsyntenic AS genes (the percentage of conserved AS genes are labeled in black).
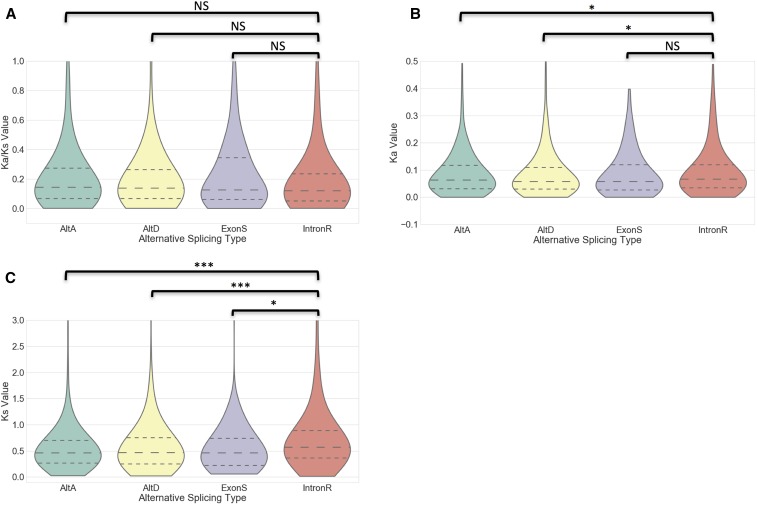
We examined selection pressure at the amino acid level on AS events conserved within grasses by performing pairwise comparisons of Ka/Ks ratios across the SASTs flanking each conserved AS event (Figure 4). Requiring Ka values of Ka < 0.5, Ks < 5, and Ka/Ks < 2, resulted in a collection of SAST alignments from 886 IntronR, 879 AltA, 424 AltD, and 169 ExonS of remained, respectively. In general, Ka/Ks values of SAST alignments flanking conserved AS events are < 1 across different AS types. There are no significant Ka/Ks differences between IntronR and other types of AS events (Figure 4). There are no single SAST alignments with Ka/Ks > 1 that pass a Fisher’s exact test for differences from neutrality, indicating that SAST alignments under positive selection were not detected. We did observe that the Ka of IntronR is significantly higher than that of AltA and AltD, but not ExonS. Also, the difference between Ka for IntronR vs. other types of AS events is smaller than the difference between Ks for IntronR vs. other types of AS events (Figure 4), which leads to differences in the Ka/Ks values between IntronR and other types of AS events being nonsignificant.
Figure 4.
Violinplot of Ka/Ks, Ka, Ks on SASTs in conserved AS events across syntenic genes within grass species. The pairwise Ka/Ks (A), Ka (B), and Ks (C) for SASTs flanking conserved AS events in five grass species were calculated and plotted. Only alignments >150 bp in length with gaps <5% of the total aligned length were considered. We only kept the Ka/Ks results if Ka < 0.5, Ks < 5 and Ka/Ks ratios < 2. The dotted lines within the violinplots represented the first, second, and third quartile. Wilcoxon Rank-Sum Tests were performed between different splicing event types (NS, Not significant; * P < 0.05, *** P < 0.001).
Conserved AS is more common among single copy genes
We tested the functional sharing hypothesis between single-copy genes vs. nonsingle-copy genes by measuring the number of genes within each category that exhibit conserved AS events; 2985 single-copy genes, including “strict” and “mostly” single copy, were described in Arabidopsis (De Smet et al. 2013). Han et al. (2014) suggested single-copy genes had increased levels of AS relative to genes from large gene families, and a decreased proportion of AS compared to genes from small gene families. Similarly, Su and Gu (2012) suggested that single copy genes had more isoforms per gene compared to the genes from large gene families, but similar average isoform counts per gene compared to the genes from small gene families.
We compared the number of genes with conserved AS events between single-copy genes vs. nonsingle-copy genes, irrespective of gene family size. Out of 2985 single-copy genes, 2717 (91.0%) are multi-exon genes, and 1113 (40.8%) of these have evidence of AS in Arabidopsis based on the Araport11 transcripts. In contrast, 6751 (34.3%) of the 19,700 nonsingle-copy multi-exon genes exhibit AS. Among multi-exon genes, the single-copy genes have significantly more AS compared to multi-exon genes (P < 0.0001, Chi-square with Yates’ correction). Among the 944 Arabidopsis genes with conserved AS events in our analysis, 173 came from the 1113 (15.5%) single-copy genes that undergo AS, while the remaining 771 came from the 6751 (11.4%) nonsingle-copy genes that undergo AS. Single-copy genes in Arabidopsis are also significantly enriched in conserved AS events (P < 0.0001, Chi-square with Yates’ correction). These results suggest that, among multi-exon genes in Arabidopsis, a greater proportion of single copy genes both undergo AS and harbor conserved AS events than do nonsingle-copy genes.
Conserved AS is more common among SR protein genes relative to hnRNP proteins
SR and hnRNP proteins are families of splicing regulatory proteins that contain RNA binding domains (reviewed in Barbazuk et al. 2008). AS events were detected in SR proteins, confirming previous observations (Kalyna et al. 2006; Rauch et al. 2014; Chamala et al. 2015). Out of 21 SR proteins in maize, 19 demonstrate AS, and 13 of these have 24 conserved AS events with other species, including 13 IntronR, four AltA, four AltD, and three ExonS events (Table S5). Out of 24 conserved AS events, 10 are present in at least five species. Zm-SCL30 (GRMZM2G065066), previously identified in maize, responds to both cold and heat (Mei et al. 2017). SCL30 has AltA, AltD, and ExonS events conserved across grass species and Amborella, as well as two IntronR events only observed within the grass lineage. Altogether, there are four maize SR proteins (two in the RS subfamily, one in the SC subfamily, and one in the SCL subfamily) that demonstrate six conserved AS events with Amborella (Table S5) supporting the existence of a deeply conserved splicing mechanism in SR proteins. Out of 40 hnRNP proteins in maize, 10 exhibit evidence of 17 conserved AS events: 10 IntronR, four AltA, two AltD, and one ExonS event (Table S5). None of these 17 AS events are conserved across more than four species. Three of the 17 conserved AS events from three hnRNP genes in maize are conserved with Amborella (Table S5), while six out of 24 AS events from four SR proteins in maize are conserved with Amborella. These results suggest that the signals for AS within SR protein genes across species are better conserved than those within the hnRNP-protein-encoding genes.
Conservation and divergence of SR proteins and their AS isoforms after whole genome duplication
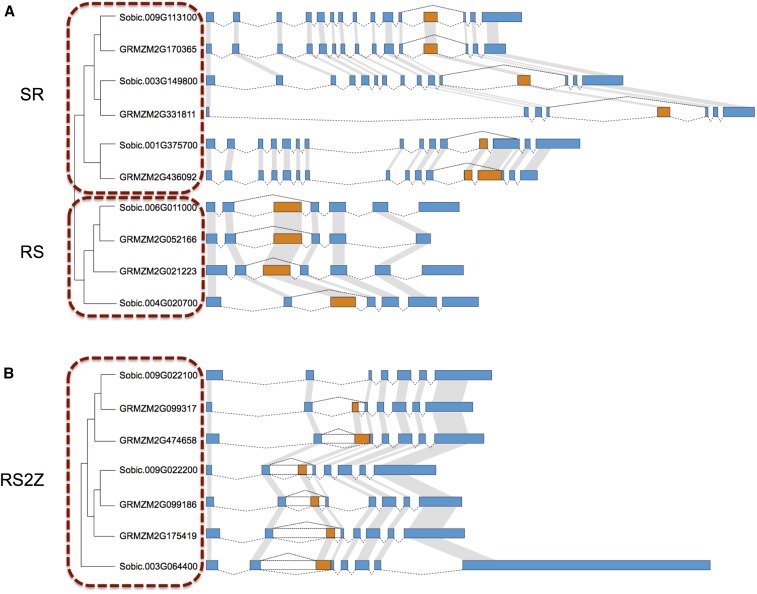
Conserved AS events across the SR protein subfamilies in maize and sorghum were identified using a phylogenetic approach. An ancestral ExonS event is conserved across all members of this subfamily in maize and sorghum (Figure 5A). Intriguingly, all three maize SR subfamily genes were reduced to one copy after whole genome duplication; one gene lost a homeologous copy from maize subgenome1, and two genes lost their homeologous copies from subgenome2. The plant-specific RS subfamily also has a shared ExonS event in both maize and sorghum (Figure 5A). The exon skipping isoform generates a complete RNA Recognition Motif (RRM) domain and is known to be conserved between green algae and some angiosperms (Kalyna et al. 2006). Our analysis confirms that this exon skipping event is conserved between maize and rice, and is also present within sorghum, Brachypodium, foxtail millet, and Amborella. Maize maintained both postwhole-genome-duplication copies of the plant-specific RS2Z subfamily genes. Remarkably, two sorghum genes Sobic.009G022100 and Sobic.009G022200 are next to each other but on opposite strands, suggesting one copy may have resulted from tandem duplication. Phylogenetic evidence suggests Sobic.009G022200 is the older copy (Richardson et al. 2011). In addition to Sobic.009G022200, and two duplicate genes in maize retaining an ExonS event, they also contain two IntronR events flanking the ExonS event (Figure 5B). Two duplicate genes in maize (GRMZM2G099317 and GRMZM2G474658) apparently gained additional IntronR and AltA events compared to Sobic.009G022100 (Figure 5B). Sobic.009G022100 preserved only the ExonS isoform, which would generate a complete RRM domain, while GRMZM2G099317 also gained the IntronR and AltA events on the right side of the ExonS event. For the second copy, GRMZM2G474658, the AltA and IntronR events on the left side are gained. There is one additional sorghum gene Sobic.003G064400 without conserved synteny in maize, which suggests that maize might have lost both syntenic copies that had represented orthologs of this sorghum gene. The AS pattern of this gene is similar to that of GRMZM2G474658 (Figure 5B). The alternative long intron in the RS2Z subfamily is also conserved from mosses to angiosperms (Kalyna et al. 2006).
Figure 5.
Phylogenetic tree and gene structure of SR, RS, and RS2Z subfamilies in maize and sorghum. (A) Phylogenetic tree representing the relationship of genes from SR and RS subfamilies in maize and sorghum; associated gene structures are illustrated by Fancygene to the right. Constitutive exons are shaded blue, and alternative regions are indicated with orange. Transcript structure is indicated by exons connected via dashed lines; orthologous exons are indicated with gray lines. (B) Phylogenetic tree and gene structure of RS2Z subfamily in maize and sorghum are illustrated by Fancygene. Annotation and shading is described above; intron retention regions are indicated in clear boxes.
R2R3-MYB is tightly connected with a plant-specific SR protein subfamily
We used a network approach to identify the degree of interactivity of TFs with conserved AS in response to five stresses (ABA, drought, cold, NaCl, and light) based on protein–protein interactions in STRING v10 (Szklarczyk et al. 2015). Several interesting relationships were identified in the network (Figure S4 in File S1). Ubiquitin genes such as UBQ10, UBQ11, and UBQ14 are centralized in the network, suggesting that AS may play a role in the susceptibility of translated isoforms to ubiquitin-mediated degradation. Ubiquitin genes are closely associated with splicing related genes (such as RS2Z33, RS41, RS2Z32, RS40, U2AF65A, SR45, and SCL33) via Glycine Rich Protein 7 (GRP7). GRP7 is part of the circadian clock and negatively autoregulates its own protein abundance by producing a nonproductive AS isoform that is subject to nonsense-mediated decay (Schöning et al. 2007, 2008). In addition, we observe an association between R2R3-MYB genes and the splicing factor network through the proline-rich spliceosome-associated protein (AT4G21660) (Figure S4 in File S1). The R2R3-MYB family has undergone expansion in plants (Du et al. 2015), and plays many important plant-specific roles. Furthermore, there are many kinase- and phosphatase-related genes in close association with genes responsible for phosphorylation and dephosphorylation of splicing regulators, such as serine-threonine protein kinase (CIPK3), calcium-dependent protein kinase 29 (CPK29), CBL-interacting protein kinase 9 (CIPK9), and calcineurin b-like protein (CBL1). CBL1 is a salt-tolerance gene and connects with components of the spliceosome (Feng et al. 2015). CBL2 and CBL3, together with CIPK3 and CIPK9, can form a calcium network to regulate magnesium levels, while CBL1 is a calcium sensor (Tang et al. 2015). Here, AS of CBL1 and CIPK3, CIPK9 could potentially contribute in regulating magnesium levels through a feedback loop sensitive to calcium concentration.
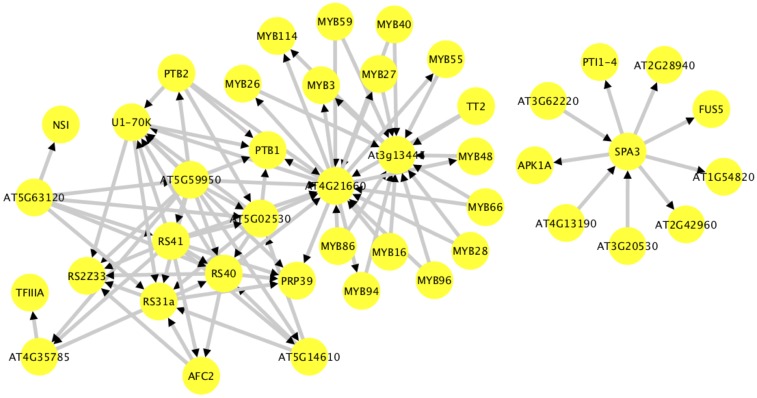
We also examined networks containing Arabidopsis genes with AS events conserved with Amborella and at least one species of monocot. Most of the genes present in the network were from the plant-specific SR protein subfamilies, such as RS2Z, RS, and SR45a. In addition, TFIIIA is connected to the splicing protein network via RNA-binding protein (AT4G35785). TFIIIA has an ExonS event that is conserved across the land plants (Fu et al. 2009; Barbazuk 2010). Similar to the network based on TFs in response to stress, we identified a close connection between the R2R3-MYB class with a splicing protein network via TFIID-1 (AT3G13445), and proline-rich spliceosome-associated family protein (AT4G21660) (Figure 6). These R2R3-MYBs are involved in many important functions, such as glucosinolate biosynthesis, phenylpropanoid pathway, conical epidermal cell outgrowth, drought, ABA- and JA-mediated pathogens, and cuticular wax biosynthesis (Table S6).
Figure 6.
Protein–protein network of Arabidopsis genes with evidence of conserved AS with Amborella and at least one species of monocot.
Discussion
In this study, we utilized a bioinformatics scheme to detect AS events and identify those that are conserved across nine plant species from publicly available RNA-Seq data. In total, across all nine species, 9804 AS events are conserved between two or more species. Newly sequenced plant genomes are published, and vast amounts of RNA-Seq data are increasingly deposited into NCBI’s Sequence Read Archive (SRA). These are treasure troves of data waiting to be mined for new discoveries. Many are already tapping into this wealth, as exemplified by Nellore et al. (2016), who examined splice junction variants in the human genome, and Chamala et al. (2015) who examined conserved AS events across several species of eudicots. The method we describe here can be used to mine to identify conserved AS events within any species with an available genome sequence and deep RNA-Seq data.
AS events are widely conserved across species but many have undergone complicated patterns of gain and loss
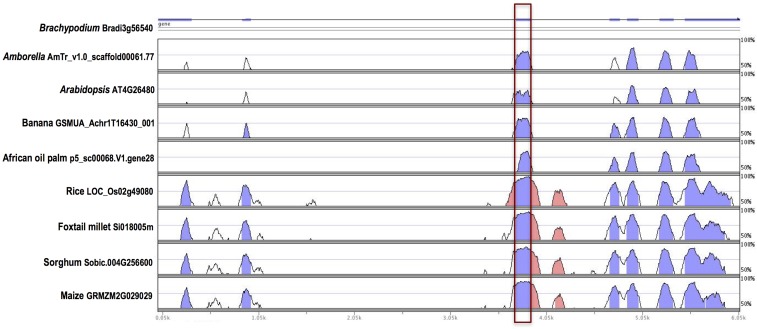
Our analysis identified Arabidopsis as having the least number of genes with conserved AS (944), which may be a result of it being the only eudicot considered in the study, while maize exhibited the greatest number of genes harboring conserved AS events (3687) (Figure 2). The smallest percentage of conserved AS genes among genes that undergo AS was found for Arabidopsis (12.0%), while foxtail millet has the greatest proportion of genes harboring conserved AS events among those genes undergoing AS (36.2%). We identified 1413 genes with conserved AS events in Amborella, which account for 18.3% of the genes in Amborella with evidence of AS. The public sequence collections used identified that between 1000 and 3000 genes undergo AS within each of the species examined. The sequence datasets have overlapping but nonidentical tissue representation and sequence depth. This likely results in a reduction in our ability to detect some events in some species, and may result in the miss-assignment of some events as nonconserved. Therefore, we expect that accuracy in identifying conserved vs. nonconserved events will improve as additional data become available. As an example, the RNA-Seq data identified an ExonS event within a gene encoding an RNA-binding KH domain-containing protein that was conserved across eight of the nine species examined. The RNA-Seq data did not detect this event in Arabidopsis; however, this isoform is reported within the AtRTD Arabidopsis transcriptome data (Zhang et al. 2015), indicating that it is broadly conserved across all species investigated here. RNA-binding KH domain proteins are vital for heat stress-responsive gene regulation (Guan et al. 2013). The alternatively spliced exon within this RNA-binding KH domain-containing protein has high sequence similarity across the nine species studied (Figure 7), and this AS event may be conserved broadly across angiosperms. We identified significant enrichment of AltA and AltD events that were conserved in at least three species relative to those conserved in only two species, and a significant enrichment of ExonS events conserved in at least four species relative to those conserved in only two species. Conserved retained introns within Arabidopsis and Brachypodium have significantly shorter lengths compared to nonconserved retained introns. Conserved retained introns in foxtail millet have shorter mean lengths but no appreciable differences in median lengths vs. nonconserved retained introns (Table 3). However, this trend is not observed across other species we examined, which suggests that other factors contribute to the lengths of conserved and nonconserved retained introns.
Figure 7.
Vista plots illustrating sequence conservation within a gene encoding an RNA-binding KH domain-containing protein that produces an exon skipping event conserved across Brachypodium, Amborella, Arabidopsis, banana, African oil palm, rice, foxtail millet, sorghum, and maize. The exon skipping in Arabidopsis is not included in the Araport11, but detected in AtRTD. The alternative exon (boxed in red) is highly conserved between Brachypodium and its orthologs across the other plant species examined. Levels of sequence identity between Brachypodium and its orthologs in the eight other plants species displayed are depicted as blue peaks. Pink bars signify segments that pass the alignment criteria of 70% identity over a 100 bp window.
The plant-specific RS and RS2Z subfamilies associate with the R2R3-MYB class within the network built from Arabidopsis genes demonstrating conserved AS with Amborella and at least one species of monocots (Figure 6). Many R2R3-MYB genes are represented in this network, and possess plant-specific functions (Table S6). A connection between R2R3-MYB family and splice factors is also detected within networks build around Arabidopsis TFs that respond to stress. Li et al. (2006) identified conserved AS events within MYB59 and MYB48. Both genes are represented within our network and associate with SR proteins via a proline-rich spliceosome-associated protein AT4G21660 (Figure 6). Together, these results suggest a conserved connection between splicing factors and R2R3-MYB TFs. These two protein classes may cooperate during developmental processes and stress response in plants. In addition, natural variation in Arabidopsis contributes to splicing changes in the proline synthesis enzyme (P5CS1) affecting drought-induced proline accumulation. This suggests that AS may play a role in adaptation to environmental challenges (Kesari et al. 2012).
Conserved AS events are enriched within grasses
Within monocots, we identified lineage-specific conserved AS events. There are 239 AS events conserved between banana, African oil palm, and at least one grass species, while an additional 204 AS events conserved between banana and African oil palm are absent in the grass species examined and were likely lost. The number of conserved AS between two species in the PACMAD clade (maize, sorghum, and foxtail millet) is similar to the number of conserved AS events between two species within the BEP clade (rice and Brachypodium). There were 224 conserved AS events found conserved across five grass species examined. In addition, the proportion of genes within syntenic blocks with AS events in the grasses is approximately twice that compared to those outside of syntenic blocks, and this trend is also mirrored across those genes with conserved AS events (Figure 3).
While KaKs analysis on regions flanking conserved AS revealed no evidence of positive selection, we did observe elevated Ks values associated with regions flanking IntronR compared to other AS types. This suggests that these regions may be functionally constrained and undergoing purifying selection, perhaps to maintain cis-regulatory signals required for AS. Some conserved intron retention events exhibit high cross-species sequence conservation based on multiple sequence alignments (Figure S1 in File S1). These retained introns may have conserved RNA structure that participates in the switch between splicing and retention analogous to the situation seen for TFIIIA (Fu et al. 2009), or may play a role in proteome plasticity. Previous studies have suggested only moderate selection pressure on the alternative vs. the constitutive regions (Xing and Lee 2005, 2006; Chen et al. 2006). However, the selection on the regions flanking conserved AS events is less explored, particularly in plants. Our study reveals the flanking exonic regions of conserved AS events are under purifying selection, perhaps preserving splicing regulatory elements.
We observed both conserved and divergent patterns of AS within SR-protein-encoding genes after the whole genome duplication event in the maize lineage. The plant-specific RS subfamily in maize and sorghum harbor conserved ExonS events, although the maize ortholog has been reduced to a single copy after whole genome duplication (Figure 5A). In the RS2Z family, maize has retained both homeologous copies of the RS2Z subfamily genes. In one case, it has maintained the conserved AS events in both maize homeologues including ExonS and IntronR events (Figure 5B); however, in another case, the AS events have diverged between the sorghum gene and its two maize homeologues (Figure 5B). Whether these divergent patters are truly indicative of sub or neofunctionalized genes needs to be experimentally confirmed.
Important functional conserved AS in SR protein in response to environmental stress
Many TFs that respond to ABA, salt (NaCl), drought, light, and cold stress have conserved AS. Two genes—AT1G78070 (Transducin/WD40 repeat-like superfamily protein) and AT4G27410 (RD26)—are expressed during all five stresses. RD26 encodes a NAC TF induced in response to desiccation. In our data, RD26 has conserved AS in seven out of the nine species we examined (except sorghum and Brachypodium). As illustrated in Figure S5 in File S1, the exonic flanking sequences of this IntronR event are the sites of the highest sequence conservation within this gene across the nine species examined. This conserved IntronR event may be broadly conserved across angiosperms.
The protein–protein interaction network based on Arabidopsis stress response TFs with conserved AS events places phosphatases, kinases, and several ubiquitin genes in the center of the network (Figure S4 in File S1). This result is in line with the recent discovery that alternative exons and exitrons (exon-like introns) have more phosphorylation sites and ubiquitination sites compared to constitutive exons (Marquez et al. 2015). In these genes with conserved AS across the angiosperms, AS may be involved in signaling pathways responsible for de/phosphorylation and protein degradation pathways, which suggests an important regulatory role in plants. Another interesting finding in the network is SPA3 (Figure 6). In moss, red and blue light photoreceptors were shown to regulate AS, and intron retention was misregulated in moss mutants defective in red light sensing phytochromes (Wu et al. 2014), which suggests an ancient connection between light regulation and conserved AS in land plants. We found both conserved alternative donor and intron retention events in SPA3, which acts as a negative regulator during light signaling via suppression of photomorphogenesis (Laubinger and Hoecker 2003). SPA3 is connected to nine additional kinase genes in the network, all of which have conserved AS across the angiosperms (Figure 6). This suggests that AS of kinases might play an important role in the regulation of the light signal, perhaps by affecting a kinase-mediated signal cascade linked to SPA3.
The conserved AS landscape in plants is complex, but likely identifies events of functional significance. Collections of evolutionarily conserved AS events provide a foundation for additional efforts to link functions and phenotypes to these ancient event and will aid in identifying sequence that participate in the regulation of these events.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300189/-/DC1.
Acknowledgments
We thank Daniel Gates, Emily Josephs, and Michelle Corey Stitzer for providing helpful comments for the manuscript. This work was supported by the Department of Biological Sciences at University of Florida, The Florida Genetics Institute, A Graduate Student Fellowship and College of Liberal Arts and Sciences Dissertation Fellowship from University of Florida awarded to W.M., and National Science Foundation grants IOS-0922742 & IOS-1547787 (W.B.B.).
Author contributions: W.M. and W.B.B. designed the work. W.M., W.B.B., L.B., G.F., and J.C.S. analyzed the data. W.M. and W.B.B. wrote the manuscript with input from L.B., G.F., and J.C.S. The authors declare that they have no competing interests.
Footnotes
Communicating editor: J. Birchler
Literature Cited
- Abdel-Ghany S. E., Hamilton M., Jacobi J. L., Ngam P., Devitt N., et al. , 2016. A survey of the sorghum transcriptome using single-molecule long reads. Nat. Commun. 7: 11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amborella Genome Project2013. The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Barbazuk W. B., 2010. A conserved alternative splicing event in plants reveals an ancient exonization of 5S rRNA that regulates TFIIIA. RNA Biol. 7: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk W. B., Fu Y., McGinnis K. M., 2008. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 18: 1381–1392. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Schmutz J., Wang H., Percifield R., Hawkins J., et al. , 2012. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30: 555–561. [DOI] [PubMed] [Google Scholar]
- Busch A., Hertel K. J., 2012. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA 3: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. A., Haas B. J., Hamilton J. P., Mount S. M., Buell C. R., 2006. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 7: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamala S., Feng G., Chavarro C., Barbazuk W. B., 2015. Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front. Bioeng. Biotechnol. 3: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.-C., Wang S.-S., Chen C.-J., Li W.-H., Chuang T.-J., 2006. Alternatively and constitutively spliced exons are subject to different evolutionary forces. Mol. Biol. Evol. 23: 675–682. [DOI] [PubMed] [Google Scholar]
- Cheng C.-Y., Krishnakumar V., Chan A., Schobel S., Town C. D., 2017. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89: 789–804. [DOI] [PubMed] [Google Scholar]
- Chuang T.-J., Yang M.-Y., Lin C.-C., Hsieh P.-H., Hung L.-Y., 2015. Comparative genomics of grass EST libraries reveals previously uncharacterized splicing events in crop plants. BMC Plant Biol. 15: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton J. L., Wysocki W. P., Clark L. G., Kelchner S. A., Pires J. C., et al. , 2015. Resolving deep relationships of PACMAD grasses: a phylogenomic approach. BMC Plant Biol. 15: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darracq A., Adams K. L., 2013. Features of evolutionarily conserved alternative splicing events between Brassica and Arabidopsis. New Phytol. 199: 252–263. [DOI] [PubMed] [Google Scholar]
- De Smet R., Adams K. L., Vandepoele K., Van Montagu M. C., Maere S., et al. , 2013. Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc. Natl. Acad. Sci. USA 110: 2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hont A., Denoeud F., Aury J.-M., Baurens F.-C., Carreel F., et al. , 2012. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488: 213–217. [DOI] [PubMed] [Google Scholar]
- Droc G., Lariviere D., Guignon V., Yahiaoui N., This D., et al. , 2013. The banana genome hub. Database 2013: bat035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Liang Z., Zhao S., Nan M.-G., Tran L. S., et al. , 2015. The evolutionary history of R2R3-MYB proteins across 50 eukaryotes: new insights into subfamily classification and expansion. Sci. Rep. 5: 11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z., 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick J., Fu A., Muppirala U., Sabharwal M., Wilkerson M. D., et al. , 2008. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 36: D959–D965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., Kelly S., 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Li J., Gao Z., Lu Y., Yu J., et al. , 2015. SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant 8: 1038–1052. [DOI] [PubMed] [Google Scholar]
- Filichkin S. A., Priest H. D., Givan S. A., Shen R. K., Bryant D. W., et al. , 2010. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Bannach O., Chen H., Teune J.-H., Schmitz A., et al. , 2009. Alternative splicing of anciently exonized 5S rRNA regulates plant transcription factor TFIIIA. Genome Res. 19: 913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S. A., Ricke D., Lan T.-H., Presting G., Wang R., et al. , 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., et al. , 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Wen C., Zeng H., Zhu J., 2013. A KH domain-containing putative RNA-binding protein is critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Mol. Plant 6: 386–395. [DOI] [PubMed] [Google Scholar]
- Haas B. J., Delcher A. L., Mount S. M., Wortman J. R., Smith R. K., Jr, et al. , 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31: 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond M. C., Wachter A., Breaker R. R., 2009. A plant 5S ribosomal RNA mimic regulates alternative splicing of transcription factor IIIA pre-mRNAs. Nat. Struct. Mol. Biol. 16: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Peng Y., Xu L., Xiao P., 2014. Identification, characterization, and utilization of single copy genes in 29 angiosperm genomes. BMC Genomics 15: 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. B., Syed N. H., Bordage S., Marshall J., Nimmo G. A., et al. , 2012. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24: 961–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.-k., Liu Y.-l., Xia E.-h., Gao L.-z., 2013. Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol. 161: 1844–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., Lopato S., Voronin V., Barta A., 2006. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 34: 4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesari R., Lasky J. R., Villamor J. G., Des Marais D. L., Chen Y.-J. C., et al. , 2012. Intron-mediated alternative splicing of Arabidopsis P5CS1 and its association with natural variation in proline and climate adaptation. Proc. Natl. Acad. Sci. USA 109: 9197–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Hoecker U., 2003. The SPA1‐like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J. 35: 373–385. [DOI] [PubMed] [Google Scholar]
- Lawrence C. J., Dong O. F., Polacco M. L., Seigfried T. E., Brendel V., 2004. MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res. 32: D393–D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Q., Dang C. C., Gascuel O., 2012. Modeling protein evolution with several amino acid replacement matrices depending on site rates. Mol. Biol. Evol. 29: 2921–2936. [DOI] [PubMed] [Google Scholar]
- Lee Y., Rio D. C., 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li X., Guo L., Lu F., Feng X., et al. , 2006. A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J. Exp. Bot. 57: 1263–1273. [DOI] [PubMed] [Google Scholar]
- Li Q., Xiao G., Zhu Y.-X., 2014. Single-nucleotide resolution mapping of the Gossypium raimondii transcriptome reveals a new mechanism for alternative splicing of introns. Mol. Plant 7: 829–840. [DOI] [PubMed] [Google Scholar]
- Mandadi K. K., Scholthof K.-B. G., 2015. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 27: 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y., Brown J. W. S., Simpson C., Barta A., Kalyna M., 2012. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22: 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y., Höpfler M., Ayatollahi Z., Barta A., Kalyna M., 2015. Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res. 25: 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo A. M., Marone D., Laidò G., De Leonardis A. M., De Vita P., 2012. Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 185: 40–49. [DOI] [PubMed] [Google Scholar]
- Mei W., Liu S., Schnable J. C., Yeh C.-T., Springer N. M., et al. , 2017. A comprehensive analysis of alternative splicing in paleopolyploid maize. Front. Plant Sci. 8: 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naika M., Shameer K., Mathew O. K., Gowda R., Sowdhamini R., 2013. STIFDB2: an updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol. 54: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellore A., Jaffe A. E., Fortin J.-P., Alquicira-Hernández J., Collado-Torres L., et al. , 2016. Human splicing diversity and the extent of unannotated splice junctions across human RNA-seq samples on the sequence read archive. Genome Biol. 17: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panahi B., Abbaszadeh B., Taghizadeghan M., Ebrahimie E., 2014. Genome-wide survey of alternative splicing in Sorghum bicolor. Physiol. Mol. Biol. Plants 20: 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., et al. , 2009. The sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., et al. , 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M., Drost H.-G., Gabel A., Ullrich K. K., Bönn M., et al. , 2012. A transcriptomic hourglass in plant embryogenesis. Nature 490: 98. [DOI] [PubMed] [Google Scholar]
- Rambaldi D., Ciccarelli F. D., 2009. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics 25: 2281–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch H. B., Patrick T. L., Klusman K. M., Battistuzzi F. U., Mei W., et al. , 2014. Discovery and expression analysis of alternative splicing events conserved among plant SR proteins. Mol. Biol. Evol. 31: 605–613. [DOI] [PubMed] [Google Scholar]
- Reddy A. S., Marquez Y., Kalyna M., Barta A., 2013. Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S. N., Rogers M. F., Richardson D. N., Hamilton M., Ben-Hur A., 2012. Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front. Plant Sci. 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D. N., Rogers M. F., Labadorf A., Ben-Hur A., Guo H., et al. , 2011. Comparative analysis of serine/arginine-rich proteins across 27 eukaryotes: insights into sub-family classification and extent of alternative splicing. PLoS One 6: e24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J. C., Freeling M., Lyons E., 2012. Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol. Evol. 4: 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schöning J. C., Streitner C., Page D. R., Hennig S., Uchida K., et al. , 2007. Auto‐regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 52: 1119–1130. [DOI] [PubMed] [Google Scholar]
- Schöning J. C., Streitner C., Meyer I. M., Gao Y., Staiger D., 2008. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 36: 6977–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severing E. I., Dijk A. D., Stiekema W. J., Ham R. C., 2009. Comparative analysis indicates that alternative splicing in plants has a limited role in functional expansion of the proteome. BMC Genomics 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhou Z., Wang Z., Li W., Fang C., et al. , 2014. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26: 996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H., Hanada K., Ushijima T., Nakashima M., Suzuki Y., et al. , 2014. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 18781–18786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Ong-Abdullah M., Low E.-T. L., Manaf M. A. A., Rosli R., et al. , 2013. Oil palm genome sequence reveals divergence of interfertile species in old and new worlds. Nature 500: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J. W., 2013. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitner C., Simpson C. G., Shaw P., Danisman S., Brown J. W., et al. , 2013. Small changes in ambient temperature affect alternative splicing in Arabidopsis thaliana. Plant Signal. Behav. 8: 11240–11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D., Malone J. H., Sun X., Smith H. E., Rabinow L., et al. , 2013. Design of RNA splicing analysis null models for post hoc filtering of Drosophila head RNA-Seq data with the splicing analysis kit (Spanki). BMC Bioinformatics 14: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Gu X., 2012. Revisit on the evolutionary relationship between alternative splicing and gene duplication. Gene 504: 102–106. [DOI] [PubMed] [Google Scholar]
- Su Z. X., Wa J. M., Yu J., Huang X. Q., Gu X., 2006. Evolution of alternative splicing after gene duplication. Genome Res. 16: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed N. H., Prince S. J., Mutava R. N., Patil G., Li S., et al. , 2015. Core clock, SUB1, and ABAR genes mediate flooding and drought responses via alternative splicing in soybean. J. Exp. Bot. 66: 7129–7149. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., et al. , 2015. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43: D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R.-J., Zhao F.-G., Garcia V. J., Kleist T. J., Yang L., et al. , 2015. Tonoplast CBL–CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: 3134–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher S. R., Zhou W., Leonard A., Wang B.-B., Beatty M., et al. , 2014. Genome-wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell 26: 3472–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher S. R., Danilevskaya O. N., Meng X., Beatty M., Zastrow-Hayes G., et al. , 2016. Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 170: 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. P., Garvin D. F., Mockler T. C., Schmutz J., Rokhsar D., et al. , 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- Walley J. W., Sartor R. C., Shen Z., Schmitz R. J., Wu K. J., et al. , 2016. Integration of omic networks in a developmental atlas of maize. Science 353: 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters B., Lum G., Sablok G., Min X. J., 2013. Genome-wide landscape of alternative splicing events in Brachypodium distachyon. DNA Res. 20: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhang Y., Zhang Z., Zhu J., Yu J., 2010. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8: 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Burge C. B., 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.-P., Su Y., Chen H.-C., Chen Y.-R., Wu C.-C., et al. , 2014. Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 15: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Lee C., 2005. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc. Natl. Acad. Sci. USA 102: 13526–13531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Lee C., 2006. Alternative splicing and RNA selection pressure—evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 7: 499–509. [DOI] [PubMed] [Google Scholar]
- Xu P., Kong Y., Song D., Huang C., Li X., et al. , 2014. Conservation and functional influence of alternative splicing in wood formation of Populus and Eucalyptus. BMC Genomics 15: 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Calixto C. P., Tzioutziou N. A., James A. B., Simpson C. G., et al. , 2015. AtRTD–a comprehensive reference transcript dataset resource for accurate quantification of transcript‐specific expression in Arabidopsis thaliana. New Phytol. 208: 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zhang N., Ma P.-F., Liu Q., Li D.-Z., et al. , 2013. Phylogenomic analyses of nuclear genes reveal the evolutionary relationships within the BEP clade and the evidence of positive selection in Poaceae. PLoS One 8: e64642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
AS events table and GFF files for the AS isoforms in seven species (Setaria italica, Oryza sativa L. ssp. Japonica, Brachypodium distachyon, Musa acuminata, and Elaeis guineensis, the eudicot Arabidopsis thaliana and the basal angiosperm Amborella) are made available to the community via figshare (https://doi.org/10.6084/m9.figshare.4789153.v2) and github (https://github.com/wenbinmei/Monocot_conserved_splicing), CoGe, the Comparative Genomics Platform. (https://www.genomevolution.org/). Scripts used in the analysis are available at the same github repository above. The Zea mays and Sorghum bicolor data are already available to the community at figshare (https://doi.org/10.6084/m9.figshare.4205079.v3).