Abstract
Eukaryotic chromosome segregation requires a protein complex known as the kinetochore that mediates attachment between mitotic spindle microtubules and centromere-specific nucleosomes composed of the widely conserved histone variant CENP-A. Mutations in kinetochore proteins of the fission yeast Schizosaccharomyces pombe lead to chromosome missegregation such that daughter cells emerge from mitosis with unequal DNA content. We find that multiple copies of Msc1—a fission yeast homolog of the KDM5 family of proteins—suppresses the temperature-sensitive growth defect of several kinetochore mutants, including mis16 and mis18, as well as mis6, mis15, and mis17, components of the Constitutive Centromere Associated Network (CCAN). On the other hand, deletion of msc1 exacerbates both the growth defect and chromosome missegregation phenotype of each of these mutants. The C-terminal PHD domains of Msc1, previously shown to associate with a histone deacetylase activity, are necessary for Msc1 function when kinetochore mutants are compromised. We also demonstrate that, in the absence of Msc1, the frequency of localization to the kinetochore of Mis16 and Mis15 is altered from wild-type cells. As we show here for msc1, others have shown that elevating cnp1 levels acts similarly to promote survival of the CCAN mutants. The rescue of mis15 and mis17 by cnp1 is, however, independent of msc1. Thus, Msc1 appears to contribute to the chromatin environment at the centromere: the absence of Msc1 sensitizes cells to perturbations in kinetochore function, while elevating Msc1 overcomes loss of function of critical components of the kinetochore and centromere.
Keywords: Msc1, CENP-A, Mis6, Mis15, Mis16/Mis18, Mis17, CCAN, lysine demethylase KDM5, RBP2, PLU-1, kinetochore, centromere
SEGREGATION of the genome at mitosis relies on the faithful association of spindle pole body-derived microtubules with regions of individual chromosomes that mediate chromosome attachment to the mitotic spindle. These chromosomal regions are epigenetically marked with a variant of canonical histone H3 known as CENP-A (Palmer et al. 1987; Allshire and Ekwall 2015). CENP-A-containing nucleosomes are found at centromeres in eukaryotic organisms ranging from unicellular budding and fission yeast, to human cells. CENP-A nucleosomes recruit complexes of proteins that form the kinetochore—the structure that mediates dynamic microtubule interactions with the mitotic spindle (Cheeseman 2014). Characteristics of the centromere vary between organisms: budding yeast chromosomes have point centromeres consisting of a single CENP-A-containing nucleosome, whereas fission yeast and human chromosomes have regional centromeres consisting of an array of CENP-A-containing nucleosomes (Westhorpe and Straight 2015). Regional centromeres of the three fission yeast chromosomes consist of distinct organizational structures: a central core (cnt) with flanking inverted repeats (imr), all of which have associated CENP-A nucleosomes, and large heterochromatic repeats to the outside of the imr region (otr) that contain histone H3 modified by methylation of lysine 9 in its N-terminal tail. Some report the presence of the histone H2A variant, H2A.Z in the otr region as well (Lawrence and Volpe 2009; Qiu et al. 2010).
Kinetochores are protein structures that associate with CENP-A-containing nucleosomes to mediate the dynamic association between chromosomes and the growing and shrinking microtubules of the mitotic spindle (reviewed in Cheeseman 2014). Kinetochores of both fission yeast and mammalian cells include a complex of conserved proteins, collectively known as the Constitutive Centromere-Associated Network or CCAN, which consists of more than a dozen proteins that tend to remain with the centromere throughout the cell cycle (McAinsh and Meraldi 2011). As individual components of the CCAN are further investigated, evidence is emerging for dynamic association of some CCAN components as cells progress through the cell cycle (Hellwig et al. 2011; Fang et al. 2015; Nagpal and Fukagawa 2016).
In fission yeast, two components of the CCAN were identified in screens for mutants that exhibit a high frequency of mini-chromosome loss: Mis6 (CENP-I) (Takahashi et al. 1994) and Mal2 (CENP-O) (Fleig et al. 1996). Mis15 (CENP-N) and Mis17 (CENP-U) were identified in a direct visual screen of temperature-sensitive (ts) mutants that missegregate chromosomes at mitosis (Hayashi et al. 2004). Another CCAN protein, Sim4 (CENP-K), was identified in a screen for mutants that are defective for silencing in the middle of the centromere, a screen that gave rise as well to an allele of CENP-A (Pidoux et al. 2003). Several other components of the CCAN complex were identified by purification of proteins associated with Sim4 and Mal2 (Liu et al. 2005). Interestingly, only a subset of the CCAN proteins are conserved in budding yeast, suggesting that the association of the CCAN with an array of CENP-A containing nucleosomes requires a distinct set of proteins as compared to those needed to form a kinetochore on the single point centromere found in budding yeast (McAinsh and Meraldi 2011).
Direct screening for mutants defective in chromosome segregation also identified Mis16 and Mis18, proteins conserved between fission yeast and human cells (Hayashi et al. 2004). While a homolog of Mis16 is found in budding yeast (Hat2), a homolog of Mis18 is not, though Mis16 and Mis18 form a complex in both fission yeast and human cells (Fujita et al. 2007; Nardi et al. 2016; Subramanian et al. 2016). Two additional proteins, Mis19/Eic1 and Mis20/Eic2, which appear to be unique to fission yeast, associate with the Mis16/18 complex as well (Hayashi et al. 2014; Subramanian et al. 2014). The Mis16/18 complex is thought to facilitate loading of CENP-A to centromeres after DNA replication when CENP-A must be reincorporated following duplication of the chromosome and consequent dilution of the CENP-A containing nucleosomes between the daughter strands (Hayashi et al. 2004; Fujita et al. 2007). CENP-A protein levels at the central core are reduced in mis16 and mis18 strains (Hayashi et al. 2004). A histone chaperone, fission yeast Scm3, which may be analogous to human HJURP, has been described as a receptor for CENP-A loading, and proposed to act with the Mis18 complex, perhaps through histone deacetylation, to facilitate loading of CENP-A (Foltz et al. 2009; Pidoux et al. 2009; Barnhart et al. 2011).
Msc1 was identified in a fission yeast genetic screen for genes that in multiple copies suppressed the DNA damage-induced lethality of cells defective for the DNA damage checkpoint (Ahmed et al. 2004). Subsequent studies of cells disrupted for the msc1 gene revealed a role for the protein in chromosome segregation (Ahmed et al. 2007), as cells lacking msc1 exhibit a chromosome loss phenotype. Further genetic analysis (Ahmed et al. 2007) revealed synthetic lethal interactions with the CCAN component mis6 (CENP-I), and with mis12, a component of the KMN protein network that is proposed to link microtubules to the kinetochore (Cheeseman et al. 2006). Double mutants of either mis6 or mis12 with a null allele of msc1 result in inviability at temperatures typically permissive for the mis mutants (Ahmed et al. 2007). Curiously, increased expression of msc1 suppresses the ts growth defect of cells with a mutation in the CENP-A encoding gene cnp1-1 (Ahmed et al. 2007).
We have demonstrated previously that Msc1 coprecipitates histone deacetylase (HDAC) activity (Ahmed et al. 2004). Given that increasing the level of Msc1 compensates for loss of cnp1-1 function, and that the Mis16/18 complex helps to maintain deacetylation of histones to facilitate CENP-A loading, we hypothesized that Msc1 might influence activity of the Mis16/18 complex. To that end, we tested whether elevating Msc1 could rescue ts mis16 or mis18 mutants, or whether deletion of msc1 exacerbates their phenotype. Indeed, at restrictive temperature, we find that extra copies of Msc1 can suppress, while deletion of msc1 exacerbates, the growth defect and chromosome missegregation phenotype of mis16 or mis18 mutants. The C-terminal PHD domains, previously shown to be necessary to precipitate HDAC activity, are necessary for Msc1 to accomplish this function. Like Msc1, the Mis16/18 complex is conserved in mammalian cells, which, like fission yeast, have regional centromeres, but Mis18 is not conserved in budding yeast, which have point centromeres. CCAN components are conserved in organisms with regional and point centromeres, and we have also examined the relationship between Msc1 and components of the CCAN. Interestingly, elevating Msc1 levels rescues ts alleles of CCAN mutants mis6, mis15, and mis17, which have been shown by others to be rescued upon increased expression of cnp1 (Hayashi et al. 2004). While elevating cnp1 or msc1 levels acts similarly to promote survival of the CCAN mutants, the rescue of mis15 and mis17 by cnp1 does not require the presence of msc1. Finally, we examine the localization of the Mis16 component of the Mis16/18 complex as well as the CCAN component Mis15 in cells lacking Msc1. Consistent with exacerbation of the mis16 phenotype by deletion of msc1, Mis16 protein is less frequently associated with the kinetochore in the absence of Msc1. In contrast, Mis15 protein is more frequently associated with the kinetochore when Msc1 is absent. We suggest that Msc1 contributes to the chromatin environment at the centromere such that in its absence, kinetochore function is compromised. On the other hand, in cells with diminished kinetochore function, elevating Msc1 can facilitate chromosome segregation and improve viability.
Materials and Methods
Strains and growth conditions
Standard Schizosaccharomyces pombe media and genetic techniques were used as described (Moreno et al. 1991). Transformation was done using the LiAC method. To construct double mutant strains, two strains with opposite mating types (h+ and h− in fission yeast) were crossed and mated on nitrogen-deficient SPA plates at 25° for at least 2 days. Asci from the crosses were incubated in 1× glusulase at 25° followed by plating on YES plates. Plasmids were transformed into strains as listed in Table 1. For spotting assays, all cells were grown at 25° in minimal media (PMA-leucine) unless otherwise noted. Cells were grown to midlog phase and fivefold serial dilutions prepared. Aliquots of each dilution were then transferred using a 48-pin Multi-Blot Replicator (VP Scientific), spotted and grown for 3–4 days at temperatures and on media as indicated in the figure legends. YES plates contain the following components per liter: 5 g yeast extract, 30 g glucose, 150 mg each of adenine, uracil, leucine, lysine and histidine, and 20 g of agar. PMA minus leucine plates contain the following components per liter: 32 g EMM (Sunrise Science Products), 150 mg each of adenine, uracil, lysine and histidine, and 20 g of agar.
Table 1. Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| SP6 | h- leu1-32 | Laboratory stock |
| NW730 | h+ msc1::kan leu1-32 | Laboratory stock |
| NW1702 | h- leu1-32 ura4+ mis6-302 | Laboratory stock |
| NW2841 | h- leu1 mis15-68 | YGRC FY20041 |
| NW2842 | h- leu1 mis16-53 | YGRC FY20042 |
| NW2843 | h- leu1 mis17-362 | YGRC FY20043 |
| NW2844 | h- leu1 mis18-262 | YGRC FY20044 |
| NW2850 | h- leu1 ura4 mis16-GFP[ura4+] | YGRC FY10474 |
| NW2851 | h- leu1 ura4 mis15-GFP[ura4+] | YGRC FY10468 |
| NW2945 | h? leu1- mis15-68 msc1::kanR | This study |
| NW2946 | h? leu1- mis16-53 msc1::kanR | This study |
| NW2947 | h? leu1- mis17-362 msc1::kanR | This study |
| NW2948 | h? leu1- mis18-262 msc1::kanR his? | This study |
| NW2949 | h- msc1::hygB mis16-GFP [ura+] leu1- | This study |
| NW2950 | h- msc1::hygB mis15-GFP [ura+] leu1- | This study |
Microscopy for mis phenotype
Cells were grown at 25° in media as indicated to midlog phase and diluted to 2.5 × 106 cells/ml. Subsequently, cells were transferred to fresh media at appropriate restrictive temperature for the period of time described in the figure legends. Methanol fixation was conducted by washing cells twice with ddH2O, then incubating cells in 100% methanol for ≤ 20 min at −20°. Cells were pelleted and incubated in 30% methanol, 1× PBS at −20°. For glutaraldehyde fixation, cells were washed with ddH2O then fixed in 0.5% glutaraldehyde for 10 min on ice. After three washes with cold ddH2O, cells were resuspended in a small volume of ddH2O. For DAPI staining, 0.4 µl of cells was mixed with 0.4 µl of 10 µg/ml DAPI solution. The DAPI-stained cell suspension was analyzed using a fluorescence microscope (Zeiss Axioplan 2) within 24 hr of fixation. Images were captured with a Zeiss AxioCam and analyzed with Openlab software.
Fluorescence microscopy
For imaging GFP signals, cells were grown at 25° to midlog phase, stained with DAPI as described above and imaged for GFP at 15–19 different focal depths through Z-stack scanning. By tracking dots throughout all individual images taken from each focal depth, we evaluated a minimum of 200 cells for each strain for the presence of discrete foci representing the centromere.
Western blot
For lysate preparation, strains were grown to midlog phase in indicated media at 25°. Cells were lysed in a buffer containing 10% 1× Protease Inhibitor (PI), 10% PMSF in 1× PBS with acid-washed glass beads in a Fastprep vortexing machine (Bio101). The following antibodies were used for Western Blot analyses: GFP B-2 (Santa Cruz), Goat anti-mouse IgG-HRP (Santa Cruz), α-tubulin T-5168 (Sigma), and polyclonal antibody to fission yeast Ded1 (Liu et al. 2002)
Data and reagent availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All strains constructed in the course of this study will be made available by request.
Results
Increased expression of msc1 improves viability of mis16/18 mutants and reduces mis16-53 chromosome missegregation
Fission yeast strains with hypomorphic alleles of kinetochore proteins encoded by mis16 and mis18 exhibit hyperacetylation of centromeric histones (Hayashi et al. 2004). Given the observed genetic interactions of msc1 with mutants defective in kinetochore function (Ahmed et al. 2007) and the observation that Msc1 coprecipitates HDAC activity (Ahmed et al. 2007), we asked whether an increase in gene dosage of msc1 can suppress the growth defect of strains with conditional, ts mutations in mis16 and mis18. The mis16-53 and mis18-262 mutant strains were transformed with a multi-copy plasmid expressing full length Msc1 (pmsc1), or with the empty vector pSP1 (Cottarel et al. 1993). Each transformed cell is expected to contain 5–10 copies of such plasmids, as the S. pombe ars1 sequence allows replication (Forsburg 1993). To assess growth, spotting assays were conducted by growing cells to midlog phase at the permissive temperature of 25° followed by plating a series of fivefold dilutions at permissive (25°), semirestrictive (33°), and restrictive temperature (36°). As shown in Figure 1A, extra copies of the wild-type msc1 gene allowed both mis16-53 and mis18-262 mutant strains to grow at 36°.
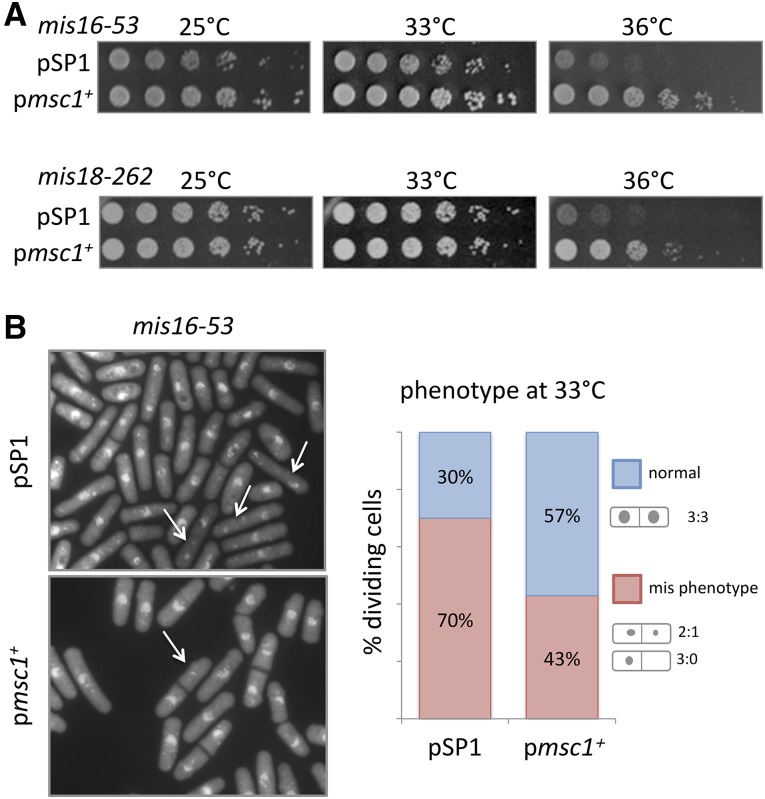
Figure 1.
Extra copies of msc1 restore viability to Mis16/18 complex mutants and decrease the frequency of the mis phenotype in mis16-53. (A) Strains with mutations in the Mis16/18 complex mis16-53 and mis18-262 were transformed with an empty vector pSP1 or plasmid carrying the msc1 gene. The indicated strains were grown to midlog phase, serially diluted (fivefold), spotted on PMA-leucine plates and incubated at 25, 33, and 36°. (B) Cells of a mis16-53 strain transformed with either empty vector (pSP1) or pmsc1+ were grown to midlog phase then shifted to 33° for 8 hr. Cells were stained with DAPI and photographed by fluorescence microscopy. Arrows indicate the mis phenotype. Right panel: quantification of the percentage of dividing cells exhibiting the mis phenotype after shift to 33° for 8 hr (n > 200).
At restrictive temperature, the mis16 and mis18 mutants exhibit chromosome missegregation such that most cells in the population arrest with an unequal distribution of chromosomes in the daughter cells (Takahashi et al. 1994; Hayashi et al. 2004). To test whether extra copies of msc1 relieve the missegregation phenotype, we grew mis16-53 and mis18-262 mutant strains containing empty vector (pSP1) or full length msc1 (pmsc1) at 25° to midlog phase. Cells were shifted to 33° for 8 hr and stained with DAPI to visualize chromosomes (Figure 1B). In normally divided daughter cells, DNA should distribute evenly as observed in wild-type cells. In contrast, mutations in kinetochore components result in a chromosome missegregation phenotype characterized by daughter cells with unequal division of chromosomes (Takahashi et al. 1994; Saitoh et al. 1997; Hayashi et al. 2004): cells either contain large and small nuclei, with an apparent DNA ratio of 2:1, or all of the chromosomes segregate to one cell resulting in a DNA distribution of 3:0 (Figure 1B). Quantification of the fraction of the binucleate cell population with the mis phenotype reveals that 70% of mis16-53 strains with the empty vector (pSP1) displayed the mis phenotype at 33° (Figure 1B). With extra copies of msc1, the fraction of cells with unequal chromosome segregation decreased to 43%. Thus, increased expression of msc1 in the mis16-53 strain improves viability and decreases the frequency of the mis phenotype.
Deletion of msc1 decreases viability of mis16/18 ts mutants and increases frequency of the mis phenotype associated with mis16-53
Given that elevated levels of Msc1 suppress the ts chromosome segregation and viability defect of the mis16/18 ts mutants, we hypothesized that loss of function of msc1 might further compromise the phenotype of mis16/18 ts mutants. Strains with a null allele of msc1 were crossed with mis16-53 or mis18-262 strains to generate double mutants. To test for the impact of loss of msc1 function on growth of the mis16 and mis18 mutants, strains were grown at 25° overnight before preparing fivefold serial dilutions to plate at 25, 33, and 36°. As shown in Figure 2A, at permissive temperature (25°), cells of both single- and double-mutant strains are viable. At semipermissive temperature (33°), mis16-53 grows well, while mis16-53 msc1Δ lost viability. At 36°, the mis16-53 single mutant retains some viability, but the double mutant is completely inviable. While the mis18-262 mutant retains viability even at 36°, the mis18-262 msc1Δ strain is inviable at that temperature. Thus, loss of msc1 lowers the restrictive temperature of mis16/18 ts mutants suggesting that viability of the kinetochore mutants is compromised when Msc1 is absent.
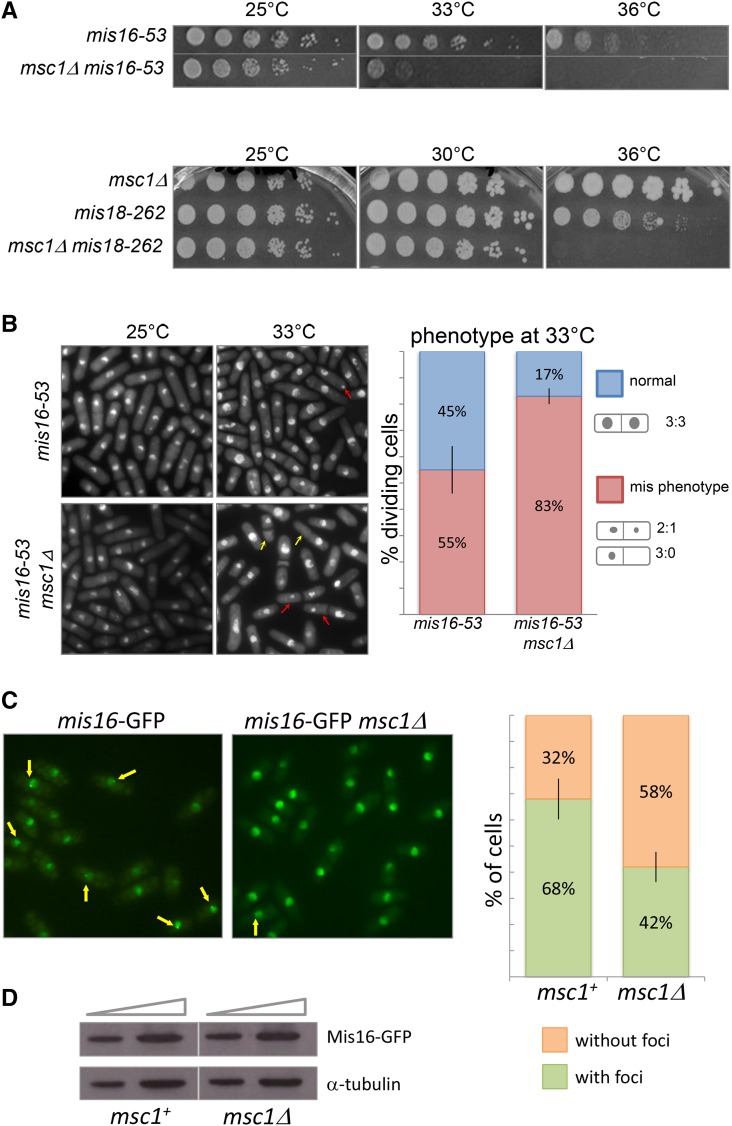
Figure 2.
In the absence of msc1, the viability of mis16/18 mutants is compromised, the frequency of the mis phenotype associated with mis16-53 is increased, and localization of Mis16 to the centromere is reduced. (A) The indicated strains were grown to midlog phase and fivefold serial dilutions were prepared, spotted on minimal media, and incubated for 3–4 days at the indicated temperatures. (B) Representative images of methanol-fixed cells grown to midlog phase at 25° (permissive temperature) and shifted to 33° (restrictive temperature) for 8 hr. Cells were stained with DAPI and photographed by fluorescence microscopy. Arrows indicate the mis phenotype: red (2:1), yellow (3:0). Right panel: quantification of the percentage of dividing cells exhibiting the mis phenotype after shift to 33° for 8 hr (n > 200). Data represent the mean of three counts of the number of cells exhibiting a normal vs. a mis phenotype; error bars indicate the SEM; the P-value for a two-tailed, unpaired t-test is 0.047. (C) The indicated strains with an integrated allele of mis16 fused to GFP were grown at 25° to midlog phase. Localization of GFP-tagged Mis16 with or without msc1 was observed by taking Z-stack pictures using a fluorescence microscope. Quantification of the frequency of cells with localization of Mis16-GFP to single foci is shown on the right. Data represent the mean of four determinations of protein localization with error bars indicating the SEM; the P-value for a two-tailed, unpaired t-test is 0.039. (D) GFP-tagged strains with wild-type msc1 and with msc1 deletion were grown to midlog phase. Two quantities of protein lysate from the indicated strains were separated by SDS-PAGE, transferred to nitrocellulose membrane and incubated with antibody to GFP to detect Mis16-GFP. Detection of α-tubulin was utilized as a loading control.
To determine whether loss of Msc1 function alters the missegregation phenotype, we examined DAPI-stained mis16-53 cells with and without msc1 by fluorescence microscopy. The mis16-53 and mis16-53 msc1Δ strains were grown to midlog phase at 25°, then shifted to 33° for 8 hr, fixed and examined by microscopy. As shown and quantified in Figure 2B, the frequency of cells with the mis phenotype at 33° is elevated in mis16-53 cells lacking msc1. Whereas 55% of mis16-53 cells exhibit the mis phenotype at 33°, 83% of mis16-53 msc1Δ show unequal chromosome segregation at this temperature. Without msc1, aberrant chromosome segregation caused by mis16-53 and mis18-262 mutants is exacerbated, consistent with the decreased viability of the double mutant cells as compared to the single mutants.
Msc1 downregulates Mis16 protein localization at the centromere without changing protein level
Other laboratories have reported that localization to the centromere of Mis16 and Mis18 proteins is codependent (Hayashi et al. 2004): when either Mis16 or Mis18 is mutated, localization to the centromere of the other is compromised. Given the observed genetic interactions of msc1 with mis16/18, we sought to determine whether Msc1 might affect localization of the Mis16 component of the Mis16/18 complex.
We acquired a strain in which the endogenous mis16+ gene is tagged with GFP (Hayashi et al. 2004). By combining the tagged mis16:GFP strain with a deletion of msc1, we could analyze whether recruitment of Mis16 protein to the centromere would be altered. As reported (Hayashi et al. 2004), Mis16-GFP appears diffusely in the nucleus as well as in discrete foci by fluorescence microscopy, representing localization of Mis16-GFP to the centromere. GFP signals were observed at different focal depths by capturing individual images through Z-stack scanning. A single representative focal plane image is presented for Mis16-GFP cells in the left panel of Figure 2C. After collecting all image slices, the frequency of Mis16-GFP foci (indicated by yellow arrows) were scored and plotted. As shown in the right panel of Figure 2C, and, consistent with previously reported results (Hayashi et al. 2004), 68% of wild-type cells have foci. In cells lacking msc1, the percentage of cells with foci is reduced to 42%. Given that Mis16 undergoes a cycle of association and dissociation from the centromere during each cell cycle, this result suggests that Msc1 either facilitates Mis16 localization to the centromere or that the absence of msc1 prolongs the period during which Mis16 is delocalized.
To determine whether the diminished localization of Mis16-GFP might be due to a change in protein abundance, we prepared cell lysates for Western blot analysis with anti-GFP antibody. Antibody to α-tubulin was used as a loading control. As shown in Figure 2D, the abundance of Mis16-GFP is equivalent in cells with and without msc1. Thus, deletion of msc1 does not change the protein expression level of Mis16.
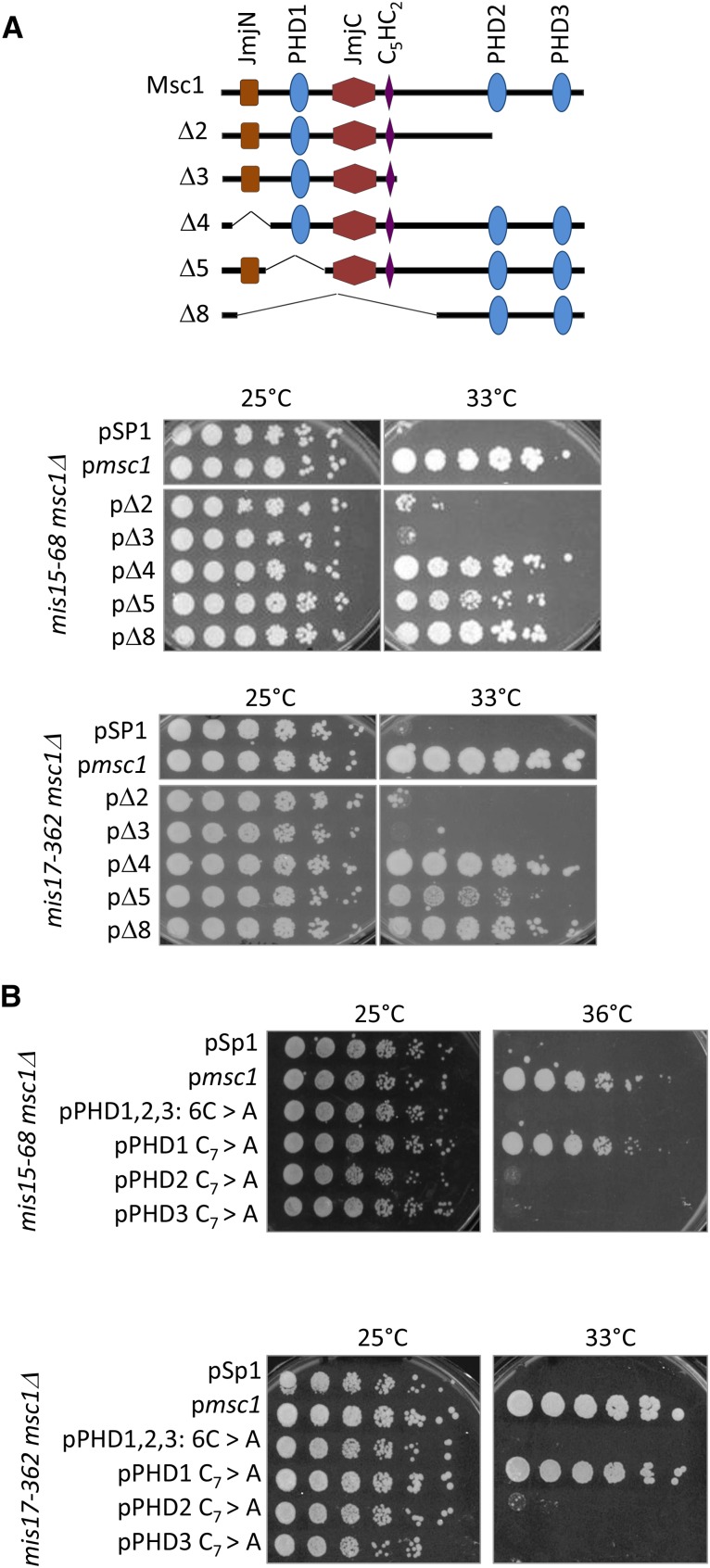
C-terminal PHDs of Msc1 are necessary for kinetochore mutant suppression
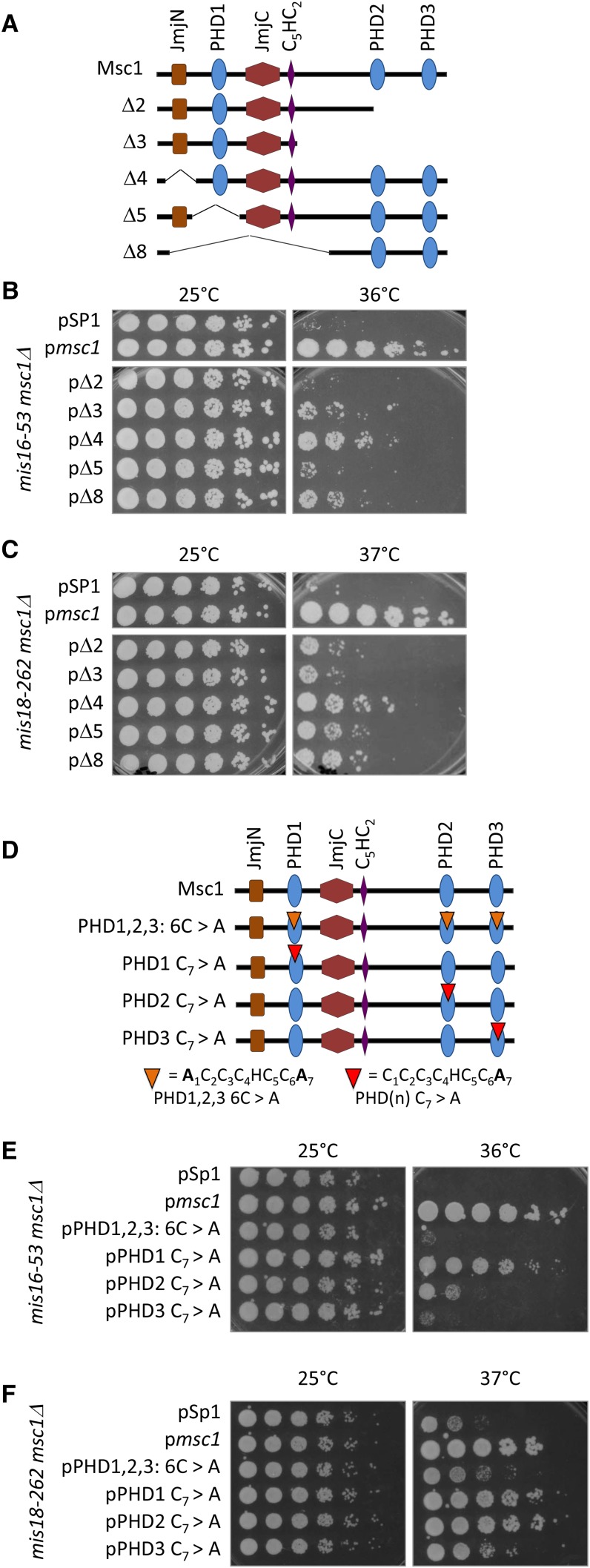
To gain insight into the domains of Msc1 necessary for supporting kinetochore function, we made use of previously constructed msc1 plasmids (Ahmed et al. 2004; Qiu et al. 2010) with deletions of one or more conserved domains (Figure 3A). Plasmids containing deletion alleles were transformed into mis16-53 msc1Δ and mis18-262 msc1Δ, along with empty vector (pSP1) and a plasmid expressing full-length msc1 as negative and positive controls, respectively. As shown in Figure 3B, when the wild-type msc1 gene is introduced, cells are able to grow on plates at 36°, whereas the empty vector does not support any growth at this temperature. A plasmid missing the jmjN domain (Δ4) or N-terminal half of msc1 (Δ8) allowed some growth. In contrast, deletion constructs lacking PHD domains (Δ2, Δ3, and Δ5) seem incapable of supporting viability of the mis16-53 msc1Δ mutant. Similar observations were found in the mis18-262 msc1Δ strain when the same plasmids were introduced (Figure 3C). Thus, we conclude that the PHD domains are necessary for Msc1 to confer viability on cells that have compromised kinetochore function due to mutations in mis16 or mis18.
Figure 3.
The C-terminal PHD domains of Msc1 are important for its function when mis16 or mis18 are compromised. (A) Schematic of Msc1 deletion mutations (Ahmed et al. 2004). (B, C) The indicated plasmids were transformed into the indicated strains. Cells were grown to midlog phase in PMA-leucine at 25°, serially diluted, then spotted on PMA-leucine plates (B), or YES plates (C), and incubated at the indicated temperatures. (D) Schematic of point mutations in Msc1 (Dul and Walworth 2007). (E, F) The indicated plasmids were transformed into the indicated strains. Cells were grown to midlog phase in PMA-leucine at 25°, serially diluted, then spotted on to PMA-leucine plates (E) or YES plates (F) and incubated at the indicated temperatures. YES plates were used for assaying mis18 growth as the temperature sensitivity is more pronounced on YES than on minimal media.
While the PHD domains seem to be critical for Msc1 function in the context of compromised kinetochore function, deletion mutants may affect protein folding. We have shown in previous studies that point mutations of conserved cysteine residues in the PHD domains of Msc1 affect their activity as E3 ubiquitin ligases (Dul and Walworth 2007). Therefore, we transformed plasmids containing point mutations within the PHD1, PHD2, and PHD3 domains into mis16-53 msc1Δ and mis18-262 msc1Δ strains to assess growth. As shown in Figure 3D, if the first and seventh cysteine in all three PHD domains of msc1 are mutated (6C > A), cells are unable to grow at 36°. With a single point mutation in the N-terminal PHD1 domain, mis16-53 msc1Δ cells grow well at 36° (Figure 3E), which indicates that PHD1 may not be important for Msc1 function in the context of compromised kinetochore function due to mutation in mis16. However, point mutations in PHD2 and PHD3 dramatically reduce the ability of Msc1 to function in the context of compromised mis16 function, as neither the PHD2C7 > A nor the PHD3C7 > A plasmids support robust growth at 36°. For the msc1Δ mis18-262 mutant, PHD3 seems to be most critical as PHD3C7 > A is most compromised for growth (Figure 3F). Previous results have shown that C-terminal PHD2 and PHD3 domains of Msc1 are important for precipitating HDAC activity (Ahmed et al. 2004). Given that mis16-53 or mis18-262 mutations increase centromeric histone acetylation levels at the centromere and reduce cell viability (Hayashi et al. 2004), our results are consistent with the hypothesis that, through its C-terminal PHD domains, Msc1 may recruit HDAC to the centromere to suppress compromised kinetochore mutants.
Msc1 restores viability to CCAN mutants
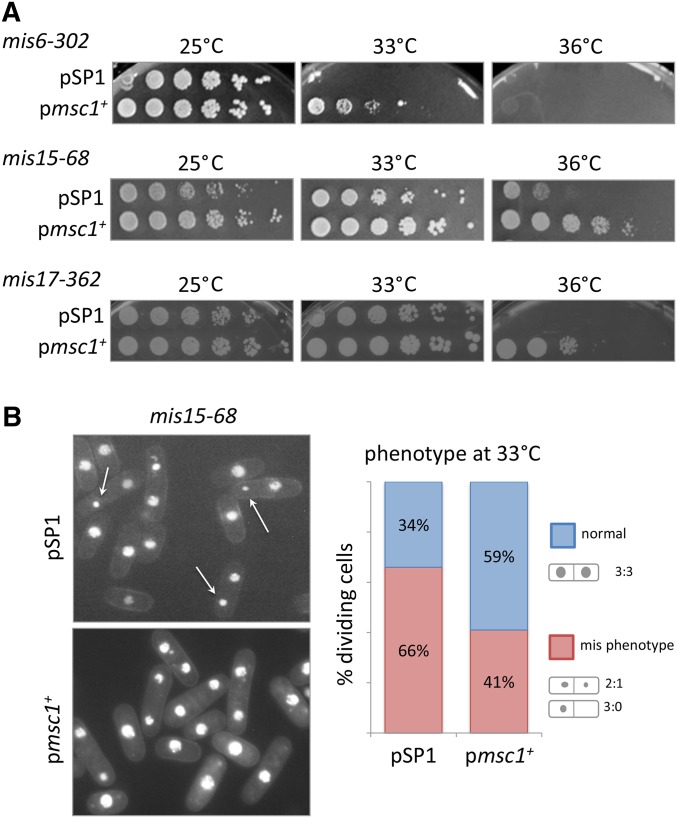
We have shown previously that increased expression of msc1 restores viability to cells with defective CENP-A due to a ts allele of cnp1 (Ahmed et al. 2007). CENP-A loading at the centromere is facilitated through a largely conserved set of proteins that associate with CENP-A nucleosomes and constitute the constitutive centromere-associated network or CCAN (Cheeseman and Desai 2008). To test whether elevating msc1 levels might rescue mutations in components of the CCAN, we transformed cells with either an empty vector or a plasmid expressing msc1 into ts mutants of mis6, mis15, and mis17, which encode CENP-I, CENP-N, and CENP-U, respectively. As shown in Figure 4A, multicopy expression of msc1 restores growth to each of these strains, though for mis6, suppression is achieved only at the restrictive temperature of 33°, and not at 36°. As shown in Figure 4B, extra copies of msc1 also alleviates the mis phenotype, increasing the number of cells undergoing a normal mitosis in the case of mis15-68, by ∼1.7-fold.
Figure 4.
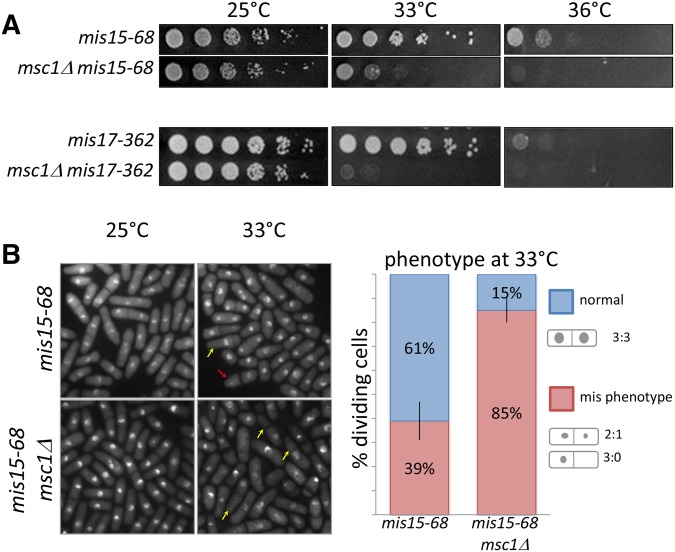
Extra copies of msc1 restore viability to CCAN mutants, and decrease the frequency of the mis phenotype. (A) Strains with mutations in CCAN kinetochore mis mutants, mis6-302, mis15-68, and mis17-362 were transformed with an empty vector pSP1 or plasmid carrying the msc1 gene. The indicated strains were grown to midlog phase, serially diluted (fivefold), spotted on PMA-leucine plates, and incubated at 25, 33, and 36°. (B) Cells of a mis15-68 strain transformed with either empty vector (pSP1) or pmsc1+ were grown to midlog phase then shifted to 33° for 8 hr. Cells were stained with DAPI and photographed by fluorescence microscopy. Arrows indicate the mis phenotype. Right panel: quantification of the percentage of dividing cells exhibiting the mis phenotype after shift to 33° for 8 hr (n > 200).
We showed previously that deletion of msc1 compromises the viability of a mis6 mutant (Ahmed et al. 2007). To test whether msc1 is important for survival of mutants with defects in the CCAN components encoded by mis15 and mis17, we generated double mutants of mis15-68 and mis17-362 with msc1Δ. As shown in Figure 5, A and B, deletion of msc1 compromises the viability of both mis15 and mis17, and exacerbates the mis phenotype in these cells at 33°: the frequency of the mis phenotype for mis15-68 at 33° is increased from 39% when Msc1 is present in the cells to 85% when cells lack msc1.
Figure 5.
In the absence of msc1, viability of CCAN mutants is compromised and the frequency of the mis phenotype is elevated. (A) The indicated strains were grown to midlog phase and fivefold serial dilutions were prepared, spotted on minimal media, and incubated for 3–4 days at the indicated temperatures. (B) Representative images of methanol-fixed cells grown to midlog phase at 25° (permissive temperature) and shifted to 33° (restrictive temperature) for 8 hr. Cells were stained with DAPI and photographed by fluorescence microscopy. Arrows indicate the mis phenotype: red (2:1 segregation), yellow (3:0 segregation). Right panel: Quantification of the percentage of dividing cells exhibiting the mis phenotype after shift to 33° for 8 h (n > 200). Data represent the mean of three counts of the number of cells exhibiting a normal vs. a mis phenotype; error bars indicate the SEM; the P-value for a two-tailed, unpaired t-test is 0.009.
To assess which domains of Msc1 are important for function when the CCAN is compromised, we introduced plasmids harboring deletions of various domains of Msc1 into double mutant cells with ts mis15 or mis17 alleles in combination with deletion of msc1. As shown in Figure 6A, truncation of the C-terminal domain of Msc1 eliminates Msc1 function in the context of mis15-68 or mis17-362, consistent with our previous report on mis6-305 (Ahmed et al. 2007). Similarly, as shown in Figure 6B, whereas a point mutation changing cysteine to alanine in position seven of PHD1 retains function in the CCAN mutants, mutating the analogous cysteine in either PHD2 or PHD3 abolishes function. Thus, as is the case for mis6-305 (Ahmed et al. 2007), PHD2 and PHD3 are important for Msc1 function when mis15 or mis17 are compromised.
Figure 6.
The C-terminal PHD domains are important for Msc1 function when CCAN function is compromised. (A) Schematic of plasmids expressing deletion mutants of Msc1 as described in Figure 3. Plasmids were transformed into the indicated strains. Cells were grown to midlog phase at 25°, serially diluted, then spotted on to PMA-leucine plates and incubated at the indicated temperatures. (B) The indicated plasmids (shown schematically in Figure 3D) were transformed into the indicated CCAN mutant strains. Cells were grown to midlog phase at 25°, serially diluted, then spotted on PMA-leucine plates and incubated at the indicated temperatures.
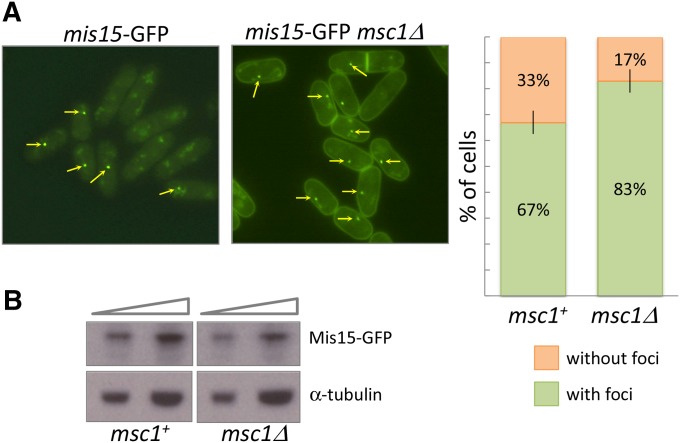
Mis15 associates with the kinetochore more frequently in cells lacking msc1
Some proteins of the CCAN, including CENP-N, the mammalian homolog of Mis15, have been shown to exhibit dynamic association with the centromere during cell cycle progression (Hellwig et al. 2011; Fang et al. 2015). To examine kinetochore association of Mis15 in cells with and without Msc1, we utilized a strain with a chromosomal copy of mis15 tagged with GFP. As shown in Figure 7A, and consistent with previously reported results (Hayashi et al. 2004), ∼70% of wild-type cells expressing Mis15-GFP show characteristic punctate foci indicative of association with the centromere. Curiously, in cells lacking msc1, >80% of cells show the presence of foci, suggesting that centromeric association of Mis15 persists when msc1 is absent. As was the case for Mis16, the abundance of Mis15-GFP as determined by Western blot analysis is unchanged in cells with and without msc1 (Figure 7B).
Figure 7.
In the absence of Msc1, the frequency at which Mis15 localizes to the centromere is elevated. (A) The indicated strains with an integrated allele of mis15 fused to GFP were grown at 25° to midlog phase. Localization of GFP-tagged Mis15 with or without msc1 was observed by taking Z-stack pictures using a fluorescence microscope. Quantification of the frequency of cells with localization of Mis15-GFP to single foci is shown on the right. Data represent the mean of four determinations of protein localization, with error bars indicating the SEM; the P-value for a two-tailed, unpaired t-test is 0.045. (B) GFP-tagged strains with wild-type msc1 and with msc1 deletion were grown to midlog phase. Two quantities of protein lysate from the indicated strains were transferred to nitrocellulose membrane, and incubated with antibody to GFP to detect Mis15-GFP. Detection of α-tubulin was utilized as a loading control.
Extra copies of cnp1 or msc1 act independently to improve viability of CCAN mutants
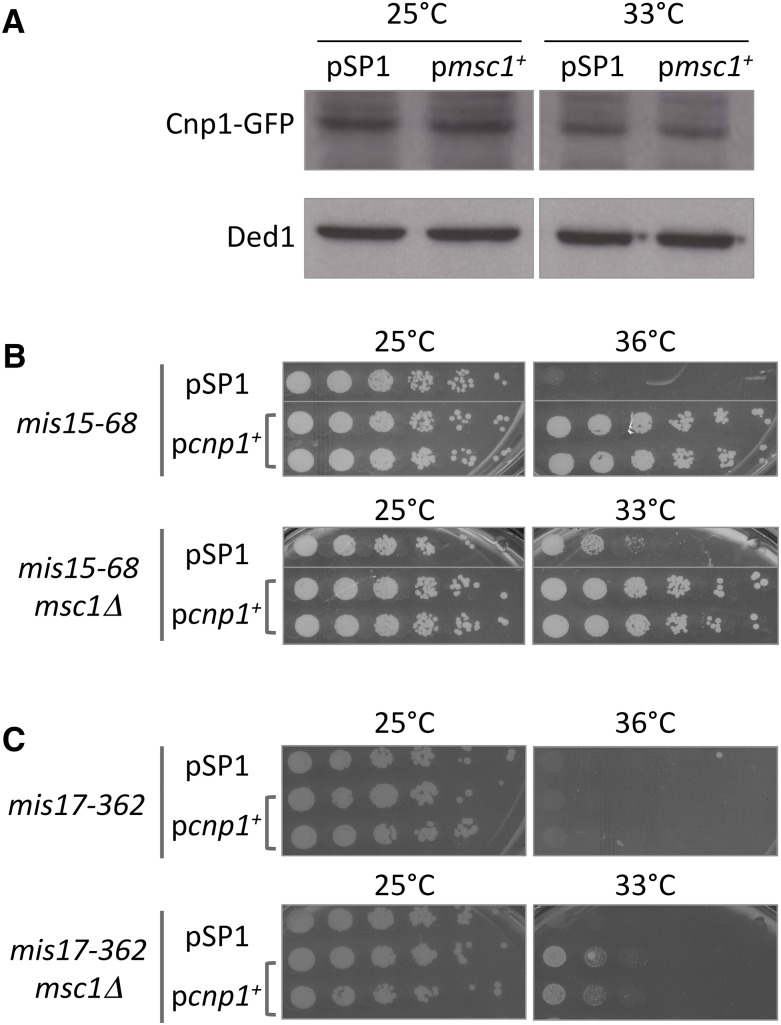
As previously reported by others, extra copies of CENP-A expressed from a plasmid encoding cnp1 can rescue the ts growth defect of several mis mutants (Hayashi et al. 2004). Similarly, extra copies of msc1 rescue mis16/18 (Figure 1) as well as mutants in CCAN components (Figure 4). We entertained the possibility that msc1 might rescue mis mutants by elevating expression of cnp1. To test this possibility, we expressed msc1 in cells with a GFP-tagged allele of cnp1 incorporated in the genome. Cells transformed with an empty vector served as a control for Cnp1-GFP protein expression. As shown in Figure 8A, there is no apparent difference in the amount of Cnp1-GFP protein expressed in cells at either 25 or 33° when cells were transformed with the empty vector (pSP1) or with pmsc1. Thus, it seems unlikely that msc1 rescues mis mutants by elevating Cnp1. To determine whether cnp1 and msc1 might be acting through different mechanisms, we asked whether cnp1 on a plasmid could rescue CCAN mutants mis15 and mis17 when msc1 is absent from cells. As shown in Figure 8, B and C, pcnp1+ rescues both the mis15-68 and mis17-362 mutants in the absence of msc1. Thus, we conclude that pcnp1+ rescue of the CCAN mutants does not require msc1.
Figure 8.
Extra copies of cnp1+ rescue viability of CCAN mutants even in the absence of msc1. (A) Elevated expression of msc1 does not alter the expression of Cnp1-GFP. A mis6-302 mutant with a chromosomally integrated Cnp1-GFP allele strain was transformed with empty vector pSP1 or pmsc1. Cells were grown to midlog phase at 25°, and then shifted to 33° for 5 hr. Cell lysates were prepared and aliquots separated on SDS-PAGE then transferred to nitrocellulose. Western blot assays were conducted by incubating with antibody to Cnp1-GFP (anti-GFP) or Ded1 (anti-Ded1) as a loading control. (B, C) CCAN mis mutants and mis msc1Δ double mutants were transformed with pSP1 or pcnp1 encoding CENP-A. Cells were grown to midlog phase at 25°, serially diluted, then spotted on PMA-leucine plates and incubated at the indicated temperatures for 3–4 days.
Discussion
Kinetochore proteins Mis16 and Mis18 form the Mis16/18 complex required for localization of newly synthesized histone H3 variant CENP-ACnp1 to the centromere (Hayashi et al. 2004; Fujita et al. 2007). By mutating either of these kinetochore proteins, unequal chromosome segregation resulting in the mis phenotype occurs (Hayashi et al. 2004). Furthermore, kinetochore mutations decrease the frequency of CENP-ACnp1 localization, and elegant mechanisms have been proposed to account for the role of Mis18 orthologs in this process (Nardi et al. 2016). Whereas fission yeast Mis18 forms a homotetramer via YIPPEE domains in the N-terminus, the conserved and related human Mis18α and Mis18β proteins form a heterotetramer (Nardi et al. 2016; Subramanian et al. 2016). In both organisms, the Mis18 oligomers serve to target Mis18BP1 and the histone chaperone HJURP to the centromere to facilitate loading of CENP-A. Notably, Mis18 is not conserved in budding yeast, which, rather than having a regional centromere composed of multiple CENP-A nucleosomes interspersed in an array of histone-H3-containing nucleosomes, has single CENP-A nucleosomes on each chromosome that form point centromeres (Sullivan et al. 2001).
Like Mis18, Msc1 is not conserved in budding yeast, though it plays a critical role in maintaining genomic stability in fission yeast (Ahmed et al. 2004). While nonessential for viability, strains harboring a deletion of msc1 exhibit elevated levels of chromosome loss, the lagging chromosome phenotype, and compromised viability when combined with conditional mutants in a number of genes encoding proteins involved in chromosome segregation. Furthermore, multi-copy expression of msc1 robustly suppresses a ts CENP-ACnp1 mutant (cnp1-1), while localization of CENP-ACnp1 to the centromere is compromised in msc1 null cells (Ahmed et al. 2007). Homologs of Msc1, which comprise the four members of the KDM5 family of proteins, are conserved in mammalian cells (Ahmed et al. 2004), and implicated in a range of nuclear functions including transcriptional repression and histone modifications (Barrett et al. 2002; Benevolenskaya et al. 2005; Liefke et al. 2010; Nishibuchi et al. 2014).
In the present study, the results of phenotypic and genetic analysis of Msc1 lead us to hypothesize that Msc1 affects chromosome stability by influencing kinetochore protein function. Extra copies of msc1 suppress loss of function of the kinetochore mutant proteins in the Mis16/18 complex, relieving the ts growth defect of mutants in either subunit, and the chromosome missegregation (mis) phenotype associated with mis16-53. Given that Mis16/18 complexes are required for CENP-ACnp1 deposition and normal chromosome segregation, we conclude that Msc1 may affect Mis16 and Mis18 protein function by either changing the environment of the centromere to facilitate Mis16/18 complex recruitment to the centromere, or supporting or compensating for functions of the Mis16/18 complex to influence CENP-ACnp1 recruitment.
Msc1 associates with the Swr1 complex, which exchanges histone H2A.Z for histone H2A (Qiu et al. 2010) at discrete locations in the genome. H2A.Z has been reported to localize to the fission yeast centromeric outer repeats (Lawrence and Volpe 2009; Qiu et al. 2010). Curiously, deletion of msc1 or swr1 relieves the mitotic arrest phenotype of cells with a cold-sensitive mutation in dis1 (Qiu et al. 2010). Dis1 encodes a protein that facilitates microtubule polymerization (Ohkura et al. 1988, 2001). Analysis of the suppression of dis1 mutants by msc1Δ suggests that deletion of msc1 alters the chromatin landscape at the centromere in such a way as to permit productive microtubule/kinetochore interactions that silence the spindle assembly checkpoint, and relieve the mitotic arrest imposed by inactivation of dis1 (George and Walworth 2015). The present study suggests that effects of Msc1 on the chromatin landscape at the centromere also influence the function of the Mis16/18 complex and the inner kinetochore CCAN complex. Increased expression of Msc1 improves viability and alleviates chromosome missegregation in ts mutants in subunits of both Mis16/18 and the discrete complexes that comprise the CCAN. Furthermore, in cells deficient for Msc1, localization is altered for representative subunits of these complexes, namely Mis16 and Mis15.
The early description of the CCAN as a stable platform upon which dynamic components of the kinetochore associate during cell cycle progression has been revised in recent years as it has become apparent that some components display cell-cycle-dependent association with the centromere. For example, CENP-N (Mis15 in fission yeast), which is thought to bind directly to the CATD domain of CENP-A, binds to kinetochores during S phase and G2 phase in mammalian cells, but is dissociated during mitosis and G1 phase (Hellwig et al. 2011). A particular region of CENP-A that becomes blocked when chromatin is compacted as cells progress through mitosis into G1 has been proposed to act as the switch for CENP-N binding (Fang et al. 2015). We observe that, in cells lacking Msc1, Mis15 exhibits increased frequency of association with the centromere (Figure 7), suggesting that dissociation may be reduced when the function of Msc1 is absent. One model to explain this observation is that, in the absence of msc1, the CATD of CENP-A may be altered in such a way that CENP-N (Mis15) has a higher propensity to remain associated with it, thereby prolonging its association with the centromere. Further studies to analyze in detail the timing of Mis15 association with the centromere in the presence and absence of Msc1 may reveal possible mechanisms to explain the observed data.
Rapid advances in identifying the protein subcomplexes that assemble to form the kinetochore, and of the mechanisms that lead to its formation, have led to elegant models for kinetochore structure and function (Cheeseman 2014; Dimitrova et al. 2016; Nardi et al. 2016; Petrovic et al. 2016; Subramanian et al. 2016). Unlike the proteins that comprise the core components of the kinetochore, Msc1 is nonessential, though it appears to modulate the ability of kinetochore proteins to execute their essential functions. As a member of the KDM5 family of proteins, Msc1 and its mammalian counterparts possesses conserved functional domains that may contribute to their cellular roles, and could serve as relevant drug targets to modulate function. The PHD domains have been shown to facilitate ubiquitylation of substrate proteins (Dul and Walworth 2007) and the JmjC domain of the mammalian protein KDM5A exhibits demethylase activity (Christensen et al. 2007; Klose et al. 2007). KDM5 family proteins are found in various protein complexes with other proteins that mediate chromatin modification. As such, they may serve regulatory roles and modulate local chromatin structure to affect a variety of nuclear functions: transcription, chromosome compaction, or chromosome segregation. While the absence of Msc1 is not lethal to the single celled organism S. pombe, genome integrity is compromised as cells exhibit an elevated rate of chromosome loss, evidenced by the appearance of lagging chromosomes indicative of merotelic chromosome attachments to the mitotic spindle. Such attachments may lead to aneuploidy. Significantly, under circumstances in which microtubule polymerization is compromised by mutation of the dis1 gene leading to mitotic arrest, the elevated frequency of merotelic attachments conferred by loss of function of Msc1 or particular HDACs, allows cells to escape from mitotic arrest (George and Walworth 2015). Thus, deciphering ways in which Msc1 and the KDM5 proteins might be modulated has implications for discovering new ways to interfere with mitosis and to appreciate how cells might overcome mitotic blocks.
Acknowledgments
The authors gratefully acknowledge the Rutgers Research in Science and Engineering (RISE) program for support of L.L. Research in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under award number R01GM53194. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions: C.G. and N.C.W. conceived and designed the experiments; C.G., L.L., and F.K. performed the experiments; C.G., L.L., F.K., A.A.G., and N.C.W. analyzed the data; C.G. and N.C.W. wrote the paper, with assistance from A.A.G.
Footnotes
Communicating editor: S. Biggins
Literature Cited
- Ahmed, S., C. Palermo, S. Wan, and N. C. Walworth, 2004 A novel protein with similarities to Rb binding protein 2 compensates for loss of Chk1 function and affects histone modification in fission yeast. Mol. Cell. Biol. 24: 3660–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S., Dul B., Qiu X., Walworth N. C., 2007. Msc1 acts through histone H2A.Z to promote chromosome stability in Schizosaccharomyces pombe. Genetics 177: 1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R. C., Ekwall K., 2015. Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 7: a018770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart M. C., Kuich P. H., Stellfox M. E., Ward J. A., Bassett E. A., et al. , 2011. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194: 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett A., Madsen B., Copier J., Lu P. J., Cooper L., et al. , 2002. PLU-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: a new cancer/testis antigen?. Int. J. Cancer 101: 581–588. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya E. V., Murray H. L., Branton P., Young R. A., Kaelin W. G., Jr, 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18: 623–635. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., 2014. The kinetochore. Cold Spring Harb. Perspect. Biol. 6: a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A., 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9: 33–46. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A., 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997. [DOI] [PubMed] [Google Scholar]
- Christensen J., Agger K., Cloos P. A., Pasini D., Rose S., et al. , 2007. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128: 1063–1076. [DOI] [PubMed] [Google Scholar]
- Cottarel G., Beach D., Deuschle U., 1993. Two new multi-purpose multicopy Schizosaccharomyces pombe shuttle vectors, pSP1 and pSP2. Curr. Genet. 23: 547–548. [DOI] [PubMed] [Google Scholar]
- Dimitrova Y. N., Jenni S., Valverde R., Khin Y., Harrison S. C., 2016. Structure of the MIND complex defines a regulatory focus for yeast kinetochore assembly. Cell 167: 1014–1027.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dul B. E., Walworth N. C., 2007. The plant homeodomain fingers of fission yeast Msc1 exhibit E3 ubiquitin ligase activity. J. Biol. Chem. 282: 18397–18406. [DOI] [PubMed] [Google Scholar]
- Fang J., Liu Y., Wei Y., Deng W., Yu Z., et al. , 2015. Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev. 29: 1058–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig U., Sen-Gupta M., Hegemann J. H., 1996. Fission yeast mal2+ is required for chromosome segregation. Mol. Cell. Biol. 16: 6169–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E., Bailey A. O., Yates J. R., III, Bassett E. A., et al. , 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137: 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21: 2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., et al. , 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell 12: 17–30. [DOI] [PubMed] [Google Scholar]
- George A. A., Walworth N. C., 2015. Escape from mitotic arrest: an unexpected connection between microtubule dynamics and epigenetic regulation of centromeric chromatin in Schizosaccharomyces pombe. Genetics 201: 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., et al. , 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Ebe M., Nagao K., Kokubu A., Sajiki K., et al. , 2014. Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP-A through interacting with NMD factors and the SWI/SNF complex. Genes Cells 19: 541–554. [DOI] [PubMed] [Google Scholar]
- Hellwig D., Emmerth S., Ulbricht T., Doring V., Hoischen C., et al. , 2011. Dynamics of CENP-N kinetochore binding during the cell cycle. J. Cell Sci. 124: 3871–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R. J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., et al. , 2007. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128: 889–900. [DOI] [PubMed] [Google Scholar]
- Lawrence R. J., Volpe T. A., 2009. Msc1 links dynamic Swi6/HP1 binding to cell fate determination. Proc. Natl. Acad. Sci. USA 106: 1163–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefke R., Oswald F., Alvarado C., Ferres-Marco D., Mittler G., et al. , 2010. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 24: 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Y., Nefsky B. S., Walworth N. C., 2002. The Ded1 DEAD box helicase interacts with Chk1 and Cdc2. J. Biol. Chem. 277: 2637–2643. [DOI] [PubMed] [Google Scholar]
- Liu X., McLeod I., Anderson S., Yates J. R., III, He X., 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24: 2919–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh A. D., Meraldi P., 2011. The CCAN complex: linking centromere specification to control of kinetochore-microtubule dynamics. Semin. Cell Dev. Biol. 22: 946–952. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Nagpal H., Fukagawa T., 2016. Kinetochore assembly and function through the cell cycle. Chromosoma 125: 645–659. [DOI] [PubMed] [Google Scholar]
- Nardi I. K., Zasadzinska E., Stellfox M. E., Knippler C. M., Foltz D. R., 2016. Licensing of centromeric chromatin assembly through the Mis18alpha-Mis18beta heterotetramer. Mol. Cell 61: 774–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi G., Shibata Y., Hayakawa T., Hayakawa N., Ohtani Y., et al. , 2014. Physical and functional interactions between the histone H3K4 demethylase KDM5A and the nucleosome remodeling and deacetylase (NuRD) complex. J. Biol. Chem. 289: 28956–28970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Adachi Y., Kinoshita N., Niwa O., Toda T., et al. , 1988. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 7: 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Garcia M. A., Toda T., 2001. Dis1/TOG universal microtubule adaptors - one MAP for all? J. Cell Sci. 114: 3805–3812. [DOI] [PubMed] [Google Scholar]
- Palmer D. K., O’Day K., Wener M. H., Andrews B. S., Margolis R. L., 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Keller J., Liu Y., Overlack K., John J., et al. , 2016. Structure of the MIS12 complex and molecular basis of its interaction with CENP-C at human kinetochores. Cell 167: 1028–1040.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A. L., Richardson W., Allshire R. C., 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A. L., Choi E. S., Abbott J. K., Liu X., Kagansky A., et al. , 2009. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Dul B. E., Walworth N. C., 2010. Activity of a C-terminal plant homeodomain (PHD) of Msc1 is essential for function. J. Biol. Chem. 285: 36828–36835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S., Takahashi K., Yanagida M., 1997. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell 90: 131–143. [DOI] [PubMed] [Google Scholar]
- Subramanian L., Toda N. R., Rappsilber J., Allshire R. C., 2014. Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open Biol. 4: 140043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L., Medina-Pritchard B., Barton R., Spiller F., Kulasegaran-Shylini R., et al. , 2016. Centromere localization and function of Mis18 requires Yippee-like domain-mediated oligomerization. EMBO Rep. 17: 496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B. A., Blower M. D., Karpen G. H., 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2: 584–596. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamada H., Yanagida M., 1994. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell 5: 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe F. G., Straight A. F., 2015. The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb. Perspect. Biol. 7: a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]