The conservation of sleep among animals suggests that this behavior serves an important adaptive function; however, a unifying function for and genetic.....
Keywords: sleep, CEP-1, p53, ultraviolet light, stress, behavior
Abstract
Stress-induced sleep (SIS) in Caenorhabditis elegans is important for restoration of cellular homeostasis and is a useful model to study the function and regulation of sleep. SIS is triggered when epidermal growth factor (EGF) activates the ALA neuron, which then releases neuropeptides to promote sleep. To further understand this behavior, we established a new model of SIS using irradiation by ultraviolet C (UVC) light. While UVC irradiation requires ALA signaling and leads to a sleep state similar to that induced by heat and other stressors, it does not induce the proteostatic stress seen with heat exposure. Based on the known genotoxic effects of UVC irradiation, we tested two genes, atl-1 and cep-1, which encode proteins that act in the DNA damage response pathway. Loss-of-function mutants of atl-1 had no defect in UVC-induced SIS but a partial loss-of-function mutant of cep-1, gk138, had decreased movement quiescence following UVC irradiation. Germline ablation experiments and tissue-specific RNA interference experiments showed that cep-1 is required somatically in neurons for its effect on SIS. The cep-1(gk138) mutant suppressed body movement quiescence controlled by EGF, indicating that CEP-1 acts downstream or in parallel to ALA activation to promote quiescence in response to ultraviolet light.
SLEEP is a highly conserved process that occurs in every animal carefully studied (Cirelli and Tononi 2008). However, the mechanisms that regulate sleep and the function of sleep are still unclear. While much work has gone into understanding circadian sleep, less is known about sleep induced by environmental stressors such as high heat and infection (Imeri and Opp 2009; Qian et al. 2015). Stress-induced sleep (SIS) is conserved from mammals to Drosophila melanogaster and Caenorhabditis elegans (Hill et al. 2014; Nelson et al. 2014; Lenz et al. 2015), which provide useful models for studying the genetic regulation of this behavior (Davis and Raizen 2016). SIS is separately regulated from other types of sleep in these organisms (Lenz et al. 2015; Trojanowski et al. 2015) and also serves to restore cellular homeostasis (Hill et al. 2014). In this study we use C. elegans to further explore the genetic regulation of SIS.
Since sleep in nonmammalian models cannot be identified via electroencephalography, we use behavioral hallmarks such as reduced responsiveness and rapid reversibility to identify sleep states. C. elegans SIS fulfills behavioral criteria for sleep in nonmammalian models (Trojanowski and Raizen 2016). The C. elegans model of SIS has similarities to sleep in response to illness in mammals (reviewed in Davis and Raizen 2016). The function of sleep as a restorative state has also been demonstrated in both mammals and nematodes. After a heat shock, worms that cannot sleep have reduced survival (Hill et al. 2014; Fry et al. 2016). Similarly, in rabbits, animals that sleep more after an infection are more likely to survive than rabbits that sleep less (Toth et al. 1993; Imeri and Opp 2009). In this study, we hope to shed light on the regulation and function of SIS by introducing a new type of stress, ultraviolet (UV) C (UVC) irradiation.
UV light is a common environmental toxin that must be dealt with by animals. Exposure to UV light in humans can lead to skin burns and fatigue (Mayo Clinic Staff 2014). By damaging DNA, UV increases the risk of skin cancer (United Nations Environment Program et al. 1994). The cellular response to UV exposure and other genotoxic stressors such as ionizing radiation largely revolves around DNA damage response. Behavioral responses to genotoxic stress are less well characterized. In patients undergoing cancer treatments involving radiation therapy (RT), fatigue is a common complaint with up to 70% of patients noting an increase in fatigue during treatment with ionizing radiation (Irvine et al. 1994; Hickok et al. 2005). Notably, fatigue is experienced by patients receiving RT far from the brain, for example in the pelvic area in patients with prostate cancer (Monga et al. 1999, 2005; Janda et al. 2000). Many patients report drowsiness with RT, suggesting that sleepiness does, in fact, contribute to their complaint of fatigue (Knapp et al. 2012). This suggests that there is cytokine-based communication between peripheral organs undergoing genotoxic stress and the central nervous system. Mice exposed to UV light also become sleepy (Van Oosterhout et al. 2012).
Exposure of C. elegans to UV light leads to acute cessation of feeding and to avoidance behavior (Liu et al. 2010; Bhatla and Horvitz 2015). These behaviors occur during UV irradiation but little is known about how the animal behaves during recovery from UV exposure. The cellular response to UV irradiation in C. elegans is homologous to the mammalian DNA damage repair pathway and includes homologs of DNA surveillance protein ataxia telangiectasia and RAD3-related kinase (ATR) and the tumor suppressor p53 (Jolliffe and Derry 2013). In this study we describe sleep behavior during recovery from UV irradiation. This sleep behavior engages the same neural pathways that are engaged after exposure to other types of stressors (Hill et al. 2014). We show that CEP-1, the C. elegans homolog of mammalian p53, plays a role in the nervous system in the behavioral response to UV light.
Materials and Methods
Animal cultivation, transgenic nomenclature, and strains used
Animals were cultivated on the surface of NGM 1.7% agar in petri dishes of 5.5 cm diameter. Unless noted otherwise, animals were cultivated in a 20° incubator. The agar used in the plates was from manufacturer Apex (catalog number 20-275). The wild-type strain was N2, variety Bristol (Brenner 1974). Animals were fed the Escherichia coli (Boyer and Roulland-Dussoix 1969) strain DA837 (Davis et al. 1995), which is a derivative of OP50 (Brenner 1974). Strains used are listed below. Recombinant DNA transgenes are designated as promoter:gene.
N2 (Bristol) (Brenner 1974).
IB16: ceh-17(np1) I (Pujol et al. 2000).
NQ602: flp-13(tm2427) IV (outcrossed 3×) (Nelson et al. 2014).
KP2018: egl-21(n476) IV (Jacob and Kaplan 2003).
MT1206: egl-21(n576) IV (Jacob and Kaplan 2003).
TJ375: gpIs1[Phsp-16.2:GFP] (Rea et al. 2005).
SJ4005: zcIs4[Phsp-4:GFP] V (Urano et al. 2002).
SJ4100: zcIs13[Phsp-6:GFP] V (Benedetti et al. 2006).
DW101: atl-1(tm853) V / nT1[unc(n754) let (qIs50)] (IV, V) (Mori et al. 2008).
TJ1: cep-1(gk138) I (Arum and Johnson 2007).
CE1255: cep-1(ep347) I (Lackner et al. 2005).
XY1054: cep-1(lg12501) I (Mateo et al. 2016).
TU3401: uIs72[pCFJ90 (Pmyo-2:mCherry); Punc-119:SID-1]; sid-1(pk3321) V (Calixto et al. 2010).
NQ918: cep-1(gk138) I; qnEx456[TransGeneOme Clone 9347172996193398G02(cep-1::gfp); Pmyo-2:mCherry].
NQ1035: qnEx550[Prgef-1:cep-1(sense); Prgef-1:cep-1(antisense); Pmyo-2:mCherry].
NQ1058: qnEx572[Pmyo-2:cep-1(sense); Pmyo-2:cep-1(antisense); Prol-6:gfp].
NQ1071: qnEx582[Psur-5:cep-1(sense); Psur-5:cep-1(antisense); Pmyo-2:mCherry].
NQ1087: qnEx589[Prgef-1:mcherry(sense); Prgef-1:mcherry(antisense); Pmyo-2:gfp].
NQ1102: cep-1(gk138) I; pha-1(e2123ts) III; him-5(e1490) V; syEx723[Phsp-16.2:LIN-3C; Pmyo2:GFP; pha-1](+)].
PS5009: pha-1(e2132ts) III; him-5(e1490) V; syEx723[Phsp16.2:LIN-3C; Pmyo2:GFP; pha-1(+)] (Van Buskirk and Sternberg 2007).
CB4037: glp-1(e2141) III (Kodoyianni et al. 1992).
Molecular biology, micro-injections, and list of oligonucleotides used
Tissue-specific RNA interference (RNAi) constructs were made using overlap extension PCR (Nelson and Fitch 2011) with the oligonucleotides listed in Supplemental Material, Table S1, and using the high fidelity enzyme Phusion (Thermo Fisher Scientific). Briefly, primers were designed to amplify the specified promoter from N2 genomic DNA or to amplify the cep-1 coding region lacking a start codon or 3′−UTR from the ORFeome RNAi feeding clone F52B5.5 (Rual et al. 2004). Promoter fragments were amplified with overlapping sequences for the cep-1 amplicon in both the sense and antisense directions. Therefore, two constructs were made (promoterX:cep-1 sense and promoterX:cep-1 antisense) and simultaneously injected to create the double-stranded RNA needed for RNAi (Fire et al. 1998). Micro-injections were performed via standard methods (Mello et al. 1991) using plasmids pCFJ90 encoding Pmyo-2:mcherry, pPD118.33 encoding Pmyo-2:gfp, or Prol-6:gfp as markers for transformation. See Table S1 for oligonucleotide sequences.
Compound genetic strain constructions
To create strain NQ1102 we crossed PS5009 males to TJ1 hermaphrodites and followed the extrachromosomal array via the GFP marker. To identify the presence of the cep-1(gk138) mutation, we used PCR with oligonucleotides oNQ1591 and oNQ1592, which detect an ∼2000-bp band in wild-type animals and an ∼500-bp band in cep-1(gk138) mutants.
To create the strain NQ1174, N2 males were crossed with CB4037 hermaphrodites. Male cross-progeny were crossed with TJ1 hermaphrodites. We detected the presence of the homozygous cep-1(gk138) mutation using PCR. To identify the presence of the glp-1(e2141) mutation, we cultivated animals at the 25° restrictive temperature and confirmed that 100% of the animals were sterile.
UV-light exposure
Animals freely moving on an NGM agar surface were placed in Spectrolinker XL-1500 (Spectroline) either on a 5.5-cm petri dish or on the WorMotel, uncovered (UV bulbs were above the sample), and irradiated with light of wavelength 254 nm. Various energies of UVC were achieved by using the energy input function, which varies the exposure time. E. Coli DA837 bacteria were transferred onto the agar surface together with the worms and were spread thinly on the agar surface so as to minimize blocking of UV rays.
Assessment of feeding
Following UVC irradiation on NGM agar plates, food was supplied by transferring bacteria from unused seeded plates to the treated plates. Feeding was assessed via visual determination of pharyngeal grinder movements (which represent pharyngeal pumps) on a stereomicroscope under 10× ocular magnification and 4–16× objective magnification. We considered any pumps in a 10-sec period of observation an indication that the animal was feeding.
Assessment of body-movement activity and quiescence
Body-movement activity and quiescence was assessed using the WorMotel (Churgin and Fang-Yen 2015; Churgin et al. 2017) at ambient temperatures ranging from 21 to 23°. Individual worms were placed in 24 or 48 separate wells and imaged under dark field illumination conditions using a red light-emitting diode light source. Images were taken at a spatial resolution of 16 μm/pixel every 10 sec for 491 min by an Imaging Source DMK 23GP031 camera. Activity values were determined via image subtraction analysis which measures number of pixels changed between frames (Raizen et al. 2008). The custom software used is available at https://github.com/cfangyen/wormotel. Due to day-to-day variability in behavior, comparisons between experimental animals and controls were only done for animals housed on the same WorMotel and they were imaged simultaneously.
Assessment of sensory responsiveness
L4 animals were put on unseeded half-peptone agar plates the day before the experiment. Bacteria were added onto the plates with a worm pick to support animal growth to adulthood. The next day, the worms were treated with 1500 J/m2 UVC irradiation and immediately transferred to plates with a full lawn of DA837 bacteria. After a period of 2 hr after treatment the worms were exposed to octanol vapor by placing an eyelash dipped in 60% octanol one pharynx length away from the animal’s nose. We measured the time it took an animal to move backward the length of at least a pharynx. Animals that were inadvertently touched during the assay were censored.
Heat exposure for SIS and transgene induction
To observe heat shock-induced sleep or transgene induction of Phsp-4:gfp, Phsp-16.2:gfp, and Phsp-16.2:lin-3c, we submerged animals housed on NGM agar plates in a 37° water bath for 30 min. The petri dishes were rendered impermeable to water by wrapping with parafilm. To heat shock directly on the WorMotel, the chip loaded with worms was placed in an empty 10-cm petri dish, which was parafilmed and submerged, also for 30 min. The WorMotel was then directly moved to the imaging setup following this treatment for measurements of body-movement quiescence and activity.
Fluorescent microscopy
After heat shock, animals were placed in 2 μl of 1 mM levamisole on a 2% agarose pad on a microscope slide and covered with a cover slip. The worms were visualized at 5× objective magnification using an upright Leica DM5500B compound microscope equipped with epifluorescence optics and a metal halide light source. Images were captured with a Hamamatsu C4742-95 camera controlled by Leica LAS software. Images were collected on the same day using the same camera exposure durations.
RNAi knockdown via feeding
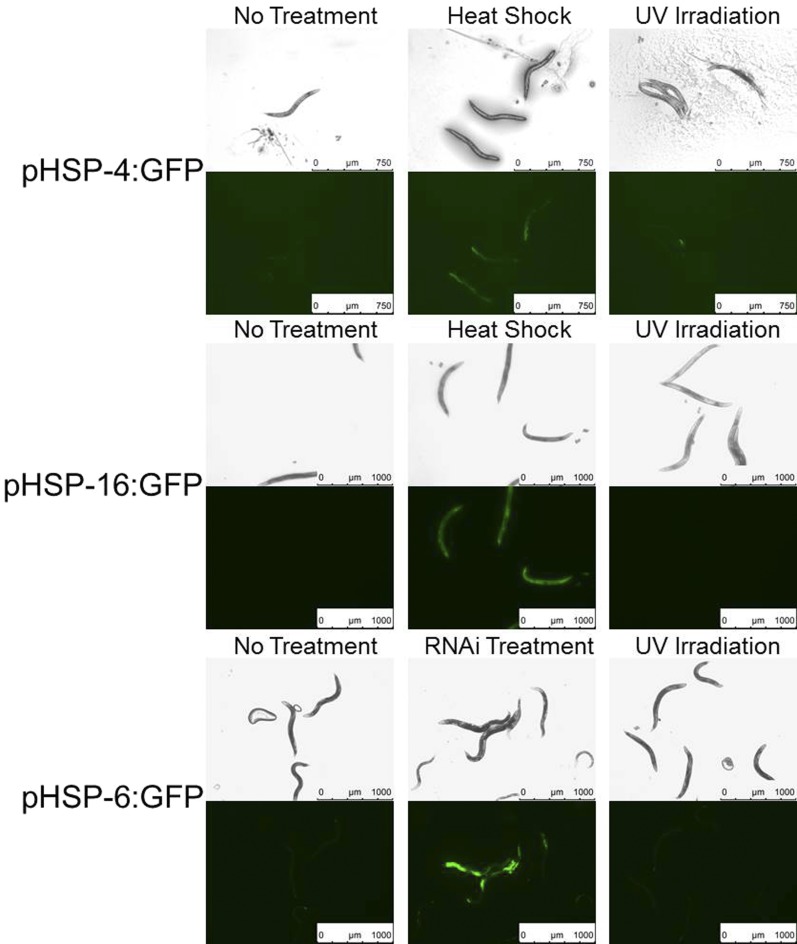
For experiments involving the use of feeding RNAi, animals were laid as eggs onto L4440 bacterial clones containing an RNAi-knockdown construct or an empty-vector control. When animals reached the L4 stage, they were transferred to a new plate containing the same RNAi clone or empty-vector control and then used as day 1 adults for experiments in which we visualized GFP induction from the hsp-6 promoter (Figure 2) or looked at the response to UVC irradiation (Figure 4B).
Figure 2.
UVC exposure does not activate proteostatic stress response pathways. Adult animals of strains SJ4005 (promoter of hsp-4 driving GFP), TJ375 (promoter of hsp16.2 driving GFP), and SJ4100 (promoter of hsp-6 driving GFP) were imaged with bright field optics (rows 1,3, and 5) and with green fluorescence optics (rows 2, 4, and 6) 4 hr after heat shock at 33° for 30 min, UVC irradiation at 1500 J/m2, after cultivation on bacteria expressing double-stranded RNA known to induce mitochondrial stress, or untreated and cultivated on DA837 bacteria. Camera exposure times and gain were kept constant within strains.
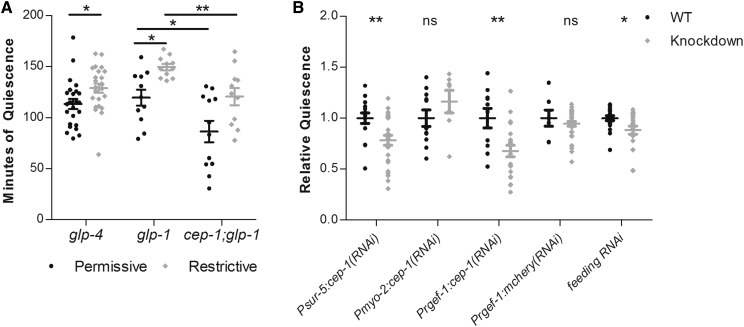
Figure 4.
CEP-1 functions in neuronal cells to promote UVC-induced movement quiescence. (A) Total minutes of body-movement quiescence following UVC irradiation of glp-4(bn2) mutant animals, glp-1(e2141) mutant animals, and cep-1(gk138) I; glp-1(e2141) III double-mutant animals cultivated at the permissive (15°) or at the restrictive (25°) temperature. The N for each group: glp-4 N = 24 permissive and 24 restrictive, glp-1 N = 11 permissive and 11 restrictive, cep-1:glp-1 N = 11 permissive and 11 restrictive. (B) Body-movement quiescence, normalized to wild-type quiescence, of animals in which either cep-1 or mCherry was knocked down via transgenic expression of double-stranded RNA in specific cell types or was knocked down via the feeding RNAi method in a strain hypersensitive to RNAi in neurons (TU3401). Each point indicates an individual animal’s total quiescence divided by the mean quiescence of wild-type control animals imaged simultaneously. The N values and mean total minutes for each group are as follows. Psur-5:cep-1: wild type (WT), N = 16, mean = 118.7; knockdown, N = 35, mean = 92.98. Pmyo-2:cep-1: WT, N = 14, mean= 93; knockdown, N = 14, mean = 52.38. Pregef:cep-1: WT, N = 10, mean = 66.87; cep-1 knockdown, N = 21, mean = 45.29. Prgef-1:mcherry: WT, N = 7, mean = 133.5; knockdown, N = 35, mean = 126.2. * P < 0.05, ** P < 0.01, Student’s t-test comparing total minutes of quiescence of each mutant group to its control. In both panels, the wide and short horizontal bars denote mean and SE, respectively. ns, not significant.
Data availability
All strains not provided by the Caenorhabditis Genetics Center are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
UV irradiation induces behavioral quiescence that requires signaling by the ALA neuron
We used UVC wavelength light as a convenient tool for rapid induction of genotoxic stress. We exposed worms to 0, 500, 1500, and 2500 J/m2 of UVC (wavelength 254 nm) irradiation. Each exposure lasted <30 sec and was administered while worms were freely moving on solid media with either a thin lawn of food or no food. Animals showed pronounced feeding and movement quiescence for several hours following UVC exposure in a dose-dependent manner (Figure 1, A–D).
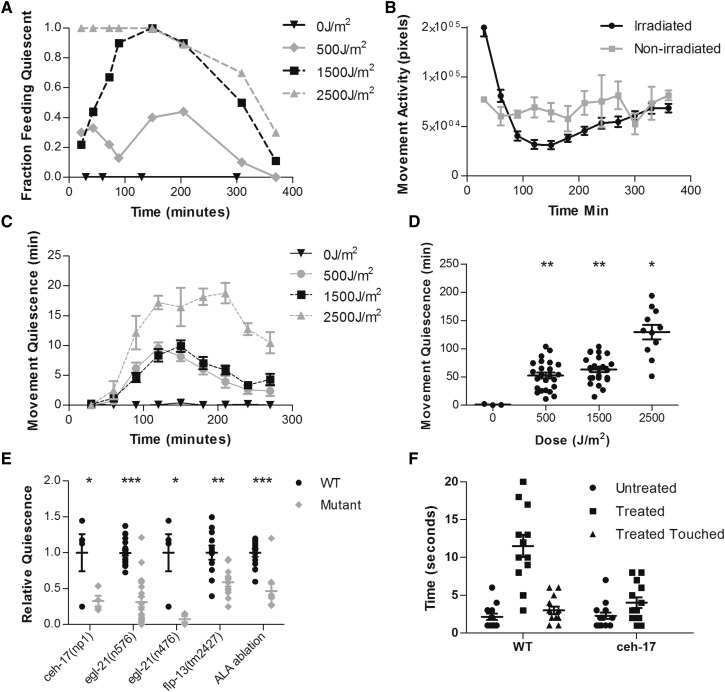
Figure 1.
UVC induces quiescence in an ALA-dependent manner. Animals on an NGM agar surface without bacteria were irradiated with 254-nm light at the specified dose (A–D) or at 1500 J/m2 (E and F). Except where noted in (E), animals used were wild type. (A) Fraction of animals quiescent for feeding in the presence of food as a function of time after UVC exposure. N = 10 for each UVC dose. (B) Activity in pixels of worms irradiated at 1500 J/m2 and of worms that were not irradiated. Irradiated group, N = 32; not irradiated group, N = 4. (C) Minutes of body-movement quiescence in each 30-min period after UVC exposure. Points denote mean and error bars denote SEM. N for each dose was as follows: 0 J/m2, N = 3; 500 J/m2, N = 24; 1500 J/m2, N = 24; 2500 J/m2, N = 5. (D) Minutes of movement quiescence during the first 6 hr after UVC irradiation. * P < 0.05, ** P < 0.01, Student t-tests comparing minutes of quiescence for each irradiated group to nonirradiated group. N for each dose was as follows: 0 J/m2, N = 4; 500 J/m2, N = 24; 1500 J/m2, N = 24; 2500 J/m2, N = 11. (E) Response latency to octanol for wild-type and ceh-17(np1) mutant animals 2 hr after no treatment (untreated) [wild type (WT), N = 13; ceh-17, N = 14], irradiation with 1500 J/m2 UVC (treated) (WT, N = 12; ceh-17, N = 13), or after irradiation followed by harsh body touch immediately prior to octanol exposure (treated touched) (WT, N = 13). (F) Quiescence relative to wild-type animals of ceh-17(np1), egl-21(n576), egl-21(n476), and flp-13(tm2427) mutant animals or of wild-type animals in which ALA was laser ablated relative to unoperated controls. Each point represents total minutes of quiescence in the 6 hr after UVC irradiation of one animal normalized to the mean of wild-type animals imaged simultaneously. The mean for total quiescence in minutes and number of animals N for each group were as follows: ceh-17(np1) (WT, N = 4, mean = 111.6; mutant, N = 4, mean = 36.67], egl-21(n576) (WT, N = 21, mean = 251.47; mutant, N = 21, mean= 130.46), egl-21(n476) (WT, N = 4, mean = 111.6; mutant, N = 4, mean = 8.42), flp-13(tm2427) (WT, N = 12, mean = 147.6; mutant, N = 12, mean = 76.37), ALA ablation (unoperated control, N = 12, mean = 126.4; ablated, N = 9, mean = 59.0). * P < 0.05, ** P < 0.01, *** P < 0.001, Student’s t-test on total minutes of quiescence, comparing mutant or ablated animals to wild-type control animals. In (D–F), the wide and short horizontal bars denote means and SE, respectively.
To measure feeding quiescence, we used bright field stereomicroscopy to inspect movements of the pharyngeal grinder, a tooth-like cuticular specialization (Raizen et al. 2012). Feeding behavior consists of pharyngeal contraction/relaxation cycles called pumps; during each pump, the grinder moves backward and forward. We counted the fraction of animals pumping at several time points up to 6 hr following UVC irradiation. All (100%) of nonirradiated control animals pumped throughout the experiment. Following exposure to either 500 or 1500 J/m2 of UVC irradiation, we observed the maximal feeding quiescent behavior at 150–200 min after UVC exposure with 40 and 100% of the animals suppressing pumping, respectively (Figure 1A). Following exposure to 2500 J/m2 irradiation, 100% of worms stopped feeding immediately and did not begin to recover feeding until ∼200 min after the UVC exposure. By 360 min after irradiation, worms exposed to either 500 or 1500 J/m2 UVC irradiation had resumed feeding behavior, while ∼25% of worms exposed to 2500 J/m2 UVC irradiation remained quiescent for feeding (Figure 1A).
To measure movement quiescence, we used a machine vision algorithm that is based on subtraction of temporally adjacent video frames (Raizen et al. 2008) taken of individually housed animals on a WorMotel (Churgin and Fang-Yen 2015). This frame-subtraction analysis gives a measure of total activity, which is reflected by the total number of pixels changed between frames; as well as of quiescence, which is quantified by summing the bouts in which there was no movement. Irradiated animals were initially more active than nonirradiated control animals (Figure 1B) and showed little quiescence for the first 30 min after UVC exposure (Figure 1B). After 30 min, there was reduced activity (Figure 1B) and increased quiescence (Figure 1C); the degree of quiescence was proportional to the dose of UVC. After each UVC exposure, most of the movement-quiescence behavior was observed in the first 4 hr and most animals irradiated with 500 or 1500 J/m2 UVC irradiation were active at 6 hr after irradiation (Figure 1C). We calculated total quiescence in minutes for the first 6 hr following exposure to UVC. Nonirradiated control animals showed minimal to no quiescence during the 6 hr of tracking (Figure 1D). Total movement quiescence following UVC irradiation increased with increasing doses of irradiation (Figure 1D). The observation of delayed onset of quiescence after irradiation, and near full recovery of movement and feeding 6 hr after UVC exposure, suggests that the quiescence following UVC exposure is not explained by acute injury to the animals; rather, it is likely to be neurally regulated, similar to sleep induced by other types of stressors in C. elegans (Hill et al. 2014).
To determine whether the quiescence following UVC irradiation is quiet wakefulness vs. sleep, we tested another behavioral hallmark of sleep, sensory depression, using an olfactory-avoidance assay. Unstressed adult awake worms nearly immediately move backward when exposed to the aversive chemical octanol (Chao et al. 2004). In contrast, during lethargus and during heat shock-induced sleep, this backing response is delayed (Raizen et al. 2008; Hill et al. 2014). We compared the response to octanol in wild-type worms 2 hr after UVC irradiation at 1500 J/m2 to that of control animals that were not irradiated. The latency to backward movement was prolonged in UVC-irradiated animals (Figure 1E). To distinguish between immobility and reduced responsiveness due to UVC-induced injury from immobility and reduced responsiveness due to sleep, we used a platinum wire to touch irradiated worms (labeled as “irradiated touched group” in Figure 1E) and then again measured the latency to respond to octanol. The octanol-response latency after applying the waking mechanical stimulus to irradiated animals was significantly shorter than the latency prior to the waking stimulus and was similar to that of nonirradiated animals (Figure 1E). The reduced responsiveness and rapid reversibility of the quiescent behavioral state demonstrate sleep properties of UVC-induced quiescence.
SIS after heat shock is dependent on the ALA neuron and signaling by neuropeptides, including those encoded by the gene flp-13 (Hill et al. 2014; Nelson et al. 2014). To test if UVC-induced quiescence is also dependent on this pathway, we measured movement quiescence after irradiating animals in which the function of the single ALA neuron was removed using the laser-ablation method (Fang-Yen et al. 2012) or by using a mutation in the gene ceh-17, which is required for ALA development and function (Pujol et al. 2000). Both approaches for eliminating ALA neuron function resulted in the same outcome: movement quiescence after UVC exposure was greatly reduced (Figure 1F). Additionally, loss of ALA in the ceh-17 mutant strain resulted in loss of sensory depression following UVC irradiation, with octanol-response times similar to those of nonirradiated controls (Figure 1E). To test whether neuropeptide signaling is required for UVC-induced quiescence, we measured movement quiescence in animals mutant for egl-21, which encodes a carboxypeptidase E required for maturation of neuropeptides (Jacob and Kaplan 2003). We also tested animals mutant for flp-13, which is expressed in the ALA neuron and encodes neuropeptides required for SIS (Nelson et al. 2014). Animals mutant for two different alleles of egl-21 and animals mutant for flp-13 showed reduced movement quiescence following UVC exposure at 1500 J/m2 (Figure 1F). The fact that egl-21 mutants had a stronger phenotype than the flp-13 mutant can be explained by additional ALA-expressed neuropeptides acting in parallel to FLP-13 (Nath et al. 2016). Taken together, these experiments demonstrate that UVC-induced sleep requires the same neural mechanisms engaged in SIS induced by heat shock (Hill et al. 2014; Nelson et al. 2014).
UVC-induced quiescence is not associated with induction of proteostatic stress pathways
Prior studies of the mechanism of SIS primarily used high heat exposure as an environmental trigger of the behavior (Hill et al. 2014; Nelson et al. 2014). High heat exposure results in protein misfolding, which triggers both HSF-1-dependent (Hajdu-Cronin et al. 2004) and endoplasmic reticulum (ER) (Link et al. 1999; Shen et al. 2001) stress response pathways, as evidenced by transcriptional upregulation of protein chaperones which serve to restore protein homeostasis (Candido 2002; Hill et al. 2014). Additionally, oxidative stress leads to upregulation of the HSP70 homolog HSP-6, an indicator of mitochondrial stress (Benedetti et al. 2006). UVC exposure generates oxygen radicals that can damage both DNA and protein (Cabiscol et al. 2000; Bhatla and Horvitz 2015). Therefore, we tested whether UVC-induced behavioral quiescence is associated with markers of proteostatic stress. We used transcriptional reporters to monitor gene expression of hsp16.2, hsp-4, and hsp-6, which encode chaperones upregulated by the HSF-1 stress response pathway, by the ER stress response pathway, and by the mitochondrial stress response pathway, respectively. We observed UVC-irradiated and heat-shocked animals for up to 8 hr and found the strongest GFP expression at 4 hr after heat shock. In contrast to heat shock, UVC exposure did not lead to upregulation of hsp-4 or hsp-16.2 (Figure 2). We also did not see upregulation of hsp-6 after UVC treatment, while as a positive control we observed induction of hsp-6:GFP following an RNAi knockdown with the clone T09B4.9, a treatment known to induce mitochondrial stress (Bennett and Kaeberlein 2014). These observations suggested that SIS is not triggered exclusively by exposures that cause proteostatic stress and led us to consider DNA damage as the relevant cellular stress following UVC exposure.
p53/CEP-1 is required for movement quiescence after UVC exposure
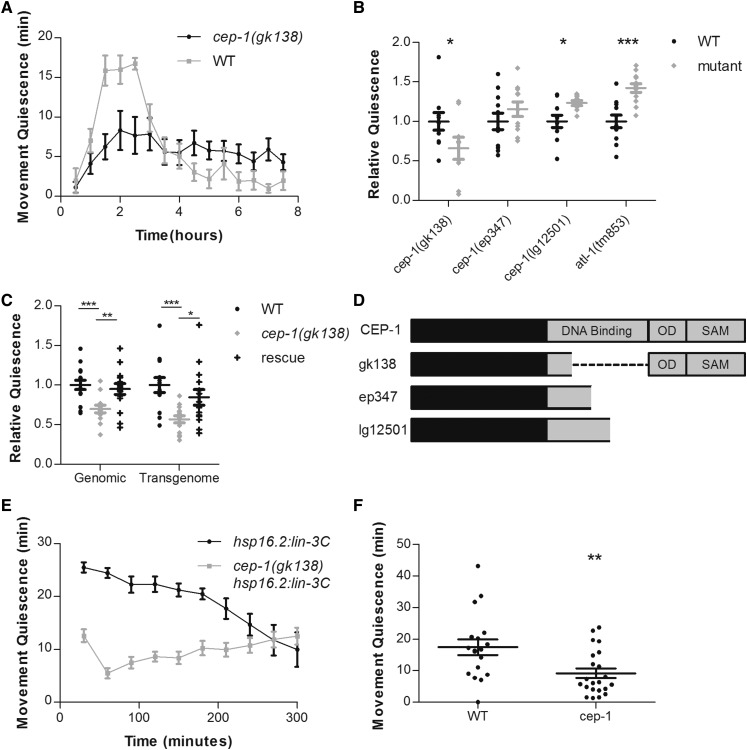
In the C. elegans germline, DNA damage induced by UVC exposure is detected by ATL-1, the C. elegans homolog of mammalian kinase ATR. ATL-1 is required to activate CEP-1 (Vermezovic et al. 2012), the C. elegans homolog of the mammalian tumor suppressor protein p53 (Derry et al. 2001; Schumacher et al. 2001). Upon activation by DNA damage, CEP-1 upregulates genes required for apoptosis and therefore promotes cell death of germline cells whose DNA is damaged by UVC irradiation. To test whether a similar DNA damage surveillance system functions in quiescent behavior after UVC exposure, we measured UVC-induced, body-movement quiescence in animals mutant for atl-1 and of animals mutant for cep-1. While atl-1 mutants showed an enhancement and not a defect in UVC-induced movement quiescence, cep-1 mutants showed a decrease in UVC-induced movement quiescence for 4 hr following 1500 J/m2 UVC irradiation (Figure 3, A and B). We observed no effect of the cep-1 mutation on feeding quiescence after UVC irradiation (2 of 48 wild-type animals and 0 of 36 cep-1 mutant animals were pumping 2 hr after UVC exposure). These results indicate that the mechanism of body-movement quiescence is partially distinct from the mechanism of feeding quiescence, as previously suggested (Trojanowski et al. 2015; Nath et al. 2016). We rescued the defect of cep-1 in movement quiescence by expressing a fosmid containing the complete cep-1 gene with 8 kb of upstream DNA, 13 kb of downstream DNA, and with a GFP tag at the C terminus of the CEP-1 protein. We also rescued the cep-1(gk138) mutant phenotype using a genomic PCR fragment containing the cep-1 coding region, 2 kb of upstream regulatory DNA and 600 bp of DNA downstream of the stop codon (Figure 3C). These data indicate that cep-1 promotes UVC-induced movement quiescence. The observation that atl-1 was not required for UVC-induced quiescence suggests that factors other than ATL-1 can activate CEP-1 following exposure to UVC.
Figure 3.
CEP-1 promotes movement quiescence. (A) Time course of body-movement quiescence per 30 min period after irradiation at 1500 J/m2. [cep-1(gk138), N = 9; wild type, N = 10]. (B and C) Each point represents an individual animal’s total minutes of body-movement quiescence for 4 hr following irradiation divided by the mean quiescence of wild-type animals imaged simultaneously. P-values from t-tests comparing minutes of quiescence for each mutant group to the wild-type control group or between groups indicated by horizontal lines: * P < 0.05, ** P < 0.01, *** P < 0.001. (B) The N and mean for total minutes of quiescence for each group is as follows. cep-1(gk138): wild type (WT), N = 10, mean = 78.35; mutant, N = 9, mean = 46.48. cep-1(ep347): WT, N = 11, mean = 75.15; mutant, N = 11, mean = 86.85. cep-1(lg12501): WT, N = 10, mean = 101.5; mutant, N = 10, mean = 134.0. atl-1(tm853): WT, N = 11, mean = 92.91; mutant, N = 12, mean = 132.2. (C) Transgenic rescues of cep-1(gk138) using PCR-amplified wild-type DNA fragment (genomic) or a TransGenome Fosmid (transgenome). The N and mean for total minutes of quiescence for each group are as follows. Genomic: WT, N = 15, mean = 24.43; cep-1(gk138), N = 13, mean = 21.64; rescue, N = 15, mean = 24.74. Transgenome: WT, N = 14, mean = 86.04; cep-1(gk138), N=14, mean = 48.02; rescue, N = 14, mean = 75.05. (D) Schematic of CEP-1 indicating position of DNA binding domain, oligomerization domain (OD), and SAM domain. Below are representations of predicted mutant peptides created by each cep-1 allele with in-frame gap represented by dashed line for the gk138 allele. (E) Time course of body-movement quiescence per 30-min period in wild-type animals and cep-1(gk138) mutants upon overexpression of the EGF LIN-3C. (F) Total body-movement quiescence in the first hour after heat shock at 35° for 30 min. WT, N = 18; cep-1(gk138), N = 22. P-values from t-tests comparing minutes of quiescence: ** P < 0.01. In (B, C, and F), the wide and short horizontal bars denote means and SE, respectively.
Three mutant alleles of cep-1 (gk138, ep347, and lg12501) affect CEP-1 function in radiation-induced apoptosis (Deng et al. 2004; Lackner et al. 2005; Schumacher et al. 2005). gk138 is the largest deletion of the DNA binding domain, removing 144 amino acids, in comparison to ep347 and lg12501, which remove 108 or 72 amino acids, respectively (Figure 3D). ep347 and lg12501 alleles are both frameshifting deletions that are predicted to eliminate the C-terminal oligomerization and sterile α-motif (SAM) domains of CEP-1, whereas the gk138 deletion is in frame, and therefore leaves these domains intact (Lackner et al. 2005; Mateo et al. 2016). There are functional differences between the alleles gk138 and lg12501 for the role of CEP-1 in genome stability, attributed to the C-terminal domains (Mateo et al. 2016). We found that among these three alleles, only the gk138 allele affected UVC-induced movement quiescence (Figure 3B).
The mechanism of SIS has been proposed to consist of stressed cells sending an epidermal growth factor (EGF) signal to the ALA neuron, which then depolarizes and releases FLP-13 peptides to induce behavioral quiescence (Hill et al. 2014; Nelson et al. 2014). In the context of this model, if CEP-1 were acting in the stressed cells upstream of ALA activation, then it should not affect the quiescent response caused by EGF overexpression. In contrast, if CEP-1 were acting downstream of ALA activation by EGF, then the gk138 mutation would be predicted to impair EGF overexpression-induced behavioral quiescence. To distinguish between these possibilities, we crossed the cep-1(gk138) allele into a strain in which the C. elegans EGF LIN-3C is controlled by a heat-inducible promoter (Van Buskirk and Sternberg 2007). cep-1(gk138) strongly suppressed movement quiescence due to overexpression of LIN-3C (Figure 3E), which suggests that CEP-1 is functioning downstream of or in parallel to EGF activation of ALA. cep-1 did not suppress the vulva-inducing effects of EGF overexpression—following EGF over expression in a wild-type or cep-1 genetic background, we observed 42% (N = 4 plates with a range of 20–100 worms per plate) and 30% (N = 9 plates with a range of 20–100 worms per plate), respectively, of animals with a multi-vulva phenotype (P = 0.19 by student t-test)—indicating that cep-1 does not reduce transgene expression or heat-shock promoter activation. Similar to its effect on UVC-induced feeding quiescence, cep-1(gk138) did not suppress feeding quiescence due to overexpression of LIN-3C (0 of 15 wild-type animals and 0 of 20 cep-1(gk138) mutant animals were feeding 2 hr after overexpression of EGF).
EGF activation of ALA is required for SIS triggered by high heat exposure (Hill et al. 2014). If CEP-1 is functioning downstream of EGF release then it should also suppress heat shock-induced quiescence. To test this, we heat shocked the cep-1(gk138) mutants at 35° for 30 min and then tracked body movements for 1 hr using the WorMotel. We found a reduction in movement quiescence following heat shock (Figure 3F).
CEP-1 is expressed in the germline as well as in somatic cells of the pharynx (Derry et al. 2001). To determine whether the germline plays a role in UVC-induced SIS, we used the mutant glp-4(bn2), which lacks a germline when cultivated at the restrictive temperature of 25° (Beanan and Strome 1992). glp-4 mutant animals cultivated at the restrictive temperature did not show reduced quiescence after UVC exposure compared to mutant animals cultivated at the permissive temperature (Figure 4A). Rather, we observed an enhancement of UVC-induced quiescence in glp-4 mutants. Since glp-4(bn2) has reduced efficiency of translation in somatic cells (Rastogi et al. 2015), which may contribute to the altered phenotype following UV irradiation, we used another temperature-sensitive mutant to eliminate the germline: glp-1(e2141). In this glp-1 background, we also saw more quiescence in response to UVC irradiation when the mutant animals were cultivated at the restrictive temperature than when they were cultivated at a permissive temperature (Figure 4A). These findings suggest that the germline is not required for UVC-induced quiescence and that the germline may act to weakly suppress quiescence. The effect of the germline on UVC-induced quiescence is not explained by an effect on activity in general since at baseline (in the absence of UVC exposure), both glp-1 and glp-4 mutants have the same overall activity after cultivation at the restrictive conditions as after cultivation at the permissive conditions (Figure S1).
To test whether the defect of cep-1 mutants in UVC-induced quiescence requires the germline, we made animals that were mutant for both cep-1(gk138) as well as for glp-1(e2141). cep-1(gk138) I; glp-1(e2141) III double mutants had reduced UVC-induced quiescence compared to glp-1(e2141) single mutants when cultivated at 15° (and therefore with an intact germline) as well as when cultivated at 25° (and therefore with an absent germline) (Figure 4A). These experiments indicate that the effect of cep-1 on movement quiescence following UVC irradiation does not require the germline (Figure 4A), and suggest that cep-1 is acting somatically for this behavioral phenotype.
Since the germline appears dispensable for UVC-induced quiescence as well as for the effect of cep-1 on UVC-induced quiescence, we hypothesized that cep-1 is acting in somatic cells to promote UVC-induced movement quiescence. We constructed tissue-specific, promoter-driven, transgenic RNAi to knock down cep-1 expression in all somatic cells using the sur-5 promoter (Yochem et al. 1998), in pharyngeal muscle cells using the myo-2 promoter (Jantsch-Plunger and Fire 1994), and in all neurons using the rgef-1 P1 promoter (Stefanakis et al. 2015). Knockdown of cep-1 either in all somatic cells or selectively in neurons resulted in a decrease in UVC-induced movement quiescence (Figure 4B). By contrast, the pharyngeal muscle-specific cep-1 transgenic RNAi had no effect on UVC-induced movement quiescence. To control for nonspecific effects of RNAi in the nervous system, we used the same transgenic RNAi strategy to knock down the irrelevant target gene mCherry in the nervous system. These control transgenic animals showed no defect in UVC-induced quiescence (Figure 4B). To test the role of neuronal cep-1 in a different fashion, we used the feeding RNAi method (Fraser et al. 2000) to target cep-1 in the strain TU3401, which expresses the RNAi importer SID-1 exclusively in neuronal cells (Calixto et al. 2010). TU3401 animals fed bacteria expressing cep-1 double-stranded RNA showed reduced UVC-induced movement quiescence relative to control animals that were fed bacteria expressing empty-vector RNAi. Taken together, these results suggest that cep-1 functions in neurons downstream of or in parallel to EGF activation of ALA to promote UVC-induced movement quiescence.
Discussion
Our results show that an intense dose of UVC irradiation will induce a quiescent behavioral state in C. elegans. The quiescent state induced by UVC irradiation is regulated by the same neural signaling pathway that regulates other forms of SIS (Hill et al. 2014; Nelson et al. 2014). This pathway includes release of FLP-13 and other neuropeptides from the ALA neuron following activation by EGF signaling. However, unlike high heat exposure, which results in proteostatic stress as evidenced by transcriptional upregulation of the chaperones HSP-4 and HSP-16.2 (Morimoto 1998; Hill et al. 2014), UVC irradiation does not induce these chaperones. This observation suggests that the type of cellular stress that triggers SIS is broader than just proteostatic stress.
A lack of proteostatic stress following UVC irradiation led us to look at genes involved in genotoxic stress, namely ATL-1 (C. elegans homolog of mammalian ATR) and CEP-1 (C. elegans homolog of P53 family). ATL-1, along with the 9-1-1 complex (HPR-9, MRT-2, and HUS-1) detects DNA lesions induced by UVC irradiation and then activates CEP-1, which in turn activates a proapoptosis transcriptional program in the germline (Jolliffe and Derry 2013). Our results show that CEP-1 is functioning to promote UVC-induced quiescence. Surprisingly, this function is independent of ATL-1 and resides in neurons downstream of or in parallel to EGF activation of ALA.
Though our transgenic rescue and RNAi experiments suggest that the gk138cep-1 allele is a reduction-of-function mutant, we did not observe a quiescence defect in other cep-1 loss-of-function alleles. Differences between the gk138 allele and other alleles of CEP-1 have been noted in CEP-1’s function in meiotic fidelity (Mateo et al. 2016). It is possible that the gk138 allele maintains some functions that may be attributed to the intact SAM and oligomerization domain at the C-terminal end of CEP-1. Alternatively, it is possible that the lg12501 and ep347 mutant alleles result in peptides that maintain enough of the CEP-1 DNA binding domain to transcriptionally regulate genes important for promoting UVC-induced movement quiescence. A final possibility is that partially reduced function of CEP-1, which we would predict to occur in the gk138 allele as well as in the transgenic RNAi knockdown, will lead to an effect on UVC-induced movement quiescence; whereas complete loss of cep-1 function, which might occur in the lg12501 and ep347 alleles due to nonsense-mediated messenger RNA degradation, is compensated by some other stress response pathway and therefore we see no effect on this particular phenotype.
It is interesting to note that while CEP-1 is required for UVC-induced movement quiescence, ATL-1 is not required. Similar to the regulation of p53 in mammals, in C. elegans, two protein kinase regulators of CEP-1 are involved in UVC-induced apoptosis and DNA damage repair. These regulators are ATL-1 and ataxia telangiectasia mutated (ATM-1). ATL-1 is largely responsible for the cellular response to UV-induced DNA damage, while ATM-1 responds to both low doses of UV and to ionizing radiation (Jolliffe and Derry 2013). Since we observed quiescence following a high dose of UVC irradiation but not at all following ionizing radiation (unpublished results), it is unlikely that ATM-1 would be involved. Importantly, these regulators of CEP-1 seem to act solely in the germline (Jolliffe and Derry 2013), which is not required for UVC-induced quiescence (Figure 4). It is likely that in somatic cells another stress regulator is required to activate CEP-1 in response to UVC irradiation. This is not surprising since mammalian p53 is activated by many stress response pathways (Horn and Vousden 2007). The discovery of a function for CEP-1 in somatic cells raises questions about how it might be differently regulated than in the germline.
Our motivation to study CEP-1 in the response to UVC-induced SIS was based on its known role in the response to genotoxic stress. Our original hypothesis was therefore that CEP-1 and its DNA damage response regulators would be required upstream of activation of the ALA neuron. To our surprise, our analysis falsified this motivating hypothesis. First, the well-characterized regulator of CEP-1 in the DNA damage response, ATL-1, is not required for UVC-induced SIS. Second, cep-1 is required only in the body-movement quiescence response to UVC irradiation and not the feeding quiescence response. Third, cep-1 is required also for SIS after heat shock, which should primarily cause proteostatic stress and not genotoxic stress. And fourth, the cep-1gk138 mutants strongly suppress the movement quiescence-inducing effects of EGF. These results suggest that CEP-1 is acting downstream of or in parallel to ALA activation by EGF. One possibility is that cep-1 is required for the development or function of ALA itself, though we and others (Derry et al. 2001), did not observe cep-1 expression in ALA. Moreover, if CEP-1 affects ALA function itself, we would expect it to affect not only body-movement quiescence but also feeding quiescence following EGF overexpression, which we did not observe. Taken together, our data suggest that cep-1 acts in neurons regulating body-movement quiescence. Trojanowski et al. (2015) showed that the regulation of body-movement quiescence in SIS occurs upstream of or at the level of motor neurons. cep-1 may be acting acutely in movement-controlling neurons in response to ALA signaling. Alternatively, cep-1 may serve a developmental function in motor neurons and may not have a signaling role in adult animal behavior.
In adult animals, CEP-1 is expressed in few somatic cells when visualized via a translational fusion to GFP. Our data suggest that CEP-1 functions in somatic cells, and specifically in neurons, to alter behavior in response to stress from UVC irradiation. The mammalian p53 family plays a role in neurogenesis and cell cycle control, which indirectly affects behavior (Killick et al. 2011; Cancino et al. 2013; Wang et al. 2014). It is unlikely that neurogenesis is the mechanism by which CEP-1 affects UVC-induced behavior since neural development is complete in adult C. elegans. Therefore, the function of CEP-1 as a modulator of behavior in adult neurons is novel.
Previous work on SIS suggested that regulation of protein homeostasis may be a core function of this sleep. Our studies with UVC-induced sleep suggest a more general function of sleep. We propose that this function is metabolic resource allocation within the organism (Schmidt 2014). When cells are damaged via genotoxic, oxidative, or proteostatic stress, the metabolic load to repair said damage is high. Therefore, the organism enters a sleep state to reallocate resources from behavioral functions to cellular repair functions. In the future, we hope to more broadly address the questions of resource allocation during sleep and quiescence.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300070/-/DC1.
Acknowledgments
We thank Michael Iannacone and Kristin Davis for technical assistance. Cheryl Van Buskirk taught us how to identify the ALA neuron for the laser ablation experiment. For comments on this manuscript, we thank Kristen Davis, Richard McCloskey, and Renske Erion. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD-010440). This research was supported by the National Institutes of Health (R01 NS-088432, principal investigator: D.M.R.; and T32 HL-07713, principal investigator: Alan Pack) and by a pilot grant from the Center for Excellence in Environmental Toxicology (P30 ES013508, principal investigator: Trevor Penning).
Footnotes
Communicating editor: D. Greenstein
Literature Cited
- Arum O., Johnson T. E., 2007. Reduced expression of the Caenorhabditis elegans p53 ortholog cep-1 results in increased longevity. J. Gerontol. A Biol. Sci. Med. Sci. 62: 951–959. [DOI] [PubMed] [Google Scholar]
- Beanan M. J., Strome S., 1992. Characterization of a germ-line proliferation mutation in C. elegans. Development 116: 755–766. [DOI] [PubMed] [Google Scholar]
- Benedetti C., Haynes C. M., Yang Y., Harding H. P., Ron D., 2006. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 174: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C. F., Kaeberlein M., 2014. The mitochondrial unfolded protein response and increased longevity: cause, consequence, or correlation? Exp. Gerontol. 56: 142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatla N., Horvitz H. R., 2015. Light and hydrogen peroxide inhibit C. elegans feeding through gustatory receptor orthologs and pharyngeal neurons. Neuron 85: 804–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D., 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41: 459–472. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabiscol E., Piulats E., Echave P., Herrero E., Ros J., 2000. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 275: 27393–27398. [DOI] [PubMed] [Google Scholar]
- Calixto A., Chelur D., Topalidou I., Chen X., Chalfie M., 2010. Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods 7: 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancino G. I., Yiu A. P., Fatt M. P., Dugani C. B., Flores E. R., et al. , 2013. p63 regulates adult neural precursor and newly born neuron survival to control hippocampal-dependent behavior. J. Neurosci. 33: 12569–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido E. P. M., 2002. The small heat shock proteins of the nematode Caenorhabditis elegans: structure, regulation and biology. Prog. Mol. Subcell. Biol. 28: 61–78. [DOI] [PubMed] [Google Scholar]
- Chao M. Y., Komatsu H., Fukuto H. S., Dionne H. M., Hart A. C., 2004. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc. Natl. Acad. Sci. USA 101: 15512–15517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churgin M. A., Fang-Yen C., 2015. An imaging system for C. elegans behavior. Methods Mol. Biol. 1327: 199–207. [DOI] [PubMed] [Google Scholar]
- Churgin M. A., Jung S.-K., Yu C.-C., Chen X., Raizen D. M., et al. , 2017. Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging. Elife 6: e26652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Tononi G., 2008. Is sleep essential? PLoS Biol. 6: e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K. C., Raizen D. M., 2016. A mechanism for sickness sleep: lessons from invertebrates. J. Physiol. DOI: 10.1113/JP273009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Somerville D., Lee R. Y., Lockery S., Avery L., et al. , 1995. Mutations in the Caenorhabditis elegans Na,K-ATPase alpha-subunit gene, eat-6, disrupt excitable cell function. J. Neurosci. 15: 8408–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hofmann E. R., Villanueva A., Hobert O., Capodieci P., et al. , 2004. Caenorhabditis elegans ABL-1 antagonizes p53-mediated germline apoptosis after ionizing irradiation. Nat. Genet. 36: 906–912. [DOI] [PubMed] [Google Scholar]
- Derry W. B., Putzke A. P., Rothman J. H., 2001. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 294: 591–595. [DOI] [PubMed] [Google Scholar]
- Fang-Yen C., Gabel C. V., Samuel A. D. T., Bargmann C. I., Avery L., 2012. Laser microsurgery in Caenorhabditis elegans. Methods Cell Biol. 107: 177–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., et al. , 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330. [DOI] [PubMed] [Google Scholar]
- Fry A. L., Laboy J. T., Huang H., Hart A. C., Norman K. R., 2016. A conserved GEF for rho-family GTPases acts in an EGF signaling pathway to promote sleep-like quiescence in Caenorhabditis elegans. Genetics 202: 1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin Y. M., Chen W. J., Sternberg P. W., 2004. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics 168: 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickok J. T., Roscoe J. A., Morrow G. R., Mustian K., Okunieff P., et al. , 2005. Frequency, severity, clinical course, and correlates of fatigue in 372 patients during 5 weeks of radiotherapy for cancer. Cancer 104: 1772–1778. [DOI] [PubMed] [Google Scholar]
- Hill A. J., Mansfield R., Lopez J. M. N. G., Raizen D. M., Van Buskirk C., 2014. Cellular stress induces a protective sleep-like state in C. elegans. Curr. Biol. 24: 2399–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H. F., Vousden K. H., 2007. Coping with stress: multiple ways to activate p53. Oncogene 26: 1306–1316. [DOI] [PubMed] [Google Scholar]
- Imeri L., Opp M. R., 2009. How (and why) the immune system makes us sleep. Nat. Rev. Neurosci. 10: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine D., Vincent L., Graydon J. E., Bubela N., Thompson L., 1994. The prevalence and correlates of fatigue in patients receiving treatment with chemotherapy and radiotherapy. A comparison with the fatigue experienced by healthy individuals. Cancer Nurs. 17: 367–378. [PubMed] [Google Scholar]
- Jacob T. C., Kaplan J. M., 2003. The EGL-21 carboxypeptidase E facilitates acetylcholine release at Caenorhabditis elegans neuromuscular junctions. J. Neurosci. 23: 2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M., Gerstner N., Obermair A., Fuerst A., Wachter S., et al. , 2000. Quality of life changes during conformal radiation therapy for prostate carcinoma. Cancer 89: 1322–1328. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V., Fire A., 1994. Combinatorial structure of a body muscle-specific transcriptional enhancer in Caenorhabditis elegans. J. Biol. Chem. 269: 27021–27028. [PubMed] [Google Scholar]
- Jolliffe A. K., Derry W. B., 2013. The TP53 signaling network in mammals and worms. Brief. Funct. Genomics 12: 129–141. [DOI] [PubMed] [Google Scholar]
- Killick R., Niklison-Chirou M., Tomasini R., Bano D., Rufini A., et al. , 2011. p73: a multifunctional protein in neurobiology. Mol. Neurobiol. 43: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp K., Cooper B., Koetters T., Cataldo J., Dhruva A., et al. , 2012. Trajectories and predictors of symptom occurrence, severity, and distress in prostate cancer patients undergoing radiation therapy. J. Pain Symptom Manage. 44: 486–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodoyianni V., Maine E. M., Kimble J., 1992. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Mol. Biol. Cell 3: 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M. R., Kindt R. M., Carroll P. M., Brown K., Cancilla M. R., et al. , 2005. Chemical genetics identifies Rab geranylgeranyl transferase as an apoptotic target of farnesyl transferase inhibitors. Cancer Cell 7: 325–336. [DOI] [PubMed] [Google Scholar]
- Lenz O., Xiong J., Nelson M. D., Raizen D. M., Williams J. A., 2015. FMRFamide signaling promotes stress-induced sleep in Drosophila. Brain Behav. Immun. 47: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link C. D., Cypser J. R., Johnson C. J., Johnson T. E., 1999. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones 4: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ward A., Gao J., Dong Y., Nishio N., et al. , 2010. C. elegans phototransduction requires a G protein-dependent cGMP pathway and a taste receptor homolog. Nat. Neurosci. 13: 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A.-R. F., Kessler Z., Jolliffe A. K., McGovern O., Yu B., et al. , 2016. The p53-like protein CEP-1 is required for meiotic fidelity in C. elegans. Curr. Biol. 26: 1148–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo Clinic Staff, 2014 Diseases and Conditions: Sunburn. Available at: http://www.mayoclinic.org/diseases-conditions/sunburn/basics/definition/con-20031065. Accessed May 1, 2016.
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monga U., Kerrigan A. J., Thornby J., Monga T. N., 1999. Prospective study of fatigue in localized prostate cancer patients undergoing radiotherapy. Radiat. Oncol. Investig. 7: 178–185. [DOI] [PubMed] [Google Scholar]
- Monga U., Kerrigan A. J., Thornby J., Monga T. N., Zimmermann K. P., 2005. Longitudinal study of quality of life in patients with localized prostate cancer undergoing radiotherapy. J. Rehabil. Res. Dev. 42: 391–399. [DOI] [PubMed] [Google Scholar]
- Mori C., Takanami T., Higashitani A., 2008. Maintenance of mitochondrial DNA by the Caenorhabditis elegans ATR checkpoint protein ATL-1. Genetics 180: 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto R. I., 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12: 3788–3796. [DOI] [PubMed] [Google Scholar]
- Nath R. D., Chow E. S., Wang H., Schwarz E. M., Sternberg P. W., 2016. C. elegans stress-induced sleep emerges from the collective action of multiple neuropeptides. Curr. Biol. 26: 2446–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, M. D., and D. H. Fitch, 2011 Overlap extension PCR: an efficient method for transgene construction. Methods Mol. Biol. 772: 459–470. [DOI] [PubMed]
- Nelson M. D., Lee K. H., Churgin M. A., Hill A. J., Van Buskirk C., et al. , 2014. FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans. Curr. Biol. 24: 2406–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N., Torregrossa P., Ewbank J. J., Brunet J. F., 2000. The homeodomain protein CePHOX2/CEH-17 controls antero-posterior axonal growth in C. elegans. Development 127: 3361–3371. [DOI] [PubMed] [Google Scholar]
- Qian S., Li M., Li G., Liu K., Li B., et al. , 2015. Environmental heat stress enhances mental fatigue during sustained attention task performing: evidence from an ASL perfusion study. Behav. Brain Res. 280: 6–15. [DOI] [PubMed] [Google Scholar]
- Raizen D. M., Zimmerman J. E., Maycock M. H., Ta U. D., You Y., et al. , 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451: 569–572. [DOI] [PubMed] [Google Scholar]
- Raizen, D., B.-M. Song, N. Trojanowski, and Y.-J. You, 2012 Methods for measuring pharyngeal behaviors (December 18, 2012), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.154.1, http://www.wormbook.org. 10.1895/wormbook.1.154.1 [DOI] [PMC free article] [PubMed]
- Rastogi S., Borgo B., Pazdernik N., Fox P., Mardis E. R., et al. , 2015. Caenorhabditis elegans glp-4 encodes a valyl aminoacyl tRNA synthetase. G3 5: 2719–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S. L., Wu D., Cypser J. R., Vaupel J. W., Johnson T. E., 2005. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat. Genet. 37: 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J.-F., Ceron J., Koreth J., Hao T., Nicot A.-S., et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14: 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. H., 2014. The energy allocation function of sleep: a unifying theory of sleep, torpor, and continuous wakefulness. Neurosci. Biobehav. Rev. 47: 122–153. [DOI] [PubMed] [Google Scholar]
- Schumacher B., Hofmann K., Boulton S., Gartner A., 2001. C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 11: 1722–1727. [DOI] [PubMed] [Google Scholar]
- Schumacher B., Hanazawa M., Lee M.-H., Nayak S., Volkmann K., et al. , 2005. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120: 357–368. [DOI] [PubMed] [Google Scholar]
- Shen X., Ellis R. E., Lee K., Liu C.-Y., Yang K., et al. , 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans. Dev. Cell 107: 893–903. [DOI] [PubMed] [Google Scholar]
- Stefanakis N., Carrera I., Hobert O., 2015. Regulatory logic of pan-neuronal gene expression in C. elegans. Neuron 87: 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth L. A., Tolley E. A., Krueger J. M., 1993. Sleep as a prognostic indicator during infectious disease in rabbits. Proc. Soc. Exp. Biol. Med. 203: 179–192. [DOI] [PubMed] [Google Scholar]
- Trojanowski N. F., Raizen D. M., 2016. Call it worm sleep. Trends Neurosci. 39: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski N. F., Nelson M. D., Flavell S. W., Fang-Yen C., Raizen D. M., 2015. Distinct mechanisms underlie quiescence during two Caenorhabditis elegans sleep-like states. J. Neurosci. 35: 14571–14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations Environment Programme. International Commission on Non-Ionizing Radiation Protection. World Health Organization , 1994. Ultraviolet Radiation: An Authoritative Scientific Review of Environmental and Health Effects of UV, with Reference to Global Ozone Layer Depletion. World Health Organization, Geneva. [Google Scholar]
- Urano F., Calfon M., Yoneda T., Yun C., Kiraly M., et al. , 2002. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J. Cell Biol. 158: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk C., Sternberg P. W., 2007. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat. Neurosci. 10: 1300–1307. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout F., Fisher S. P., Van Diepen H. C., Watson T. S., Houben T., et al. , 2012. Report ultraviolet light provides a major input to non-image-forming light detection in mice. Curr. Biol. 22: 1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermezovic J., Stergiou L., Hengartner M. O., d’Adda di Fagagna F., 2012. Differential regulation of DNA damage response activation between somatic and germline cells in Caenorhabditis elegans. Cell Death Differ. 19: 1847–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. B., Kinoshita C., Kinoshita Y., Morrison R. S., 2014. p53 and mitochondrial function in neurons. Biochim. Biophys. Acta 1842: 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Gu T., Han M., 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains not provided by the Caenorhabditis Genetics Center are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.