Abstract
The evolution of complex body plans in land plants has been paralleled by gene duplication and divergence within nuclear auxin-signaling networks. A deep mechanistic understanding of auxin signaling proteins therefore may allow rational engineering of novel plant architectures. Toward that end, we analyzed natural variation in the auxin receptor F-box family of wild accessions of the reference plant Arabidopsis thaliana and used this information to populate a structure/function map. We employed a synthetic assay to identify natural hypermorphic F-box variants and then assayed auxin-associated phenotypes in accessions expressing these variants. To more directly measure the impact of the strongest variant in our synthetic assay on auxin sensitivity, we generated transgenic plants expressing this allele. Together, our findings link evolved sequence variation to altered molecular performance and auxin sensitivity. This approach demonstrates the potential for combining synthetic biology approaches with quantitative phenotypes to harness the wealth of available sequence information and guide future engineering efforts of diverse signaling pathways.
Keywords: synthetic biology, auxin-induced degradation, natural variation
AUXIN controls many aspects of plant development and environmental adaptation. Natural and synthetic auxins have been used to control plant growth in fields, greenhouses, and laboratories for nearly a century. In recent years, the gene families of biosynthetic and metabolic enzymes, transporters, and perception machinery that determine the spatial, temporal, and developmental specificity of auxin signals have been identified (Enders and Strader 2015). Recent work has just begun to determine how functionally robust the auxin signaling machinery is to mutation (Yu et al. 2013, 2015; Dezfulian et al. 2016) and to measure the propensity for mutations to produce novel plant phenotypes that result in evolutionary innovation (Delker et al. 2010; Rosas et al. 2013). As auxin effects are so wide ranging, it is not surprising to find that significant variation exists in auxin sensitivity and auxin-induced transcription across Arabidopsis thaliana accessions (Delker et al. 2010), perhaps contributing to morphological diversity. As such, mapping evolutionary trajectories in auxin signaling could facilitate the engineering of numerous plant traits, such as root architecture, shoot branching, or leaf venation—all traits associated with crop yield (Mathan et al. 2016).
Auxin is perceived by a coreceptor complex consisting of an F-box protein transport inhibitor response 1/auxin signaling F-boxes (TIR1/AFB), hereafter referred to as AFBs, an auxin molecule, and a member of a transcriptional coreceptor/corepressor family auxin/indole-3-acetic acid proteins (Aux/IAAs). The F-box domain of the AFB associates with a Skp/Cullin/F-box (SCF) ubiquitin ligase complex that facilitates ubiquitination of the Aux/IAA proteins, targeting them for degradation (Lavy and Estelle 2016). In low auxin conditions, Aux/IAA proteins interact with and repress a family of transcription factors, the auxin response factors (ARFs) (Guilfoyle and Hagen 2007). Auxin response genes are turned on when local auxin accumulation triggers degradation of Aux/IAAs thereby relieving the repression on ARFs.
A. thaliana has six AFB genes, TIR1 and AFB1–AFB5 (Dharmasiri et al. 2005a). The N-terminal F-box domain is modular and functionally conserved in TIR1 and AFB2, both of which form functional E3 ubiquitin ligase complexes with components in yeast and animals (Nishimura et al. 2009; Zhang et al. 2015). The C-terminal domain of the AFBs is a leucine-rich repeat (LRR). LRR domains offer a highly evolvable scaffold for binding small molecules and proteins and perform diverse functions across all domains of life (Bella et al. 2008). The AFB LRR domain allows auxin sensing by interacting with both auxin and the Aux/IAA transcriptional repressor/co-receptor proteins (Dharmasiri et al. 2005a; Tan et al. 2007; Calderón Villalobos et al. 2012). The identity of the subunits and their affinity for one another governs the rate of Aux/IAA degradation which, in turn, governs transcriptional dynamics, cell fate, and morphological change (Dreher et al. 2006; Pierre-Jerome et al. 2014; Galli et al. 2015; Guseman et al. 2015; Winkler et al. 2017).
Here, we paired an examination of the natural coding sequence variation in the AFB family with quantification of functional variation. We used a synthetic auxin-induced degradation assay in yeast to assess the function of natural variants in isolation from the rest of the auxin response network. Variants with altered function were then evaluated in their native context by quantifying auxin-associated root growth inhibition in accessions containing these polymorphisms. Finally, we directly measured the contribution to auxin sensitivity of the most hypermorphic TIR1 allele by generating transgenic plants expressing this variant under a constitutive promoter. Through this work, we have generated a higher resolution structure/function map of the AFB family and highlighted the challenge of identifying functional divergence in highly buffered signaling pathway components using intact plants.
Materials and Methods
Materials, media composition, and general growth conditions
PCRs were performed with Phusion (cloning reactions; New England Biolabs, Ipswich, MA), GoTaq (diagnostics; Promega, Madison, WI) or GemTaq (genotyping; MGQuest, Lynnwood, WA) with primers from IDT (Coralville, Iowa). Media were standard formulations as described in Pierre-Jerome et al. (2017). Plants were grown on 0.5× LS media (Caisson Laboratories, Smithfield, UT) containing 0.5% sucrose and 0.7% phytoagar (Plantmedia; Dublin, OH). Seeds were obtained from the Arabidopsis Biological Resource Center (Columbus, OH).
Analysis of sequence variation
A reference data set of the genome locations of the TIR1/AFB family and COI1 was assembled from the TAIR10 database on July 28, 2015. Transcript and coding sequences were identified using the ENSEMBL biomart version of TAIR10. The 1001 genomes Salk data set (June 28, 2010) was obtained from http://1001genomes.org/. SNPs and 1-bp deletions with a quality (PHRED) score of ≥25 (i.e., “quality_variant_filtered” files) were used for the following analysis using custom R scripts unless otherwise specified. SNPs located in genes of interest were isolated and mapped to their respective gene structures using the VariantAnnotation package (Obenchain et al. 2014). Coding variants were identified and assembled for each gene and each accession. Nucleotide diversity, Watterson’s theta, and Tajima’s D were calculated using the PopGenome package (Pfeifer et al. 2014).
In identifying polymorphisms, TIR1/AFB genes were split into F-box and LRR domains, with the F-box defined as the N terminus of the protein to I50 of TIR1 and the corresponding residues of the other genes according to the alignment generated by Tan et al. (2007). The N-terminal extension of AFB4 and -5 were excluded.
For functional analysis, nonsynonymous polymorphisms in TIR1 and AFB2 were isolated. As domain swap experiments revealed that the F-box regions of TIR1 and AFB2 confer highly similar or identical function in yeast (Supplemental Material, Figure S3), we focused our analysis on variants within the LRR domain. Highly represented and potentially functionally divergent nonsynonymous polymorphisms were then identified by creating a dN/dS matrix of all-by-all pairs of accessions for each gene using the kaks function within the seqinr R package (Charif and Lobry 2007), which implements the method of Nei and Gojobori (1986). Incalculable and infinite values were excluded from these matrices prior to extraction of outlier pairs and associated nonsynonymous polymorphisms. This set of TIR1 and AFB2 polymorphisms was then cloned into yeast expression vectors and functionally characterized as described below. The remaining polymorphisms were subsequently cloned and characterized (Figure S4 and Figure S5). Annotated code and supplemental data are in File S1.
Strain construction
Plasmids were designed using j5 (Hillson et al. 2012) and constructed by aquarium (http://klavinslab.org/aquarium.html). TIR1 and AFB2 were separately inserted into pGP8G (Havens et al. 2012) downstream of a GPD promoter and followed by 3X-FLAG–6X-HIS tandem affinity purification tag, via Golden Gate cloning (Engler et al. 2009). Mutations were introduced into the parent vectors via two-fragment Gibson assembly (Gibson et al. 2009). The coding sequence of the gene of interest was confirmed by sequencing (Genewiz; South Plainfield, NJ).
Plasmids were digested with PmeI before Lithium PEG transformation (Gietz and Schiestl 2007) into W303-1A ADE2+ yeast (MATa, leu2-3,112 trp1-1 can1-100 ura3-1 his3-11,15 ybp1-1). Correct integration of transformed colonies was confirmed by diagnostic PCR across the 3′ boundary of homologous recombination, relative to the gene of interest. Similarly, pGP4GY-IAA1 and -IAA17 (Havens et al. 2012) were transformed into W814-29B yeast (MATα ade2-1 trp1-1 can1-100 ura3-1 leu2-3,112 his3-11,15). Confirmed transformants were struck to isolation on YPAD plates. AFB strains were individually mated with each Aux/IAA strain using standard methods (Pierre-Jerome et al. 2016).
Auxin-induced degradation assays in yeast
Assays were performed as described in Pierre-Jerome et al. (2017) using a Becton Dickinson (BD) special order cytometer with a 514-nm laser exciting fluorescence that is cutoff at 525 nm prior to photomultiplier tube collection (BD, Franklin Lakes, NJ). Events were annotated, subset to singlet yeast, and normalized to initial levels of fluorescence using the flowTime R package (Wright et al. 2017). The full data set is available via FlowRepository (http://flowrepository.org/id/FR-FCM-ZZPB). Additional detail is provided in File S1.
Western blot analyses
Yeast cultures that had been incubated overnight in synthetic complete (SC) media were diluted to OD600 = 0.6 and incubated until cultures reached OD600 ∼1. Cells were harvested by centrifugation from 4 ml of culture. Cells were then lysed by vortexing for 5 min at 4° in the presence of 200 µl of 0.5-mm diameter acid washed glass beads and 200 µl SUMEB buffer (1% SDS, 8 M urea, 10 mM MOPS pH 6.8, 10 mM EDTA, 0.01% bromophenol blue) per one OD unit of original culture. Lysates were then incubated at 65° for 10 min and cleared by centrifugation prior to electrophoresis and blotting (Sambrook and Russell 2001). Mouse anti-FLAG M2 monoclonal primary antibodies (Sigma-Aldrich, St. Louis, MO) were used at a 1:1000 dilution per the manufacturer’s directions.
Root growth inhibition assays
After sterile seeds were stratified on plates oriented vertically at 4° in the dark for 3 days (or 1 week for wild accessions), they were transferred to long day conditions at 20° for 4 days. Ten plants each of four different genotypes were then transferred in two rows to plates containing either DMSO carrier or 2,4-dichlorophenoxyactic acid (2,4-D) with root tips aligned to a reference mark for each row. Plants were scanned after an additional 3 days of incubation. Plates were regularly rotated during incubation to avoid position effects. Root growth was measured using ImageJ (Rasband 1997) and an Intuos Pro drawing pad (Wacom, Portland, Oregon). Additional detail can be found in File S1. The experiment in Figure 3, A–C was repeated on two different days. The experiments in Figure 3D were repeated on 3 different days for T2 lines and 5 different days to T3 lines.
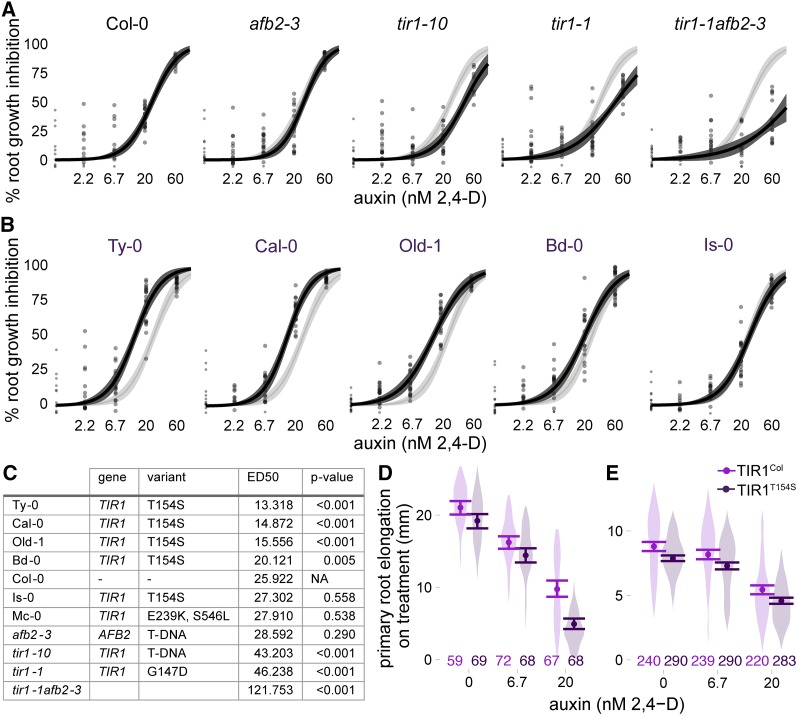
Figure 3.
Auxin sensitivity varies only subtly in wild accessions. (A and B) The impact of auxin on root growth (normalized to mock-treated controls) was measured in 8-day-old seedlings. Results from two biological replicates, each containing 10 plants per treatment level, are shown. Each measurement is shown as a transparent gray point. Solid lines represent log-logistic dose response model fits with a lighter ribbon indicating 95% confidence intervals. The Col-0 curve is reproduced in light gray in each panel to facilitate comparisons. (A) Assays on the reference accession Col-0, and mutants in the Col-0 background, are shown. (B) Auxin sensitivity of accessions containing the hypermorphic TIR1T154S allele. (C) Estimated ED50 values for selected accessions and controls. Parameters were compared ratiometrically to Col-0 and one-sample t-tests were used to estimate the likelihood that the ratio of parameters equals one. P-values were corrected for multiple testing using the Benjamini–Hochberg method. (D and E) A natural polymorphism was sufficient to alter auxin sensitivity in plants. Mean root growth (large points) and 95% confidence intervals (error bars) are shown on top of a violin plot representing the distribution of all measurements. All transgenes were expressed under the pUBQ10 promoter. The number of plants measured for each condition is shown above the x-axis. (D) Three experimental replicates were performed with T2 plants from four independent lines of Col-0 expressing the reference allele and three independent lines expressing TIR1T154S. (E) Five experimental replicates were performed with T3 plants from five independent lines of Col-0 expressing the reference allele and six independent lines expressing TIR1T154S.
Construction and analysis of transgenic plants
Genes of interest were inserted via Golden Gate cloning (Engler et al. 2009) into pGreenII (Hellens et al. 2000) with a pUBQ10 promoter (Grefen et al. 2010) and 3X-FLAG–6X-HIS tandem affinity purification tag. Plasmids were transformed into Agrobacterium tumefaciens GV3101 containing pSOUP (Hellens et al. 2000) via electroporation, and transformants were selected on plates with 50 µg/ml gentamicin and 25 µg/ml kanamycin. Plants were transformed by floral dip (Zhang et al. 2006), and transformants were selected on plates with 30 µg/ml hygromycin at 4 days postgermination after an initial light exposure for 7 hr. Root growth inhibition phenotypes were quantified in T2 generation of at least three independent transformants and at least five independent transformants in the T3 generation as described above. Each T2 plant was genotyped for the presence of the hygromycin resistance gene after the growth assay, using the forward primer (GATGTTGGCGACCTCGTATT) and the reverse primer (GTGCTTGACATTGGGGAGTT). Expression levels in T3 lines were measured by quantitative PCR. RNA was isolated from the tissue from young leaves of T3 plants using illustra RNAspin Mini RNA Isolation Kit (GE Healthcare, Little Chalfont, United Kingdom) and reverse transcribed using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative PCR was performed using iQ SYBR Green Supermix (Bio-Rad) and primers for TIR1 (f-CACGGAACAAGAAGACATCCAAAGG, r-TGAGGAAACTAGAGATAAGGGACTGC) or PP2A (f-AACGTGGCCAAAATGATGC, r-AACCGCTTGGTCGACTATCG) in a CFX96 Touch Real-Time PCR detection system (Bio-Rad).
Plasmids, strains, and sequence files are available upon request or via Addgene. All code used to perform analysis and visualization is provided in File S1. All data including raw images are available upon request.
Results
We identified polymorphisms across the entire AFB gene family in the 170 A. thaliana accessions of the SALK subset of the 1001 Genomes Project (Schmitz et al. 2013). The AFB gene family is highly conserved, relative to other auxin signaling gene families (Delker et al. 2010). We found 1631 total polymorphisms within coding regions and 175 segregating sites across the whole family (Figure S1, Figure S2, and Table S1). AFB3 had the highest ratio of per-site diversity at nonsynonymous sites relative to synonymous sites. AFB4, critical for response to the synthetic auxin picloram (Prigge et al. 2016), had the highest nonsynonymous diversity (>10× that of TIR1) and the only two nonsense polymorphisms identified in this data set. In contrast, AFB1, which is largely incapable of forming a functional SCF complex (Yu et al. 2015), had a similar ratio of nonsynonymous-to-synonymous diversity to TIR1. Many of the accessions contained nonsynonymous polymorphisms in multiple members of the AFB family (Table S2). These additional polymorphisms occurred more frequently in TIR1/AFB1 and AFB2/AFB3 sister pairs than expected (permutation analysis, P < 0.05, File S1 section Assessing covariation within and between sister pairs of AFBs).
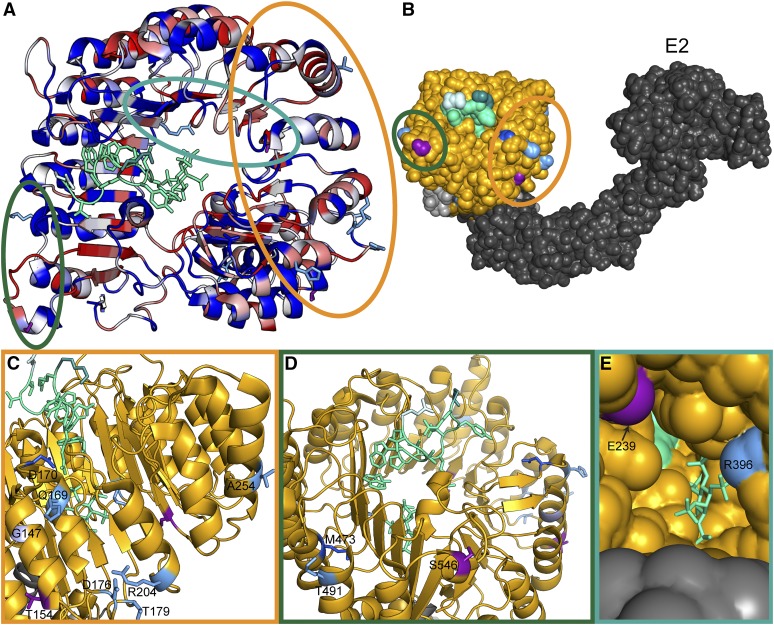
None of the identified accessions had nonsynonymous polymorphisms in both TIR1 and AFB2 (Table S2), an unlikely pattern to occur by chance (permutation analysis, P < 0.05, File S1 section Assessing covariation within and between sister pairs of AFBs). This may reflect the fact that AFB2 and TIR1 are the major auxin receptors and serve partially redundant functions, a conclusion supported by genetic analysis (Dharmasiri et al. 2005a; Parry et al. 2009). The majority of the nonsynonymous polymorphisms in TIR1 and AFB2 occurred in positions of high diversity across the Col-0 AFB family, and most were located in surface residues of the LRR domain (Figure 1A). The majority of these polymorphisms spanned the exterior helices and loops of the fourth through eighth LRRs, which face the Cullin subunit (Figure 1, B and C). This region was recently identified as being responsible for SCFTIR1 dimerization (Dezfulian et al. 2016) and is also proximal to the S-nitrosylation site (Terrile et al. 2012). A pair of polymorphisms exists on the surface spanning the final three LRRs and the C-terminal cap (Figure 1D). This region may interact with the KR motif known to strongly affect auxin-induced degradation rates (Dreher et al. 2006; Moss et al. 2015). A final pair of polymorphisms was found on the interior surface of the LRR domain horseshoe (Figure 1E).
Figure 1.
Clusters of natural variation in TIR1 and AFB2. (A) Identified nonsynonymous polymorphisms tend to occur in residues of high diversity within the Arabidopsis AFB family. A top down view of the LRR domain of the TIR1 structure [Protein Data Bank (PDB): 2P1Q] is shown with the F-box domain in the bottom right and the LRR domain spiraling counterclockwise. The backbone of the TIR1 structure (Tan et al. 2007) was colored according to protein sequence diversity with conserved residues in blue and diverging residues in red. Diversity was calculated as Shannon entropy using an alignment of the protein sequences of the Arabidopsis AFB family (TIR1, AFB1–5). All nonsynonymous polymorphisms are shown as sticks. AFB2 variants are in light blue and TIR1 variants are in purple. Previously identified TIR1 mutations are in dark blue (Ruegger et al. 1998; Yu et al. 2013). The IAA7 degron is shown as a light green ribbon with sidechains as sticks. The N-terminal residue of the IAA7 degron is in lighter green and the C-terminal residue is darker green. Circles around polymorphisms match the detailed views shown in C–E. (B) Polymorphisms face the Cullin subunit of the predicted SCFTIR1 structure. ASK1 (light gray) was aligned with SKP1 from the human SKP2-SKP1-Cul1-RBX1 structure (PDB: 1LDK, shown in dark gray), docking with TIR1 (gold). Putative E2 location is labeled. (C) The dimerization domain on the N-terminal side of the LRR horseshoe contains the majority of natural variation in TIR1 and AFB2. The tir1-1 allele (tir1G147D) is in light purple. (D) Two variants were located on the C-terminal side of the LRR close to the N terminus of the degron. (E) Two additional variants were located inside the LRR horseshoe, near the inositol–hexakisphosphate cofactor.
Synthetic yeast assays reveal functional variation in TIR1 and AFB2
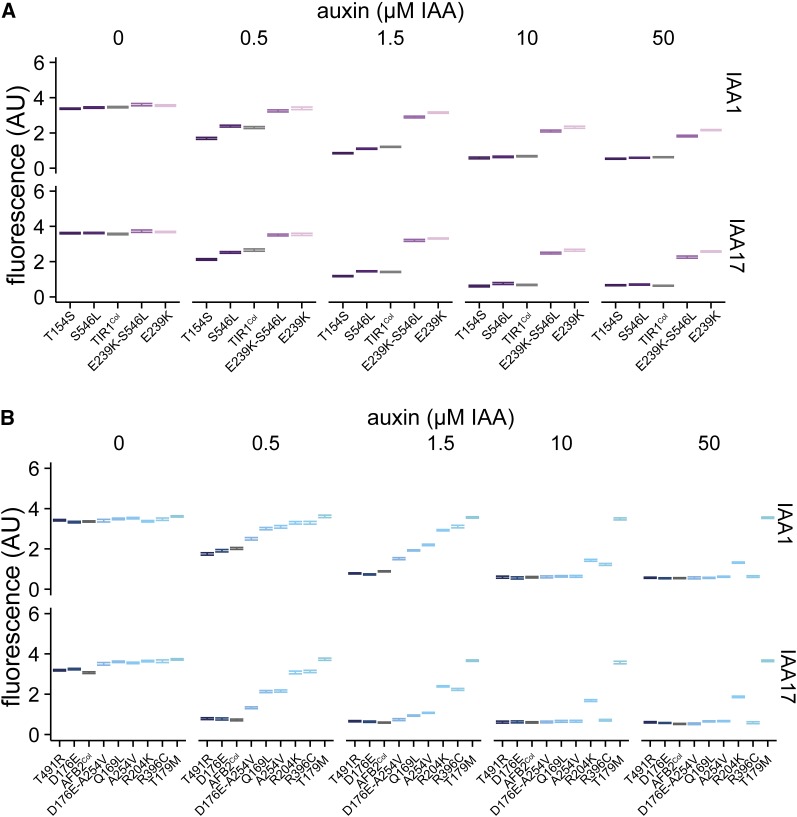
An auxin-induced degradation assay has been established in yeast using heterologous expression of either TIR1 or AFB2 (Havens et al. 2012). We used this synthetic assay to quantify the function of AFB natural variants in the absence of the potentially confounding effects of feedback from the auxin pathway itself or from modulation by other integrating pathways. Natural variants were engineered into the Col-0 reference sequence with co-occurring polymorphisms cloned individually and in combination. Each AFB was then constitutively co-expressed in yeast with fluorescently labeled Aux/IAA targets. Auxin-induced degradation was measured for two targets, IAA1 and IAA17, as these substrates show distinct patterns of behavior when assayed with Col-0 TIR1 and AFB2. TIR1Col induces degradation of IAA1 and IAA17 at similar rates, while AFB2Col causes IAA17 to degrade much faster than what is observed for IAA1 (Havens et al. 2012). We focused on polymorphisms in the LRR domain that were predicted to be functionally divergent (having any pairwise dN/dS value >1), but analysis of the few additional polymorphisms is shown in Figure S4 and Figure S5.
Some natural variants increased function compared to the Col-0 reference, while others decreased or nearly abrogated function (referred to hereafter as hypermorphs, hypomorphs, and amorphs, respectively) (Figure 2). Of the TIR1 polymorphisms, T154S was hypermorphic and E239K-S546L was strongly hypomorphic (Figure 2A). E239K alone was nearly amorphic, and adding S546L only slightly restored activity. These polymorphic TIR1 variants are expressed at similar levels to TIR1Col (Figure S6). Among the AFB2 polymorphisms, T491R was the only clear hypermorph identified (Figure 2B). D176E was slightly hypermorphic, whereas A254V was a moderate hypomorph. In combination, these two polymorphisms were largely additive, giving a response quite similar to AFB2Col. AFB2Q169L was also a moderate hypomorph. Two AFB2 alleles, R396C and R204K, were strong hypomorphs, and T179M was amorphic in our assays. Interestingly, the two most highly represented variants, TIR1T154S (present in five accessions) and AFB2R204K (six accessions), show strong functional divergence from their respective wild-type proteins.
Figure 2.
Synthetic assays reveal significant functional variation in naturally occurring AFB polymorphisms. Nonsynonymous polymorphisms in the LRR domains of TIR1 (A) and AFB2 (B) were synthesized and co-expressed in yeast with fluorescently labeled IAA1 or IAA17. Degradation was assessed using flow cytometry on cultures exposed to different concentrations of the auxin indole-3-acetic acid (IAA) for 1 hr. Error bars represent 95% confidence intervals around the median fluorescence calculated from three independent experiments. In many cases, intervals are small enough that they appear as a single line. The reference Col-0 variant is shown in gray.
Accessions containing a hypermorphic TIR1 allele are hypersensitive to auxin
We next assessed whether the functional variation observed in the synthetic assays was manifested as phenotypic differences in the respective accessions. As there are many polymorphisms between accessions and previous genome-wide association studies of auxin response have not identified the AFB family (Rosas et al. 2013; Meijón et al. 2014), we did not expect a strong correlation between genotype and phenotype from our analysis. To increase the sensitivity and precision of the test, we measured inhibition of primary root growth in the presence of exogenous auxin and fit a log-logistic dose response model to the data. Similar bioassays have been used extensively to identify and characterize mutants in the AFB gene family (Gray et al. 1999; Dharmasiri et al. 2005a,b; Parry et al. 2009). The effective dose of auxin required to elicit 50% of the maximum root growth inhibition (ED50) was the most effective parameter in our model for differentiating among genotypes. Two tir1 mutants in the Col-0 background (a point mutation tir1-1 and a T-DNA insertion tir1-10) were also included in our analysis. Both mutants had significantly higher ED50 values than Col-0, as expected (Figure 3, A and C). A loss of function afb2 allele did not significantly affect the root growth response in our assays, although tir1-1 afb2-3 double mutants had a much larger ED50 than the tir1-1 single mutant.
Four of five accessions carrying TIR1T154S were hypersensitive to auxin, following the pattern predicted by the hypermorphic behavior of that variant in yeast (Figure 3, B and C). In three of these accessions, ED50 values were quite close to one another and significantly lower than the ED50 measured for Col-0 or any other accession. However, Mc-0, which contains the strong hypomorph TIR1E239K-S546L, had auxin responses that were only subtly different Col-0. Consistent with the modest phenotype of afb2-3 in our assays, auxin responses of most accessions with AFB2 polymorphisms were essentially similar to those of Col-0 (File S1, pp. 38–40).
A common TIR1 allele confers auxin hypersensitivity to Col-0
The aberrant auxin responses in yeast and the majority of accessions led us to hypothesize that TIR1T154S is a natural gain-of-function allele with the capacity to impact organ-level auxin responses. To test this, we generated transgenic Col-0 lines expressing TIR1Col or TIR1T154S under a constitutive promoter. Most transgenic lines had relatively similar expression levels, although the Col-0 allele was expressed on average at modestly higher levels than the T154S variant (Figure S7). Expression level and root growth phenotypes were not strongly correlated—the lines with the lowest expression levels showed essentially similar auxin responses as lines with higher transgene expression. In the T3 generation, silencing of the endogenous TIR1 and transgenes was observed in three lines. Two of these lines were from the family with the highest transgene expression (Figure S7, TIR1-7).
Overall, plants expressing TIR1T154S had shorter roots than plants expressing TIR1Col even in the absence of auxin treatment, consistent with the expectation that the T154S polymorphism conferred increased sensitivity to endogenous auxin. In the T2 generation, auxin treatments in root inhibition assays confirmed that TIR1T154S had increased auxin sensitivity relative to TIR1Col (Figure 3D) (transgene:treatment effect F = 9.3, P = 0.0001, full statistical analysis shown in File S1). In the T3 generation, TIR1T154S expressing plants consistently had shorter roots than TIR1Col-expressing plants (transgene effect F = 100.4, P < 10−16); however, the difference in sensitivity to exogenous auxin when compared with TIR1Col-expressing plants was diminished. This may be because the auxin response is near saturation even in the TIR1Col-expressing plants in this generation. The roots of homozygous T3 plants with either transgene were much shorter and had a significantly reduced auxin response when compared with the T2 generation (Figure 3D vs. Figure 3E, note especially the difference in the y-axis).
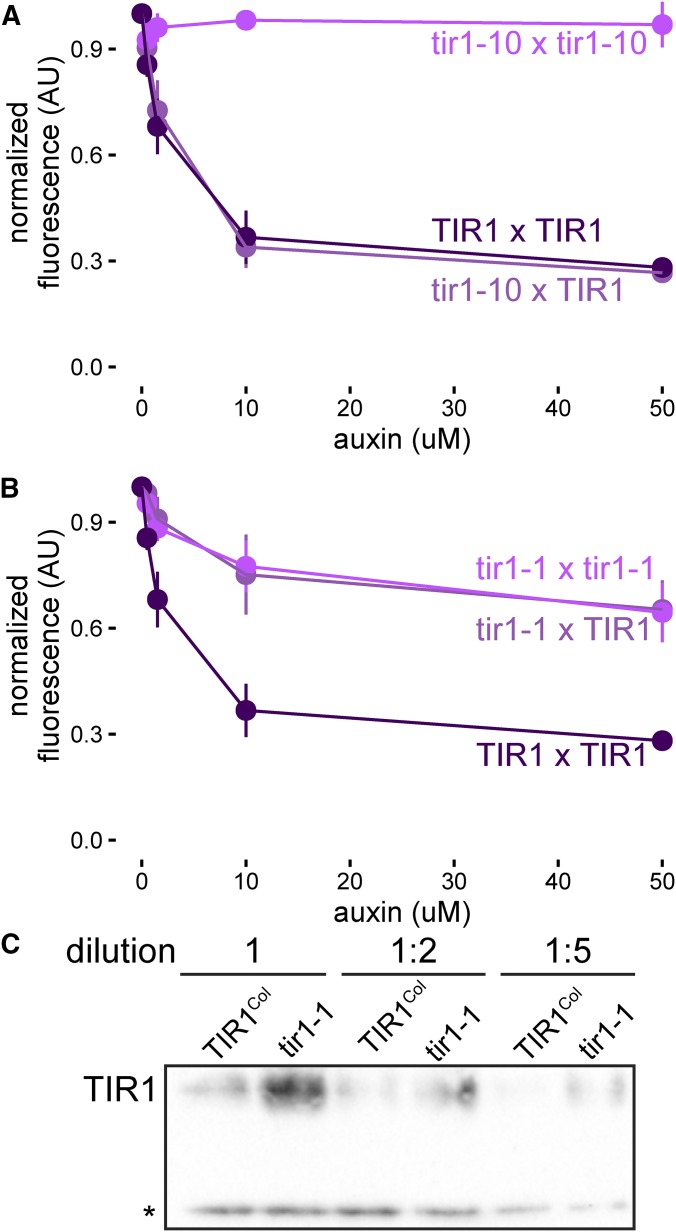
Dimerization domain variation affects dominance relations between TIR1 alleles
One of the unexpected findings in our analysis of auxin response across genotypes was a subtle but highly reproducible difference between the two induced alleles of tir1 in the Col-0 background (Figure 3, A and C). The point mutation tir1-1 showed a consistently stronger loss-of-auxin sensitivity than the T-DNA insertion tir1-10, raising the possibility that tir1-1 might be acting as a dominant negative or antimorph rather than as a simple loss of function. In support of that interpretation, tir1-1 mutants are semidominant (Ruegger et al. 1998), and the tir1-1 allele (G147D) and several other mutations in nearby residues negatively affect SCFTIR1 dimerization and activity (Dezfulian et al. 2016).
We turned to the yeast synthetic system to further investigate this question. By transforming a single copy of each allele into haploid yeast strains of each mating type, we created all pairs of alleles via mating. We also created tir1K159* a mimic of the tir1-10 T-DNA insertion allele. As expected, tir1K159* was an amorph, behaving similarly to an empty expression cassette (Figure S8). TIR1 dosage had little effect on auxin response in these assays, as TIR1/tir1-10 heterozygotes responded similarly to TIR1 homozygotes (Figure 4A). In contrast, expression of tir1-1 nearly completely abrogated TIR1 activity (Figure 4B), providing strong evidence that tir1-1 is a dominant negative allele. In addition to having a greatly reduced ability to induce Aux/IAA degradation, it is likely that tir1-1 is also outcompeting TIR1 for SCF complex formation and substrate binding as tir1-1 protein accumulated to much higher levels in yeast than TIR1 (Figure 4C).
Figure 4.
tir1-1 is a dominant negative allele. Yeast expressing YFP-IAA17 and pairwise combinations of (A) TIR1 and tir1-10 (tir1K159*) or (B) TIR1 and tir1-1 (tir1G147D) alleles were treated with various concentrations of auxin for 1 hr before YFP-IAA17 fluorescence was measured by flow cytometry. Mean fluorescence ± SE was calculated from four experiments. Some error bars are within the points. (C) Dilutions of lysates of IAA17 and FLAG-tagged TIR1-variant expressing yeast strains were subjected to Western blotting. A nonspecific band (*) is included as a loading control.
Discussion
The analysis of intraspecific variation in auxin sensitivity presented here critically extends previous work on the evolution of this pathway by focusing on protein level functional variation. Synthetic assays allowed for direct quantification of differences in the ability of TIR1 and AFB2 variants to facilitate ubiquitin-mediated degradation of their substrates. The creation of a structure/function map of natural variation revealed several areas of the F-box–LRR protein scaffold that can accommodate mutations, while modulating auxin sensitivity. For example, this analysis further underscored the importance of the AFB dimerization domain (Dezfulian et al. 2016) to regulate SCF activity.
The AFB family provides a test case for genome evolution after gene duplication, as there is evidence of both significant novelty and redundancy between family members (Dharmasiri et al. 2005a; Walsh et al. 2006; Parry et al. 2009; Hu et al. 2012). Analysis of intraspecific coding sequence polymorphisms in this study has identified a subset of the tolerated polymorphisms within the AFB family. This natural variation has also revealed potential differences in evolutionary rates across the gene family and redundancies within sister pairs. In the future, quantitative phenotyping and precision genetics will allow us to test related hypotheses and accurately partition the novel and redundant effects of individual AFB genes on developmental phenotypes. Accessions containing highly represented polymorphisms and having phenotypes not predicted by our synthetic functional analysis, should facilitate future examination of evolutionary robustness and plasticity in nuclear auxin signaling and downstream gene networks.
Our analysis demonstrates that functional diversification is occurring within the Arabidopsis TIR1 lineage and clarified the role of induced variants that are commonly used for auxin studies. The integrated biochemical and phenotypic analysis of natural variants refined the map of functionally relevant residues in TIR1 and AFB2. Synthetic analysis of the chemically induced tir1-1 allele, which is in proximity to many of the natural polymorphisms found in TIR1 and AFB2, has established tir1-1 as a dominant negative allele and highlighted the potential importance of interactions among family members to the auxin response.
The auxin pathway in Arabidopsis, like many critical signaling pathways across eukaryotes, has high levels of redundancy at each node and numerous modes of feedback. Together these factors act as strong buffers, masking functional changes in any one component. This effect likely explains the discrepancies between synthetic and plant phenotypes in this study. Similar factors may also contribute to the lack of TIR1/AFB genes identified in genome-wide association studies. Candidate gene approaches incorporating isolated functional assays as demonstrated here can complement genome-wide approaches by removing feedback and other compensatory effects. Future efforts that combine synthetic assays with higher throughput allelic replacement technologies in plants (Čermák et al. 2017) would substantially increase the ability to precisely compare the impact of a given variant in isolation and in a common plant context.
Extending this pipeline for structure/function and genotype/phenotype mapping to additional auxin signaling genes and developmental phenotypes will improve our understanding of how plant form is shaped by this small molecule. This information, along with the general evolvability of the LRR scaffold (Bella et al. 2008), make the AFBs potential candidates for engineering novel traits in crops (Sun et al. 2016).
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300092/-/DC1.
Acknowledgments
We thank Doug Fowler, Adam Leaché, and Eric Klavins for guidance on methods, analysis, and interpretation of our findings; members of the J.L.N., E. Klavins, and T. Imaizumi laboratories for helpful discussions; and Brenda Martinez for technical assistance. This work was supported by the National Institutes of Health (R01-GM107084), the National Science Foundation (MCB-1411949), and the Howard Hughes Medical Institute. R.C.W. received fellowship support from the National Science Foundation (DBI-1402222).
Author contributions: R.C.W. and J.L.N. conceived of the project and designed experiments. R.C.W., M.L.Z., and S.R.G. acquired data. R.C.W., M.L.Z., and J.L.N analyzed and interpreted data. R.C.W., M.L.Z., S.R.G., and J.L.N drafted, revised, and approved the manuscript.
Footnotes
Communicating editor: S. Poethig
Literature Cited
- Bella J., Hindle K. L., McEwan P. A., Lovell S. C., 2008. The leucine-rich repeat structure. Cell. Mol. Life Sci. 65: 2307–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón Villalobos L. I. A., Lee S., De Oliveira C., Ivetac A., Brandt W., et al. , 2012. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T., Curtin S. J., Gil-Humanes J., Čegan R., Kono T. J. Y., et al. , 2017. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29: 1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D., Lobry J. R., 2007. SeqinR 1.0–2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis, pp. 207–232 in Structural Approaches to Sequence Evolution: Molecules, Networks, Populations, Biological and Medical Physics, Biomedical Engineering, edited by Bastolla U., Porto M., Roman H. E., Vendruscolo M. Springer-Verlag, New York. [Google Scholar]
- Delker C., Pöschl Y., Raschke A., Ullrich K., Ettingshausen S., et al. , 2010. Natural variation of transcriptional auxin response networks in Arabidopsis thaliana. Plant Cell 22: 2184–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezfulian M. H., Jalili E., Roberto D. K. A., Moss B. L., Khoo K., et al. , 2016. Oligomerization of SCF TIR1 is essential for Aux/IAA degradation and auxin signaling in Arabidopsis. PLoS Genet. 12: e1006301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., et al. , 2005a Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M., 2005b The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Dreher K. A., Brown J., Saw R. E., Callis J., 2006. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders T. A., Strader L. C., 2015. Auxin activity: past, present, and future. Am. J. Bot. 102: 180–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Gruetzner R., Kandzia R., Marillonnet S., 2009. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M., Liu Q., Moss B. L., Malcomber S., Li W., et al. , 2015. Auxin signaling modules regulate maize inflorescence architecture. Proc. Natl. Acad. Sci. USA 112: 13372–13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., 2007. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 1–4. [DOI] [PubMed] [Google Scholar]
- Gray W. M., del Pozo J. C., Walker L., Hobbie L., Risseeuw E., et al. , 1999. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13: 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., et al. , 2010. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T. J., Hagen G., 2007. Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Guseman J. M., Hellmuth A., Lanctot A., Feldman T. P., Moss B. L., et al. , 2015. Auxin-induced degradation dynamics set the pace for lateral root development. Development 142: 905–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens K. A., Guseman J. M., Jang S. S., Pierre-Jerome E., Bolten N., et al. , 2012. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 160: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R. P., Edwards E. A., Leyland N. R., Bean S., Mullineaux P. M., 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hillson N. J., Rosengarten R. D., Keasling J. D., 2012. j5 DNA assembly design automation software. ACS Synth. Biol. 1: 14–21. [DOI] [PubMed] [Google Scholar]
- Hu Z., Keçeli M. A., Piisilä M., Li J., Survila M., et al. , 2012. F-box protein AFB4 plays a crucial role in plant growth, development and innate immunity. Cell Res. 22: 777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., Estelle M., 2016. Mechanisms of auxin signaling. Development 143: 3226–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan J., Bhattacharya J., Ranjan A., 2016. Enhancing crop yield by optimizing plant developmental features. Development 143: 3283–3294. [DOI] [PubMed] [Google Scholar]
- Meijón M., Satbhai S. B., Tsuchimatsu T., Busch W., 2014. Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat. Genet. 46: 77–81. [DOI] [PubMed] [Google Scholar]
- Moss B. L., Mao H., Guseman J. M., Hinds T. R., Hellmuth A., et al. , 2015. Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol. 169: 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T., 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M., 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6: 917–922. [DOI] [PubMed] [Google Scholar]
- Obenchain V., Lawrence M., Carey V., Gogarten S., Shannon P., et al. , 2014. VariantAnnotation: a bioconductor package for exploration and annotation of genetic variants. Bioinformatics 30: 2076–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Calderon-Villalobos L. I., Prigge M., Peret B., Dharmasiri S., et al. , 2009. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. USA 106: 22540–22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B., Wittelsbürger U., Ramos-Onsins S. E., Lercher M. J., 2014. PopGenome: an efficient swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31: 1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E., Jang S. S., Havens K. A., Nemhauser J. L., Klavins E., 2014. Recapitulation of the forward nuclear auxin response pathway in yeast. Proc. Natl. Acad. Sci. USA 111: 9407–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E., Moss B. L., Lanctot A., Hageman A., Nemhauser J. L., 2016. Functional analysis of molecular interactions in synthetic auxin response circuits. Proc. Natl. Acad. Sci. USA 113: 11354–11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E., Wright R. C., Nemhauser J., 2017. Characterizing auxin response circuits in Saccharomyces cerevisiae by flow cytometry. Methods Mol. Biol. 1497: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M. J., Greenham K., Zhang Y., Santner A., Castillejo C., et al. , 2016. The Arabidopsis auxin receptor F-box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 6: 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband W., 1997. ImageJ. U. S. National Institutes of Health, Bethesda, MD. [Google Scholar]
- Rosas U., Cibrian-Jaramillo A., Ristova D., Banta J. A., Gifford M. L., et al. , 2013. Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root systems architecture. Proc. Natl. Acad. Sci. USA 110: 15133–15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M., Dewey E., Gray W. M., Hobbie L., Turner J., et al. , 1998. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12: 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W., 2001. Molecular Cloning: A Laboratory Manual. CSHL Press, New York. [Google Scholar]
- Schmitz R. J., Schultz M. D., Urich M. A., Nery J. R., Pelizzola M., et al. , 2013. Patterns of population epigenomic diversity. Nature 495: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Wang B., Wang X., Hu K., Li K., et al. , 2016. Genome-wide association study dissecting the genetic architecture underlying the branch angle trait in Rapeseed (Brassica napus L.). Sci. Rep. 6: 33673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L. I. A., Sharon M., Zheng C., Robinson C. V., et al. , 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Terrile M. C., París R., Calderón-Villalobos L. I. A., Iglesias M. J., Lamattina L., et al. , 2012. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70: 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. A., Neal R., Merlo A. O., Honma M., Hicks G. R., et al. , 2006. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic Picolinate auxins and not to 2,4-Dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 142: 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M., Niemeyer M., Hellmuth A., Janitza P., Christ G., et al. , 2017. Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat. Commun. 8: 15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. C., N. Bolten, and E. Pierre-Jerome, 2017 FlowTime: annotation and analysis of biological dynamical systems using flow cytometry. R package version 1.0.0. Available at: http://bioconductor.org/packages/flowTime/. Accessed July, 2017.
- Yu H., Moss B. L., Jang S. S., Prigge M., Klavins E., et al. , 2013. Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 162: 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Zhang Y., Moss B. L., Bargmann B. O. R., Wang R., et al. , 2015. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat. Plants 1: 14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ward J. D., Cheng Z., Dernburg A. F., 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142: 4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S., Niu Q., Chua N., 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.