Abstract
Large-scale forward genetic screens have been instrumental for identifying genes that regulate development, homeostasis, and regeneration, as well as the mechanisms of disease. The zebrafish, Danio rerio, is an established genetic and developmental model used in genetic screens to uncover genes necessary for early development. However, the regulation of postembryonic development has received less attention as these screens are more labor intensive and require extensive resources. The lack of systematic interrogation of late development leaves large aspects of the genetic regulation of adult form and physiology unresolved. To understand the genetic control of postembryonic development, we performed a dominant screen for phenotypes affecting the adult zebrafish. In our screen, we identified 72 adult viable mutants showing changes in the shape of the skeleton as well as defects in pigmentation. For efficient mapping of these mutants and mutation identification, we devised a new mapping strategy based on identification of mutant-specific haplotypes. Using this method in combination with a candidate gene approach, we were able to identify linked mutations for 22 out of 25 mutants analyzed. Broadly, our mutational analysis suggests that there are key genes and pathways associated with late development. Many of these pathways are shared with humans and are affected in various disease conditions, suggesting constraint in the genetic pathways that can lead to change in adult form. Taken together, these results show that dominant screens are a feasible and productive means to identify mutations that can further our understanding of gene function during postembryonic development and in disease.
Keywords: zebrafish, dominant screen, mapping, postembryonic, pigment, skeletogenesis
THE use of systematic forward genetic screens has been instrumental in uncovering genes and pathways involved in a multitude of developmental processes (e.g., Brenner 1974; Nüsslein-Volhard and Wieschaus 1980; Mayer et al. 1991; Driever et al. 1996; Haffter et al. 1996a). This phenotype-driven approach allows for the unbiased analysis of gene function through generation of random mutations throughout the genome using chemicals or irradiation as mutagens.
Many genes that are found to be essential for early development and functional alterations are often not compatible with viability. Such lethality hinders the study of gene function during postembryonic stages. The establishment of tissue-specific or inducible knock-out lines circumvents this problem, and enables analysis in tissues of interest or at specific time-points during development. However, these methods do not lend themselves to broad unbiased screening in development, as often only a few loci can be feasibly tested at any one time. Different types of mutations such as partial loss-of-function or dominant mutations can help in elucidating functions in late development even in genes with key roles in embryogenesis. Dominant mutations, in particular, can be revealing of the full range of molecular and developmental gene functions, as increased and novel actions of a gene can result in unexpected phenotypes. These dominant mutations can also exhibit dosage-dependent effects, showing graded phenotypic differences between heterozygous and homozygous individuals. Thus, unique mutations apart from complete loss-of-function alleles can be informative about molecular regulation of gene function in postembryonic development.
The zebrafish is a well-established genetic model. Screens have focused on the identification of genes important for early developmental processes, with mutants showing recessive inheritance of the phenotype (e.g., Driever et al. 1996; Haffter et al. 1996a). Screens for recessive mutations require breeding the induced mutations to homozygosity and therefore multiple generations need to be raised before a phenotype is visible in the F3 generation. Thus, recessive screens require the ability to raise and screen a large number of fish in order to screen the function of genes affecting a specific developmental process in sufficient depth.
The majority of the identified recessive mutants display early phenotypes and are embryonic or larval lethal. Only ∼3% of mutants identified in these screens for early larval phenotypes led to viable adults with observable phenotypes (Haffter et al. 1996a). Thus, much of the genetic regulation of late development remains undescribed. Few screens have looked for genes affecting late development and most have been restricted in depth and phenotypic breadth (Haffter et al. 1996b; Bauer and Goetz 2001; Fisher et al. 2003; Andreeva et al. 2011; Saito et al. 2011). However, larger screens looking specifically for genes necessary for normal patterning and growth of the adult fish, demonstrated that a large number of mutants could be identified, supporting that late developmental processes can be investigated by classic genetic approaches (ZF models; www.zf-health.org/zf-models). In comparison, the largest of these screens from the ZF models consortium scored ∼1000 genomes for adult traits, ∼1/6 of the total predicted number of genomes analyzed for larval phenotypes in this screen, as well as the number of combined genomes screened in the early zygotic screens (Driever et al. 1996; Haffter et al. 1996a). Intriguingly, many of the mutations identified from these adult screens affect genes whose orthologs are associated with disease in humans (fgfr1a, col1a1a, bmp1a, and edar) (Fisher et al. 2003; Harris et al. 2008; Rohner et al. 2009; Asharani et al. 2012).
While isolating mutant lines through genetic screens has been very successful, identification of the causative mutations underlying mutant phenotypes was previously difficult, limiting the broad analysis of large classes of mutants and preventing detailing of their cognizant genetic pathways. The advent of next-generation sequencing techniques in combination with targeted capture and multiplexing of samples has allowed for cost-effective and efficient identification of mutations (Bowen et al. 2012; Leshchiner et al. 2012; Obholzer et al. 2012; Voz et al. 2012; Henke et al. 2013; Kettleborough et al. 2013; Ryan et al. 2013). This has opened the potential for analysis of genetic networks regulating specific developmental processes, as linkage and potential causative mutations can be quickly defined in whole sets of mutants.
We sought to take advantage of these new sequencing technologies to explore the utility of zebrafish genetic screens to investigate the genes controlling postembryonic development. Toward this end, we initiated a screen for mutations affecting the form of the adult zebrafish. To facilitate depth of screening, we focused on mutants with a dominant effect on morphology. Importantly, unlike most alleles identified or created using genome editing techniques, our focus on dominant mutations was centered on the ability to provide insight into the molecular action of a gene other than simple loss-of-function alleles through identification of potential gain-of-function and neomorphic alleles. Here, we show the feasibility of large-scale dominant screens in zebrafish for postembryonic development, including methods to systematically identify linkage and causative mutations underlying dominant mutant phenotypes. Results from this screen define important disease models in the zebrafish. Furthermore, by clustering analysis of similar phenotypes, we show enrichment for mutations affecting extracellular matrix formation as a regulator of late development. Importantly, this screen sets the stage for the use of zebrafish as an experimental tool to investigate genetic regulation through modifier analysis and efficient identification of gene networks regulating late development.
Materials and Methods
Husbandry and management of identified mutant lines
Zebrafish (Danio rerio) were raised and maintained under standard conditions (Nüsslein-Volhard and Dahm 2002) in compliance with internal regulatory review at Boston Children’s Hospital. Mutant lines were named following the rules set out by ZFIN, where “mh” is the designation of the founding lab (Harris lab) and “d” indicates dominant inheritance of the allele.
Mutagenesis and screen design
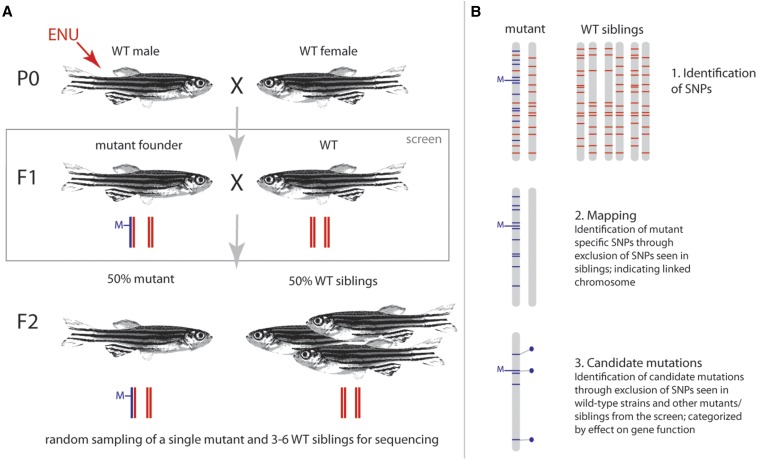
To identify dominant mutations that affect the adult form of the zebrafish, mutations were induced by treatment of 30 wild-type Tübingen males with N-ethyl-N-nitrosourea (ENU) treatment following an optimized protocol (Rohner et al. 2011). The surviving 14 mutagenized males were mated twice a week with wild-type females, and over 14,000 progeny were raised. At 10–12 weeks postfertilization (wpf), F1 fish were anesthetized in 0.02% MS-222 and screened under a dissecting scope for morphological changes. Potential mutants were isolated and crossed to wild-type fish. Similarly, F2 progeny were screened at 10–12 wpf. Crosses re-expressing the phenotype in roughly 50% of progeny were maintained and further analyzed. Varied wild-type lines, such as AB, Tübingen, and albino mutants, were used for outcrossing.
Next-generation sequencing library preparation and exome capture
Exome sequencing was performed as previously described (Kettleborough et al. 2013) with minor modifications. Briefly, 1–2 μg of DNA from individual mutants and pooled siblings was fragmented to enable the construction of 150–200 bp insert libraries according to standard Illumina protocols. Following genomic library construction, 500 ng of DNA was hybridized for 24 hr to the Agilent SureSelect Zv9.2 biotinylated whole exome RNA baits. The hybridization was then enriched with streptavidin-coated beads, and the rest of the RNA baits were digested. Libraries were amplified and barcoded through PCR (10 cycles) and libraries were paired-end sequenced (50 bp) using HiSeq2000v4 chemistries. As this version of the zebrafish exome capture is based on the Zv9 genome assembly, genes and exons annotated subsequent to the Zv9 release were not be captured by this method.
Mapping by association and candidate gene identification
Sequencing reads were aligned to the zebrafish genome (GRCz10) using Novoalign software (http://www.novocraft.com/products/novoalign/), with default settings and including 3′-adaptor trimming. PCR duplicates were removed using the MarkDuplicates command in Picard (http://picard.sourceforge.net/). Variants were called using SAMtools and BCFtools (Li et al. 2009; Li 2011). To identify SNPs useful for mapping, we used the GATK VariantFiltrationWalker (McKenna et al. 2010) to exclude the following variants: (1) SNPs lying in low-complexity regions or interspersed repeats, classified by RepeatMasker; (2) SNPs lying in a cluster of more than three SNPs per 10 bp; (3) SNPs with a quality score <30; (4) SNPs with a root-mean-square mapping quality of covering reads <40; (5) SNPs with a total read depth <20 or >100; (6) SNPs with a total read depth <40. Furthermore, SNPs with an allele frequency of 1 were excluded. For exclusion of common variants, SNPs present in at least two reads in siblings or mutants were identified from the individual variant files using a custom python script. After removing SNPs present in the siblings, the mutant chromosome was identified by counting the remaining SNPs per chromosome.
To identify candidate mutations, the mutant variant file was filtered using GATK VariantFiltrationWalker to exclude the following variants: (1) SNPs lying in a cluster of more than three SNPs per 10 bp; (2) SNPs with a quality score <30; (4) SNPs with a root-mean-square mapping quality of covering reads <40; and (5) SNPs with a total read depth <3 or >60. In addition, all SNPs that were called as having more than one nonreference allele were removed. SNPs present in at least two reads in wild-type strains, siblings, or other mutants from the screen were excluded using a custom python script. The remaining SNPs were analyzed and annotated using the Ensembl Variant Effect Predictor (McLaren et al. 2016). This stringent filtering protocol, while needed to exclude false positive calls, can lead to the exclusion of real variants. For example, the most likely candidate mutation in col2a1a in the dmh28 mutant was computationally excluded as a candidate mutation due to a low Phred-scaled quality score. While we identified a closely linked SNP in a neighboring gene used for the initial linkage analysis, reanalysis of variants in candidate genes in the region by eye led to the identification of the mutation in col2a1a.
Mapping and candidate gene data were visualized using PhenoGram (Wolfe et al. 2013).
Confirmation of linkage
To confirm linkage of the phenotype to a specific candidate mutation, genomic DNA from at least 10 heterozygous mutants and four wild-type siblings was Sanger sequenced to confirm presence or absence of the mutation, respectively. As a measure for linkage, we calculated the logarithm of the odds (LOD) score for each tested variant.
Generally, a LOD score of three or above is considered as proof of linkage of two loci. In our case, if no recombination event was detected in 10 heterozygous mutants analyzed (10 meiosis), the LOD score would equal 3.
Skeletal staining and microcomputed tomography
Fish were killed in 22% MS-222 and fixed in 3.7% formaldehyde for 24 hr at room temperature. For staining of mineralized tissues, fish were transferred into a 0.005% alizarin red/0.5% KOH solution overnight. Excess stain was removed with several washes in 0.5% KOH. Specimens were transferred through a glycerol series into 80% glycerol for storage.
For scanning, fish were washed in PBS or water following fixation, and then placed in sample tubes embedded in 1% agarose to reduce movement during scanning. Images were obtained using a Skyscan 1173 (Bruker), 240-degree scan with 0.2 rotational step. X-ray source voltage set to 70 kV and current set to 80 μA. Exposure time was 1500 msec. Resolution of scan was 7.14 μm per pixel. Images were processed using Paraview (http://www.paraview.org/overview/) or Amira software package, version 6.0 (FEI).
Data availability
Sperm from the 25 mutant lines described in this paper is preserved to secure the lines. Mutant lines are available on request from the author. Supplemental Material, File S1 contains total and mutant specific SNP counts for each chromosome for the 23 mutants mapped with the method described in this manuscript. File S2 contains numbers of unique nonsynonymous mutations per chromosome for the 23 mutants. A list of primers used for confirmation of linkage is provided in Table S1 in File S4. Raw sequence data were submitted to the European Nucleotide Archive (ENA) under study PRJEB13615 (http://www.ebi.ac.uk/ena/data/view/PRJEB13615). Accession numbers for individual sequencing files can be found in File S3. Custom scripts for data analysis are available for download from the laboratory website (www.fishyskeleton.com) or upon request.
Results
Screen design
To facilitate a broad sampling of genetic variation throughout the zebrafish genome, we used ENU mutagenesis and an F1 dominant screen design (Figure 1A). We screened 10,424 fish, representing a similar number of haploid genomes, for phenotypes affecting adult form. We identified 269 potential mutants affecting a wide diversity of traits. These mutants all showed specific, qualitatively distinct, adult phenotypes. We focused recovery and isolation toward mutants that exhibited differences in the formation and patterning of the adult skeleton, including fins and scales, as well as changes in pigmentation (Table 1). Screening for changes in reproductive ability—another key adult trait—was not possible, as reduction of fertility would preclude recovery of the mutant line in an F1 screen design. Other auxiliary phenotypes include defects in development of the eyes, represented by a large number of fish exhibiting cloudy lenses. These were not collected nor included in the screen results.
Figure 1.
Screen and mapping strategy (A). Wild-type males were mutagenized with ENU to induce random mutations in the genome. Mutagenized males were outcrossed to wild-type females, and the subsequent generation (F1) was screened for dominant mutations affecting adult form. Isolated mutant founders were outcrossed, and the phenotype and penetrance was assessed in their progeny (F2). (B) For mapping and candidate mutation identification, one mutant and a pool of three to six wild-type siblings from the same cross were sequenced. Sequence data were analyzed in three steps: (1) all SNPs in the mutant and sibling pool were identified (blue and red lines; gray bars represent individual chromosomes); (2) to identify the chromosome carrying the mutation (M), mutant specific SNPs (blue bars) were identified by removing all SNPs present in the wild-type siblings from the set of previously identified SNPs; (3) candidate mutations are then identified by excluding all background SNPs from wild-type strains or other mutants/siblings sequenced. The remaining SNPs are classified as synonymous or nonsynonymous, and ranked by their predicted consequence on gene function (deleterious/tolerated). Blue dots indicate nonsynonymous changes on the linked chromosome.
Table 1. Screen results.
| F1 Screen | |
|---|---|
| #Fish screened | #Potential mutants |
| 10,424 | 269 |
| F2 confirmation screen | |
| #Successful matings | #Confirmed mutants |
| 135 | 72 |
| Phenotype | #Mutants |
| Fin | 23 |
| Dermal skeleton | 4 |
| Craniofacial | 12 |
| Stature | 21 |
| Pigment | 12 |
To confirm true monogenic inheritance of a phenotype, isolated F1 mutant founders were outcrossed to wild-type fish, and the F2 generation was screened for the parental phenotype. Out of the 269 potential mutants isolated in the F1 generation, 135 produced offspring. In these 135 crosses, 72 showed the expected phenotype in 50% of the offspring, suggesting a monogenic trait. These mutants were grouped into five major classes based on their most prominent phenotype. A group of 23 mutants showed changes in the development of fins; 21 mutants are characterized by changes in stature; the craniofacial skeleton was affected in 12 mutants; four mutants showed a broader change in the dermal skeleton, and 12 mutants displayed changes in pigmentation (Table 1). A select group of 25 mutants from these classes were maintained for mapping, mutation identification, and further phenotypic analysis.
Systematic mapping of dominant mutations from the screen
A candidate gene approach was used to successfully identify mutations in two of the 25 mutants isolated, dm1 and dmh3 (see mutant phenotypes section below). For linkage analysis and mutation identification of the remaining 23 mutants, DNA was isolated from a single mutant carrier, and three to six wild-type siblings for each mutant (Table S2 in File S4). Most progeny stemmed from heterozygous mutant outcrosses to wild-type strains, for six mutants progeny from heterozygous mutant incrosses were analyzed. Sequencing libraries were prepared for each single mutant and pooled wild-type siblings. These pools were enriched for coding regions using exome capture. Paired end Illumina sequencing was performed on eight libraries per lane (four mutant/sibling pairs), resulting in 15–29 million reads per sample, and average exome coverage of 23× (Table S2 in File S4).
Two overlapping but distinct methods were used to rough map and refine potential causative mutations underlying the dominant phenotypes. First, to localize the genetic location of the mutation in the genome, the chromosome carrying mutant-specific SNPs was identified (Figure 1B). Second, through identification of unique nonsynonymous, mutations in the mutant, candidate mutations potentially underlying the mutant phenotype were defined for subsequent confirmation of linkage.
Identification of mutant-specific haplotypes:
Given the high level of heterogeneity of the zebrafish genome even within strains (Stickney et al. 2002; Guryev et al. 2006; Bradley et al. 2007; Coe et al. 2009; Bowen et al. 2012), causative mutations should be associated with discernable haplotype blocks of variants unique to the mutant. We defined mutant-specific haplotypes solely by the presence of SNPs unique to the mutant. These SNPs can be identified by excluding all SNPs seen in the sibling pool from SNPs present in the mutant (Figure 1B). If all haplotypes present in the cross analyzed are sampled and sequenced to sufficient depth in the sibling pool (Figure S1), only the chromosome carrying the phenotype-causing mutation should have SNPs left after this analysis.
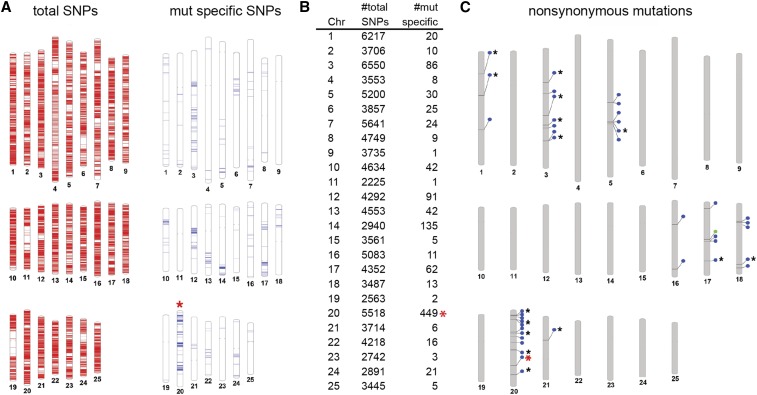
To this end, sequencing reads were aligned to the current assembly of the zebrafish genome (GRCz10), and high-quality SNPs were identified that showed at least 20× coverage in both the mutant and corresponding sibling pool (see Methods for details). Although we achieved average exome coverage of 23×, only ∼50% of the exome is covered by >20 reads and thus is used in the analysis (Table S2 in File S4). This is likely due to unequal capture efficiency of different exons and preferential sequencing of certain regions. SNPs that were homogeneous in both the mutant and sibling pool were excluded from the analysis as uninformative. This resulted in the identification of between 450 and 7000 SNPs per chromosome (Figure 2, A and B and File S1). In the mutants analyzed here, removal of SNPs found in the corresponding sibling pool resulted in reduction of total SNPs to between 0 and 1469 SNPs per chromosome. The highest number of mutant specific SNPs on a chromosome within the genome varies significantly between different mutant lines analyzed, and ranges from 106 to 1469 (File S1). This variation highlights the different levels of background heterogeneity in the crosses analyzed. Although one would expect no mutant specific SNPs for unlinked chromosomes, most chromosomes showed a relatively low level of mutant-specific SNPs. For example, analysis of the sequence data for dmh35 and the corresponding sibling pool resulted in the identification of 1–135 mutant specific SNPs per chromosome for unlinked chromosomes (Figure 2, A and B). Chromosome 20 showed with 449 SNPs the highest number of mutant specific SNPs and was subsequently shown to be linked to the mutant phenotype, as described below.
Figure 2.
Mapping and candidate gene identification in dmh35. To identify mutant specific haplotypes, first all SNPs in the genome are identified. After exclusion of all SNPs that are present in the sibling pool, only mutant specific SNPs are left. The highest number of mutant specific SNPs on a chromosome should indicate the chromosome carrying the mutation. (A) The red bars in the diagram on the left represent SNPs identified in the mutant and sibling pool on each of the 25 chromosomes. Blue bars in the diagram on the right represent mutant-specific SNPs on each of the 25 chromosomes. (B) Summary of the number of total and mutant-specific SNPs per chromosome. (C) In a second step, all nonsynonymous changes are identified in the genome. Blue circles indicate missense mutations; green circles mark nonsense mutations; black asterisks mark mutations predicted to be deleterious; the red asterisk indicates the position of the G138D missense mutation in connexin 43, shown to be linked to the dmh35 mutant phenotype.
To test the bounds of sampling, for two mutants, dmh15 and dmh4, we sequenced two pools of three siblings to a depth of 20× each. In both cases, the remaining numbers of mutant-specific SNPs per chromosome were significantly lower when compared to other mutants where only one pool of three siblings or a pool of six siblings was sequenced to 20× depth (File S1 and Table S3 in File S4). This is most likely due to unequal sequencing of haplotypes present in the pool, as well as insufficient sequencing depth to cover all haplotypes. Differences in sampling of parental haplotypes were obvious when running the analysis, with only one of the sibling pools. For example, for the dmh15 mutant, chromosome 5 has the highest number of mutant specific SNPs with 534 when only using sibling pool 1 (Table S3 in File S4). However, the chromosome harboring the highest number of SNPs switches to chromosome 7 with 866 mutant-specific SNPs if only the second sibling pool is used in the analysis (Table S3 in File S4). Both chromosomes show almost no mutant-specific SNPs when both sibling pools are used in combination, indicating that the haplotype present in the mutant for these two chromosomes was sampled in only one of the sibling pools.
In addition to a second sibling DNA pool, we also sequenced a second mutant individual for dmh15. By repeating the experiment with a different mutant, we noticed that not all chromosomes showed the same trend as seen with the first comparison (Table S3 in File S4). More specifically, chromosome 4 showed an extremely high number of mutant specific SNPs in the second mutant, whereas the first mutant sequenced had almost no mutant-specific SNPs on chromosome 4. This indicates that the chromosomal haplotype of the second mutant was insufficiently sampled in the two sibling pools. It should be noted that, in this example, the chromosome carrying the phenotype-causing mutation (chromosome 19), did not have a high number of mutant-specific SNPs. This suggests that the phenotype-causing mutation does not lie within a discernable haplotype in this specific example.
Identifying candidate mutations through filtering related and common SNPs:
Since whole exomes are sequenced by this method, the sequencing data can, at the same time, be used for mapping as well as the identification of potential candidate mutations affecting gene function. First, all variants with at least 3× coverage are identified in the mutant (Table 2). This coverage was consistently seen over, on average, 88% of the exome in all sequencing libraries (Table S2 in File S4). Due to the high rate of variation within the zebrafish genome, this analysis results in a high number of potential phenotype-causing mutations. To refine this list of potential candidate mutations further, all SNPs that are present in the corresponding wild-type siblings are excluded. This reduces the number of potential candidate mutations significantly (Table 2). Furthermore, taking advantage of existing sequencing data from wild-type strains (Bowen et al. 2012), all wild-type variants observed in this dataset are excluded as potential candidate mutations as well. In addition, as a large number of mutants from the same dominant screen were sequenced and share a common set of background alleles from the founders, we used this data to further refine potential candidates by excluding all variants detected in wild-type sibling pools or mutants with phenotypes different from the mutant under analysis. This way of filtering shared SNPs reduces the number of nonsynonymous mutations observed genome-wide to a small number that can then be prioritized based on potential linkage, predicted gene function and the nature of the mutation (File S2 and Table 2). For the dmh35 mutant, our analysis identified 41 nonsynonymous mutations genome wide (Figure 2C); 12 of these mutations are located on chromosome 20, the chromosome with the highest number of mutant-specific SNPs (Figure 2, A and B).
Table 2. Candidate gene identification.
| SNPs Identified in dmh35 | ||||
|---|---|---|---|---|
| Genome Wide | Chromosome 20 | |||
| Coding | Nonsynonymous | Coding | Nonsynonymous | |
| #Total | 159,882 | 52,562 | 6262 | 1934 |
| #Not present in siblings | 7,804 | 3,138 | 676 | 262 |
| #Not present in siblings or WT strains | 1,345 | 619 | 97 | 41 |
| #Not present in siblings, WT strains or other mutants and their siblings | 62 | 41 | 16 | 12 |
To confirm linkage of the phenotype to a specific SNP, regions containing the variant were amplified from genomic DNA from a number of individual heterozygous mutants and wild-type siblings, and then Sanger sequenced to assess presence of the variant. With this approach, we were able to identify closely linked mutations for >80% of the mutants sequenced (20/23; Table 3). In 14 out of the 20 mutants (70%), linkage was confirmed to the chromosome with the highest, or second highest, number of mutant specific SNPs in the genome (Table S4 in File S4). For several mutants the number of mutant-specific SNPs on the linked chromosome was very low (0–56) (File S1 and Table S3 in File S4), suggesting that there was not a discernable haplotype associated with the mutation. But even in cases where our mapping approach did not correctly predict the chromosome containing the mutation, the most likely candidate mutation could be predicted for six of the mutants by analyzing the genome-wide candidate mutation list after filtering (Table 3). As variants found in siblings are excluded as potential candidates, this technique relies heavily on the ability to phenotypically distinguish mutants from wild-type siblings. For mutants with incomplete penetrance, it can be difficult to identify candidate mutations if a mutant was improperly scored as a wild-type sibling, as both phenotype-linked SNPs and the causative mutation will be removed during the analysis. Therefore, for mutants with weak phenotypes, or incomplete penetrance, it may be helpful to reanalyze potential variants without exclusion of sibling SNPs, comparing against common SNPs from wild types and unrelated mutants.
Table 3. Linked variants.
| Mutant Allele | #Nonsyn Genome | # Linked Chr | Location; bp Change | Gene | AA Change | LOD Score |
|---|---|---|---|---|---|---|
| Stature | ||||||
| dmh14 | 29 | 3 | 3:23104458 G/A X | col1a1a | G1144E | 5.41 |
| dmh13 | 26 | 2 | 3:23104092 G/A | col1a1a | G1093R | 3.61 |
| dmh29 | 303 | 17 | 12:3070126 C/T | col1a1b | G1123D | 7.22 |
| dmh15 | 17/12 | 1/2 | 19:41411721 G/A | col1a2 | G882D | 12 |
| dmh27 | 27 | 3 | 8:21181959 G/T | col2a1a | G1174X | 3.91 |
| dmh28 | 22 | 6 | 8:21181691 G/A X | col2a1aa | G1141D | 4.8 |
| dmh21b | 66 | 8 | 8:21303580 G/A X | itpr3 | G2135D | 8.4 |
| dmh30b | 10 | 5 | 8:21303580 G/A | itpr3 | G2135D | 1.5 |
| dmh16 | 17 | 4 | 12:13080359 T/G X | cmn | M10R | 5.11 |
| dmh31 | 9 | 5 | 5:31610564 G/A X | myhz2 | M543I | 5.97 |
| dmh4 | 10 | 3 | 24:39229412 T/A X | CU929145.1 | I156L | 3.61 |
| Fin | ||||||
| dmh35 | 41 | 12 | 20:40820378 C/T X | cx43 | G138D | 3.01 |
| dmh20 | 38 | 12 | 20:28348113 C/A X | dll4 | H190N | 8.4 |
| dmh32 | 28 | 3 | 20:28350489 T/A X | dll4 | C263X | 7.22 |
| dmh33 | 16 | 5 | 20:28352804 C/A X | dll4 | Y449X | 6.02 |
| dmh34 | 32 | 17 | 20:28350403 A/T X | dll4 | R235X | 1.5 |
| Dermal skeleton | ||||||
| dmh3c | — | — | 9:56069237 G/T | edar | D409Y | 14.8 |
| dmh19 | 23 | 1 | 8:17607022 A/G X | prkcz | S113P | 5.7 |
| dmh18 | 49 | N/A | ||||
| dmh22 | 66 | N/A | ||||
| Pigment | ||||||
| dmh8 | 23 | 3 | 1:46725420 G/T X | cx41.8 | N63K | 4.82 |
| dmh7 | 35 | 5 | 1:46725024 T/A X | cx41.8 | R195S | 4.52 |
| dmh9 | 25 | 6 | 15:40379722 G/A | kcnj13 | T128M | 3.01 |
| dmh1d | 102 | 6 | 15:40373740 A/T | kcnj13 | Y325X | N/D |
| dmh11 | 12 | N/A | ||||
Mutation in col2a1a was filtered out as a candidate due to a low Phred-scaled quality score of 5.46 of the alternative allele.
Mutants are most likely clonal; N/A, for these mutants, no linkage was found at time of publication; N/D, not determined; X, linkage to predicted chromosomes was confirmed.
Identified by candidate gene approach.
Identified by whole exome sequencing of a homozygous mutant pool.
For three out of 23 mutants analyzed by mapping, a linked chromosome was not identified at time of publication. Further analysis will be needed to assess if alternative SNPs identified in our analysis show linkage, or if there was sampling bias stemming from the exome design. Regardless, over 85% of mutants analyzed were mapped.
Identified mutant phenotypes and underlying genetic changes
For mutant characterization, we first clustered mutants into broad phenotypic classes. Following this clustering, we then compare the suite of candidate genes identified in each class to assess affected genetic pathways leading to change in form. Here, we detail four classes of mutants containing 25 mutants in total. For each mutant, we have performed linkage analysis and identification of potentially causative mutations. The remaining mutants from the screen are no longer available, and were therefore excluded from the manuscript.
Mutants with vertebral defects
A group of 11 dominant mutants, identified by their stature, were kept for further analysis. These vertebral mutants were grouped into five classes described below.
Class I mutants:
Four of these mutants (dmh13, dmh14, dmh15, and dmh29) were characterized by short stature, with no apparent larval phenotype at 5 days post fertilization (dpf). Analysis by microcomputed tomography (μCT) revealed strong deformations of the vertebrae, especially of the hemal and neural arches (Figure 3, B–D). In addition, we found excessive bone formation, with the formation of osteophytes, especially in the centrum of the vertebrae. Interestingly, mapping of the causative change in each of these mutants indicated that all four mutants carry missense mutations in components of Collagen type 1, changing a conserved glycine in the triple-helical domain to a different amino acid (Table 3). The Collagen 1 protein consists of three protein chains, two Collagen1a1 (Col1a1) and one Collagen1a2 (Col1a2) chain that form a triple helix. Proper helix formation requires a glycine residue in every third position of the helical domain of the protein. Missense mutations in these glycine residues are known to act in a dominant fashion (Gajko-Galicka 2002). Stemming from the whole genome duplication in the teleost common ancestor, zebrafish have two paralogs of the collagen1a1 gene: col1a1a and col1a1b. Both the dmh13 and dmh14 mutants carry mutations in the col1a1a gene (G1093R; G1144E), which are closely linked, while the dmh29 mutants exhibit mutations in a glycine residue of the col1a1b gene (G1123D), leading to a very similar phenotype. The observed phenotype of these mutations are similar to, but more severe than, previously identified col1a1a mutants (Fisher et al. 2003; Asharani et al. 2012). The dmh15 mutants exhibit the strongest phenotype in the group, as heterozygous mutants show the greatest reduction in body length (Figure 3D). In these mutants, we detected a mutant-specific nonsynonymous mutation in col1a2 (G882D) linked to the phenotype. The phenotypic similarity, taken together with the predicted functional consequence of the identified changes, makes these mutations in collagen 1 the most likely candidates for causing the phenotype. Interestingly, in this phenotypic class, we were able to identify dominant mutations in all components of Collagen I, expanding our knowledge about phenotypic consequences of mutations in collagen1 over the previously published col1a1a alleles.
Figure 3.
Mutants with vertebral defects. Representative photographs (left) and μCT images of the vertebral column (right) of the five different subgroups of mutants with vertebral defects. (A) wild-type zebrafish, regularly patterned and shaped vertebrae, and vertebral spines. (B–D) Heterozygous dmh13, dmh29 and dmh15 mutants have a shorter trunk, and show strongly deformed vertebral bodies and spines with excessive bone formation. (E) Heterozygous dmh27 fish are also shorter than wild-type fish, but show mostly normal patterned vertebral bodies with the exception of a couple of fused vertebrae. Vertebral spines show slight changes in angle. (F) In addition to a shorter body, heterozygous dmh16 mutants have strong deformation of their vertebrae. In addition to fusion of vertebral bodies, some vertebrae are split up into multiple hemi-segments. (G) The body of dmh31 mutants seems to be proportionally normal until the region in front of the caudal peduncle. Here, vertebrae are strongly deformed, leading to a bend in the body axis. (H) Strong curvature of the spine in dorsal, ventral, and lateral direction leads to an overall shorter body length of the dmh4 mutant. The vertebral bodies seem to be mostly normally patterned.
Class II mutants:
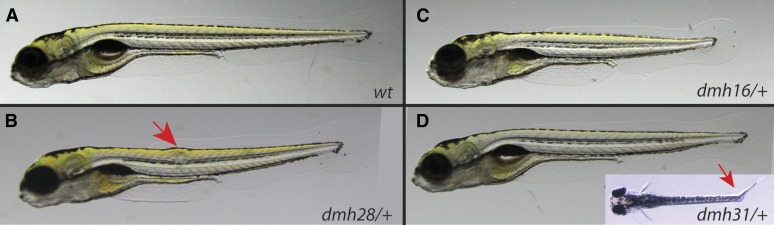
Unlike the first class of vertebral mutants, which primarily exhibited phenotypes arising late in development, a second group of four mutants (dmh21, dmh27, dmh28, and dmh30) showed altered length already as larvae at 5 dpf (Figure 4B). A fraction of the mutant progeny showed deformations of the notochord at 5 dpf in addition to a reduction in length. Analysis of the adult spine of the mutants in this class revealed mostly normal vertebral morphology, with the exception of a few fused centra (Figure 3E). We found that all four mutants are linked to chromosome 8. For two of the mutants, dmh27 and dmh28, we identified missense mutations in a gene encoding a component of type II Collagen. Similar to col1a1, zebrafish have two copies of the col2a1 gene, col2a1a, and col2a1b. Both dmh27 and dmh28 were found to harbor mutations in col2a1a. While the mutation in dmh28 changes a conserved glycine residue to aspartic acid (G1141D), similar to col1a mutants we identified, the dmh27 allele leads to a truncation of the C-terminal part of the protein (G1174X) (Table 3) that may act in a dominant negative manner. dmh21 and dmh30 are likely clonal mutations as they show the same haplotype pattern surrounding a linked variant in itpr3 on chromosome 8. As the precise mutation for these alleles is not known, we maintain their separate allele designation. Itpr3 is in close proximity to col2a1a. The dmh21 and dmh30 mutants, however, do not show any obvious nonsynonymous or splice site mutations in the predicted col2a1a gene. Given the similarity in phenotype to dmh27 and dmh28, it is likely that these too are alleles of col2a1a, and that the mutations are regulatory, or in not well annotated isoforms of the gene. Further biochemical analysis of Col2a1a in these alleles will need to be carried out to test the potential causative effect of these unidentified mutations.
Figure 4.
Larval phenotypes of mutants with vertebral defects. Representative photographs of wild-type (A) and mutant (B–D) phenotypes at 5 dpf. Mutants show a shorter body length when compared to wild-type larvae. A fraction of heterozygous dmh28 and dmh31 mutants, in addition show deformations of the notochord (red arrows). Inset in (D) shows a dorsal view of a dmh31/+ mutant larva, illustrating the kink in the trunk coinciding with deformations in the notochord.
Class III mutants:
The dmh16 mutant is superficially similar to the mutants linked to col2a1a, displaying shorter adult body length early at 5 dpf and in the adult (Figure 3F and Figure 4C). However, μCT analysis revealed that the vertebral bodies in this mutant are highly deformed, with composite vertebrae phenotype. Several vertebrae were found to have two or more hemi-segments. Fused vertebrae were also commonly noted (Figure 3F). For this mutant, we show linkage to a mutation in calymmin (cmn) on chromosome 12 (Table 3). This gene was discovered in a cDNA library screen through its expression in the zebrafish notochord, and is thought to be an extracellular matrix protein (Cerdà et al. 2002). The identified mutation, M10R, resides within the signal peptide, and could potentially interfere with proper protein localization.
Class IV and V mutants:
Two further mutants show deformation in the spine beyond axial shortening. The dmh31 mutant (class IV) seems to be proportionally normal in length until right before the caudal peduncle, where the vertebrae are highly deformed and bend (Figure 3G). This bend in the body can already be seen at 5 dpf as a kink in the notochord (Figure 4D). The dmh31 phenotype shows strong linkage to a missense mutation in a myosin heavy chain gene, myosin, heavy polypeptide 2, fast muscle specific (myhz2). The myhz2 gene encodes a fast-muscle-specific myosin heavy chain, and is expressed in muscles of the posterior part of the trunk at 3 dpf (Peng et al. 2002; Nord et al. 2014), coinciding with the position of the deformations of the trunk seen in dmh31 mutants (Figure 4D). The identified mutation substitutes a highly conserved methionine in the myosin tail 1 domain for an isoleucine (M543I; Table 3) and is predicted to be deleterious.
The class V mutant, dmh4, is characterized by a strong curvature in the spine. Whereas the embryos appear to be perfectly normal at 5 dpf, multiple bends in dorsal, ventral, and lateral orientation can be detected in the adult (Figure 3H). Apart from the strong curvature, the vertebral bodies seem to be patterned correctly when analyzed by μCT imaging. We did identify a closely linked predicted deleterious mutation in an uncharacterized protein, CU929145.1, with sequence similarity to a predicted myloid differentiation marker-like protein 2 (XP_005174257.1; Table 3). As for genes in which just a single allele was identified, further experiments will be needed to show causality of the identified change.
Mutations broadly affecting the dermal skeleton
Four mutants were isolated with broad defects on the formation and/or patterning of the dermal skeleton. One of the hallmark characters that differentiates these mutants from other classes such as fins, which are also constrained components of the dermal skeleton, is their effect on scales and/or dermal bones of the skull, such as the bone covering the gills, the opercle, subopercle, and branchiostegal rays.
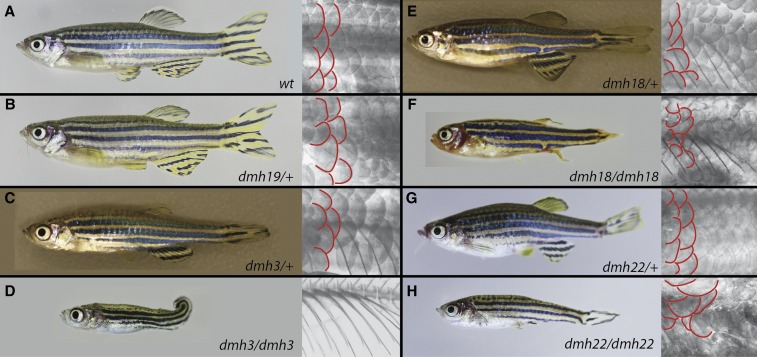
The first mutant in this class, dmh19, shows a very mild scale phenotype characterized by irregularly sized and patterned scales (Figure 5B). In addition, the mouth is positioned slightly more superiorly in mutants when compared to wild-type siblings. Our mapping approach indicated linkage to chromosome 8 and identified a single unique variant on this chromosome in a gene encoding Protein kinase C zeta (prkcz). Sequencing of individual mutant carriers showed strong linkage to this change; however, the mutation is predicted to be tolerated. Further analysis of the region will be needed to prove causality of this change, or to identify alternative potential candidate mutations.
Figure 5.
Mutants with defects of the dermal skeleton. Representative photographs of wild type and mutants (left), with close ups of the flank stained with alizarin red to visualize scales (right). For better visualization, two rows of scales are outlined in red. (A) In wild-type fish, the scales are similar in size and are very regularly patterned. (B–H) All mutants in this class show differences in size, pattern, and some even in orientation of scales. (D, F, H) These differences are even more obvious in homozygous mutants. In addition, all mutants show changes in the patterning and length of their fin rays to varying degree.
The dmh3 mutant is characterized by shorter and deformed fin rays, and has fewer but larger scales (Figure 5C). Dmh3 homozygous mutants show a strongly enhanced phenotype with a complete loss of the dermal component of the fins, and complete loss of scales (Figure 5D). Due to the similarities in phenotype of the dmh3 mutant to previously published mutants with defects in ectodysplasin signaling (Harris et al. 2008), we choose a candidate gene approach to identify potential phenotype-causing mutations. We isolated cDNA from heterozygous dmh3 mutants and sequenced the coding region of ectodysplasin pathway genes. This led to the isolation of a linked missense mutation in the ectodysplasin A receptor (edar) gene, changing a conserved aspartic acid in the death-domain to a tyrosine (D409Y).
Heterozygous dm18 mutants show characteristic defects in the fronto-nasal aspect of the skull leading to the formation of a notch right behind the maxilla (Figure 5E). The scales are irregularly sized, and show changes in pattern and orientation. Some of the fin rays appear to be truncated, while other rays in the same fin appear normal in size. These phenotypes are strongly enhanced in the homozygous mutants (Figure 5F). Here the scales are generally smaller than in wild type and grow in no obvious pattern or orientation. The skull shows extreme fronto-nasal shortening generating a very small and round shape. In a large percentage of mutants, the gill cover is very small and fails to cover the full extent of the gills. In addition, the fin rays seem to converge distally.
The last mutant in this group, dmh22, shows mild irregularities in scale size and patterning, mostly located toward the front of the trunk (Figure 5G). The fins display varying degrees of defects, ranging from a reduction in the number of rays to deformed rays and ray segments. Here again, the homozygous mutant phenotype is more extreme. The scales in homozygous dmh22 mutants grow in all orientations and do not have any pattern or consistent size (Figure 5H). The fin phenotypes range from complete loss of fins to a strong decrease in fin ray and ray segment number.
For both the dmh18 and dmh22 mutants, a variant linked to the phenotype could not be identified at time of publication.
Mutants with specific defects in fin formation
Five mutants were further analyzed that show specific alterations in the fins when compared to the entire skeleton. Mutants in this class could generally be grouped into two subclasses: (1) mutants with normal patterned but shorter fins, and (2) mutants with deformed fins. No long-finned mutants were found.
From the first subclass, we further analyzed and sequenced one mutant with shorter fins, dmh35. In dmh35 both the paired and median fins are normally patterned but shorter due to shortening of the fin ray segments (Figure 6B). Sequencing analysis of dmh35 revealed a missense mutation in connexin 43 (cx43) on chromosome 20 (Table 3). Connexin 43 is a component of gap junctions and mutations in cx43 have been shown previously to cause the short fin phenotype (Iovine et al. 2005).
Figure 6.
Mutants with fin deformations. Representative photographs of mutants with wild type (A), short (B), and deformed (C) fins. (B) Both the median and paired fins of dmh35 heterozygous mutants are shorter. Alizarin red stained pelvic fins of wild type (D) and dmh33/+ mutant (E) fish. Dmh33 heterozygous mutants show shorter fin rays with deformed segments of unequal length.
From the second subclass class of fin mutants, we analyzed four mutants dmh20, dmh32, dmh33, and dmh34. Here, the pelvic fins are most severely affected in that they show partial truncations of fin rays and generally unequal length of segments within a fin (Figure 6, C and E). Fins in general show bending of fin rays in varying extent. All four mutants in this class show strong linkage to missense mutations in the notch ligand delta-like 4 (dll4) (Table3). The change in dmh20 mutates a conserved histidine at position 190 in the delta serrate ligand (DSL) domain in dll4 to an asparagine, while the mutations in dmh32, dmh33, and dmh34 lead to early truncations of the protein, partially deleting the epidermal growth factor (EGF) domains as well as the predicted transmembrane domain. These data support previous findings that haploinsufficiency in dll4 can lead to fin deformations in adult zebrafish (Leslie et al. 2007).
Pigment mutants
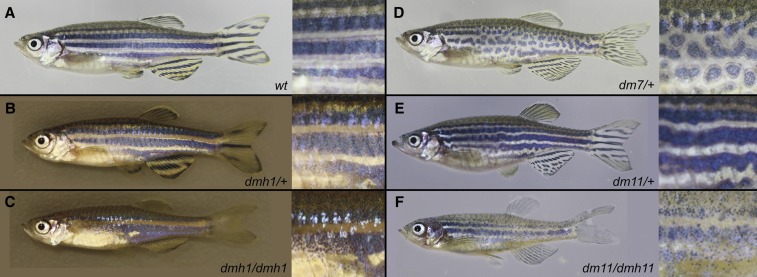
The adult pigment pattern is specified early during postembryonic development. The majority of pigmentation mutants we identified exhibited altered stripe patterns (Figure 7), some of which closely resembled phenotypes identified in previous screens (Haffter et al. 1996b). Two mutants, dmh1 and dmh9, displayed fewer melanophore stripes and wider interstripe regions (Figure 7B), resembling the dominant phenotype previously described for the obelix/jaguar mutants altering kcnj13/kir7.1 (Iwashita et al. 2006). To identify potential candidate mutations in dmh1, a pool of nine homozygous mutants was sequenced, and all homozygous mutations in the genome were identified. After excluding known background variants as described before, the list of genes was scanned for potential candidate mutations revealing a mutation in kcnj13/kir7.1. Similarly, mapping of dmh9 by the approach described before also revealed a missense mutation in kcnj13/kir7.1 that is closely linked to the phenotype (Table 3). The dmh7 and dmh8 mutants were identified by the formation of pigment spots instead of stripes (Figure 7D), resembling the phenotype previously described for the leopard mutant (Watanabe et al. 2006). Here, we found missense mutations in connexin 41.8 (cx41.8) (Table 3)—a gap junction protein shown to cause the leopard phenotype when mutated. Thus, these phenotypes are likely caused by these mutations, and represent additional dominant obelix/jaguar or leopard alleles, respectively.
Figure 7.
Pigment mutants. Representative photographs of wild type (A) or pigment mutants on the left panels, and a close-up of their flank on the right, highlighting the differences in pigmentation. Homozygous phenotypes are shown for dmh1 (C) and dmh11 (F). (B) dmh1 mutants are characterized by a reduced number of melanophore stripes on the fins and wider stripes on the trunk. (C) Homozygous dmh1 mutants have only a single xanthophore stripe on their trunks. (D) Stripes in dmh7 mutants are disrupted, and show a spot-like pattern. (E) Heterozygous dmh11 mutants show mild disruptions of the melanophore stripes, leading to a wave appearance. (F) In contrast, homozygous dmh11 mutants show a strong reduction in melanophore number, while mostly maintaining a stripe like pattern.
The other pigmentation mutant, dmh11, remains unmapped as linkage has not yet been identified. The phenotype is characterized by mild irregularities in the melanophore stripe, leading to a wavy appearance of the stripes (Figure 7E). Similar to the other pigment mutants, this mutant shows a dose dependent effect as homozygotes exhibit a strikingly enhanced phenotype (Figure 7, C and F).
Discussion
Genetic approaches to understand the developmental regulation of the adult form are limited by the essential function of many genes during early development. To expand forward genetic approaches for investigating late development, we show that dominant screens in zebrafish are efficient in identifying a wide spectrum of mutants that retain viability in adults. To make full use of these mutant collections, it is essential to be able to identify the underlying mutations in a fast and efficient manner.
Establishing high-throughput mapping strategies to support systematic analysis of dominant mutations
The current strategy for identifying mutations in zebrafish by massively parallel sequencing relies on bulked segregant analysis, in which a locus containing the mutation is identified due to retained homogeneity after recombination (Bowen et al. 2012; Leshchiner et al. 2012; Obholzer et al. 2012; Voz et al. 2012). As many dominant mutants do not have a discernable homozygous phenotype or are homozygous lethal, typically, the wild-type locus is mapped to define a linked interval by homogeneity (reverse mapping) (Smith et al. 2016). This can be problematic when working with highly polymorphic, noninbred genomes, like that of the zebrafish, as the wild-type locus will likely be heterogeneous, and thus noninformative for homogeneity mapping. To eliminate the need to perform multiple and specific crosses to reduce the potential pitfalls of polymorphic wild-type loci, we devised a new strategy to facilitate identification of dominant mutations. Our protocol uses identification of phenotype-associated haplotypes rather then homogeneity, to define a linked chromosome in combination with genome wide analysis of potential candidate mutations. This strategy is fast, and permits systematic assessment of large numbers of mutants within phenotypic classes.
Although we use exome sequencing, this strategy can easily be applied to whole genome sequencing data. The higher level of polymorphisms in noncoding regions may bolster the ability to identify mutant specific haplotypes, and could therefore increase the accuracy of the approach. However, getting comparable sequencing depth critical for this analysis will substantially increase the cost per mutant analyzed. Performing additional whole genome sequencing on just the mutant sample may be beneficial in cases where linkage is detected but viable candidate mutation(s) remain elusive. Some phenotype-causing mutations may reside within a noncoding region, or in a gene or exon not included in the exome capture design, therefore not recovered. Since the approach described here does not define a small, linked region on a chromosome, the high level of polymorphisms in noncoding regions will lead to the identification of a large number of potentially phenotype-causing variants with hard to predict functional consequences. Therefore, identification of a phenotype-causing noncoding mutation with this approach would not be straightforward, and would require substantial additional analysis. Other techniques such as RNASeq may prove valuable in these circumstances, as they will identify putative affected loci for further analysis.
Using our new strategy on a subset of 23 mutants from our screen, we find a bias for mutations within specific classes of genes and biological mechanisms, suggesting constraint in the genes that can be altered while retaining viability.
Molecular pathways of postembryonic development
Constraint on gene function:
Screens for mutations affecting postembryonic development will be biased toward genetic changes that are consistent with viability. Thus, genes that are nonessential are more likely to be identified. Similarly, the probability to identify mutations that uncover haploinsufficiency, or cause dominant negative or gain-of-function changes, is higher. Even given the modest sampling of genomes done in this study, we found in certain phenotypes that mutations in particular genes were overrepresented. For example, we identified four distinct mutations of dll4, representing the highest number of alleles within one gene in our screen. In this case, haploinsufficiency of alleles increased the frequency of this gene being identified, as generation of a loss-of-function allele is more common than generation of gain-of-function or dominant negative alleles. But, in most cases, only one or two alleles were identified for different genes, suggesting that the screen has not reached saturation.
When we extend these findings to previous screens, it is clear that several gene classes, and specific genes, are found repeatedly. This is clear for pigmentation mutants, where additional dominant kcnj13/kir7.1/obelix and connexin 41.8/leopard alleles were identified. Similarly, in fin and dermal skeleton development, additional cx43.1/sof and edar/fls alleles were identified. All these genes were identified as dominant mutations both in the initial Tübingen and ZF Model screens. These frequently identified loci probably represent key nodes in which variation in form can arise.
Phenotypic variability and genetic background effects:
Notably, no dominant long-finned mutants were observed in this screen, although these have been previously identified as byproducts of recessive screens (Haffter et al. 1996b; van Eeden et al. 1996). This suggests that, even though we screened a significant number of genomes, we have not fully sampled potential unique mutations in genes that can affect late development. The lack of long-finned mutants in our screen could also be attributed to genetic background effects. For many of the phenotypes, we observed significant variability in phenotypic strength when mutants were crossed to different wild-type strains. This suggests that some genetic backgrounds are able to buffer effects of genetic mutations.
Systematic screening and identifying genetic networks:
With use of targeted capture and next-generation sequencing technologies, it is now feasible to identify causative mutations in large sets of mutants. If substantial numbers of genomes are screened, it is now possible in the zebrafish to systematically approach identification of genetic networks similar to screens routinely performed in Drosophila, Caenorhabditis elegans or yeast. We identified 21 stature mutants in our screen. Of the 10 mutants we analyzed, six had mutations in fibril-forming collagens. Fibril-forming collagens have large helical domains with conserved glycine residues at every third position. Mutations of any of these residues will lead to structural changes of the protein that act in a dominant fashion, and therefore represent a large number of targets for the creation of phenotype-causing mutations. When these mutants are combined with several mutants previously identified through their recessive phenotype such as bmp1a (Asharani et al. 2012), hsp47/serpinh1b (Bhadra and Iovine 2015), and plod2 (Gistelinck et al. 2016)—a component of known genes of the collagen network—including genes regulating collagen protein function and processing, are identified. Genetic analysis of similar phenotypic classes of mutants should allow further refinement of this network.
Constraint on identified alleles and identification of disease mechanisms:
Mutation recovery in adult screens in the zebrafish could be reflective of a broader evolutionary restriction in which genes can be altered and still be consistent with viability. Thus, a prediction of performing a screen on postembryonic development is that the results will be biased toward changes that also would be found underlying disease states in humans.
We recovered mutants that have similar or analogous effects in human orthologs that are known to be associated with disease. Sixteen out of the 22 mutants with potential phenotype-causing mutations identified from the 25 mutants analyzed harbor mutations in known disease genes (Table 4). This supports the utility of forward genetic screens in zebrafish to predict the genetic cause of human diseases in cases where the underlying gene mutations have not been deduced. Further, this increases the potential for genetic screening in zebrafish to uncover modifiers of gene function that can serve as a basis for discovery in medicine. The systematic mapping of dominant mutations as described here will greatly facilitate this approach.
Table 4. Human disease genes.
| Gene | Human Disease |
|---|---|
| col1a1 | Caffey disease |
| Ehlers-Danlos syndrome, classic and type VIIA | |
| Osteogenesis imperfecta, type I, type II, type III, type IV | |
| col1a2 | Ehlers-Danlos syndrome, cardiac valvular form and type VIIB |
| Osteogenesis imperfecta, type II, type III, type IV | |
| col2a1 | Achondrogenesis, type II or hypochondrogenesis |
| Avascular necrosis of the femoral head | |
| Czech dysplasia | |
| Epiphyseal dysplasia, multiple, with myopia and deafness | |
| Kniest dysplasia | |
| Legg-Calve-Perthes disease | |
| Osteoarthritis with mild chondrodysplasia | |
| Otospondylomegaepiphyseal dysplasia | |
| Platyspondylic skeletal dysplasia, Torrance type | |
| SED congenita | |
| SMED Strudwick type | |
| Spondyloepiphyseal dysplasia, Stanescu type | |
| Spondyloperipheral dysplasia | |
| Stickler syndrome, type I, nonsyndromic ocular and type I | |
| Vitreoretinopathy with phalangeal epiphyseal dysplasia | |
| cx43 | Atrioventricular septal defect 3 |
| Craniometaphyseal dysplasia, autosomal recessive | |
| Erythrokeratodermia variabilis et progressiva | |
| Hypoplastic left heart syndrome 1 | |
| Oculodentodigital dysplasia | |
| Oculodentodigital dysplasia, autosomal recessive | |
| Palmoplantar keratoderma with congenital alopecia | |
| Syndactyly, type III | |
| dll4 | Adams-Oliver syndrome 6 |
| edar | Ectodermal dysplasia 10A, hypohidrotic/hair/nail type, autosomal dominant |
| Ectodermal dysplasia 10B, hypohidrotic/hair/tooth type, autosomal recessive | |
| kcnj13/kir7.1 | Leber congenital amaurosis 16 |
| Snowflake vitreoretinal degeneration | |
| cx41.8/GJA5 | Atrial fibrillation, familial, 11 |
| Atrial standstill, digenic (GJA5/SCN5A) |
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300187/-/DC1.
Acknowledgments
We would like to thank Althea James, Jason Best, and Christian Lawrence at the Boston Children’s Hospital aquatics program for help with zebrafish husbandry. The authors would also like to thank Kristin Radcliffe for help with screening fish and maintaining mutant lines. This project was funded in part through National Institute of Dental and Craniofacial Research (NIDCR) U01DE024434 grant to M.P.H., and generous support from the Children’s Orthopedic Surgery Foundation.
Footnotes
Communicating editor: D. Parichy
Literature Cited
- Andreeva V., Connolly M. H., Stewart-Swift C., Fraher D., Burt J., et al. , 2011. Identification of adult mineralized tissue zebrafish mutants. Genesis 49: 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asharani P. V., Keupp K., Semler O., Wang W., Li Y., et al. , 2012. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am. J. Hum. Genet. 90: 661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M. P., Goetz F. W., 2001. Isolation of gonadal mutations in adult zebrafish from a chemical mutagenesis screen. Biol. Reprod. 64: 548–554. [DOI] [PubMed] [Google Scholar]
- Bhadra J., Iovine M. K., 2015. Hsp47 mediates Cx43-dependent skeletal growth and patterning in the regenerating fin. Mech. Dev. 138: 364–374. [DOI] [PubMed] [Google Scholar]
- Bowen M. E., Henke K., Siegfried K. R., Warman M. L., Harris M. P., 2012. Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics 190: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley K. M., Elmore J. B., Breyer J. P., Yaspan B. L., Jessen J. R., et al. , 2007. A major zebrafish polymorphism resource for genetic mapping. Genome Biol. 8: R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdà J., Gründ C., Franke W. W., Brand M., 2002. Molecular characterization of calymmin, a novel notochord sheath-associated extracellular matrix protein in the zebrafish embryo. Dev. Dyn. 224: 200–209. [DOI] [PubMed] [Google Scholar]
- Coe T. S., Hamilton P. B., Griffiths A. M., Hodgson D. J., Wahab M. A., et al. , 2009. Genetic variation in strains of zebrafish (Danio rerio) and the implications for ecotoxicology studies. Ecotoxicology 18: 144–150. [DOI] [PubMed] [Google Scholar]
- Driever W., Solnica-Krezel L., Schier A. F., Neuhauss S. C., Malicki J., et al. , 1996. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123: 37–46. [DOI] [PubMed] [Google Scholar]
- Fisher S., Jagadeeswaran P., Halpern M. E., 2003. Radiographic analysis of zebrafish skeletal defects. Dev. Biol. 264: 64–76. [DOI] [PubMed] [Google Scholar]
- Gajko-Galicka A., 2002. Mutations in type I collagen genes resulting in osteogenesis imperfecta in humans. Acta Biochim. Pol. 49: 433–441. [PubMed] [Google Scholar]
- Gistelinck C., Witten P. E., Huysseune A., Symoens S., Malfait F., et al. , 2016. Loss of type I collagen telopeptide lysyl hydroxylation causes musculoskeletal abnormalities in a zebrafish model of Bruck syndrome. J. Bone Miner. Res. 31: 1930–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V., Koudijs M. J., Berezikov E., Johnson S. L., Plasterk R. H. A., et al. , 2006. Genetic variation in the zebrafish. Genome Res. 16: 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P., Granato M., Brand M., Mullins M. C., Hammerschmidt M., et al. , 1996a The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123: 1–36. [DOI] [PubMed] [Google Scholar]
- Haffter P., Odenthal J., Mullins M. C., Lin S., Farrell M. J., et al. , 1996b Mutations affecting pigmentation and shape of the adult zebrafish. Dev. Genes Evol. 206: 260–276. [DOI] [PubMed] [Google Scholar]
- Harris M. P., Rohner N., Schwarz H., Perathoner S., Konstantinidis P., et al. , 2008. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. 4: e1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke K., Bowen M. E., Harris M. P., 2013. Perspectives for identification of mutations in the zebrafish: making use of next-generation sequencing technologies for forward genetic approaches. Methods 62: 185–196. [DOI] [PubMed] [Google Scholar]
- Iovine M. K., Higgins E. P., Hindes A., Coblitz B., Johnson S. L., 2005. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev. Biol. 278: 208–219. [DOI] [PubMed] [Google Scholar]
- Iwashita M., Watanabe M., Ishii M., Chen T., Johnson S. L., et al. , 2006. Pigment pattern in jaguar/obelix zebrafish is caused by a Kir7.1 mutation: implications for the regulation of melanosome movement. PLoS Genet. 2: e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettleborough R. N. W., Busch-Nentwich E. M., Harvey S. A., Dooley C. M., de Bruijn E., et al. , 2013. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496: 494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchiner I., Alexa K., Kelsey P., Adzhubei I., Austin-Tse C. A., et al. , 2012. Mutation mapping and identification by whole-genome sequencing. Genome Res. 22: 1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie J. D., Ariza-McNaughton L., Bermange A. L., McAdow R., Johnson S. L., et al. , 2007. Endothelial signalling by the notch ligand delta-like 4 restricts angiogenesis. Development 134: 839–844. [DOI] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U., Torres Ruiz R. A., Berleth T., Miseéra S., Juürgens G., 1991. Mutations affecting body organization in the Arabidopsis embryo. Nature 353: 402–407. [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S. E., Riat H. S., Ritchie G. R. S., et al. , 2016. The ensembl variant effect predictor. Genome Biol. 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord H., Burguiere A.-C., Muck J., Nord C., Ahlgren U., et al. , 2014. Differential regulation of myosin heavy chains defines new muscle domains in zebrafish. Mol. Biol. Cell 25: 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Dahm R., 2002. Zebrafish: A Practical Approach, Ed. 1 Oxford University Press, New York. [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. [DOI] [PubMed] [Google Scholar]
- Obholzer N., Swinburne I. A., Schwab E., Nechiporuk A. V., Nicolson T., et al. , 2012. Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development 139: 4280–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M.-Y., Wen H.-J., Shih L.-J., Kuo C.-M., Hwang S.-P. L., 2002. Myosin heavy chain expression in cranial, pectoral fin, and tail muscle regions of zebrafish embryos. Mol. Reprod. Dev. 63: 422–429. [DOI] [PubMed] [Google Scholar]
- Rohner N., Bercsényi M., Orbán L., Kolanczyk M. E., Linke D., et al. , 2009. Duplication of Fgfr1 permits Fgf signaling to serve as a target for selection during domestication. Curr. Biol. 19: 1642–1647. [DOI] [PubMed] [Google Scholar]
- Rohner N., Perathoner S., Frohnhöfer H. G., Harris M. P., 2011. Enhancing the efficiency of N-ethyl-N-nitrosourea-induced mutagenesis in the zebrafish. Zebrafish 8: 119–123. [DOI] [PubMed] [Google Scholar]
- Ryan S., Willer J., Marjoram L., Bagwell J., Mankiewicz J., et al. , 2013. Rapid identification of kidney cyst mutations by whole exome sequencing in zebrafish. Development 140: 4445–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Siegfried K. R., Nüsslein-Volhard C., Sakai N., 2011. Isolation and cytogenetic characterization of zebrafish meiotic prophase I mutants. Dev. Dyn. 240: 1779–1792. [DOI] [PubMed] [Google Scholar]
- Smith H. E., Fabritius A. S., Jaramillo-Lambert A., Golden A., 2016. Mapping challenging mutations by whole-genome sequencing. G3 (Bethesda) 6: 1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickney H. L., Schmutz J., Woods I. G., Holtzer C. C., Dickson M. C., et al. , 2002. Rapid mapping of zebrafish mutations with SNPs and oligonucleotide microarrays. Genome Res. 12: 1929–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden F. J., Granato M., Schach U., Brand M., Furutani-Seiki M., et al. , 1996. Genetic analysis of fin formation in the zebrafish, Danio rerio. Development 123: 255–262. [DOI] [PubMed] [Google Scholar]
- Voz M. L., Coppieters W., Manfroid I., Baudhuin A., Von Berg V., et al. , 2012. Fast homozygosity mapping and identification of a zebrafish ENU-induced mutation by whole-genome sequencing. PLoS One 7: e34671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Iwashita M., Ishii M., Kurachi Y., Kawakami A., et al. , 2006. Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep. 7: 893–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe D., Dudek S., Ritchie M. D., Pendergrass S. A., 2013. Visualizing genomic information across chromosomes with phenoGram. BioData Min. 6: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sperm from the 25 mutant lines described in this paper is preserved to secure the lines. Mutant lines are available on request from the author. Supplemental Material, File S1 contains total and mutant specific SNP counts for each chromosome for the 23 mutants mapped with the method described in this manuscript. File S2 contains numbers of unique nonsynonymous mutations per chromosome for the 23 mutants. A list of primers used for confirmation of linkage is provided in Table S1 in File S4. Raw sequence data were submitted to the European Nucleotide Archive (ENA) under study PRJEB13615 (http://www.ebi.ac.uk/ena/data/view/PRJEB13615). Accession numbers for individual sequencing files can be found in File S3. Custom scripts for data analysis are available for download from the laboratory website (www.fishyskeleton.com) or upon request.