Abstract
Y chromosome function, structure and evolution is poorly understood in many species, including the Anopheles genus of mosquitoes—an emerging model system for studying speciation that also represents the major vectors of malaria. While the Anopheline Y had previously been implicated in male mating behavior, recent data from the Anopheles gambiae complex suggests that, apart from the putative primary sex-determiner, no other genes are conserved on the Y. Studying the functional basis of the evolutionary divergence of the Y chromosome in the gambiae complex is complicated by complete F1 male hybrid sterility. Here, we used an F1 × F0 crossing scheme to overcome a severe bottleneck of male hybrid incompatibilities that enabled us to experimentally purify a genetically labeled A. gambiae Y chromosome in an A. arabiensis background. Whole genome sequencing (WGS) confirmed that the A. gambiae Y retained its original sequence content in the A. arabiensis genomic background. In contrast to comparable experiments in Drosophila, we find that the presence of a heterospecific Y chromosome has no significant effect on the expression of A. arabiensis genes, and transcriptional differences can be explained almost exclusively as a direct consequence of transcripts arising from sequence elements present on the A. gambiae Y chromosome itself. We find that Y hybrids show no obvious fertility defects, and no substantial reduction in male competitiveness. Our results demonstrate that, despite their radically different structure, Y chromosomes of these two species of the gambiae complex that diverged an estimated 1.85 MYA function interchangeably, thus indicating that the Y chromosome does not harbor loci contributing to hybrid incompatibility. Therefore, Y chromosome gene flow between members of the gambiae complex is possible even at their current level of divergence. Importantly, this also suggests that malaria control interventions based on sex-distorting Y drive would be transferable, whether intentionally or contingent, between the major malaria vector species.
Keywords: vector genetics, hybrid incompatibility, gene flow, Y chromosome, malaria
SEX chromosomes often play an important role in speciation, though the molecular factors that influence this process remains an area of active investigation (Ellegren 2011). Y chromosome sequence content in heterogametic animals is transmitted in a clonal manner due to the lack of crossing over with the X across some or all of its length. This absence of recombination promotes a progressive genetic degeneration including the accumulation and rapid turnover of repetitive sequences (Charlesworth 1991; Rice 1996; Charlesworth and Charlesworth 2000; Bachtrog 2013). It is generally thought that remaining Y-linked genes represent the remnants of an inexorable process of inactivation, degradation, and gene loss, and only genes with a selectable function, such as the male-determining factor, are likely to survive on the Y chromosome. However, while this view appears to hold true for mammals, the birth of new genes on the Y has been described in Drosophila, and may even dominate the evolution of its Y chromosome (Vibranovski et al. 2008; Carvalho et al. 2015). This suggests an evolutionary dynamic that is particular to the Y of different phylogenetic groups.
Sex chromosomes play an important role in reproductive isolation; however, it is mostly the gene-rich X chromosome that has been implicated with three interrelated patterns of hybrid incompatibility: Haldane’s rule, the large X-effect, and the asymmetry of hybrid viability and fertility in reciprocal crosses (Wu et al. 1996; Masly and Presgraves 2007; Turelli and Moyle 2007). In a few cases, a link between the Y and hybrid incompatibilities has been demonstrated. For example, it has been suggested that the Y chromosome contributes to reproductive barriers between rabbit subspecies (Geraldes et al. 2008). This can be explained by interactions involved in determining male dimorphism or fertility, between genes that diverged from allele pairs on the Y and X, and that break down in the heterospecific context. Such X–Y chromosome incompatibilities have been demonstrated to contribute to hybrid male sterility in house mice (Campbell and Nachman 2014). Additionally, the introgression of heterospecific Y chromosomes in Drosophila was found to affect male fertility, and alters the expression of 2–3% of all genes in hybrids (Sackton et al. 2011).
The varied picture of the biological role of Y emerging form work in mammals and Drosophila suggests the need for additional studies using other model systems. The Anopheles genus, which contains all human malaria-transmitting mosquito species, has received much attention in recent years, not only due to its stark medical importance but also as a model system for studying speciation and chromosome evolution. In particular the Anopheles gambiae species complex of eight sibling species, including the most widespread and potent vectors of malaria in sub-Saharan Africa, offers an excellent platform to further our understanding of the biology of the Y and its possible role in reproductive isolation.
To this end, we decided to focus on the two species with prime medical importance, A. gambiae and A. arabiensis, as they are the most anthropophilic members of the complex with the widest distributions. A. gambiae predominates in zones of forest and humid savannah, whereas A. arabiensis prevails in arid savannahs and steppes, including those of the South-Western part of the Arabian Peninsula. In the sympatric areas, changes in seasonal prevalence are observed showing an increase in the relative frequency of A. arabiensis during the dry season. In areas where the distribution of A. gambiae and A. arabiensis overlaps, hybrids are detected at extremely low frequency (0.02–0.76%) (Temu et al. 1997; Toure et al. 1998; Mawejje et al. 2013). However, a recent study conducted in Eastern Uganda to investigate hybridization between these species showed that 5% of the samples analyzed were hybrid generations beyond F1 (Weetman et al. 2014). F1 male sterility and other postzygotic isolation mechanisms have been studied in A. gambiae and A. arabiensis hybrids. In addition to mapping multiple loci contributing to male sterility, Slotman et al. (2004) demonstrated, by observing the absence of particular genotypes in backcross experiments, that inviability is caused by recessive factors on the X chromosome of A. gambiae incompatible with at least one factor on each autosome in A. arabiensis.
Y chromosomes of A. gambiae and A. arabiensis have recently been shown to differ dramatically (Hall et al. 2016). This study, which, to date, remains the only in depth analysis of Y chromosome content in A. gambiae and its sibling species, revealed that the Y chromosomes of A. gambiae and A. arabiensis differ dramatically both in the content and abundance of sequences on the Y chromosome. Despite using long sequence reads, a high quality assembly of the estimated ∼26 Mbp Y chromosome (∼10% of the genome) was not achieved, but the genic and repetitive content were sufficiently characterized to paint a picture of Y chromosome evolution and provide key data for the present study. Of the five genes that were confirmed on the Y chromosome of A. gambiae, only one, YG2, recently shown to act as a male-determining factor (Krzywinska et al. 2016), was found to be present on the Y chromosome of all species of the gambiae complex. YG1, whose function remains unknown, but which physically flanks YG2 and is homologous to it, is also shared between the Y chromosome of A. gambiae and A. arabiensis. Surprisingly, the most abundant A. gambiae Y chromosome sequences that represent 92% of its total sequence content, namely the satellite DNAs AgY477 and AgY373 and the Zanzibar transposon, which appears to have expanded through satellite-like processes, are not abundant on the Y chromosome of A. arabiensis, or are completely absent, further demonstrating the rapid turnover and expansions of sequences on these Y chromosomes. Overall previous studies suggest the rapid evolution of Y chromosome in this highly dynamic genus of malaria vectors.
In the present study, we wanted to establish whether the introgression of the A. gambiae Y chromosome into an A. arabiensis genetic background is possible when selected for in a controlled laboratory setting, whether the Y contributes to reproductive isolation, and whether a heterospecific Y, as has been reported in Drosophila, would markedly modulate gene expression patterns, fertility, or behavior of Y hybrid males. In addition to basic biological insights, our attempt to better understand the biology of the mosquito Y is key for both the development of male-specific traits for genetic control as well as predicting the behavior of such traits in the field.
Materials and Methods
Mosquito strains and rearing
Wild-type A. gambiae and A. arabiensis mosquitoes of strains G3 and Dongola, respectively, were used. Strain G3 was originally isolated from West Africa (MacCarthy Island, The Gambia) in 1975, and obtained from the MR4 (MRA-112). It is considered a hybrid stock with mixed features derived from both A. gambiae s.s. and Anopheles coluzzii. The Dongola strain of A. arabiensis was obtained from the MR4 (MRA-856), and was originally isolated from Sudan. For the introgression experiments, we used two independently generated Y-linked A. gambiae transgenic strains GY1 [referred to as YattP in Bernardini et al. (2014)] carrying a Pax-RFP marker and GY2 (R.G., unpublished data). Strain GY2 contains a Y linked insertion of construct pBac[3xP3-DsRed]β2-eGFP::I-PpoI-124L (Galizi et al. 2014) also carrying a Pax-RFP marker (no expression from the inactive β2-eGFP::I-PpoI-124L locus is detectable in this strain). Inverse PCR suggests position 17757 on the Y_unplaced collection as a likely insertion site of this construct; however, no assembly of the repetitive A. gambiae Y chromosome exists. Strain AY2 contains the A. gambiae Y chromosome from strain GY2 within an A. arabiensis background as described in the Results section. All mosquitoes were reared under standard condition at 28° and 80% relative humidity, with access to fish food as larvae and 5% (weight/volume) glucose solution as adults. For egg production, young adult mosquitoes (2–4 days after emergence) were allowed to mate for at least 6 days, and then fed on mice. Two days later, an egg bowl containing rearing water (dH2O supplemented with 0.1% pure salt) was placed in the cage. One or 2 days after hatching, larvae were placed into trays containing rearing water. The protocols and procedures used in this study were approved by the Animal Ethics Committee of Imperial College in compliance with United Kingdom Home Office regulations.
Genetic crosses and fertility assays
Crosses were set up in BugDorm-1 cages with size 30 × 30 × 30 cm. Generally 100 female and 100 male mosquitoes were crossed during the introgression experiment, although the number of males varied after the F3 bottleneck and was dependent on the number of male progeny that could be recovered from the previous generation. To assay fertility of the Y-introgressed males, after 11 generation of backcrossing in cage, single crosses in cups were set up. Y-Introgressed males were singularly introduced into a cup together with one A. arabiensis female. In parallel, the same number of cups was set up for A. arabiensis males and females as a control. After 6 days of mating and blood feeding of females, eggs were collected from every cup, and the hatching rate (number of larvae/number of eggs) relative to every cross was calculated.
DNA sequencing
Samples for DNA sequencing were 10 adult male AY2, and two wild-type control A. arabiensis males were used individually for WGS. AY2 males had been mated to A. arabiensis females prior to DNA extraction, allowing us to establish fertility for seven of the 10 AY2 males. The DNA libraries were prepared in accordance with the Illumina Nextera DNA guide for Illumina Paired-End Indexed Sequencing. AMPure XP beads were used to purify the library DNA and for size selection, after which the resulting libraries were validated using the Agilent 2100 bioanalyzer and quantified using a Qubit 2.0 Fluorometer. Sequencing runs were performed on six lanes (two samples per lane) of an Illumina flowcell (v3) on the HiSeq1500 Illumina platform, using a 2 × 100 bp PE HiSeq Reagent Kit according to the manufacturer’s recommendations. Raw reads were processed using FastQC (Andrews 2010, available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and trimmomatic (Bolger et al. 2014).
Variant calling and read coverage analysis
Reads were aligned to the A. gambiae PEST reference genome assembly (AgamP4) using BWA mem (Li 2013, bio-bwa v0.7.5a) and sorted using Samtools (v1.2). We used the MarkDuplicates module from Picard tools (v1.9) to remove PCR duplicates and the genome analysis tool kit (GATK, v3.3) to realign reads around indels (McKenna et al. 2010). First we used the GATK modules HaplotypeCaller and UnifiedGenotyper to call raw SNPs and merged them across the 10 AY2 and two A male samples using GenotypeGVCFs. Biallelic SNPs were selected using GATK VariantFiltration and SelectVariants modules following the GATK best practices guideline. For the coverage analysis we used bowtie (v1.1.1), exclusively reporting alignments for reads having only a single reportable alignment and displaying no mismatches. We used the bedtools (v2.25) makewindows tool to generate a sliding window bed file with 5 kb windows overlapping by 2.5 kb. We then used Samtools bedcov to generate per-window read counts, and calculate the group means for the seven fertile AY2 and two A male control samples.
Analysis of Y signature elements
Paired WGS reads were mapped using Bowtie2 (Langmead and Salzberg 2012) with standard parameters (bowtie2 -x [consensus_build] -a -1 [ x_R1] -2 [x_R2] -S [x.sam]) against a collection of consensus sequences of all known Y chromosome loci of A. gambiae (Hall et al. 2016). Read counts at every locus were generated with Samtools (Li et al. 2009), and normalized by library size and locus length in fragments per kilobase of transcript per million mapped reads (FPKM). WGS data in the form of paired Illumina reads corresponding to the control samples from A. gambiae males (NCBI SRA: SRR534285) and females (NCBI SRA:SRR534286) and A. arabiensis females (NCBI SRA: SRR1504792) were taken from the Hall et al. (2016) study. We also performed a separate analysis to evaluate Y chromosome satellite DNA abundance in the introgressed male samples, in part because satellites Ag53A, B and D, due to their short length, could not be appropriately assessed using Bowtie2. We used jellyfish (Marçais and Kingsford 2011) to generate unique 25mers (kmers of 25 bp long) from each of the six known Y satellites consensus sequences (jellyfish count -C -m 25 [stDNA-locus-x.fasta] -o [output] -c 5 -s 1000000000 -t [cores]). Using the same approach, we then generated and counted unique 25mers from each of the aforementioned WGS samples (jellyfish count -C -L 5 -m 25 [x.fastq] -o [output] -c 5 -s 1000000000 -t [cores]), and assessed the abundance of each of the stDNA specific kmers within the sample-specific kmers list, resulting in a table providing abundance of each kmer in each sample (grouped by stDNA locus). Raw kmer counts were normalized by library size (sequencing depth), and, for the WGS generated in this study (control and introgressed males), we calculated the median abundance for each kmer in these two groups.
RNA and small RNA sequencing experiments
Samples for RNA sequencing were generated using the following experimental design. Four cages were set up with 40 A. arabiensis wild-type males and 40 wild-type females, and four cages with 40 AY2 males and their nontransgenic sibling females. After mating and blood feeding, progeny was collected from the cages. 80 freshly hatched larvae were collected from each of both sets of cages and combined in a single tray for rearing. At the pupal stage, males were sexed and screened for the fluorescent marker linked to the Y chromosome. From every tray, 18 RFP positive and 18 RFP negative males were collected and placed in separate cages to allow emergence. Three days after emergence males were dissected in order to separate three different tissues the head, the abdominal segments harboring the reproductive tissues, and the remainder of the carcass. This experiment was performed twice, resulting in a total number of eight replicates for both controls and experimental samples. Libraries for total RNA sequencing were prepared using the TruSeq RNA sample preparation kit by Illumina, and sequenced on three lanes of an Illumina HiSequation 2500 using 2 × 100 paired end reads. The samples described above were also used for the construction of libraries for small RNA sequencing. Libraries were prepared using the NEBNext Multiplex Small RNA kit for Illumina and sequenced on three lanes of a MiSeq using the 1 × 42 single-read mode.
Differential expression analysis
Reads were aligned to the A. arabiensis genome supplemented with the 25 contigs corresponding to known A. gambiae Y loci (Hall et al. 2016) using HISAT2 (Pertea et al. 2016). We then used Stringtie (Pertea et al. 2016) in conjunction with the Anopheles arabiensis reference transcriptome version AaraD1.3 to predict novel genes and novel isoforms of known genes, which were merged across samples into a combined geneset using Cuffmerge (Ghosh and Chan 2016). Expression of all transcripts was then quantified using Stringtie -B. We used the Ballgown (Pertea et al. 2016) suite to determine transcripts showing significantly different expression levels using a cutoff of P-value <0.05 adjusted for multiple testing, a mean expression >1 FPKM across the samples in the tissue analyzed and a log2 fold-change >1 between the introgressed and control groups. To assign a repetitiveness score, the sequence of each transcript was blasted against the A. gambiae PEST RepeatMasker library provided by Vectorbase.org. For the small RNA dataset. we generated a count matrix using Seqbuster and SeqCluster suites (Pantano et al. 2011). and used DESeq2 (Love et al. 2014) for differential expression analysis and log2 transformation of the count data. Only putative small RNA loci with a mean expression of >5 counts across samples were taken into account for this analysis.
Competition experiments
Matings were performed in BugDorm-1 cages. Females and competing males were allocated in cage with a 1:1:1 ratio. Every experiment was run in triplicate. Crosses were set up as follows: 50 A females × 50 A males + 50 GY2 males, 50 G females × 50 G males + 50 AY2 males, 50 A females × 50 A males + 50 AY2 males, 50 G females × 50 A males + 50 AY2 males. After 6 days mating and blood feeding, females were collected from each experimental replicate and allowed to lay singularly in cups. The number of eggs and hatched larvae was calculated for every cup in order to estimate the hatching rate values. Progeny was screened for 3×P3 RFP to assess paternity, and transgene ratio was calculated in order to identify any occurring secondary mating.
Data availability
All sequence data has been submitted to the National Center for Biotechnology Information (NCBI) short read archive, and are available under BioProject IDs PRJNA381402, PRJNA381403 and PRJNA381033. A detailed list of per-sample accession numbers is given in Supplemental Material, File S1.
Results
Experimental introgression of the A. gambiae Y chromosome into A. arabiensis
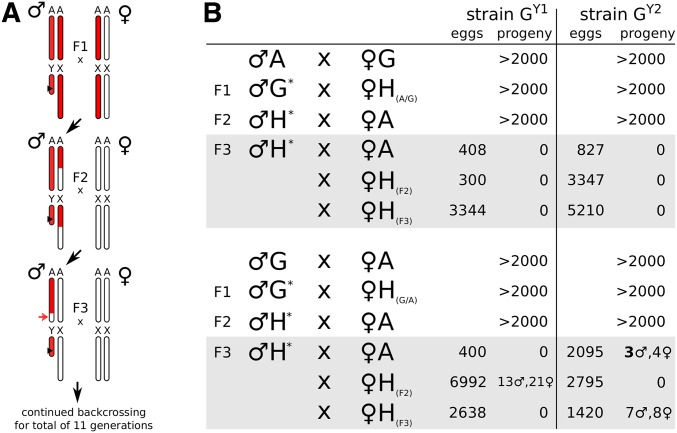
We have previously established a number of transgenic strains in which different fluorescent transgenes were inserted onto the A. gambiae Y chromosome (Bernardini et al. 2014; R.G., unpublished data). Limited recombination between the A. gambiae sex chromosomes has been suggested to occur in specific genetic backgrounds (Wilkins et al. 2007), but is generally believed to occur at very low frequencies (Mitchell and Seawright 1989; Bernardini et al. 2014; Hall et al. 2016). We concluded that, for our purpose, a Y-linked fluorescent transgene would be a reliable tool to track the Y chromosome throughout a multi-generational introgression experiment. It has previously been shown that F1 male hybrids between A. gambiae and A. arabiensis suffer from complete male sterility, which precludes Y chromosome introgression experiments in a straightforward manner. Our pilot experiments confirmed this finding (Table S1A). We employed an F1 × F0 crossing strategy (we define this as a scheme where first generation female hybrids are crossed to pure species males) in which transgenic Y gambiae males were backcrossed to F1 hybrid females, which were in turn generated using either wild-type gambiae or arabiensis mothers (Figure 1A). We used two different transgenic Y strains, herein referred to as GY1 and GY2, that both express an RFP reporter gene driven by the neuronal 3×P3 promoter from different insertion sites on the Y chromosome. In the F2 cross, the hybrid males containing the labeled Y are crossed again to wild-type arabiensis females, and then third generation hybrid males are crossed to both hybrid females and wild-type arabiensis females in the attempt to recover offspring. Using this crossing scheme, we encountered a severe bottleneck at generation F3 when males are predicted to have inherited a predominantly A. gambiae autosomal genome from their fathers in conjunction with pure A. arabiensis genome (including the X chromosome) from their mothers (Figure 1A). We recovered no larvae from >13,000 eggs when backcrossing of either GY1 or GY2 males was carried on from F1 hybrid females originated from A. gambiae mothers. In the reverse cross, using F1 hybrid females from A. arabiensis mothers, we managed to recover seven larvae (three males and four females) with strain GY2 in a direct cross to wild-type A. arabiensis females (Figure 1B). These three hybrid males obtained were used to progress the introgression, and we continuously maintained backcross purification by crossing hybrid males recovered each generation to wild-type arabiensis females for a total of 11 generations to establish the introgressed strains AY2 used for all subsequent experiments in this study. The occurrence of fertile phenotypes in these crosses is a rare event. In additional experiments where either GY1 or GY2 males were crossed to F2 or F3 hybrid females (Figure 1B) the majority of the resulting male progeny (15 larvae, of which seven were male, and 34 larvae, of which 13 were male) failed to develop into fertile adults.
Figure 1.
F0 × F1 hybrid crossing scheme and resulting progeny. (A) Crossing scheme indicating the A. gambiae (red) or A. arabiensis (white) genomic contributions in generations F1–F3, with autosomes represented as a single pair labeled A. In the F1 cross, A. gambiae males of strains GY1 or GY2 are crossed to hybrid F1 females. Each generation, the resulting males are backcrossed to A. arabiensis wild-type females. In the bottleneck generation F3 hybrid males harbor an A. arabiensis X with a fraction (∼25% on average) of autosomal regions expected to be homozygous for the arabiensis background (red arrow). (B) Observed number of eggs and progeny arising for each indicated cross where the F1 hybrid females had either an A. gambiae mother (top) or an A. arabiensis mother (bottom). The asterisk indicates strain GY1 or GY2, respectively. In the bottleneck generation (F3), males were crossed to either pure species arabiensis females (♀A), or crossed to female hybrids with decreasing levels of A. gambiae genome content (♀H(F2), ♀H(F3)).
Fertility of males carrying a heterospecific Y chromosome
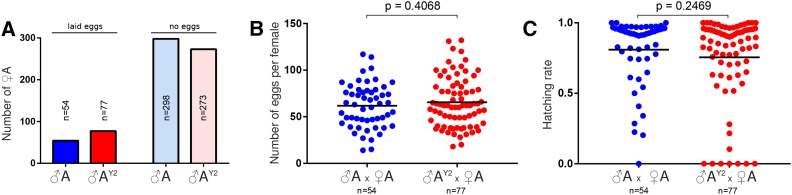
We first asked whether AY2 males showed reduced levels of fertility when compared to wild-type A. arabiensis males due to the presence of the heterospecific Y chromosome. In order to assay individual males, rather than a population average, we performed single-copula mating experiments of strain AY2 or wild-type males mated to A. arabiensis females, measuring the mating rates, and oviposition and egg hatching rates of single females. Every generation, the males of the largest family were used for establishing the next round of single-copula backcrosses, and, over the course of seven generations, a total of 344 wild-type A. arabiensis and 350 AY2 males were assayed. The rationale for this design was to exclude the possibility of A. gambiae fertility loci having been retained by selection in introgressed males, because such loci would be expected to segregate in this design. Figure 2 shows a summary of these experiments. Single copula matings are inefficient, in fact <25% of females would mate and oviposit under these condition (Figure 2A). No significant difference in the rate of mating was observed between AY2 (22%) and control A. arabiensis males (15.3%). Power analysis suggested that the sample size of the successfully mated males would allow for the reliable detection of an effect of medium size (P = 0.79 for a two-sided t-test, P-value = 0.05, d = 0.5). We observed that females mated to AY2 males and females mated to A. arabiensis wild-type males laid a comparable numbers of eggs (Figure 2B) that had comparable hatching rates (Figure 2C). This analysis indicates that, under laboratory conditions, and, in the absence of mate choice and male–male competition, and taking into account the above considerations on power, AY2 males show no significant difference in fertility when compared to wild-type A. arabiensis males that retain their native Y chromosome. As an additional control, we back-crossed AY2 males to A. gambiae females. Despite the presence of the A. gambiae Y chromosome in these males, we expected this experiment to recreate hybrid incompatibility in the form of male infertility in the resulting progeny. Indeed, we found hybrid males to be fully sterile (Table S1B), thus confirming that X-A incompatibilities are sufficient to explain this phenotype (Slotman et al. 2004).
Figure 2.
Single copula mating experiments. (A) Number of females ovipositing following single matings with A or AY2 males. Counts of eggs (B) and hatching larvae (C) for each individual family.
Genomic analysis of males with a heterospecific Y chromosome
After n = 11 generations of backcrossing, assuming no selection for sections of the A. gambiae genome, the expected autosomal genome proportion of the A. gambiae donor would be 1/2n or <0.05%. Given that gambiae genomic regions contributing to hybrid incompatibilities would be selected against in males, this is likely an underestimation due to such detrimental haplotypes being selectively removed. To confirm that our backcrossing scheme had eliminated the A. gambiae autosomal genome but retained the A. gambiae Y chromosome, we performed DNA WGS from 10 AY2 males and two wild-type control males of our A. arabiensis laboratory colony. To determine whether introgressed males were fertile, they were singularly mated to wild-type arabiensis females before their genomic DNA was extracted. WGS reads were mapped to the A. gambiae genome to identify genomic regions with fixed allele differences between introgressed and control groups by confining our analysis to biallelic SNPs. No assembly of the A. gambiae Y exists; however, the PEST assembly includes the Y_unplaced sequence collection that includes ∼230 kb of unscaffolded contigs that have been assigned to Y. Our analysis (Table 1) showed that the autosomes of introgressed-Y and pure species contained a small number of differentially represented SNPs comparable in number to the X chromosome, which, since it is replaced in every backcross generation, serves as a background control. In contrast, the majority of differentially represented SNPs (74.7% of the total number of differential fixed SNP and 35.6% of the total number of SNPs on the Y) arose from reads mapping to the Y_unplaced portion of the A. gambiae genome that represents <0.1% of the total genome assembly. Within the Y_unplaced collection, we found that most SNPs mapped to the largest contig, which also contains the male-determine gene (Figure S1). In addition 12.7% of the total number of SNPs arose from the UNKN collection (unassigned contigs) that is also expected to contain a number of unassigned Y sequences and repetitive elements. We performed an additional sliding-window analysis, where we considered only reads mapping uniquely to the A. gambiae genome and allowed for no mismatches. The rationale was that, given the observed levels of divergence between these genomes, perfectly matching reads are expected to predominantly map to the genome of origin. When comparing the mean number of reads of AY2 fertile introgressed males and the samples of the A. arabiensis control group, we find that the Y_unplaced collection experiences significant coverage only in AY2 males, as do parts of the UNKN collection. For the autosomes and the X, few windows accrue a significant number of reads, and we find no substantial differences between the groups in the direction of AY2, with the possible exception of an intergenic region on chromosome 2L (Figure S2). This suggests that, apart from the Y, both groups have a similar A. arabiensis background, and we concluded that the transgenic A. gambiae Y had been successfully purified in an A. arabiensis genomic background, although our data cannot rule out that some fraction of A. gambiae genomic DNA other than the Y chromosome persists in the AY2 strain.
Table 1. Analysis of single nucleotide polymorphisms between A and AY2 males.
| Chromosome | Base pairs | Total number of variants | Biallelic SNPs without missing data | Differentially fixed SNPs | |
|---|---|---|---|---|---|
| Number | % | ||||
| X | 24,393,108 | 1,040,639 | 583,061 | 27 | 0.0046 |
| 2L | 49,364,325 | 2,180,463 | 1,464,616 | 42 | 0.0029 |
| 2R | 61,545,105 | 2,233,941 | 1,573,367 | 12 | 0.0008 |
| 3L | 41,963,435 | 1,702,574 | 1,159,627 | 27 | 0.0023 |
| 3R | 53,200,684 | 2,480,254 | 1,653,367 | 6 | 0.0004 |
| Mt | 15,363 | 23 | 21 | 0 | 0 |
| Unknown | 42,389,979 | 749,098 | 351,870 | 115 | 0.0327 |
| Y_unplaced | 237,045 | 4,180 | 1,902 | 677 | 35.5941 |
Analysis of Y chromosome sequence content in introgressed males
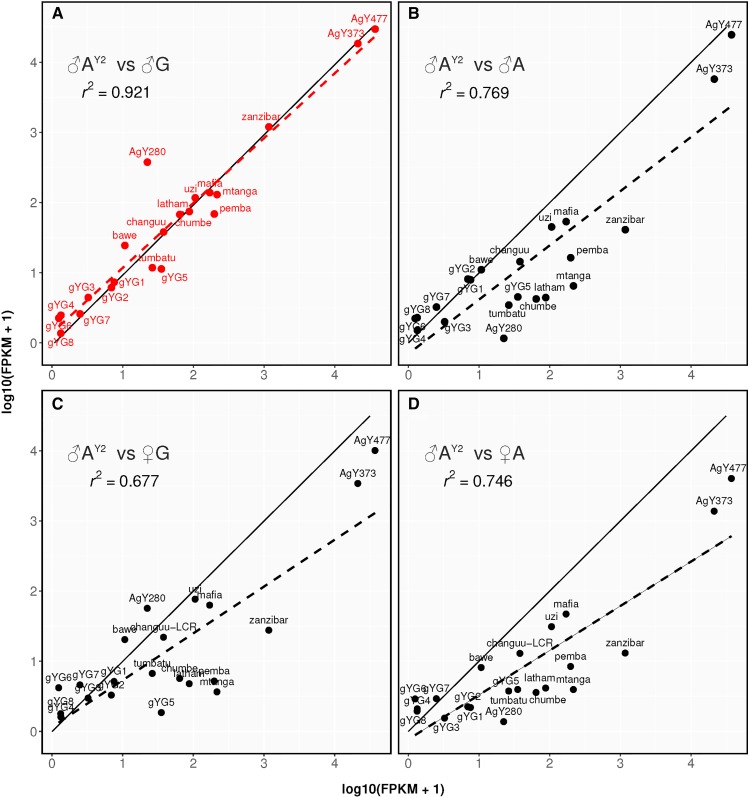
To assess whether introgression of the A. gambiae Y chromosome into the A. arabiensis genome coincided with any detectable structural rearrangements of the Y, for example, the selective elimination of sequences that would be detrimental in hybrids, we mapped DNA reads from all AY2 individuals as well as pooled control datasets from Hall et al. (2016) against a collection of known Y chromosome genes and repeats of A. gambiae. This collection includes consensus sequences of all known and putative genes and repetitive elements, as well as satellite DNA. We assessed normalized read depth at each locus (FPKM) as a proxy for copy number of each sequence element in a given background. This analysis is complicated by the occurrence of autosomal copies of many of these elements, as well as possible variation between Y chromosome isolates. Figure 3 shows normalized read counts for these elements in strain AY2 plotted vs. males and females of both A. gambiae and A. arabiensis. We observe an excellent correlation in the representation of these Y signature sequence elements between A. gambiae males and AY2 males (Figure 3A). Because these signature elements are derived from, and, to some degree, specific to the A. gambiae Y chromosome, they were under-represented in A. arabiensis control males and in females of both species. Interestingly, the AgY280 satellite shows an ∼15× higher coverage in A. gambiae males compared to the AY2 strain. However this satellite is known to be highly variable between Y isolates (Hall et al. 2016). In order to better assess the content of DNA satellites, in particular satellites Ag53A, B, and C, which are too short for standard read mapping, we performed an additional analysis based on kmer counts in the read libraries. This analysis showed that AY2 males resemble A. gambiae males with regards to the content of DNA satellites attributed to the Y (Figure S3). Together, these analyses therefore confirm the presence of the A. gambiae Y chromosome in the introgressed strain, and suggest that the introgression experiment did not impact in any evident way the original content of this Y chromosome.
Figure 3.
Analysis of the content of the introgressed Y chromosome. The plots show the number of normalized reads mapping to the A. gambiae Y chromosome reference loci calculated as log10 transformed FPKM values for AY2 males on the x-axis compared to either wild-type A. gambiae males (A), wild-type A. arabiensis males (B), A. gambiae females (C) and wild-type A. arabiensis females (D) on the y-axis. The dashed linear regression line and associated r2 coefficient indicate the best correlation in read counts of signature Y elements between AY2 and A. gambiae males.
Transcriptomic analysis of males carrying a heterospecific Y chromosome
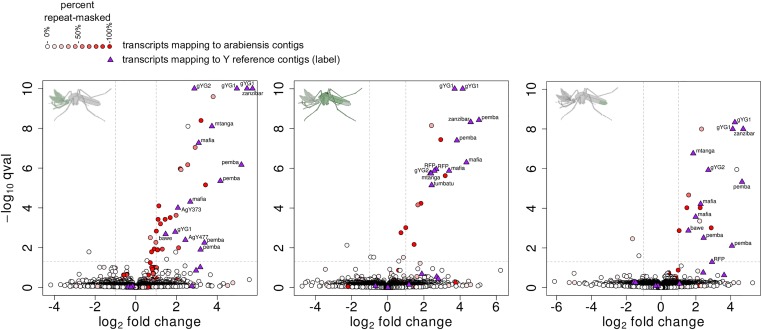
We next asked to what extent the presence of a heterospecific Y chromosome would alter the expression of autosomal or X-linked genes of the A. arabiensis background. This could occur by (i) the action of gambiae Y trans-acting factors or absent A. arabiensis Y trans-acting factors, (ii) by Y sequences that recruit cellular trans-acting factors and/or modulate the chromatin structure and epigenetic state of other chromosomes, or (iii) by the activation of mobile genetic elements present on the A. gambiae Y that trigger a cellular response in X or autosomal sequences. We focused our analysis on three tissues, the head, the terminal abdominal segments containing the reproductive tissues and the remainder of the carcass. We performed RNA sequencing of a total of eight control and eight experimental samples for each tissue from AY2 and wild-type males that had been reared in the same larval tray and that were sexed and separated at the pupal stage. Paired-end reads were mapped against the 1214 A. arabiensis genomic scaffolds supplemented by 25 consensus sequences corresponding to known A. gambiae Y loci previously mentioned, as well as the reporter gene construct. Due to the incomplete annotation of the A. arabiensis genome we performed an isoform level analysis, where we first predicted novel genes and novel isoforms of known genes across all samples. Gene-level expression of the experimental groups as well as sample relationships are summarized in Figure S4 and File S1. Finally, in order to indicate whether differentially expressed transcripts potentially represented known mobile elements or repetitive DNA arising from the Y (but matching paralogous sequences present on the A. arabiensis scaffolds, which could thus be misreported as differentially expressed) we blasted each predicted transcript to the A. gambiae repeat library and assigned a repetitiveness score. We then predicted differential expression of transcripts between AY2 and wild-type males (Figure 4 and File S2). Few transcripts were expressed significantly lower in AY2 males. This is partially expected because A. arabiensis genomic scaffolds are derived from the DNA of females (Neafsey et al. 2015), and it is thus not possible to identify any A. arabiensis Y-linked genes that would have fallen into this class. However, it also indicates that few, if any, endogenous genes are downregulated as a result of the presence of the heterospecific Y chromosome. In contrast, a number of transcripts displayed significantly higher levels of expression in AY2 males. However, the majority of these correspond to the known A. gambiae Y loci (purple triangles in Figure 4). The expression of these genes, not present on the A. arabiensis Y, and hence absent from A. arabiensis wild-type males, is also summarized in Figure S5. Of the remaining upregulated A. arabiensis transcripts (colored circles in Figure 4) the majority had a high repetitiveness score i.e., significant homology to A. gambiae repeats. A manual homology search using all A. arabiensis transcript sequences passing the significance threshold and cut-off for expression confirmed that virtually all differentially expressed transcripts are related to repetitive DNA. In addition to the above analysis, we also measured small RNA expression in these tissues, finding no evidence for the differential expression of small noncoding RNAs between AY2 and wild-type males (Figure S6). While it is possible that such effects could occur in other developmental stages or under specific environmental conditions, we find little evidence that the heterospecific Y-chromosome markedly affects expression of the nonrepetitive, autosomal, or X-linked gene repertoire of the A. arabiensis genome.
Figure 4.
Differential expression analysis by RNA-seq. Volcano plots showing log2 fold-change values (x-axis) by −log10 corrected P-values (y-axis) for all transcripts between introgressed AY2 males and A. arabiensis control males. The analysis was performed separately for the head (left panel), the carcass (middle panel), and the abdominal segments containing the reproductive tract (right panel). Transcripts derived from A. arabiensis scaffolds are represented as circles, and colored based on the percentage of their sequence masked by sequences in the A. gambiae repeats library. Transcripts from the reference set of A. gambiae Y loci are indicated by purple triangles and the name of the signature locus. Dashed lines represent the thresholds used for adjusted P-value (P < 0.05) and log2 fold change (>1.0).
Female mate-choice and male competition experiments
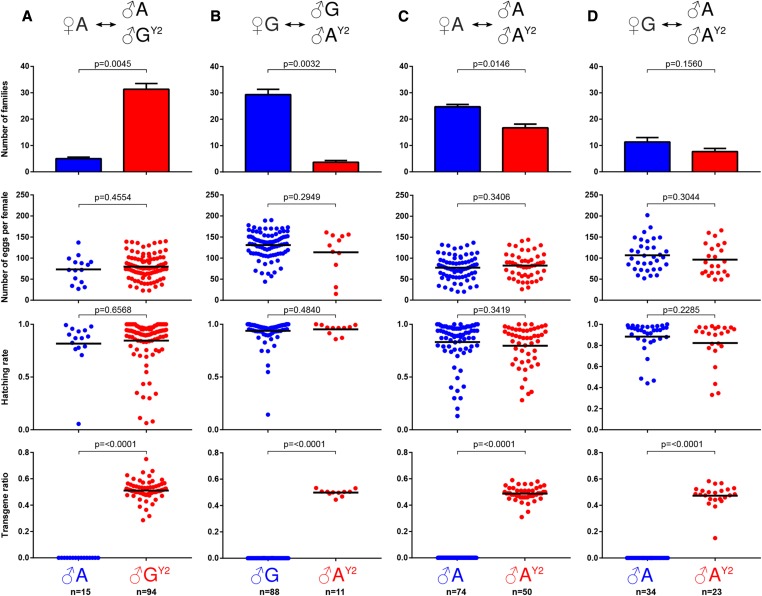
Although we find no strong effect of the heterospecific Y chromosome on the transcriptome and fertility of individual AY2 males, it is possible that Y-linked sequences do play a role in male fitness or female choice that would also limit the practical use of introgressed Y-linked traits e.g., for vector control. In order to test this hypothesis, we set up a panel of competitive mating experiments, where two strains of males were allowed to compete for mating with females in population cages (Figure 5). Both wild-type A. arabiensis and AY2 males performed substantially worse (winning only 13.7 and 11.1% of matings, respectively) than A. gambiae males in competition experiments for either A. gambiae or A. arabiensis females. These findings confirms previous observations (Schneider et al. 2000), and demonstrate that, irrespective of the female type, gambiae males are superior to A. arabiensis males in the laboratory setting. In contrast, AY2 males were only slightly less competitive compared to wild type arabiensis males, winning ∼40% of matings with arabiensis females (P = 0.0146), and no significant difference was observed when they competed against arabiensis males for gambiae females (P = 0.156). The particular set up of the experiments also allowed to score for secondary mating measured by the percentage of transgenic male progeny. With the possible exception of one case, remating (showing a significant deviation from a 50% transgene ratio in the progeny) was not observed in these experiments. Overall, a slight reduction in male competitiveness was observed which could relate, in addition to the effect of the heterospecific Y, to inbreeding or fitness costs associated with expression of the fluorescent marker gene. Importantly, in all cases no significant difference to the wild-type A. arabiensis males was observed in terms of the number of laid and hatching eggs from matings with AY2 males, confirming our previous analysis.
Figure 5.
Mating competition experiments. The genotypes of the females and the two types of competing males is indicated at the top for each experiment (A–D). The second, third, and fourth row of panels show the number of eggs laid by individual females, the hatching rate for each family, as well as the ratio of transgenic to wild-type larvae respectively. P-values were calculated using Welch’s two-tailed t-test.
Discussion
In a classic study, Slotman et al. (2004) mapped quantitative trait loci related to male sterility in hybrids between A. gambiae and A. arabiensis, and at least five or six sterility factors were detected in each of the two species. The X chromosome was found to have a disproportionately large effect on male hybrid sterility (Slotman et al. 2004), which is likely related to divergent alleles present within multiple fixed chromosomal rearrangements on the X. A possible role of the Y chromosome in hybrid incompatibilities was suggested by the authors but not followed up on experimentally. Using an F1 × F0 crossing scheme, we generated F3 males with an arabiensis X chromosome, and a set of arabiensis autosomes, as well as an A. gambiae Y chromosome. The second set of autosomes is expected to contain, on average, 75% A. gambiae sequences. The majority of such F3 males were expected to be sterile; however, we hypothesized that it should be possible to select a small fraction of fertile males that lacked the A. gambiae incompatibility loci that cause sterility when interacting with the A. arabiensis background. Indeed, we recovered 56 larvae out of a total 29,776 eggs laid (0.18%) from pooled backcrosses of ∼600 F3 males sampled.
Surprisingly, after multiple generations of backcross purification, we found that the A. gambiae Y chromosome does not markedly influence male fertility, fitness, or gene expression in Y hybrids. This rules out the Y chromosome as a major factor contributing to hybrid incompatibilities, and, more importantly, it is in stark contrast to findings in the Drosophila model (Sackton et al. 2011). Despite the relative paucity of genes in the fly Y, even intraspecific Y chromosome variants profoundly affect the expression a substantial number of genes located on the X or the autosomes. The introgression of heterospecific Y chromosomes in Drosophila, to the extent that it is even possible (Johnson et al. 1993), has consequently been found to markedly affect male fertility and gene expression in interspecific hybrids. In a D. simulans background, the D. sechellia Y has little effect on viability, but it reduced male fecundity by 63% as well as sperm competitiveness (D. simulans/sechellia divergence time has been estimated at only 0.25 MY). Y introgression differentially affected genes involved in immune function and spermatogenesis, suggesting a trade-off in investment between these processes. However, it has also been suggested that a significant part of the observed effect in Drosophila may be attributed to the ribosomal DNA (rDNA) clusters that are abundant on both the X and the Y in the fly. Paredes et al. (2011) have shown that deletions within the Y-linked rDNA arrays of Drosophila result in the differential expression of hundreds to thousands of unlinked genes due to a decreased heterochromatic composition of the genome. The affected genes significantly overlap with those affected by natural polymorphisms on Y chromosomes, suggesting that rDNA copy number variation is an important determinant of gene expression diversity in natural populations, and contributes to biologically relevant phenotypic variation (Paredes and Maggert 2009; Paredes et al. 2011). Importantly, in the A. gambiae strain G3 we used for this study [unlike some strains such as those used by Wilkins et al. (2007)] and in A. arabiensis, the rDNA is located exclusively on the X-chromosome, and thus does not confound introgression experiments the same way. Our results may thus be more indicative than similar experiments in Drosophila of the true effect a gene-poor heterospecific Y exerts on the genome of a related species.
It is possible that more subtle effects of the introgressed Y, not detectable in our experimental setup, exist. Future work could involve hybrid performance testing in mating swarms or under semi-field conditions. However, the fact that, despite its radically different structure and an estimated divergence time of ∼1.85 MY or >7 times that between D. simulans and D. sechellia (Fontaine et al. 2015), the A. gambiae Y seems to be able to fully replace the A. arabiensis Y, suggests that, in Anopheles, the Y either carries no functionally important factors or that such factors have not undergone functional divergence between these two species. Although, early work had implicated the Anopheline Y chromosome in mating behavior in a study using A. labranchiae and A. atroparvus species (Fraccaro et al. 1977), our work is thus more in line with a recent study that leveraged long single-molecule sequencing to determine the content and structure of the Y chromosome of the primary African malaria mosquito, A. gambiae in comparison to its sibling species in the gambiae complex (Hall et al. 2016).
The role of gene flow between A. gambiae and A. arabiensis in leading to adaptive introgression, and the implications for vector control has been highlighted by Weetman et al. (2014). Post-F1 gene flow occurs between A. gambiae and A. arabiensis, and, especially for traits under strong selection, could readily lead to adaptive introgression of genetic variants relevant for vector control. Introgression of the Y chromosome between species is generally viewed as unlikely, and markers found on the Y generally show restricted gene flow relative to other loci. However, this contrasts with the recent hypothesis (Neafsey et al. 2015; Hall et al. 2016) of Y gene flow between A. gambiae and A. arabiensis. Contrary to the established species branching order, the YG2 gene tree suggested that Y chromosome sequences may have crossed species boundaries between A. gambiae and A. arabiensis (Hall et al. 2016) and the authors suggested that Y chromosome introgression could have predated the development of male F1 hybrid sterility barriers that exist between this pair of species (Hall et al. 2016). Our study lends support to this hypothesis as it experimentally demonstrates the possibility of a cross-species Y chromosome transfer, and shows that the Y is functional in this context.
Our findings thus also suggest that Y-linked genetic traits generated in A. gambiae could be transferred to its sister species. For example, strain AY1 carries a site-specific docking site that now also allows the generation of male-exclusive genetic traits in A. arabiensis. Recent progress toward the elusive goal of efficient sex-ratio distortion by a driving Y chromosome (Bernardini et al. 2014; Galizi et al. 2014) could lead to the development of invasive distorter traits in A. gambiae that may then be transferred to sibling species. This could be done deliberately in the laboratory as we have demonstrated here, but it could also occur contingently in the wild after a large-scale release of transgenic males. While one should always be wary extrapolating laboratory studies to conditions in the field, the notion that the Y chromosome does not represent a genetic barrier for gene flow between two members of the A. gambiae species complex (A. gambiae and A. arabiensis) should inform the design and implementation of genetic control interventions based on transgenic mosquitoes.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300221/-/DC1.
Acknowledgments
This study was funded by the European Union’s Seventh Framework Programme (FP7 2007-2013) Marie Curie Actions cofund (Project I-Move) under GA no. 267232. This study was funded by a grant from the Foundation for the National Institutes of Health through the Vector-Based Control of Transmission: Discovery Research (VCTR) program of the Grand Challenges in Global Health initiative of the Bill & Melinda Gates Foundation. P.A.P. was supported by a Rita Levi Montalcini award from the Ministry Education, University and Research (MIUR – D.M. no. 79 04.02.2014). This study was funded by the European Research Council under the European Union’s Seventh Framework Programme ERC grant no. 335724 awarded to N.W. M.K.N.L. was supported by an MRC Career Development Award (G1100339) and by the Wellcome Trust (098051).
Author Contributions: F.B. and N.W. contributed to the experimental design of the study with help from M.L. Experiments were performed by F.B., M.R.W., C.T., K.K. and A.H. Analyses were performed by F.B., M.L., P.A.P. and N.W. The paper was written by F.B. and N.W. with help from P.A.P, T.N.,A.C. and M.L.
Footnotes
Communicating editor: M. Hahn
Literature Cited
- Bachtrog D., 2013. Y chromosome evolution: emerging insights into processes of Y chromosome degeneration. Nat. Rev. Genet. 14: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini F., Galizi R., Menichelli M., Papathanos P.-A., Dritsou V., et al. , 2014. Site-specific genetic engineering of the Anopheles gambiae Y chromosome. Proc. Natl. Acad. Sci. USA 111: 7600–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P., Nachman M. W., 2014. X-Y interactions underlie sperm head abnormality in hybrid male house mice. Genetics 196: 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A. B., Vicoso B., Russo C. A. M., Swenor B., Clark A. G., 2015. Birth of a new gene on the Y chromosome of [i]Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 112: 12450–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1991. Evolution of sex chromosomes. Science 251: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., 2011. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Genetics 12: 157–166. [DOI] [PubMed] [Google Scholar]
- Fontaine M. C., Pease J. B., Steele A., Waterhouse R. M., Neafsey D. E., et al. , 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347: 1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraccaro M., Tiepolo L., Laudani U., Marchi A., Jayakar S. D., 1977. Y chromosome controls mating behaviour on Anopheles mosquitoes. Nature 265: 326–328. [DOI] [PubMed] [Google Scholar]
- Galizi R., Doyle L. A., Menichelli M., Bernardini F., Deredec A., et al. , 2014. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun. 5: 3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A., Carneiro M., Delibes-Mateos M., Villafuerte R., Nachman M. W., et al. , 2008. Reduced introgression of the Y chromosome between subspecies of the European rabbit (Oryctolagus cuniculus) in the Iberian Peninsula. Mol. Ecol. 17: 4489–4499. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Chan C.-K. K., 2016. Analysis of RNA-seq data using TopHat and Cufflinks, pp. 339–361 in Plant Bioinformatics: Methods and Protocols, edited by Edwards D. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Hall A. B., Papathanos P.-A., Sharma A., Cheng C., Akbari O. S., et al. , 2016. Radical remodeling of the Y chromosome in a recent radiation of malaria mosquitoes. Proc. Natl. Acad. Sci. USA 113: E2114–E2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. A., Hollocher H., Noonburg E., Wu C. I., 1993. The effects of interspecific Y chromosome replacements on hybrid sterility within the Drosophila Simulans Clade. Genetics 135: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinska E., Dennison N. J., Lycett G. J., Krzywinski J., 2016. A maleness gene in the malaria mosquito Anopheles gambiae. Science 353(6294): 67–9. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Kingsford C., 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly J. P., Presgraves D. C., 2007. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5: 1890–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawejje H. D., Wilding C. S., Rippon E. J., Hughes A., Weetman D., et al. , 2013. Insecticide resistance monitoring of field-collected anopheles gambiae s.l. populations from jinja, eastern Uganda, identifies high levels of pyrethroid resistance. Med. Vet. Entomol. 27: 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S. E., Seawright J. A., 1989. Recombination between the X and Y chromosomes in Anopheles quadrimaculatus species A. The Journal of Heredity 80: 496–499. [DOI] [PubMed] [Google Scholar]
- Neafsey D. E., Waterhouse R. M., Abai M. R., Aganezov S. S., Alekseyev M. A., et al. , 2015. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347: 1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano L., Estivill X., Martí E., 2011. A non-biased framework for the annotation and classification of the non-miRNA small RNA transcriptome. Bioinformatics 27: 3202–3203. [DOI] [PubMed] [Google Scholar]
- Paredes S., Maggert K. A., 2009. Ribosomal DNA contributes to global chromatin regulation. Proc. Natl. Acad. Sci. USA 106: 17829–17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes S., Branco A. T., Hartl D. L., Maggert K. A., Lemos B., 2011. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA–sensitive” genes and natural variation. PLoS Genet. 7: e1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L., 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. R., 1996. Evolution of the Y sex in animals: Y chromosomes evolve through the degeneration of autosomes. Bioscience 46: 331–343. [Google Scholar]
- Sackton T. B., Montenegro H., Hartl D. L., Lemos B., 2011. Interspecific Y chromosome introgressions disrupt testis-specific gene expression and male reproductive phenotypes in Drosophila. Proc. Natl. Acad. Sci. USA 108: 17046–17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider P., Takken W., McCall P. J., 2000. Interspecific competition between sibling species larvae of Anopheles arabiensis and An. gambiae. Med. Vet. Entomol. 14: 165–170. [DOI] [PubMed] [Google Scholar]
- Slotman M., Della Torre A., Powell J. R., 2004. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis. Genetics 167: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temu E. A., Hunt R. H., Coetzee M., Minjas J. N., Shiff C. J., 1997. Detection of hybrids in natural populations of the Anopheles gambiae complex by the rDNA-based, PCR method. Ann. Trop. Med. Parasitol. 91: 963–965. [DOI] [PubMed] [Google Scholar]
- Toure Y. T., Petrarca V., Traore S. F., Coulibaly A., Maiga H. M., et al. , 1998. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia 40: 477–511. [PubMed] [Google Scholar]
- Turelli M., Moyle L. C., 2007. Asymmetric postmating isolation: Darwin’s Corollary to Haldane’s Rule. Genetics 176: 1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski M. D., Koerich L. B., Carvalho A. B., 2008. Two new Y-linked genes in Drosophila melanogaster. Genetics 179: 2325–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D., Steen K., Rippon E. J., Mawejje H. D., Donnelly M. J., et al. , 2014. Contemporary gene flow between wild An. gambiae s.s. and An. arabiensis. Parasit. Vectors 7: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins E. E., Howell P. I., Benedict M. Q., 2007. X and Y chromosome inheritance and mixtures of rDNA intergenic spacer regions in Anopheles gambiae. Insect Mol. Biol. 16: 735–741. [DOI] [PubMed] [Google Scholar]
- Wu C. I., Johnson N. A., Palopoli M. F., 1996. Haldane’s rule and its legacy: why are there so many sterile males? Trends Ecol. Evol. 11: 281–284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data has been submitted to the National Center for Biotechnology Information (NCBI) short read archive, and are available under BioProject IDs PRJNA381402, PRJNA381403 and PRJNA381033. A detailed list of per-sample accession numbers is given in Supplemental Material, File S1.