Selection during evolution, whether natural or artificial, is evidenced through the phenotype. For complex phenotypes like plant and inflorescence.....
Keywords: domestication, maize, teosinte, tga1, tb1
Abstract
Selection during evolution, whether natural or artificial, acts through the phenotype. For multifaceted phenotypes such as plant and inflorescence architecture, the underlying genetic architecture is comprised of a complex network of interacting genes rather than single genes that act independently to determine the trait. As such, selection acts on entire gene networks. Here, we begin to define the genetic regulatory network to which the maize domestication gene, teosinte branched1 (tb1), belongs. Using a combination of molecular methods to uncover either direct or indirect regulatory interactions, we identified a set of genes that lie downstream of tb1 in a gene network regulating both plant and inflorescence architecture. Additional genes, known from the literature, also act in this network. We observed that tb1 regulates both core cell cycle genes and another maize domestication gene, teosinte glume architecture1 (tga1). We show that several members of the MADS-box gene family are either directly or indirectly regulated by tb1 and/or tga1, and that tb1 sits atop a cascade of transcriptional regulators controlling both plant and inflorescence architecture. Multiple members of the tb1 network appear to have been the targets of selection during maize domestication. Knowledge of the regulatory hierarchies controlling traits is central to understanding how new morphologies evolve.
PLANT biologists have been remarkably successful at using genetic analysis to identify a long list of major domestication genes that have contributed substantially to the transformation of wild plants into cultivated crop species (Olsen and Wendel 2013). For example, crop geneticists have identified individual genes controlling fruit size in tomato (Cong et al. 2008), grain size and shape in rice (Shomura et al. 2008; Li et al. 2011), grain number in barley (Komatsuda et al. 2007), seed shattering in rice and sorghum (Konishi et al. 2006; Li et al. 2006; Lin et al. 2012), erect plant growth habit in rice (Jin et al. 2008; Tan et al. 2008), grain covering in wheat and barley (Simons et al. 2006; Taketa et al. 2008), and many more. Some of these genes were independently selected in multiple species such as Shattering1 in sorghum, rice, and maize (Lin et al. 2012). This body of research has firmly established that allele substitutions of large effect in single genes were a key mechanism contributing to evolution under domestication.
Over the past several decades, plant developmental biologists have discovered and described the networks of interacting genes that control how plants proceed from fertilization through development to produce complex adult processes and structures. Examples include floral organ identity (Coen and Meyerowitz 1991), leaf determinacy (Sinha et al. 1993; Bharathan et al. 2002), root structure (Malamy and Benfey 1997), inflorescence branching (Gallavotti et al. 2010), plant branching (Waldie et al. 2014), embryo polarity (Smith and Long 2010), and meristem maintenance (Fletcher et al. 1999). Gene networks also modulate differences in the size and shape of organs, as shown by the control of floral symmetry in snapdragon (Costa et al. 2005), leaf complexity in a broad array of species (Ichihashi et al. 2014), and leaf lobing in multiple species (Blein et al. 2008). Perhaps the best-understood developmental network in plants is that describing floral induction in Arabidopsis (Posé et al. 2012). This rich body of research has established how complex networks of genes control adult phenotypes.
Selection, whether natural or artificial, during evolution acts on phenotypes that affect the fitness of individuals. Given that adult phenotypes are the “read outs” of complex gene networks, selection targets the entire network. The importance of selection on the joint effects of all members of a gene network as opposed to the individual effects of single genes has long been recognized in evolutionary biology (Wright 1929, 1982). Comparing the structure of gene networks among species has shown that both membership and interactions among network members change over time and these changes correlate with complex phenotypic differences among species and higher level taxa (Peter and Davidson 2011). For example, Zhang et al. (2015) report a dynamic process by which mammalian gene networks acquire new members which themselves acquire an increasing number of gene–gene interactions within the network over time.
Maize (Zea mays subsp. mays) was domesticated from its wild progenitor, teosinte (Z. mays subsp. parviglumis), ∼9000 years ago in the Balsas river drainage in southwestern Mexico (Matsuoka et al. 2002; Piperno et al. 2009). Morphological change during maize domestication has generated broad interest because maize and teosinte differ more profoundly in plant and inflorescence architecture than any other crop–progenitor pair. As with other crop plants, research on maize has witnessed some success at identifying by genetic analysis multiple genes for which the maize allele confers a domesticated phenotype distinct from the teosinte allele which confers a wild phenotype. At teosinte branched1 (tb1), the teosinte allele confers a highly branched plant and the maize allele a less-branched plant (Doebley et al. 1997). At teosinte glume architecture1 (tga1), the teosinte allele confers covered grains and the maize allele, uncovered grains (Wang et al. 2005). At grassy tillers1 (gt1), the teosinte allele confers multiple ears per branch and the maize allele a single ear per branch (Wills et al. 2013).
In this article, we investigate the regulatory connections of two known maize domestication genes: tb1 and tga1. We show that tb1, a plant architecture gene, directly regulates tga1, an inflorescence architecture gene, and that tb1 also regulates two cell cycle genes, suggesting how it may in part control branch outgrowth. We also show that tga1 regulates multiple MADS-box transcription factors, a class of genes known to control inflorescence morphology. From the literature, two other maize domestication genes, gt1 and enhancer of tb1.2 (etb1.2), act downstream of tb1 in the network we describe. Our results, combined with published work in maize and other cereals, provides a first view of the network of genes that underlies multiple phenotypes which changed dramatically during maize domestication. This network was the target of selection during maize domestication.
Materials and Methods
Plant materials and phenotyping
All plant materials used for this research have been described previously. Briefly, tb1-teosinte is an introgression line that contains a teosinte chromosomal segment, which includes the tb1 locus, in a maize W22 background (IS3 in Studer and Doebley 2012). T249 (tga1-teosinte) is an isogenic line to W22 with tga1 replaced with a teosinte allele (H. Wang et al. 2015). To measure glume length, we used an F2 population that segregated for the maize and teosinte alleles at both tb1 and tga1. The plants were genotyped at these two genes with a molecular marker in each gene. We phenotyped plants that represent all three genotypic classes at tb1 but were all heterozygous at tga1. Heterozygosity at tga1 confers a slightly enlarged glume, which enhances the ability to accurately measure this organ. To measure glume length, mature cobs were broken in the middle and calipers were used to measure all of the glumes in the exposed row (∼12). The average glume length for each ear was calculated.
Quantitative real-time PCR
Immature ears of tb1-teosinte for quantitative real-time PCR (qPCR) were collected from plants grown at the West Madison Agricultural Research Station during the summer of 2005. Immature ears of T249 for qPCR were collected in the greenhouse at a length of 2.5 cm. qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and an ABI Prism 7000 Sequence Detection System (Applied Biosystems). Relative gene expression was calculated by normalizing to actin (GRMZM2G126010). Two-tailed t-tests were performed on expression data to test for significant differences between the maize allele and either tb1-teosinte or T249 using statistical packages in R (R Development Core Team 2010). Primers for qPCR are listed in Supplemental Material, Table S1A. qPCR with tb1-teosinte was performed with 10 independent biological replicates and at least two technical replicates. qPCR with T249 was done with 15 biological replicates. Normalized expression data can be found in Table S2.
TB1 antibody generation
The TB1 antibody was generated following the same procedure previously described by Wang et al. (2005). Briefly, a C-terminal region of the TB1 coding sequence was amplified using the PCR primers 5′-caccatgAGAAAATCGGCCAATAACGCAC-3′ and 5′-acccggGTAGTGTTCAGTAGAAGCGTGA-3′. The amplicon was cloned into pET-151/D-TOPO (Invitrogen, Carlsbad, CA) and expressed in Escherichia coli. The protein was purified using a His-column and 12% SDS-PAGE gel. The TB1-specific C-terminal antigen was injected into rabbits (Invitrogen Custom Antibodies).
PCR-assisted binding site selection and electrophoretic mobility shift assays
Full length Tb1 complementary DNA was cloned into a pVP-GW vector to produce soluble TB1 protein (Singh et al. 2005) using the primers 5′-acccggGTAGTGTTCAGTAGAAGCGTGA-3′ and 5′-caccgaaaacctgtacttccagtccATGTTTCCTTTCTGTGATTCCT-3′. After E. coli expression and purification, TB1 was used for PCR-assisted binding site selections. PCR-assisted binding site selection was used to determine the TB1 binding site following the method described in H. Wang et al. (2015). Succinctly, a 76-mer [α-32P]dATP-labeled probe [5′-actcgaggaattcggcaccccgggt(n)26tggatccggagagctcccaacgcgt-3′] containing a core of 26 randomized nucleotides was used for the electrophoretic mobility shift assay (EMSA) with TB1 protein. Five sequential EMSAs were used to enrich for probes containing the TB1 binding site. Purified sequences were cloned into pCR2.1-TOPO cloning vector (Invitrogen), and 52 independent clones were sequenced to determine the consensus binding site. EMSAs were performed as described previously (H. Wang et al. 2015).
Chromatin immunoprecipitation assays
Bulked young developing ears (1–5 cm in length) were used for chromatin immunoprecipitation (ChIP) assays with anti-TB1 and anti-TGA1 respectively. Wild-type inbred W22 ears were used for anti-TB1 ChIP assays and ears from Not1-Mu were used for anti-TGA1 ChIP. Neighbor of Tga1 (Not1) is a duplicate of Tga1 (H. Wang et al. 2015). Because of the high homology between NOT1 and TGA1, we used a null allele of not1 that contains a Mutator transposable element. This ensures that the anti-TGA1 ChIP assays enriched for only TGA1 binding sites. ChIP assays were performed as described previously (Gendrel et al. 2002; H. Wang et al. 2002, 2015). Four independent anti-TB1 ChIP replicates and six independent anti-TGA1 ChIP replicates were performed. Input and immunoprecipitation (IP) fractions were compared using qPCR to amplify promoter sequences containing TB1 or TGA1 binding sites. Primers are listed in Table S1B. One-sided, paired t-tests were performed to test ChIP enrichment using statistical packages in R (R Development Core Team 2010). Normalized enrichment data can be found in Table S3.
Data availability
File S1 contains the putative promoter sequences and binding sites of TB1 and TGA1. Figure S1 shows the consensus sequence result from the PCR-assisted bind site selection assay. Table S1A contains the primer sequences for gene expression, and Table S1B contains the sequences for ChIP-enrichment quantification. Table S2 contains the normalized expression qPCR data. Table S3 contains the normalized ChIP-enrichment qPCR data.
Results
tb1 regulates two cell cycle genes
tb1 functions as a repressor of branching such that a loss-of-function allele of tb1 leads to excessive branch outgrowth (Doebley et al. 1997). The molecular mechanism by which tb1 exercises this function is unknown. Previously, it was reported that the maize allele of tb1 is expressed approximately two-fold higher than the teosinte allele, indicating that the more highly expressed maize allele confers reduced branching in comparison with the lower expressed teosinte allele (Doebley et al. 1997). We confirmed the approximately two-fold higher expression of the maize allele using qPCR by comparing isogenic stocks carrying the maize (tb1-maize) vs. teosinte (tb1-teosinte) alleles of tb1 in a maize (W22) genetic background (Figure 1).
Figure 1.
Real-time PCR shows decreased levels of cell cycle genes with increased expression of tb1. The relative gene expression of tb1 and select cell cycle genes was assayed from immature ears of wild-type maize (tb1-maize) and a tb1-teosinte introgression line (tb1-teosinte). The mean of the biological replicates and SE are shown. Expression of each gene was measured using qPCR and normalized to actin.
tb1 belongs to the TCP family of transcriptional regulators and other members of this family are known to regulate cell proliferation and organ growth both through the control of, and response to, hormone signaling as well as through direct regulation of cell cycle genes (Nicolas and Cubas 2016). To determine if tb1, like some other TCP genes, regulates cell cycle genes, we assayed the relative level of expression of two cell cycle genes, proliferating cell nuclear antigen2 (pcna2) and minichromosome maintainence2 (mcm2; an ortholog of mcm7/Prolifera in Arabidopsis), using the same isogenic stocks mentioned above. We observed a significant increase in the expression of both pcna2 (t = 3.9112, P = 0.0027) and mcm2 (t = 2.4054, P = 0.0337) in the stock carrying the teosinte allele as compared to the one carrying the maize allele (Figure 1). Thus, when tb1 expression is higher with the maize allele, there is a corresponding reduction in the expression of pcna2 and mcm2. This reduction in pcna2 and mcm2 expression with tb1-maize is consistent with tb1 acting as a repressor of organ growth by the regulation of genes that function in cell division (Springer et al. 1995; López et al. 1997).
TB1 binds to GGNCCC motifs upstream of pcna2
To identify targets of maize tb1, the consensus binding site needed to be determined. This was done using a PCR-assisted binding site selection assay. Many of sequenced clones (23/52) contained a GGGCCC motif, and 40 of the 52 sequenced clones had a GGNCCC binding motif (Figure S1). This motif is consistent with the known binding of class II TCP transcription factors, GTGGNCCC, found in rice (Kosugi and Ohashi 2002). Our data suggest that the maize TB1 binding requirement is limited to the core GGNCCC sequence shared by both class I and II TCP transcription factors (Kosugi and Ohashi 2002).
To further investigate the direct binding of TB1 to target gene promoters, we produced a TB1-specific antibody. This was done by making an antigen using the C-terminal domain of the TB1 protein. By using the C-terminal end of TB1, we avoided the conserved TCP domain. A BLAST search of the maize genome revealed that this region of TB1 has only ∼75% sequence homology to one other gene, TCP18 (AC190734.2_FG003), located on chromosome 5. The specificity of the antibody was tested using a Western blot, which produced a single band of the expected size (data not shown).
Given our expression data for pcna2, and the previous reports that TCP-family transcription factors regulate cell division (Li et al. 2005), we searched for putative TB1 binding motifs upstream of pcna2. A single GGGCCC TB1 binding motif is located in the first 1000 bp upstream of the pcna2 start codon (Figure 2A and Figure S1). To test for a direct interaction between TB1 and the putative pcna2 promoter in vitro, we used EMSAs. When the pcna2 putative promoter fragment containing the GGGCCC binding motif was incubated with purified TB1 protein, an upshift was observed on the gel (Figure 2C). This demonstrates the binding of TB1 to the pcna2 putative promoter. To further show that the TB1 binding motif is in fact the GGGCCC sequence, the putative binding motif in the probe was mutated to GGGTTC. No upshift was observed on the gel with the mutated probe, indicating that TB1 failed to bind the GGGTTC motif. This result is consistent with the consensus TB1 binding site that was identified through the PCR-assisted binding site selection assay.
Figure 2.
TB1 binds to pcna2 and tga1 promoters in vitro and in vivo. (A and B) Sequences upstream of pcna2 and tga1 where the probes bind the GG(G/C)CCC motifs. (C) EMSAs show that TB1 binds to the pcna2 and tga1 promoters via GG(G/C)CCC motif. Mutating the TB1 binding motif in the probe eliminates binding as seen by the lack of shifted bands. Each condition was replicated as paired lanes on the gel. (D and E) ChIP confirms that TB1 binds to the pcna2 and tga1 promoters in vivo. Input, total input chromatin DNA before precipitation; ChIP, chromatin DNA precipitated with anti-TB1 antibody PI, DNA precipitated with pre-immune serum; Mock, no antibody or serum added. (D) Agarose gel image of semi-qPCR amplification of the enriched promoter fragments. (E) qPCR with error bars indicating SEs.
The binding of TB1 to the pcna2 putative promoter in vivo was tested using ChIP. The pcna2 putative promoter fragments showed enrichment after IP with the TB1 antibody (Figure 2D). The enrichment was quantified using qPCR, and a highly significant (t = −10.207, P = 7.747e−05) fourfold enrichment of the pcna2 putative promoter sequence was observed in the IP fraction when compared to the input control (Figure 2E). These data suggest that TB1 directly binds the pcna2, through direct binding to a GGGCCC motif located 260 bp upstream of the start codon, and represses pcna2 expression. Therefore, the selection for increased tb1 expression during domestication likely reduced axillary bud outgrowth in part by the direct binding and repression of pcna2, which is necessary for cell division. The cell cycle gene mcm2 is similarly downregulated in association with the tb1-maize allele, however we did not test whether this was due to direct or indirect regulation.
tga1 regulates multiple MADS-box genes
We also investigated genes regulated by tga1 to better understand how it controls covered vs. uncovered grains. tga1 is a member of the Squamosa promoter binding protein (SBP) family of transcription factors which have a known role in directly regulating MADS-box genes (Theissen et al. 2000). Therefore, we identified eight MADS genes and investigated whether they are regulated by tga1, and whether the nature of this regulation differs for tga1-maize vs. tga1-teosinte. We used qPCR to investigate MADS gene expression with isogenic lines carrying tga1-maize vs. tga1-teosinte. Seven of the eight MADS genes were upregulated in the line carrying the teosinte allele as compared to the maize allele (Figure 3A), and five of these differences were significant (P < 0.05). The higher expression of these MADS genes in the presence of tga1-teosinte as compare to tga1-maize is consistent with our prior report that the single amino acid substitution (ASP > LYS) which distinguishes these two alleles causes the maize allele to act as repressor of its targets (H. Wang et al. 2015).
Figure 3.
TGA1 regulates the expression of some MADS-box genes. (A) qPCR results showing the relative gene expression of eight MADS-box genes in wild-type maize (tga1-maize) and an isogenic line T249, which carries a tga1-teosinte allele. RNA was isolated from immature ears. The mean of the biological replicates and SE are shown. Expression of each gene was normalized to actin. (B) ChIP confirms that TGA1 binds to the promoter of some MADS-box genes in vivo. Bulked immature ears were collected and fixed for ChIP. The mean of the biological replicates and SE are shown.
To test for direct binding of TGA1 to these MADS gene promoters, ChIP was performed using the TGA1 antibody. We had previously identified GTAC as the binding site for TGA1 (H. Wang et al. 2015). Although the number of TGA1 binding sites in the putative promoter of each MADS gene varied (File S1), given the number of genes being tested only one putative binding site per gene promoter was selected for ChIP analysis. Of the eight genes tested by ChIP, five showed an enrichment in the IP fraction compared to the input control (Figure 3B). These results indicate that TGA1 binds and regulates multiple MADS genes. Given the known roles of MADS genes in regulating floral and inflorescence architecture (Medard and Yanofsky 2001), these MADS genes are likely involved in developmental control of the grain covering or “fruitcase” of teosinte.
Among the five MADS genes with different expression levels associated with the teosinte and maize alleles of tga1, zmm19 is of particular interest because this gene has been identified as being the Tunicate locus (Han et al. 2012; Wingen et al. 2012). Tunicate is a dominant mutant of maize that has enlarged glumes on the ear that completely surround the individual kernels. The molecular lesion in Tunicate (zmm19) that causes the phenotype involves a structural rearrangement and duplication of the locus which leads to strong overexpression of zmm19 in the developing glume. Our expression data show that zmm19 is more highly expressed in the isogenic stock with the teosinte allele of tga1 than the stock containing the maize allele (Figure 3A). Thus, as with the Tunicate mutant allele, higher expression of ZMM19 in the isogenic stock containing tga1-teosinte as compared to tga1-maize might partially explain the development of the large glumes in teosinte that cover the grains.
tb1 regulates tga1
Since tb1 regulates organ growth and maize has reduced growth of the fruitcase that covers the grain, tb1 represents a candidate for a contribution to the difference between covered vs. uncovered grains. In this regard, tb1 has been shown to be expressed in the ear glumes, one of two organs that compose the fruitcase (Hubbard et al. 2002). If tb1 functions as a repressor of organ growth, then increased tb1 expression in the glume as conferred by tb1-maize will function to reduce the size of the glume and the extent to which the glume covers the grain.
To investigate the possibility of direct regulation of tga1 by tb1, we searched for putative TB1 binding motifs upstream of tga1. Two putative TB1 binding sites are located in the first 1000 bp upstream of the tga1 start codon. One of these is a GGGCCC sequence identical to the TB1 binding site in pcna2, and the other sequence motif is GGCCCC (Figure S1). Given that our data suggest that TB1 binds to a GGNCCC motif, it is possible that TB1 binds at least twice, upstream of tga1. To test for a direct interaction between TB1 and the tga1 putative promoter, we performed EMSA using two different probes, each one specific to a single binding motif (Figure 2B). When each probe was incubated with purified TB1 protein, an upshift was observed on the gel (Figure 2C). The binding of TB1 to the GGGCCC and GGCCCC sequence motifs was further tested by mutating the putative binding sites to GGGTTC and GGCTTC, respectively. The TB1 protein failed to bind either of the mutated probes (Figure 2C).
The binding of TB1 to the tga1 putative promoter in vivo was tested by ChIP using a pair of primers that spanned both the binding sites in the tga1 promoter. The tga1 putative promoter fragments that were tested in vitro also showed enrichment in vivo after IP with the TB1 antibody (Figure 2D). The enrichment was quantified using qPCR and a significant enrichment in the IP fraction as compared to the input control was observed (t = −4.4362, P = 0.01065; Figure 2E). This fivefold enrichment strongly supports the direct binding of TB1 to the GGGCCC and GGCCCC motifs upstream of tga1 in vivo.
Next, we asked whether tb1 acts as a positive or negative regulator of tga1. Comparing tga1 expression for isogenic stocks carrying the maize vs. teosinte alleles of tb1, we observed a significant reduction in the expression of tga1 in the stock with tb1-teosinte relative to tb1-maize (Figure 1; t = −2.8103, P = 0.01228). This result suggests that tb1 functions as a direct activator of tga1. Overall, our results suggest that tb1 functions as an activator of tga1, but as a repressor of pcna2. Previous studies have suggested that specific TCP genes can function as a direct activator of one gene and direct repressor of another (Hervé et al. 2009).
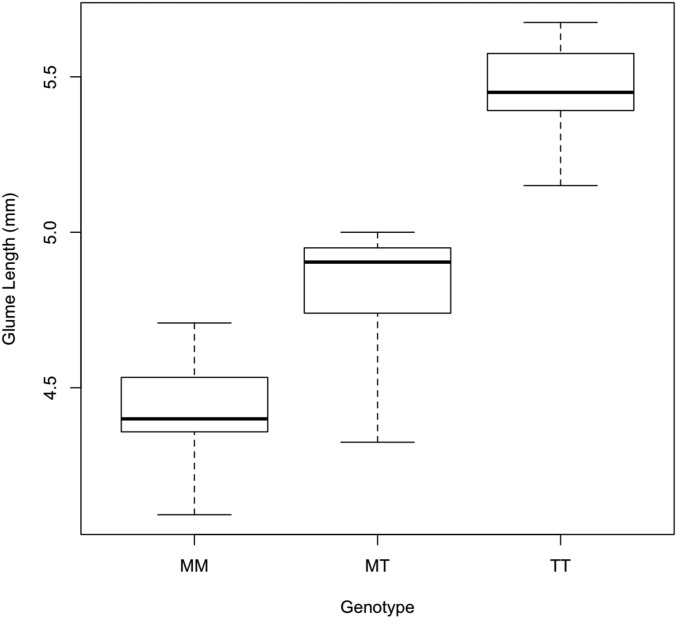
As a final question regarding the regulatory relationship between tb1 and tga1, we asked how the maize vs. teosinte allele at tb1 affects glume length. If tb1 regulates (activates) tga1, then we hypothesize that tb1 should affect glume length. Specifically, tb1-maize should give higher expression of tga1, which in turn should more strongly repress glume elongation. We compared glume length in isogenic lines with one of three genotypes at tb1 (maize homozygous, heterozygous, or teosinte homozygous), but all were heterozygous at tga1. tga1 heterozygous plants have partially enlarged glumes which enhance the phenotype. We observed a strong effect of tb1 on glume length and, as anticipated, tb1-maize confers shorter glumes relative to tb1-teosinte (Figure 4). These results are consistent with the prior report that tb1 is expressed in the glume (Hubbard et al. 2002), and they support the hypothesis that tb1 functions to control the grain covering via its role in regulating tga1.
Figure 4.
Glume length is affected by both tb1 and tga1. Quantitative effects of the tb1-teosinte allele on glume length. The y-axis indicates glume length. The x-axis indicates the allele of tb1; M denotes the tb1-maize allele and T denotes the tb1-teosinte allele. Each line was heterozygous for the tga1-maize/tga1-teosinte allele. The boxes show the first and third quartile with the line indicating the mean and the whisker showing the data extremes.
Discussion
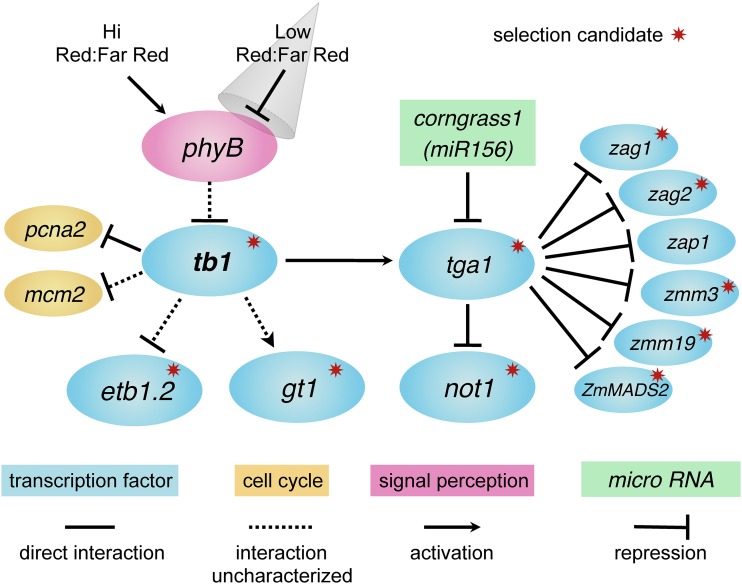
Figure 5 summarizes the regulatory interactions documented in this article or known from the literature. Here, we have shown that the maize domestication gene tb1 directly regulates another maize domestication gene tga1. We have also identified two cell cycle genes that are regulated by tb1, and six MADS-box genes that are direct targets of tga1. From the literature, multiple other interacting genes are known. Yang et al. (2016) demonstrate that tb1 lies upstream of etb1.2 and that these two genes interact epistatically on phenotype (ear structure). Moreover, tb1 acts as a repressor of etb1.2 expression. Whipple et al. (2011) showed that in sorghum phyB acts upstream of tb1 in response to the ratio of red:far-red light to mediate branch outgrowth, a component of the shade-avoidance response of plants (Kebrom et al. 2006, 2010). Whipple et al. (2011) have also shown that gt1 lies downstream of tb1, with tb1 acting as an activator of gt1 expression and thereby suppressing bud outgrowth. H. Wang et al. (2015) identified a paralog of tga1 called not1 and showed that tga1 directly regulates not1 with the tga1-maize allele acting to repress not1 expression. not1 has no known phenotypic effects. Finally, Chuck et al. (2007) demonstrated that the dominant maize mutant Corngrass1, which affects both ear and plant architecture, encodes a member of the microRNA156 family, which targets and downregulates tga1.
Figure 5.
Maize domestication gene network. Regulatory relationships among genes contributing to differences in plant and inflorescence architecture between maize and teosinte which are reported in this article or known from the literature. Solid lines and arrows indicate direct regulatory interactions supported by ChIP and gene expression assays or RNA–DNA complementary binding for corngrass1. Dotted lines and arrows indicate interactions based on gene expression assays alone and thus uncharacterized as to whether they are direct or indirect. The gray cone represents shading or a reduction in the ratio of red:far-red light. Red starbursts demark genes that show evidence of selection during domestication.
Change in a gene network for plant architecture improved harvest quality
A key aspect of the domestication of many crops was to convert highly branched wild species with multiple small fruits into less-branched crops with fewer larger fruits. Harvest efficiency is greater for crops with a single large fruit or inflorescence, as compared to a wild species with dozens of smaller fruits (Tanksley 2004). This change is accomplished by a reduction in branch number, giving fewer branches with a smaller number of larger fruits. Annual plant species may be preadapted to evolve in this manner given their innate shade-avoidance response, which favors reduced branching by suppression of axillary buds to promote growth of the main stalk under the low ratio of red:far-red light that characterizes shade (Franklin 2008; Rameau et al. 2015).
The network in Figure 5 suggests how the innate shade-avoidance pathway downstream of phyB was coopted to evolve a less-branched maize plant from a more-branched teosinte ancestor (Figure 6A). The upregulation of tb1 in maize would directly downregulate the cell cycle in buds, maintaining bud dormancy or reducing branch outgrowth. tb1 would also work through activating gt1, which promotes reduced branching as a component of the shade-avoidance response (Whipple et al. 2011). Finally, tb1 may control plant architecture through regulation of tga1, an unexpected possibility given that tga1 was identified as a gene controlling the grain covering or fruitcase (Wang et al. 2005). H. Wang et al. (2015) demonstrated that an RNA interference (RNAi) knock down of tga1 results in plants with a larger number of longer branches. In Figure 5, upregulation of tb1 would increase the expression of tga1, and increased expression of tga1 should have the opposite effect of the RNAi knock down, i.e., it should confer a more maize-like plant with fewer, shorter branches.
Figure 6.
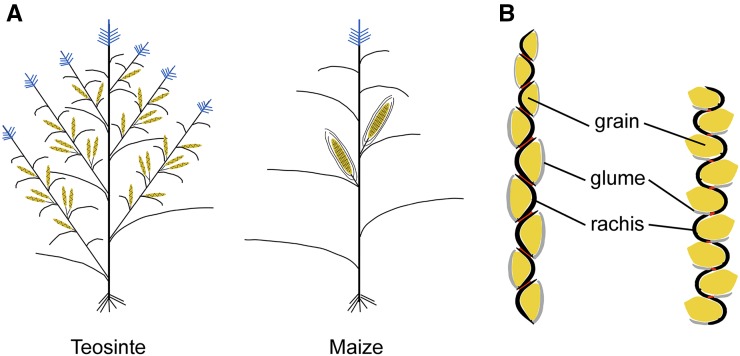
Phenotypes of maize and teosinte. (A) Schematic drawings of teosinte and maize plants showing differences in plant architecture. Female inflorescences (ears) are yellow and male inflorescences (tassels) are blue. (B) Schematic drawings of longitudinal cross sections through a teosinte ear (left) and expectation for a modified teosinte ear carrying maize alleles at tga1, tb1, and etb1.2 (right). Glumes are gray, rachis (internodes) are black, grains are yellow, and nodes (abscission layers) are red.
The network in Figure 5 suggests how the change from a larger number of small ears in teosinte to a smaller number of larger ears in maize evolved, i.e., how harvest quality was improved (Figure 6A). gt1 was identified as a major QTL for the production of secondary ears along the branches (Wills et al. 2013). Teosinte produces many secondary ears, but maize does not. The mechanism to evolve fewer secondary ears is a gain of gt1 expression in the nodes of the upper branches of maize, where this expression blocks the outgrowth of buds to form secondary ears. The suppression of secondary ear formation may also be influenced by tga1. Plants with an RNAi knock down of tga1 not only have longer branches, but on some plants, the secondary ears along these branches develop (figure 6C in H. Wang et al. 2015). As shown in Figure 5, increased expression of tb1 would drive higher expression of tga1, which should have the opposite effect of the RNAi knock down, i.e., it should confer fewer secondary ears.
Changes in organ size and identity contributed to the naked grains of maize
The gene network shown in Figure 5 suggests three mechanisms by which the change from covered grains in teosinte to naked grains in maize evolved through changes in organ size and identity. Teosinte grains are encapsulated in a stony fruitcase comprised of an ear internode (rachis) and a glume (bract) (Figure 6B, left). Maize grains are uncovered on the exterior of a cob, which is comprised of the ear internodes and glumes. This change involved a switch from elongated internodes that form a cup-like structure in which the kernel sits, to shortened (collapsed) internodes that are too small to house the kernels but form the sturdy central axis of the maize ear (the cob). There is also a change in the glume from the specialized hardened (silicated, lignified) organ in teosinte to a glume that is more leaf-like (Dorweiler and Doebley 1997).
The network in Figure 5 suggests reduction in the size of the glume could be accomplished in part through the downregulation of the cell cycle genes (pcna2 and mcm2) by tb1. Hubbard et al. (2002) showed that tb1 is expressed in the glume. We have shown that tb1 directly represses the cell cycle genes, and the anticipated effect of such repression would be to reduce the size of the glume (Figure 6B, right). The higher expression of tb1-maize relative to tb1-teosinte would cause greater repression of cell division and thus less growth of the glume. Consistent with this model, we showed that the glumes of plants carrying tb1-maize are smaller than those carrying tb1-teosinte.
The network in Figure 5 also suggests that the reduction in ear internode length could be accomplished through the downregulation of both the cell cycle genes (pcna2 and mcm2) by tb1, as well as through the downregulation of etb1.2 by tb1, as previously suggested by Yang et al. (2016). etb1.2, which encodes a YABBY-class transcription factor, acts as a positive regulator of ear internode elongation. Maize alleles of etb1.2 have either reduced or no expression and confer shorter internodes than the teosinte allele (Figure 6B right). Moreover, tb1 acts as a repressor of etb1.2 so that the reduction in internode length is reinforced with the more highly expressed tb1-maize that more strongly represses etb1.2.
Finally, the ear glumes of maize and teosinte have distinct identities. Teosinte has highly lignified and silicated glumes, while the glumes of maize are more leaf-like, being less lignified and silicated (Dorweiler et al. 1993; Dorweiler and Doebley 1997). MADS-box genes are known regulators of reproductive organ identity in plants, including grasses (Bommert et al. 2005; Sablowski 2015). Our observation that tga1 directly regulates a set of MADS-box genes invites the hypothesis that these MADS genes play a role in determining the identity of the teosinte ear glume by activating programs to promote their lignification and silication. As shown by H. Wang et al. (2015), an amino acid substitution in the maize allele of tga1 relative to the teosinte allele converts the TGA protein into a repressor of its targets. Thus, the maize allele may interfere with the specification of teosinte glume identity, causing the glumes to revert to a more leaf-like identity intermediate between the hardened glumes of teosinte and the chaffy glumes of most other grasses. A presumption of this model is that the hardened glumes of teosinte, which are unique among the grasses, evolved via complex changes in MADS genes and their targets.
The gene network as the target of selection
Evidence that the network just described was a target of selection during maize domestication comes from the literature. Those members of the network that were initially identified as domestication QTL have all been reported to show signatures of selection during domestication; tb1 (Studer et al. 2011), tga1 (Wang et al. 2005), and gt1 (Wills et al. 2013). etb1.2, which was identified as a QTL that interacts epistatically with tb1, also exhibits evidence of past selection (Yang et al. 2016). The remaining 11 genes were all identified because they interact with one of the aforementioned domestication QTL. Of these 11 genes, 6 have previously been shown to exhibit evidence for selection during maize domestication (Table 1). Interestingly, genes involved in signal perception and the cell cycle do not show signatures of selection, whereas selection pressure seemed to have acted on most of the transcription factors that control the developmental processes.
Table 1. Evidence for the signature of selection for genes in the defined network.
| Gene name | Gene identification no. | Zhao et al. (2011) | Hufford et al. (2012) | Other |
|---|---|---|---|---|
| tb1 | AC233950.1_FG002 | Studer et al. (2011) | ||
| phyB | GRMZM2G124532 | |||
| pcna2 | GRMZM2G108712 | |||
| mcm2 | GRMZM2G112074 | |||
| etb1.2 | GRMZM2G085873 | Yang et al. (2016) | ||
| gt1 | GRMZM2G005624 | Wills et al. (2013) | ||
| tga1 | GRMZM2G101511 | ✓ | Wang et al. (2005) | |
| corngrass1 | GRMZM2G022489 | |||
| not1 | AC233751.1_FG002 | ✓ | ||
| zag1 | GRMZM2G052890 | ✓ | ✓ | |
| zag2 | GRMZM2G160687 | ✓ | ✓ | |
| zap1 | GRMZM2G148693 | |||
| zmm3 | AC197699.3_FG001 | ✓ | ||
| zmm19 | GRMZM2G370777 | ✓ | ||
| ZmMADS2 | GRMZM2G316366 | ✓ |
Overview
The network of genes shown in Figure 5 included several genes previously implicated in the shade-avoidance response of plants (Franklin 2008; Kebrom et al. 2010; Rameau et al. 2015). This overlap between shade-avoidance genes and domestication genes supports an interpretation that maize domestication “hijacked” a preexisting developmental gene network to create a crop that has a constitutive shade-avoidance phenotype, in that maize has fewer and shorter branches than teosinte. In this context, other gene members of the shade-avoidance gene network should be considered as candidate domestication genes.
The network of genes shown in Figure 5 may also be related to the network controlling “phase change” through the juvenile to adult to reproductive stages of plant development (Hansey et al. 2011; Yang et al. 2013). The Corngrass1 mutant has been interpreted as promoting a constitutive juvenile identity, as overexpression of miR156 suppresses the transition to the adult phase (Chuck et al. 2007). tga1 can also be blended into this model in that RNAi knock-down lines of tga1 exhibit one of the hallmarks of the juvenile stage in maize, extension of nodes with prop roots vertically up the plant (H. Wang et al. 2015). Finally, tb1 has been identified as a gene that interacts with the phase-change program in maize (Poethig 1990). In this context, domestication may be partially explained as coopting the phase-change network to promote an accelerated transition to the adult phase for some phenotypes as compared to their progenitors (L. Wang et al. 2015).
The gene network depicted in Figure 5 contains at best a small fraction of the genes that interact to control plant and inflorescence architecture as related to maize domestication. Research in other species suggests additional genes that may act in this network in maize, and these genes are strong candidates for maize domestication genes. Ideal plant architecture1 (IPA1) is a rice Squamosa promoter binding protein that controls plant architecture and has an allele that substantially enhances grain yield (Jiao et al. 2010). The maize ortholog of IPA1 appears to be GRMZM2G160917, a gene that shows evidence of selection during maize domestication (Hufford et al. 2012), and for which allele-specific expression assays show that the maize allele is expressed at twice the level of the teosinte allele (Lemmon et al. 2014). Interestingly, IPA1 is a direct regulator of tb1 in rice (Lu et al. 2013). Hexokinase (hex1; GRMZM2G104081) is also an interesting candidate as it has been implicated in sugar signaling as related to branching (Yang et al. 2013), and it shows both evidence for selection (Hufford et al. 2012) and upregulation in maize as compared to teosinte (Lemmon et al. 2014).
Finally, tb1 holds a central position in Figure 5, being downstream of the shade-signaling but upstream of all other transcription factors, and upstream of all the identified domestication genes. In Arabidopsis, the tb1 ortholog (BRC1) has been proposed to act as the integrator of multiple signals to modulate branching via control of cell division and growth (Aguilar-Martínez et al. 2007). tb1 may have played a key position in the restructuring of plant and inflorescence architecture during maize domestication.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300071/-/DC1.
Acknowledgments
We thank Bao Kim Nguyen, Jesse Rucker, and Tina Nussbaum Wagler for technical assistance and R. Scott Poethig for insightful comments on the manuscript. This work was supported by the National Science Foundation, funding IOS1025869 and IOS1238014, and a United States Department of Agriculture Hatch grant MSN169062.
Footnotes
Communicating editor: J. Birchler
Literature Cited
- Aguilar-Martínez J. A., Poza-Carrion C., Cubas P., 2007. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan G., Goliber T. E., Moore C., Kessler S., Pham T., et al. , 2002. Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860. [DOI] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., et al. , 2008. A conserved molecular framework for compound leaf development. Science 322: 1835–1839. [DOI] [PubMed] [Google Scholar]
- Bommert P., Satoh-Nagasawa N., Jackson D., Hirano H. Y., 2005. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46: 69–78. [DOI] [PubMed] [Google Scholar]
- Chuck G., Cigan A. M., Saeteurn K., Hake S., 2007. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39: 544–549. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Meyerowitz E. M., 1991. The war of the whorls. Nature 353: 31–37. [DOI] [PubMed] [Google Scholar]
- Cong B., Barrero L. S., Tanksley S. D., 2008. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40: 800–804. [DOI] [PubMed] [Google Scholar]
- Costa M. M. R., Fox S., Hanna A. I., Baxter C., Coen E., 2005. Evolution of regulatory interactions controlling floral asymmetry. Development 132: 5093–5101. [DOI] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L., 1997. The evolution of apical dominance in maize. Nature 386: 485–488. [DOI] [PubMed] [Google Scholar]
- Dorweiler J., Doebley J., 1997. Developmental analysis of teosinte glume architecture1: a key locus in the evolution of maize (Poaceae). Am. J. Bot. 84: 1313–1322. [PubMed] [Google Scholar]
- Dorweiler J., Stec A., Kermicle J., Doebley J., 1993. Teosinte glume architecture1: a genetic locus controlling a key step in maize evolution. Science 262: 233–235. [DOI] [PubMed] [Google Scholar]
- Fletcher J. C., Brand U., Running M. P., Simon R., Meyerowitz E. M., 1999. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914. [DOI] [PubMed] [Google Scholar]
- Franklin K. A., 2008. Shade avoidance. New Phytol. 179: 930–944. [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Long J. A., Stanfield S., Yang X., Jackson D., et al. , 2010. The control of axillary meristem fate in the maize ramosa pathway. Development 137: 2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A., Lippman Z., Yordan C., 2002. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873. [DOI] [PubMed] [Google Scholar]
- Han J.-J., Jackson D., Martienssen R., 2012. Pod corn is caused by rearrangement at the Tunicate1 locus. Plant Cell 24: 2733–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansey C. N., Johnson J. M., Sekhon R. S., Kaeppler S. M., de Leon N., 2011. Genetic diversity of a maize association population with restricted phenology. Crop Sci. 51: 704–715. [Google Scholar]
- Hervé C., Dabos P., Bardet C., Jauneau A., Auriac M. C., et al. , 2009. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149: 1462–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L., McSteen P., Doebley J., Hake S., 2002. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162: 1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford M. B., Xu X., van Heerwaarden J., Pyhäjärvi T., Chia J.-M., et al. , 2012. Comparative population genomics of maize domestication and improvement. Nat. Genet. 44: 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Aguilar-Martínez J. A., Farhi M., Chitwood D. H., 2014. Evolutionary developmental transcriptomics reveals a gene network module regulating inter-specific diversity in plant leaf shape. Proc. Natl. Acad. Sci. USA 111: E2616–E2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., et al. , 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544. [DOI] [PubMed] [Google Scholar]
- Jin J., Huang W., Gao J. P., Yang J., Shi M., et al. , 2008. Genetic control of rice plant architecture under domestication. Nat. Genet. 40: 1365–1369. [DOI] [PubMed] [Google Scholar]
- Kebrom T. H., Burson B. L., Finlayson S. A., 2006. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 140: 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom T. H., Brutnell T. P., Finlayson S. A., 2010. Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 33: 48–58. [DOI] [PubMed] [Google Scholar]
- Komatsuda T., Pourkheirandish M., He C., Azhaguvel P., Kanamori H., et al. , 2007. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 104: 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S., Izawa T., Lin S. Y., Ebana K., Fukuta Y., et al. , 2006. An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396. [DOI] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y., 2002. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30: 337–348. [DOI] [PubMed] [Google Scholar]
- Lemmon Z. H., Bukowski R., Sun Q., Doebley J. F., 2014. The role of cis regulatory evolution in maize domestication. PLoS Genet. 10: e1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colon-Carmona A., Gutierrez R. A., Doerner P., 2005. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhou A., Sang T., 2006. Rice domestication by reducing shattering. Science 311: 1936–1939. [DOI] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., et al. , 2011. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43: 1266–1269. [DOI] [PubMed] [Google Scholar]
- Lin Z., Li X., Shannon L., Yeh C., Wang M., et al. , 2012. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44: 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López I., Khan S., Vázquez J., Hussey P. J., 1997. The proliferating cell nuclear antigen (PCNA) gene family in Zea mays is composed of two members that have similar expression programmes. Biochim. Biophys. Acta 1353: 1–6. [DOI] [PubMed] [Google Scholar]
- Lu Z., Yu H., Xiong G., Wang J., Jiao Y., et al. , 2013. Genome-wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J. E., Benfey P. N., 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y., Vigouroux Y., Goodman M. M., Sanchez G. J., Buckler E., et al. , 2002. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 99: 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medard N., Yanofsky M. F., 2001. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2: 186–195. [DOI] [PubMed] [Google Scholar]
- Nicolas M., Cubas P., 2016. TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33: 33–41. [DOI] [PubMed] [Google Scholar]
- Olsen K. M., Wendel J. F., 2013. A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 64: 47–70. [DOI] [PubMed] [Google Scholar]
- Peter I., Davidson E., 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144: 970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno D. R., Ranere A. J., Holst I., Iriarte J., Dickau R., 2009. Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. USA 106: 5019–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R. S., 1990. Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923–930. [DOI] [PubMed] [Google Scholar]
- Posé D., Yant L., Schmid M., 2012. The end of innocence: flowering networks explode in complexity. Curr. Opin. Plant Biol. 15: 45–50. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2010 R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: https://www.r-project.org/.
- Rameau C., Bertheloot J., Leduc N., Andrieu B., Foucher F., et al. , 2015. Multiple pathways regulate shoot branching. Front. Plant Sci. 5: 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R., 2015. Control of patterning, growth, and differentiation by floral organ identity genes. J. Exp. Bot. 66: 1065–1073. [DOI] [PubMed] [Google Scholar]
- Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., et al. , 2008. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028. [DOI] [PubMed] [Google Scholar]
- Simons K. J., Fellers J. P., Trick H. N., Zhang Z., Tai Y. S., et al. , 2006. Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Cornilescu C. C., Tyler R. C., Cornilescu G., Tonelli M., et al. , 2005. Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein. Protein Sci. 14: 2601–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N. R., Williams R. E., Hake S., 1993. Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7: 787–795. [DOI] [PubMed] [Google Scholar]
- Smith Z. R., Long J. A., 2010. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464: 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer P. S., Mccombie W. R., Sundaresan V., Martienssen R. A., 1995. Gene trap tagging of PROLIFERA, an essential MCM2-3-5–like gene in Arabidopsis. Science 268: 877–880. [DOI] [PubMed] [Google Scholar]
- Studer A. J., Doebley J. F., 2012. Evidence for a natural allelic series at the maize domestication locus teosinte branched1. Genetics 191: 951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J., Doebley J., 2011. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43: 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketa S., Amano S., Tsujino Y., Sato T., Saisho D., et al. , 2008. Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc. Natl. Acad. Sci. USA 105: 4062–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Li X., Liu F., Sun X., Li C., et al. , 2008. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364. [DOI] [PubMed] [Google Scholar]
- Tanksley S. D., 2004. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16: S181–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Becker A., Di Rosa A., Kanno A., Kim J. T., et al. , 2000. A short history of MADS-box genes in plants. Plant Mol. Biol. 42: 115–149. [PubMed] [Google Scholar]
- Waldie T., McCulloch H., Leyser O., 2014. Strigolactones and the control of plant development: lessons from shoot branching. Plant J. 79: 607–622. [DOI] [PubMed] [Google Scholar]
- Wang H., Tang W., Zhu C., Perry S. E., 2002. A chromatin immunoprecipitation (ChlP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. Plant J. 32: 831–843. [DOI] [PubMed] [Google Scholar]
- Wang H., Nussbaum-Wagler T., Li B., Zhao Q., Vigouroux Y., et al. , 2005. The origin of the naked grains of maize. Nature 436: 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Studer A. J., Zhao Q., Meeley R., Doebley J. F., 2015. Evidence that the origin of naked kernels during maize domestication was caused by a single amino acid substitution in tga1. Genetics 200: 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sun S., Jin J., Fu D., Yang X., et al. , 2015. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA 112: 15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple C. J., Kebrom T. H., Weber A. L., Yang F., Hall D., et al. , 2011. Grassy Tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 108: E506–E512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills D. M., Whipple C. J., Takuno S., Kursel L. E., Shannon L. M., et al. , 2013. From many, one: genetic control of prolificacy during maize domestication. PLoS Genet. 9: e1003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingen L. U., Münster T., Faigl W., Deleu W., Sommer H., et al. , 2012. Molecular genetic basis of pod corn (Tunicate maize). Proc. Natl. Acad. Sci. USA 109: 7115–7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1929. Fisher’s theory of dominance. Am. Nat. 63: 274–279. [Google Scholar]
- Wright S., 1982. Character change, speciation, and the higher taxa. Evolution 36: 427–443. [DOI] [PubMed] [Google Scholar]
- Yang C. J., Kursel L. E., Studer A. J., Bartlett M. E., Whipple C. J., et al. , 2016. A gene for genetic background in Zea mays: fine-mapping enhancer of teosinte branched1.2 to a YABBY class transcription factor. Genetics 204: 1573–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xu M., Koo Y., He J., Scott Poethig R., 2013. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Landback P., Gschwend A. R., Shen B., Long M., 2015. New genes drive the evolution of gene interaction networks in the human and mouse genomes. Genome Biol. 16: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Weber A. L., McMullen M. D., Guill K., Doebley J. F., 2011. MADS-box genes of maize: frequent targets of selection during domestication. Genet. Res. 93: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S1 contains the putative promoter sequences and binding sites of TB1 and TGA1. Figure S1 shows the consensus sequence result from the PCR-assisted bind site selection assay. Table S1A contains the primer sequences for gene expression, and Table S1B contains the sequences for ChIP-enrichment quantification. Table S2 contains the normalized expression qPCR data. Table S3 contains the normalized ChIP-enrichment qPCR data.