Abstract
Selection experiments and experimental evolution provide unique opportunities to study the genetics of adaptation because the target and intensity of selection are known relatively precisely. In contrast to natural selection, where populations are never strictly “replicated,” experimental evolution routinely includes replicate lines so that selection signatures—genomic regions showing excessive differentiation between treatments—can be separated from possible founder effects, genetic drift, and multiple adaptive solutions. We developed a mouse model with four lines within a high running (HR) selection treatment and four nonselected controls (C). At generation 61, we sampled 10 mice of each line and used the Mega Mouse Universal Genotyping Array to obtain single nucleotide polymorphism (SNP) data for 25,318 SNPs for each individual. Using an advanced mixed model procedure developed in this study, we identified 152 markers that were significantly different in frequency between the two selection treatments. They occurred on all chromosomes except 1, 2, 8, 13, and 19, and showed a variety of patterns in terms of fixation (or the lack thereof) in the four HR and four C lines. Importantly, none were fixed for alternative alleles between the two selection treatments. The current state-of-the-art regularized F test applied after pooling DNA samples for each line failed to detect any markers. We conclude that when SNP or sequence data are available from individuals, the mixed model methodology is recommended for selection signature detection. As sequencing at the individual level becomes increasingly feasible, the new methodology may be routinely applied for detection of selection.
Keywords: behavior, experimental evolution, exercise, F statistics, population differentiation
Complex traits, such as most behaviors, are affected by alleles segregating at multiple loci. Mapping quantitative trait loci (QTL) for such traits can be difficult, often requiring a large sample. In general, two approaches are used to map QTL, involving the use of a designed line cross experiment (Lander and Botstein 1989) or selectively bred populations (Wurschum 2012; Cui et al. 2015). Use of a line cross experiment requires a large sample to avoid the Beavis effect (Beavis 1994; Xu 2003), in which reported QTL effects are often biased and the amount of bias is inversely proportional to the sample size. Moreover, the inference space of QTL parameters is narrow, only applicable to the lines initiating the cross, and the result cannot be extended to crosses derived from other lines (Xu 1996). Using selectively bred populations for QTL mapping takes advantage of existing resources with no need to create a line cross (Chan et al. 2012; Lo et al. 2016). It is also possible to use selected lines to make a mapping cross. QTL detected from a set of selected populations can be directly applied to the same populations to further improve breeding efficiency (Wurschum 2012), and results can also be applied to the original starting (base) population from which the selected lines were derived. Another advantage of using selected populations for QTL mapping is that the sample size does not have to be very large because allelic data are used instead of the phenotypic values of a selected trait (Cui et al. 2015). The reason for this is that mapping QTL in selected populations takes advantage of the shifts of allele frequencies away from expected Mendelian ratios, i.e., equivalent to detection of segregation distortion, which does not require large sample sizes (Luo and Xu 2003; Luo et al. 2005).
Statistical methods for QTL mapping in selected populations often involve a χ2 test. When replicated lines of a selection experiment are available, a t-test or F test can be used to detect QTL via comparison of allele frequencies of the selected population with expected allele frequencies, an approach called detection of segregation distortion (Vogl and Xu 2000; Luo and Xu 2003; Luo et al. 2005). If multiple selected populations are involved, then allele frequencies among the populations may be compared, which is called population differential analysis (Weir and Cockerham 1984; Balding and Nichols 1995).
Differential analysis is an important area in population genetics and molecular evolution. Wright (1950) proposed three F statistics to describe population differentiation, where a whole population is subdivided into two or more subpopulations. The F statistics (not the F tests) describe the correlation of alleles at different levels in the population. For example, the correlation coefficient between two alleles from the same individual is called the inbreeding coefficient (FIT), the correlation coefficient between two alleles from different individuals in the same subpopulation is called FST, and the correlation coefficient between the two alleles from the same individual within the same subpopulation is called FIS. In Cockerham and Weir’s (Cockerham 1969; Weir and Cockerham 1984) notation, , and The three F statistics are related by The key parameter in population differentiation analysis is Wright (1950) only proposed the concept of F statistics and did not address how to estimate them from samples. It was Weir and Cockerham (1984) who developed a systematic approach to estimate these F statistics—ANOVA—by treating the binary indicator (0 or 1) of a reference allele as the response variable. Prior to Weir and Cockerham (1984), much confusion surrounded the relationship between the F statistics and correlation coefficients of alleles at different levels of the population hierarchy. Cockerham (1969) discovered that these F statistics can actually be expressed as various intraclass correlations (variance ratios) from the ANOVA. More importantly, one can perform a statistical test for the significance of using a nonparametric method, such as the Jackknife, the bootstrap, or the permutation test (Weir and Cockerham 1984). An estimated significantly different from that expected from neutrality means that the population differentiation may be caused by some sort of evolutionary forces beyond random genetic drift, e.g., selection. When markers of the entire genome are tested this way, selection signatures can be detected, where a selection signature is defined as a genomic region subject to selection (Brookfield 2001). Conventionally, detection of selection signatures is conducted via population differentiation analysis, and rarely have these applications included replicated lines within the differentiated populations. Without replications, it is difficult to separate selection from drift, and thus false positives may be high.
An alternative and more effective way to investigate selection acting at particular locus is through experimental evolution (Garland and Rose 2009; Baldwin-Brown et al. 2014; Schlotterer et al. 2015; Franssen et al. 2017), in which a replicated bidirectional selection experiment or a unidirectional selection experiment with nonselected control(s) is conducted. In experimental evolution, each treatment population often has multiple replicated lines that allow separation of selection effects from genetic drift, and thus reduce false positives. Although the F statistic approach can be applied to selection signature detection from experimental populations, each population represents a treatment level purposely chosen by investigators and is not a randomly selected level out of a large pool of populations. As a result, the F statistics that are based on random selection of populations may not be appropriate. Instead, a mixed model approach may be more appropriate by treating the selection (treatment) effects as fixed (e.g., high-selected, control, or low-selected) and effects of replicated lines (subpopulations) as random. Such a mixed model analysis may be more powerful than the F statistics. The purpose of the present study is to develop such a mixed model for detection of selection signatures using genome-wide markers in selected and control populations with multiple replicated lines within each population.
To distinguish population differentiation analysis from selection signature detection in experimental evolution, we now use “selection treatments” to represent “populations” and use “replicated lines” within each treatment to represent “subpopulations.” When only two levels of selection treatments (selection and control) are available for comparison, Baldwin-Brown et al. (2014) proposed a regularized t-test to compare their allele frequencies. Because this approach requires pooled DNA sequences, it was also called “evolve and resequence,” initially proposed by Turner et al. (2011) and then by Baldwin-Brown et al. (2014). The method depends on replicated lines within each selection treatment to correct allele frequency variation caused by genetic drift (or possibly founder effects). The idea was very simple, using the allele frequency of each replicated line as the original observed data point to test the mean difference in allele frequency between the two levels of selection treatments. Their main contribution was the addition of a regularization factor to the test to prevent some unexpected behavior of the test (see Discussion). The regularized factor is particularly useful when the number of replicated lines within each selection treatment is small because, by chance, the variances of allele frequency among replicates may be extremely small, leading to false detection of small difference in allele frequency between selection treatments. Many other methods are also available for detecting selection signatures, as reviewed by Schlotterer et al. (2015), but the regularized t-test is the state-of-the-art method for replicated selection experiments. The Cochran–Mantel–Haenszel test (Mantel 1963) is optimal for a replicated 2 × 2 χ2 test with replication (stratification), but not suitable for replications within each level of the treatments. If DNA sequences are available at the individual level, then using pooled allele frequency data may lead to loss of essential information and reduced power of detecting causally-related single nucleotide polymorphisms (SNPs). Information on the allelic composition of individual organisms in the population hierarchy may be very important in boosting the statistical power, and incorporation of such information into the selection model is the main goal of the present study. Although the F statistics (Weir and Cockerham 1984) already deal with genes at the level of individual organisms, a mixed model approach to detecting selection signatures in artificially manipulated populations may be more appropriate. In this study, we propose to use the minimum variance quadratic unbiased estimation (MIVQUE) procedure (Rao 1971b) for mixed models to estimate variance components and test differentiation among selection treatments that contain replicate lines.
To validate the efficacy of the mixed model methodology, we used mouse populations under long-term artificial selection for high amounts of voluntary wheel-running behavior (Swallow et al. 1998; Careau et al. 2013). The selection experiment includes two treatments, selection for high running (HR) and unselected control (C), each treatment with four replicate lines. These lines were developed as a model system to study correlated evolution and coadaptation of behavior (exercise) physiology (Wallace and Garland 2016). They are also viewed as relevant to human voluntary exercise behavior, which is very important in human health (Garland et al. 2011b). Detected selection signatures from this study will indicate that these genomic regions harbor genes responsible for voluntary wheel running. In subsequent reports, the biological functions of the identified genomic regions will be considered in detail, but that is beyond the scope of the present study.
While preparing this manuscript, we found a very similar study in rats to detect selection signatures for alcohol preference (Lo et al. 2016). That experiment included bidirectional selection for high- and low-alcohol preference, with each treatment replicated twice (four lines in total; no nonselected control lines). They collected 10 rats from each line at generation 60, where the first 30 generations were continuously selected and the last 30 generations were relaxed (no selection applied). Although their sample size was only 40 rats, they were able to detect many regions harboring genes that may be causally related to alcohol preference. Lo et al. (2016) directly estimated the (FST) parameters under the random model methodology and used a permutation test to detect that significantly deviated from the null model. Results from our mouse selection experiment are expected to be more powerful because of the larger number of lines (eight), larger sample size (80), and the use of the mixed model methodology.
Materials and Methods
Experimental material
As described in the original publication (Swallow et al. 1998, 2009), replicated within-family selection for increased voluntary wheel running in outbred laboratory house mice (Mus domesticus; Hsd:ICR strain: base population was 112 males and 112 females) was applied with four high-selected (HR) and four nonselected control (C) lines (10 families/line were carried forward each generation, with average litter size at weaning of ∼10 pups). As young adults, mice were housed individually with access to activity wheels for a period of 6 days, and selection was based on the mean number of revolutions run on days 5 and 6. Animal model analyses indicated that at least three of the four HR lines reached plateaus between generations 17 and 27 of the selection experiment, depending on sex and line. At the apparent selection limits, mice from the HR lines ran approximately threefold more than did those from the control lines (Careau et al. 2013). Various correlated responses to selection have been observed, including reduced body mass and body length, decreased body fat as a percentage of total mass, increased endurance at maximal aerobic capacity, and various alterations related to neurobiology, motivation, and brain reward system, as reviewed in (Rhodes and Kawecki 2009; Swallow et al. 2009; Garland et al. 2011a; Wallace and Garland 2016).
As outlined elsewhere (Swallow et al. 1998; Carter et al. 1999), the outbred Hsd:ICR mice used as the base population were originally bred from Swiss–Webster albino house mice in the early 1950s, including a period during which they were selected for large litters and perfect weaning success (Hauschka and Mirand 1973). Our mice were purchased from the Indianapolis, IN facility of Harlan Sprague Dawley. Levels of allozyme variation in Hsd:ICR mice are similar to those reported in wild populations of house mice [Carter et al. (1999) and references therein].
The selection experiment has been ongoing for almost 80 generations. For the present analyses, we collected DNA samples from 80 female mice at generation 61, 10 mice from each replicate line. Lines 1, 2, 4, and 5 were the nonselected C lines and lines 3, 6, 7, and 8 were the HR-selected lines. Given that the HR lines had been at selection limits (Careau et al. 2013) for many generations at the time of sampling, random genetic drift is likely to have caused further differentiation that may have obscured many SNPs affected by the selection protocol. In the future, we plan to analyze earlier generations by use of historical tissue samples, as described in Didion et al. (2016). Thus, the present data should be viewed as an exemplar to illustrate the utility of the proposed new statistical methods, not definitive with respect to signatures of selection in this particular selection experiment.
We used the Mega Mouse Universal Genotyping Array, which provides up to 77,800 single SNP markers and is built on the Illumina Infinium platform (Morgan et al. 2016). The SNP markers are distributed throughout the mouse genome (average spacing of 33 kb) and with a slight excess of probes in the telomeric regions of each autosome to facilitate detection of recombination events throughout the chromosomes. Eight mice were eliminated from the analysis because of low-quality SNP callings (one from line 2, one from line 5, two from line 3, one from line 6, and three from line 8). Of the 77,808 SNPs in the panel, 52,490 SNPs (7137 SNPs with at least one missing genotype, 45,339 monomorphic SNPs, and 14 SNPs on P and M elements) were deleted. After this quality control, the data set subject to analysis has 72 female mice with 25,318 SNPs. In contrast to genome-wide association studies (GWAS), population differentiation analysis does not use minor allele frequency and Hardy–Weinberg disequilibrium as criteria for quality control. The 25,318 selected SNPs in the analysis were evenly distributed across 19 autosomes and the X chromosome. The SNP alleles were numerically coded as 1 for the reference allele and 0 for the alternative allele. As a result, there were observations (one per allele) for each locus analyzed.
Mixed model analysis
The allelic model:
We first introduce the random model methodology for the F statistic (Weir and Cockerham 1984). As the response variable is the allelic value represented by a binary variable, the maximum likelihood method is not appropriate, unless a generalized linear mixed model (GLMM) is used (discussed later). Instead, we used the MIVQUE, denoted by MIVQUE(0), for variance component estimation (Rao 1971b). The basic idea is to construct a hierarchical model to perform ANOVA using allelic indicator (0 or 1) as the response variable and the hierarchical structures of selection treatments and replicate lines within treatments as the design matrices, where the hierarchical structure is represented by alleles within individuals, individuals with replicate lines, and lines within selection treatments. When the data are balanced, MIVQUE(0) generates equivalent results to ANOVA (Rao 1971b). We now consider two selection treatments only, one being the control treatment and the other the HR selection treatment. In this experiment, the number of treatments was two, the number of replicate lines within each treatment was four, the number of individuals within each line was 10 (but varied after deletion of eight mice with low-quality SNP callings), and the number of alleles within each individual was two (diploid organism).
Let be the indicator variable (0 or 1) for the lth allele of the kth individual from the jth line within the ith treatment, where for the two alleles of each individual, for the 10 individuals within each line, for the four lines within each treatment, and for the two treatments. Let be the reference allele and be the alternative allele of a locus under consideration. Denote the whole population frequency of by The allelic indicator variable for reference allele is
| , | (5) |
which is a Bernoulli variable, and thus the expectation is identical to the frequency of the reference allele. We now use Cockerham’s (1969) linear model to describe
| , | (6) |
where is the overall mean (frequency of for the whole experimental population), is the allele frequency of treatment i expressed as deviation from that of the whole population, is the allele frequency of the jth line expressed as deviation from the ith treatment, is the allele frequency of the kth individual expressed as deviation from the jth line within the ith treatment, and is the residual error. Note that the allele frequency of an individual is defined as which only takes three possible values, 0, 0.5, and 1. The two selection treatments were not randomly sampled but designed by the investigators prior to the experiment. Therefore, should be treated as a fixed effect. However, the Cockerham’s model is random and thus we will take the random model approach as review of the background of population differentiation. The model contains only one fixed effect () and thus it is called the random model. All other effects are random with mean zero and different variances. The variances are denoted by for effect for effect for effect , and for residual The expectation of is and the variance of is
| . | (7) |
Cockerham (1969) defined three Wright’s F statistics (Wright 1951) based on these variance components. For the four-level hierarchical model, there are four F statistics, which are defined as described in Yang (1998),
| , | (8) |
| , | (9) |
| , | (10) |
| . | (11) |
These F statistics are different from the F statistics developed by Weir and Cockerham (1984) but they have a nice property of
| (12) |
If we ignore the treatments by treating all lines as populations, then we have
| , |
which leads to
| . | (13) |
This is the typical in the three-level hierarchical population subdivision model, where all lines are promoted to populations and represents the variance of the promoted populations.
As the two levels of treatments were not randomly sampled from a universe of all possible selection treatments, it is more appropriate to treat as a fixed effect. Therefore, the model defined in Equation 6 is a mixed model, under which the expectation of is
| (14) |
and the variance of is
| . | (15) |
Our purpose of detecting selection signals is to test the null hypothesis
| , | (16) |
which does not require the F statistics but needs the original variance components to facilitate the hypothesis test. We proposed to use the MIVQUE(0) method of Rao (1971b) to estimate the variance components because normal distributions of the random effects and the residual errors are not required with MIVQUE(0).
It is much more convenient to use a matrix notation to derive the MIVQUE(0) procedure, as shown below:
| , | (17) |
where is an vector of unity, is an vector whose elements are 1 for individuals in HR and −1 for individuals in C, is the difference of allele frequencies between C and HR, is an incidence matrix representing the eight replicate lines, is a vector of allele frequencies for the eight lines, is an incidence matrix for the 72 mice (38 from C and 34 from HR), is an vector for individual effects, and is an vector of residuals. All random effects have expectations of zero and a variance for a variance for , and a variance for
The expectation and variance of the model are
| (18) |
and
| . | (19) |
The MIVQUE of the three variance components are obtained using the following linear equation system the details of which are
| , | (20) |
where the right hand sides of the equations are various quadratic forms of y and the left hand sides are the expectations of the quadratic forms. Let us define as column bind of the two matrices in the brackets and as the fixed effects. Further define , and The six unique elements of the H matrix are:
| , |
| , |
| , |
| , |
, and
| . |
The remaining three elements of H take the three corresponding elements with flipping subscripts because the matrix is symmetrical. The three elements of the Q matrix are
| . |
The MIVQUE estimate of the parameter vector is Note that the MIVQUE estimate of a variance component can be negative because of the unbiased nature of the estimate. If that happens, it is simply set to zero.
The estimated variance components, denoted by are then used to estimate the fixed effects and perform hypothesis tests. The estimated variance matrix of y is
| . | (21) |
The best linear unbiased estimate (BLUE) of the fixed effect is
| (22) |
and the variance matrix of this estimate is
| (23) |
Note that
The F test for is
| (24) |
with d.f. 1 (numerator) and 6 (denominator). The P-value is calculated using
| , | (25) |
where is a random variable of F distribution with 1 and 6 d.f. The P-value is then converted into −log10(p), which is used in the Manhattan plots.
The genotypic model:
Our interest here is not to estimate the F statistics; rather, we are interested in a statistical test for the difference between HR and C. Therefore, we can use a model that takes individual genotypes as input data. Such a model is called the genotypic model, in which the response variable for each individual mouse is the average of the two allelic values (assuming the entire population only includes two alleles at each locus). If there are more than two alleles in the experimental population, then the biallelic model still applies by treating all nonreference alleles as the “other” allele, as suggested by Weir (1996). Let be the numerically coded genotypic value for the kth individual within the jth line within the ith treatment and it is defined as
| . | (26) |
The genotypic model is
| , | (27) |
where is the residual effect with variance where and are variances defined in the allelic model. Under the mixed model, the expectation of is
| (28) |
and the variance of is
| . | (29) |
This genotypic model has reduced the model size by half and only involves two variance components. Therefore, it is computationally much more efficient than the allelic model. Parameter estimation and significance test are the same as the allelic model, except that the sample size has been reduced by half.
The gene frequency model
Baldwin-Brown, Long, and Thornton’s regularized F test:
Baldwin-Brown et al. (2014) recently developed a regularized t-test for detecting loci responsible for the phenotypic response to artificial selection or in experimentally evolved populations. The square of the regularized t-test is the regularized F test. The test uses arcsine square root-transformed allele frequency data. The test statistic is defined as
| , | (30) |
where
is a coefficient of regularization set by the investigator (0.1 is the default value),
is the number of lines within each treatment,
is the allele frequency of the HR population,
is the allele frequency of the C population,
is the variance of the allele frequencies over the four selected lines,
is the variance of the allele frequencies over the four control lines,
and is the average within treatment variance in allele frequency averaged over the two treatments and over all m loci.
When is set, the method is the usual F test without regularization. The second term in the denominator of the test, borrows information from all loci under investigation. Baldwin-Brown et al. (2014) interpreted as an empirically motivated Bayesian prior on allowable variances in allele frequencies and has the effect of stabilizing the denominator of the F test. They claimed that such a regularization is important in experimental evolution studies in which a SNP could differentially fix in the experimental vs. control replicates purely due to drift alone, and thus be associated with a traditional F test of infinity. Under the null model, the regularized F test follows an F distribution with 1 and d.f.
Regularized F test using linear regression:
The regularized F test can be achieved using a general linear model (regression analysis). The general linear model has an advantage of being able to handle multiple treatments. For example, if there are three selection treatments and multiple replicated lines are available within each treatment, then the regularized F test cannot test the difference among the three selection treatments. In the present study, we extend the regularized F test using a general linear model approach. The response variable (y) is the arc-sine square root-transformed allele frequency with eight observations for the mouse data. The linear model is
| , | (31) |
where is an vector of unity, is the intercept, an vector coded as −1 for C and 1 for HR, is the regression coefficient representing the difference in allele frequencies between the two selection treatments, and is an vector of residual errors with an unknown variance Let be the row bind of and be the column bind of and The estimated parameters are
| (32) |
and
| . | (33) |
Incorporating the regularized parameter, the variance matrix of is
| , | (34) |
where and is the average estimated across all loci in the neighborhood of the current locus or in the entire genome. The variance is a matrix with elements defined as
| (35) |
The regularized F test from this regression analysis is
| . | (36) |
One can verify that is the difference of the allele frequencies between the two selection treatments and is identical to the denominator of Equation 30 if is replaced by the average within-population variance of the current locus.
Permutation test
As the response variable in the mixed model analysis is a binary variable, the F test statistic does not follow the expected F distribution. In addition, multiple tests were involved in the analysis and the nominal 0.05 criterion of Type 1 error for the P-value cannot be used. To control the genome-wide Type 1 error at 0.05, we used the permutation test (Churchill and Doerge 1994) by randomly shuffling the mouse identification numbers so that any association of a locus with the treatment label would be a false positive. For each permuted data set, all 25,318 SNPs were analyzed, and the single largest F statistic was recorded. The permutation was replicated 1000 times and then the 95th percentile of the empirical distribution of F statistics from permuted data were compared with the 25,318 real F tests to determine the significance for each SNP. Any SNPs for which the F test was greater than the 95th percentile of the empirical distribution of F from the permuted data were considered significant at P < 0.05. This procedure thus controls the genome-wide Type 1 error rate at 5%. In the Manhattan plot, we presented the −log(p) test statistics of all loci against the genome positions. The empirical critical value of the F statistic was converted into an empirical critical value of p using d.f. of 1 and 6, which reflects the experimental design with one fixed effect (selection) and four replicate lines (random effects (nested within linetype). That empirical critical value in p was further converted into the empirical critical value in the −log(p) scale. This critical value is sample-specific, and thus is more appropriate than the Bonferroni correction, which is often too conservative (Gao et al. 2010).
In summary, we have presented four methods for detection of selection signatures. The mixed model approach under the allelic model (ALLELIC MODEL), the mixed model under the genotypic model (GENOTYPIC MODEL), the regularized F test using allele frequency (REGULARIZED F TEST), and the regularized F test using regression (REGRESSION F TEST). Except the REGULARIZED F TEST, all other models can handle more than two treatment levels. All four methods were used to analyze the SNP data of the selection experiment. A working example is provided in Supplemental Material, Note S1 in File S1, using data presented in File S8, File S9, and File S10. The R code for each method is provided in Note S2 in File S1. Users familiar with SAS programs can directly call PROC MIXED with the Method = MIVQUE0 option to perform the mixed model analysis. However, if the number of markers is large, looping over all markers in SAS can be extremely slow.
Data availability
All data are included as supplementary files. File S2 shows a marker map of the 25,318 SNPs used in the mouse data analysis. File S3 outlines mouse population information including treatments (0 and 1), lines (1, 2, 4, 5, 3, 6, 7, and 8), mouse ID (1, 2, …, 72), and allele (1 and 2). File S4 presents allelic data of 144 alleles from 72 mice for 25,318 SNP loci, where 1 and 0 represent the presence and absence of the reference allele. File S5 shows genotypic data of 72 mice for 25,318 SNP loci, where each genotypic value takes one of the three values: 0, 0.5, and 1. File S6 gives gene frequencies of eight lines (p1, p2, …, p8) of the mouse population for 25,318 SNP loci, where yi is the count of the reference alleles and ni is the total number of alleles for the ith line. File S7 presents the significant loci and their test statistics, where the column with header “Mixed” shows the 152 significant loci identified by the permutation test of the mixed model procedure. File S8 shows allelic information for SNP UNC2173488 used in the working example. File S9 provides genotypic information for SNP UNC2173488 used in the working example. File S10 outlines gene frequencies of the eight lines for SNP UNC2173488 used in the working example.
Results
Mouse data analysis
The genetic map of 25,318 markers and information about the mouse populations are provided in File S2 and File S3, respectively. The SNP data coded as binary allelic states are provided in File S4. The corresponding SNP data coded as genotypic values are provided in File S5. Each SNP data set has 25,318 rows (one row per marker), but the allelic data set has 144 columns (one column per allele) and the genotypic data set has 72 columns (one column per mouse). The data have no missing values and the number of individuals per line varied due to deletion of eight mice with low-quality SNP callings. The mice in the population information file and the mice in the allelic and genotypic data files are arranged in the same order. The allele frequency data taken by the regularized F test are given in File S6 with 25,318 rows and eight columns (one column per line). The average heterozygosities across loci are 0.1404 for C and 0.1382 for HR, and the difference has a P-value of 0.01 from a paired t-test.
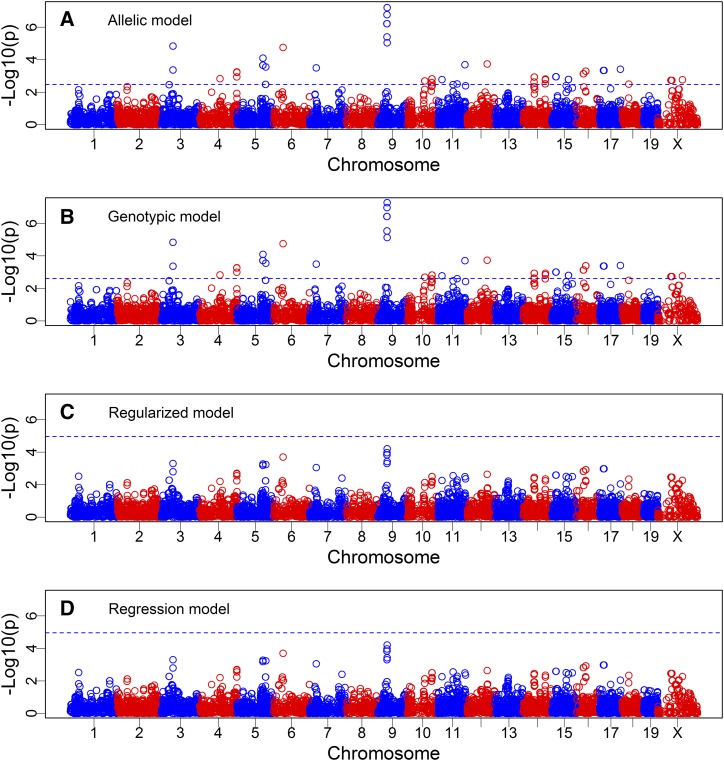
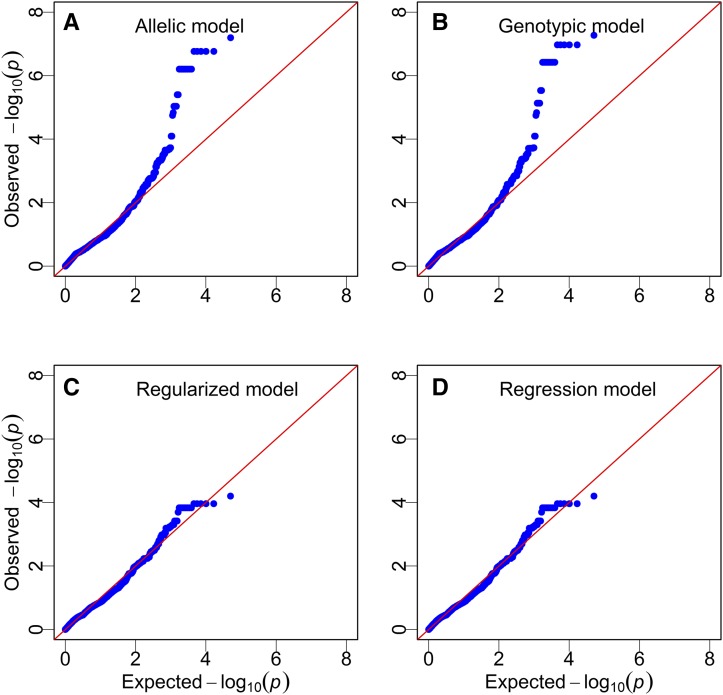
All four approaches described in the Materials and Methods section (allelic model, genotypic model, regularized model, and regression model) were used for the data analysis. The first two methods are mixed model-based methods (new methods) and collectively named MIVQUE(0), while the last two are based on gene frequencies (existing methods) and collectively named REGULAR (regularized F test). The Manhattan plots of the −log(p) test statistics are shown in Figure 1 for all four methods. The critical value of −log10(p) from 1000 permutation analyses is 2.4644 for the allelic model, 2.6405 for the genotypic model, and 4.95 for the two methods using gene frequency data. These critical values are shown in Figure 1 as the blue dashed horizontal lines. The allelic and the genotypic models are visually indistinguishable (Figure 1, A and B). The regularized F test and the regression F test are identical (Figure 1, C and D). Compared to the permutation-generated thresholds, MIVQUE(0) identified 152 markers, but REGULAR failed to identify any markers. The 152 loci and their test statistics are listed in File S7, where the column with header “Mixed” shows the significant loci identified by the permutation test of the mixed model procedure. The more stringent threshold calculated from Bonferroni correction is −log(0.05/25318) = 5.70. If we had used this threshold, MIVQUE(0) would still detect 21 markers in the middle of chromosome 9. These observations imply that MIVQUE(0) is more powerful than REGULAR (see result of simulation studies). Figure 2 shows qq-plots of the four methods, where a qq-plot is the plot of the observed test statistics against the expected test statistic calculated under the null model. The allelic and genotypic models (both are mixed models) behave as expected; the majority of markers fall on the diagonal lines and some markers deviate from the diagonal (Figure 2, A and B). The regularized and regression models (both use frequency data) show that all markers are around the diagonal lines (Figure 2, C and D).
Figure 1.
Manhattan plots of genome-wide selection signals from the mouse selection experiment (Swallow et al. 1998) at generation 61 using four different methods. The top two panels are the mixed model approach under the allelic model (A) and the genotypic model (B). The bottom two panels show the plot from the regularized F test (Baldwin-Brown et al. 2014) (C) and the regression model (D). The dashed horizontal line (blue) is the empirical threshold obtained from analysis of 1000 permuted samples.
Figure 2.
QQ-plots of genome-wide loci of the mouse selection experiment using four different methods. In each qq-plot, the y-axis is the observed test statistic and the x-axis is the expected test statistic under the null model. The upper two panels are the mixed model approach under the allelic model (A) and the genotypic model (B). The lower two panels show the plots from the regularized F test (C) and the regression model (D). Both of the mixed model approaches show more data points deviating from the diagonal lines than the other approaches, thus indicating higher statistical power.
From one permuted sample, we generated Manhattan plots (Figure S1 in File S1) for the four methods. None of the markers shows any extreme values of the test statistic for the mixed models, but many markers show very large test statistics for the frequency models. This explains why the permutation-generated critical values for the frequency models are high. Detail is provided in the Discussion section. For the same permuted sample, we drew qq-plots (Figure S2 in File S1) and observed that the test statistics of the mixed model approaches do not fall on the expected diagonal lines, whereas the frequency models behave as expected. The F tests from the mixed models do not follow the expected F distribution; therefore, if one relied on the standard F distribution, the tests would be too conservative. However, the F tests of the arc-sine square root-transformed frequency data do follow the expected F distribution.
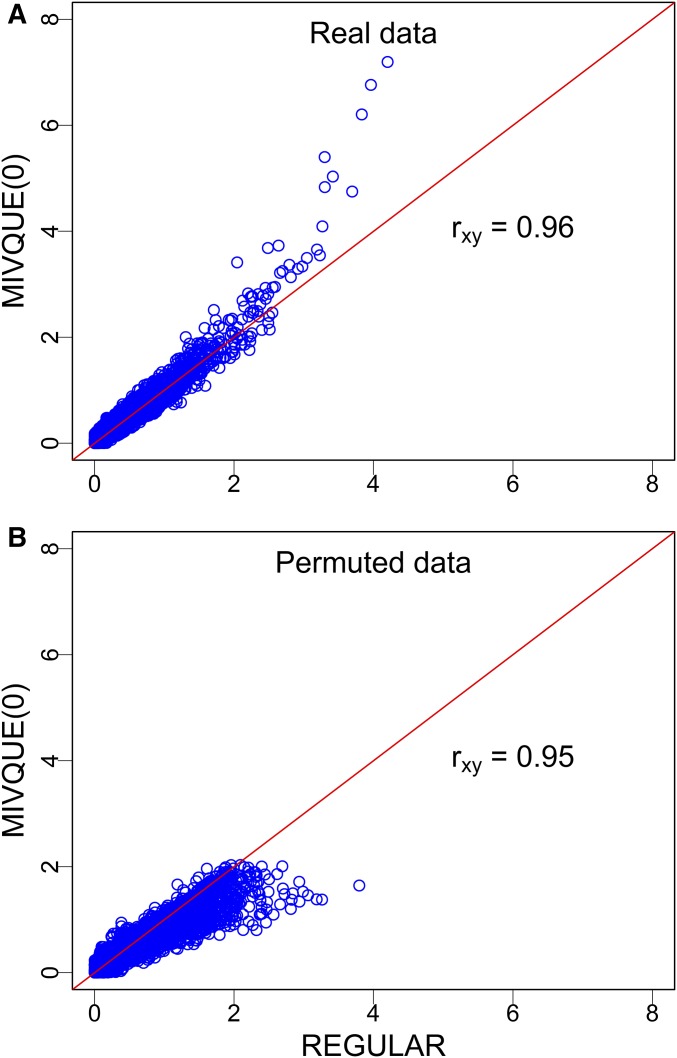
Although the regularized F test failed to identify any markers, the −log10(p) test statistic is highly correlated with that of the mixed model (), as illustrated in Figure 3A, which shows that the test statistic of MIVQUE(0) is higher than that of REGULAR. From a single permuted sample, the correlation is 0.95 (Figure 3B) and REGULAR has a higher statistic than MIVQUE(0) (i.e., the behavior is opposite to the real data analysis). We then selected the top 152 markers from REGULAR to see how many of them overlap with the 152 detected marker from MIVQUE(0). We assume that the top 152 markers from REGULAR are “significant.” We found that 118 markers overlapped (detected by both methods) and 34 markers were uniquely identified by one of the two methods. The 152 + 34 = 186 markers detected by both methods are listed in File S7 along with the test statistics and allele frequencies for each of the eight lines. Except chromosomes 2, 8, 13, and 19, each chromosome (including chromosome X) caries at least one significant marker.
Figure 3.
Comparison of the −log10(p) test statistics of MIVQUE(0) (minimum variance quadratic unbiased estimation) with REGULAR (regularized F test) from the real data analysis (A) and from the analysis of a permuted sample (B). The Pearson correlation coefficients between the test statistics of the two methods are represented by These plots demonstrate that the test statistic of the mixed model is highly correlated with the test statistic of the regularized F test, but the patterns are different for the real data and the permuted data.
The 152 significant markers occurred on all chromosomes except 2, 8, and 19, and show a variety of patterns in terms of fixation (or the lack thereof) in the four HR and four C lines (File S7). Although a number of alleles were fixed within lines, none were fixed between the two selection treatments. For example, marker UNC10025993 on chromosome 5 had frequencies of 1, 1, 0.55, and 1 in HR lines 3, 6, 7, and 8, respectively, vs. zero in all four C lines. In contrast, marker UNC12559756 on chromosome 7 had frequencies of zero in all HR lines vs. 0.45, 0.667, 0.65, and 0.389 in the C lines. Others showed intermediate frequencies, such as UNC24564099 on chromosome 14, with frequencies of 0.1875, 0.2222, 0, and 0.2857 in the HR lines vs. 0.65, 0.5556, 0.75, and 1 in the C lines.
Intuitively, if this region is under selection due to the artificial selection protocol, then populations with such small sizes after 61 generations of selection should have some loci that are completely fixed in all four HR lines. The lack of this pattern for this region may be in part related to the within-family selection scheme (Swallow et al. 1998), which is known to slow the fixation process (Falconer and Mackay 1996).
Interestingly, a total of 21 loci in the middle of chromosome 9 were detected even using the most stringent Bonferroni correction criterion. These loci are within a 901-kb region on chromosome 9. The P-values from the allelic model and the allele frequencies of the eight lines are given in Table 1. Three lines in C and one line in HR were completely fixed in allele frequency for all loci in this region. There appeared to be two recombination breakpoints taking place within this region.
Table 1. Markers detected on chromosome 9 of the mouse genome that show significant differentiation between the C and HR-selected lines.
| Ca | C | C | C | HR | HR | HR | HR | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker (Chr 9) | Position (bp) | P-Value | p1 | p2 | p4 | p5 | p3 | p6 | p7 | p8 |
| UNC16231229 | 41,246,129 | 6.24E−07 | 0 | 0 | 0 | 0 | 0.8125 | 1 | 0.9 | 0.857143 |
| JAX00170437 | 41,266,019 | 6.24E−07 | 0 | 0 | 0 | 0 | 0.8125 | 1 | 0.9 | 0.857143 |
| UNC16231874 | 41,301,221 | 6.24E−07 | 0 | 0 | 0 | 0 | 0.8125 | 1 | 0.9 | 0.857143 |
| UNC16232212 | 41,326,208 | 6.24E−07 | 0 | 0 | 0 | 0 | 0.8125 | 1 | 0.9 | 0.857143 |
| UNC16232585 | 41,353,991 | 6.24E−07 | 0 | 0 | 0 | 0 | 0.8125 | 1 | 0.9 | 0.857143 |
| UNC16232919 | 41,381,162 | 6.24E−07 | 0 | 0 | 0 | 0 | 0.8125 | 1 | 0.9 | 0.857143 |
| JAX00691456 | 41,473,757 | 6.24E−07 | 0 | 0 | 0 | 0 | 0.8125 | 1 | 0.9 | 0.857143 |
| UNC16235286 | 41,547,967 | 6.24E−07 | 0 | 0 | 0 | 0 | 0.8125 | 1 | 0.9 | 0.857143 |
| JAX00170461 | 41,592,916 | 6.24E-07 | 0 | 0 | 0 | 0 | 0.8125 | 1 | 0.9 | 0.857143 |
| UNC16236699 | 41,636,184 | 1.73E−07 | 0 | 0 | 0 | 0 | 0.875 | 1 | 0.9 | 0.857143 |
| UNC16237066 | 41,656,313 | 1.73E−07 | 0 | 0 | 0 | 0 | 0.875 | 1 | 0.9 | 0.857143 |
| UNC16237562 | 41,689,627 | 1.73E−07 | 0 | 0 | 0 | 0 | 0.875 | 1 | 0.9 | 0.857143 |
| UNC16238010 | 41,729,317 | 3.98E−06 | 0 | 0 | 0 | 0.166667 | 0.875 | 1 | 0.9 | 0.857143 |
| UNC16238418 | 41,767,394 | 1.73E−07 | 0 | 0 | 0 | 0 | 0.875 | 1 | 0.9 | 0.857143 |
| UNC16240425 | 41,877,786 | 1.73E−07 | 0 | 0 | 0 | 0 | 0.875 | 1 | 0.9 | 0.857143 |
| UNC16241644 | 41,948,973 | 3.98E−06 | 0 | 0 | 0 | 0.166667 | 0.875 | 1 | 0.9 | 0.857143 |
| UNC16242398 | 41,992,897 | 9.28E−06 | 1 | 1 | 1 | 0.777778 | 0 | 0 | 0 | 0.142857 |
| UNC16242829 | 42,013,727 | 9.28E−06 | 1 | 1 | 1 | 0.777778 | 0 | 0 | 0 | 0.142857 |
| UNC090061659 | 42,067,067 | 9.28E−06 | 1 | 1 | 1 | 0.777778 | 0 | 0 | 0 | 0.142857 |
| UNC16243882 | 42,070,360 | 9.28E−06 | 1 | 1 | 1 | 0.777778 | 0 | 0 | 0 | 0.142857 |
| UNC16244740 | 42,147,771 | 6.38E−08 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0.857143 |
Chr, chromosome; C, control; HR, high running.
The last eight columns are the allele frequencies of the eight lines (four from C and four from HR).
Power analysis from a simple simulation study
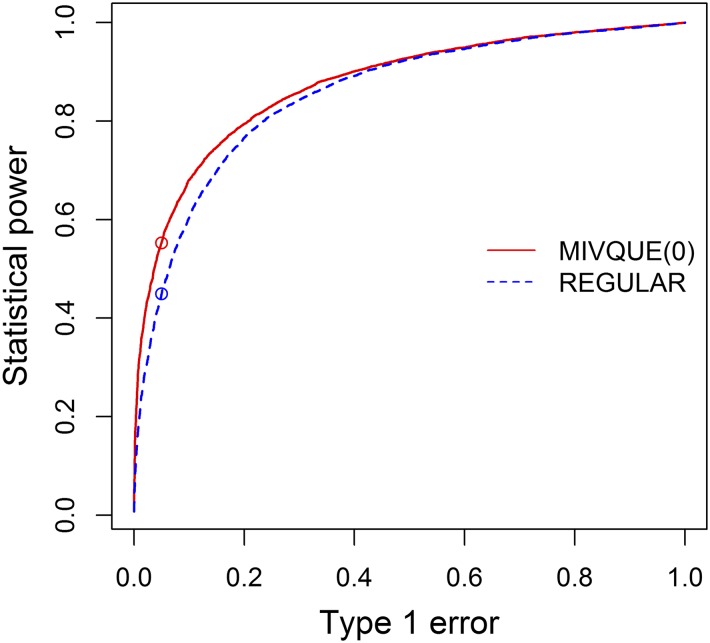
We mimicked the mouse experiment with eight lines and 10 mice in each line to examine the statistical power of the methods. We simulated 10 independently-segregating loci to investigate the powers using 10 independent neutral loci to control the Type 1 error. We used two β distributions to simulate the allele frequencies of the eight lines. For the four control lines, the β distribution was where and leading to an average allele frequency of For the four HR lines, the allele frequencies were generated from where and leading to an average allele frequency of Therefore, the average difference in allele frequency between the HR and C populations was 0.2. Once the allele frequencies were simulated for all lines, we then simulated the allele of each line from a Bernoulli distribution with the simulated allele frequency as the parameter. The actual count data (allele presences) for each line were drawn from a β-Binomial distribution. Such a simulation was replicated 1000 times. The number of loci detected over the total number of loci simulated was the empirical power for the methods compared. The criterion of a locus being detected was determined from another 1000 simulated samples under the null model where the allele frequencies of all lines from the C and HR selection treatments were generated from where The critical value of the −log10(p) test statistics under the Type 1 error of 0.05 from the 1000 null samples was 0.83 for the mixed model and 1.23 for the regularized F test. Based on these critical values, the empirical power was 0.5541 for the mixed model method and 0.4465 for the regularized F test. The new method was indeed more powerful than the regularized F test (see Figure 4). We then changed the Type 1 error and monitored the change of the empirical statistical power from the simulated samples to perform a sensitivity analysis. The receiver operating characteristic curves of the two methods are shown in Figure 4. The curve of the mixed model is consistently higher than that of the regularized F test method, indicating that the power of MIVQUE(0) is always higher than the power of REGULAR F for all levels of Type 1 error.
Figure 4.
Comparison of the receiver operating characteristic (ROC) curves of the mixed model method [MIVQUE(0) (minimum variance quadratic unbiased estimation)] and the regularized F test (REGULAR). The x-axis is the Type 1 error and the y-axis is the statistical power. The curve for the mixed model is consistently higher than that of the regularized F test method, indicating that power of the former is always higher than or equivalent to the power of the latter for all levels of Type I error. Distance (0.5528−0.4495 = 0.1033) between the two points on the plot represents the gain in statistical power of the mixed model (0.5528) over the regularized F test (0.4495) when the Type 1 error is set at 0.05.
It is difficult to relate the 0.55 − 0.45 = 0.10 power increase to the 152 more significant markers detected. The simulations were merely to demonstrate that the MIVQUE(0) method is qualitatively more powerful than the regularized F test, but there is no quantitative comparison. Among the 152 detected markers, we do not know how many are true and how many are false.
Discussion
When the two selection treatments are treated as random effects, there are four variance components for each locus: for treatments (TRT), for lines (LINE) within treatments, for individuals within lines within treatments, and for residuals. We estimated these variance components for all loci and took the ratios to obtain the F statistics for each of the 25,318 loci. We then pooled the variance components over loci and obtained overall F statistics (over all loci) using the following equations (Weir 1996),
| (1) |
| (2) |
| (3) |
| . | (4) |
The four genome-wide F statistics for the mouse populations are , and Thus, the two selection treatments were not differentiated, but the eight lines were significantly differentiated, which may be caused by random genetic drift and possibly also by different adaptive responses, called multiple solutions (Garland et al. 2011a), in the HR lines. The average inbreeding coefficient within lines (0.0316) was very small due to the use of within-family selection scheme.
The regularized F test proposed by Baldwin-Brown et al. (2014) is the state-of-the-art method for the detection of selection signatures in selection experiments with multiple replicated lines. The method is extremely simple, yet performs very well based on their simulation studies. The key issues addressed in that study are (i) replications and (ii) regularization. (i) Replications mean that there must be replicated lines within each selection treatment to separate the effect of selection from genetic drift. However, replications per se rarely happen in natural populations [for notable exceptions, both involving natural parallel adaptations of fish, see Rogers and Bernatchez (2005) and Jones et al. (2012)], and thus detection of selection signatures from natural populations is more difficult because of the confounding effect between selection, possible multiple solutions, and drift (Muir 1986). (ii) Regularization refers to a process in which a small positive number is added to the denominator of the F test statistic. Regularization is an intelligent way to deal with a special case where the within-population variances of allele frequencies are extremely small (e.g., due to drift), so that the F test is severely inflated even if the difference in allele frequency between selection treatments is small. The regularized F test borrows the average within-treatment variance from other loci and incorporates it into the within-treatment variance of the current locus to smooth the test statistics and thus prevents such an inflation in test statistics. The regularization procedure can also prevent reckless changes in test statistic between consecutive loci.
Of the 25,318 loci analyzed in the mouse data, 69 have allele frequency of exactly 0.5 for each of the eight lines. The usual F test (without regularization) statistic is not defined for these loci because the denominator is zero. The fact that the numerator of the test for these loci is also zero means that the test statistics should be zero (the two selection treatments are not different in allele frequency). The regularized F test correctly gives a zero test statistic value for all the 69 loci. Another example comes from marker UNC30702889 on chromosome X. The allele frequencies of the four C lines are 0.45, 0.4444, 0.45, and 0.4444, while the allele frequencies of the four HR lines are all 0.5. Although the difference in allele frequency between C and HR is very small (∼0.05), the unregularized F test is 1075.95 with a P-value of 5.36E−08 and a −log10(p) = 7.2712, which is the highest test value across the entire genome. This test statistic is severely inflated due to the extremely small variance within treatments. However, the regularized F test gives a test statistic of 0.2012 with a P-value of 0.6695 and a −log10(p) of 0.1743. Thus, as desired, the regularization factor has corrected such an inflation.
The most obvious advantage of the regularized F test is that it takes pooled DNA samples as input data. Each pooled DNA sample represents a replicate line within a given selection treatment. For the eight replicate lines in the mouse selection experiment, only eight pooled samples are required to perform tests. This represents a tremendous cost saving. Unfortunately, such an advantage can turn into a disadvantage if DNAs are sequenced at the individual level because this F test cannot handle allelic data. Clearly, if all individuals are sequenced, and individual variation within lines exists, then pooling the DNA samples will lead to information loss. This is the very reason for us to develop the mixed model approach when DNAs from multiple individuals are separately sequenced in a selection experiment.
The higher empirical threshold value for the test statistic of the regularized F test caused the lower power of this method compared with MIVQUE(0). Here is a tentative explanation for the high critical value. The regularized F test is very sensitive to the variance of allele frequencies between replicates within treatment, defined in the text, where is the variance of the allele frequencies of the four replicates in the control and is the variance of the allele frequencies of the four replicates in the treatment. In the original data (25,318 loci), the average value of across loci was 0.1389, but it was 0.0144 in a randomly permuted sample. Permutation randomly shuffled the individual labels so that an individual from one subpopulation could shift to another subpopulation. This is why the variance between replicates within treatments (control and selection) has shrunken so much. Since this (and only this) variance appeared in the denominator of the regularized F test statistic, its reduction has increased the test statistics genome-wide. This explains why the test statistic under the null model becomes high for the regularized F test. However, the MIVQUE(0) method implicitly uses two (genotypic model) or three (allelic model) variance components in the test statistic. For example, in the genotypic model, the two variance components are and Both variance components are involved in the test statistic in the MIVQUE(0) method. For example, in the original data analysis for the MIVQUE(0) method, the average across loci is 0.1203 and the average across loci is 0.0323, leading the sum of the two of 0.1526. For the particular permuted sample presented in the text, the corresponding values are 0.0017 and 0.1351, respectively, with a sum of 0.1368. The sums of the two variances in the original sample and the permuted sample are very close to each other. Therefore, the critical value for the MIVQUE(0) method is not sensitive to because both and contribute to the test statistic.
The regularized F test in the current form can only test the difference in allele frequency between two treatment levels because it is a squared t-test, which is only suitable for comparing two groups. We have extended this method to handle multiple treatment levels using a general linear model approach (regression method). When applied to two treatments, the regularized regression method and the regularized t square method generate identical results (see Figure 1, C and D). The regression method has an option to incorporate the sample size information of replicated lines into the model. For example, the sample sizes (n) were 10, 9, 10, 9, 8, 9, 10, and 7, respectively, for lines 1, 2, 4, and 5 (C) and 3, 6, 7, and 8 (HR). Such information can be easily incorporated into the regression model through a weight variable that is defined as the total number of alleles (two times the sample size) of that line. The exact weight value for each line should be the inverse of pq/(2n). However, when or the weight is infinity. Therefore, simply using as the weight is justifiable. The regularized regression analysis conducted here is not the weighted method because we wanted to demonstrate the equivalence of this method to the regularized t square test.
Current DNA sequencing technology is sufficiently inexpensive so that sequencing can be easily conducted at the individual level. When individuals are sequenced, pooling DNA sequences of all individuals within a line may represent a tremendous information loss. In this study, the difference between selection treatments is treated as a fixed effect, and effects of replicate lines within treatments are treated as random. There are two versions of the mixed models: the allelic model and the genotypic model. The allelic model is the classical model of Weir and Cockerham (1984), where each entry of the response variable is an allele. The hierarchical structure of the alleles is preserved and such a test captures maximum information from the populations. The genotypic model simply takes the “allele frequencies of individuals” as the response variable. Given that every diploid individual only carries two alleles, the “allele frequency” of an individual only takes three possible values: 0, 0.5, and 1. No information is lost by pooling the two alleles of each individual together. Therefore, the genotypic model generates identical results as the allelic model (see Figure 1, A and D). The genotypic model is computationally much more effective than the allelic model because the number of entries has been reduced to half. Hence, the genotypic model is recommended for GWAS for selection signatures.
An interesting feature of the mixed model approach (both the allelic and genotypic models) is that no regularization is required in the test. For example, the SNP named UNC30702889 on chromosome X discussed early in this section requires regularization for the F test because the within-treatment variance is too small. However, the allelic model without any regularization gives a test statistic of 0.2134, a P-value of 0.6604, and a −log10(p) of 0.1802, which are comparable to the regularized F test.
The fact that the response variable of the mixed model analysis is the allelic state (binary) may challenge the validity of the mixed model methodology and lead someone to think that a GLMM may be more appropriate. However, there are two justifications for the current mixed model methodology. (1) When the response variable is the allelic state (binary variable), different variance components and variance ratios have special biological meanings, covariance and correlation between alleles at different levels of the hierarchy (various types of inbreeding coefficients). Such a treatment also preserves the original natures of Wright’s F statistics. (2) The mixed model analysis with the allelic state as the response variable is computationally more effective than the GLMM analysis, which requires iterations and often faces convergence issues. If the purpose of the analysis is just to test the difference between two populations, the GLMM analysis may be considered if computational complexity is not a concern. Following the suggestion from one reviewer, we did perform GLMM analysis with the logit link function and the binary distribution using both the glmer() function in the lme4 package of R and the GLIMMIX procedure of SAS Institute Inc. (2009). The likelihood function under GLMM was approximated using the Laplace algorithm in both R and SAS. Manhattan and qq-plots of the GLMM analyses are shown in Figure S3 and Figure S4, respectively, in File S1. To our surprise, the GLMM results are not very appealing. The bad news is that the results of SAS and R are not consistent. The worst news is that, of all the 25318 loci, the programs failed to converge for 4405 loci. The SAS and R do have one thing in common, they all failed for exactly the same 4405 loci. In addition, both R and SAS were extremely slow for the GLMM analysis. For example, it took over 2 hr to scan all 25318 loci for R and half a day to complete the analysis for SAS. With such a slow speed, it was too difficult to perform permutation analysis with 1000 shuffled samples to draw empirical critical values for the test statistics. We examined some of the loci that failed to converge and found that many of them have a pattern like “all 72 individuals, except one or two, are heterozygotes.” These loci typically would fail in the t-test and thus require the regularized t-test to generate a meaningful result. However, the MIVQUE(0) method developed here worked smoothly without any problems for these loci. We set the test statistics of these problematic loci to zero and drew Manhattan plots for GLMM(R) and GLMM(SAS) along with MIVQUE(0) and REGUALR for comparison (Figure S3 in File S1). If the critical value from MIVQUE(0) were used here, GLMM(R) would detect seven significant loci, one of which is the major one detected by MIVQUE(0) on chromosome 9 (C of Figure S3 in File S1). The remaining six loci have all been detected by MIVQUE(0). Using the same critical value, GLMM(SAS) would detect 137 significant loci (D of Figure S3 in File S1). Some of the loci overlap with the ones detected by MIVQUE(0), but others do not. Interestingly, the one on chromosome 9 was also detected. Since the critical values are not known, we are not certain about these detected loci by the GLMM methods. If the Bonferroni-corrected threshold (5.7) were used, GLMM(R) would detect none but GLMM(SAS) would detect four loci (including the one on chromosome 9). We also drew qq-plots for the GLMM analyses in comparison with MIVQUE(0) and REGULAR (Figure S4 in File S1). Contrary to our common belief, the test statistics of GLMM(R) and GLMM(SAS) do not show the usual behaviors of test statistics observed in MIVQUE(0) and REGULAR. No points of the qq-plots for the GLMM methods fall on the diagonal line, indicating that the test statistics do not have the expected F distribution, probably due to the failure in convergence of many SNPs for the GLMM methods. The GLMM methods perhaps require large samples to generate meaningful results. The sample sizes in experimental evolutions are often too small to meet the requirement.
Although GLMM automatically complies with the natural boundaries in the data [0,1], out of boundaries for the proposed MIVQUE(0) is not our concern because we do not predict the response variable. The estimated variance components have no such boundaries except that they must not be negative. The MIVQUE(0) method is equivalent to the ANOVA for estimation of variance components when the data are balanced (Rao 1971a,b). The ANOVA with binary allelic indicator as the response variable has been used for more than half a century (Cockerham 1969), and has never been questioned for its validity. We originally hoped to show some advantages of GLMM over the seemingly ad hoc MIVQUE(0). Unfortunately, we failed to demonstrate any advantages of GLMM over MIVQUE(0). We would be surprised if other people had not tried to use GLMM to analyze allelic data. They might have found the same problems as we did here and just never reported the undesirable results.
The mixed model analysis of GWAS studies for selection signature detection is similar to the GWAS for quantitative trait analysis (Hirschhorn and Daly 2005; Yu et al. 2006), except that there is no specific trait associated with the genetic analysis. Therefore, this method is also called GWAS without traits (Lo et al. 2016). Unlike the regular quantitative trait GWAS, where we can control the polygenic background by incorporating a marker-inferred kinship matrix into the covariance structure, GWAS for selection signature detection does not have an obvious way to control the polygenic background. Therefore, the Type 1 error may not be controlled properly. To mimic GWAS in quantitative trait analysis, we may treat the population structure as the response variable and the allelic state as an independent variable. This treatment may be easily modified to incorporate the “polygenic effect” into the model, just like the regular mixed model GWAS (Yu et al. 2006). It is straightforward to do so if there are only two populations, where the response variable is binary. For multiple populations, a multinomial response may be used to indicate the population entries. However, hierarchical population structures may not be easily handled this way. GWAS and QTL mapping for selection signatures is a relatively new area, with large room for improvement. The present study is one of the first attempts to merge quantitative genetics, selection experiments, and genetic information in the genomic era [for other examples with rodent models, see Chan et al. (201), Ren et al. (2013), Konczal et al. (2016), and Lo et al. (2016)]. We have adopted the mixed model in our selection signature detection but have not yet incorporated the kinship matrix into the selection model. A complete unification of GWAS and selection is possible but still has a long way to go. In future studies, it will also be important to identify the presumably smaller number of haplotypes that contain the statistically significantly differentiated SNPs analyzed herein, but doing so accurately will likely require whole-genome sequence data.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300102/-/DC1.
Acknowledgments
We thank Fernando Pardo-Manuel de Villena, Daniel Pomp, and Liran Yadgary for providing the data analyzed here and for many useful discussions. We also thank Layla Hiramatsu and David Hillis for helpful discussions, and two anonymous reviewers for comments on the manuscript. This work was supported by a National Science Foundation (NSF) Collaborative Research grant (DBI-1458515) to S.X. and an NSF grant IOS-1121273 to T.G.
Footnotes
Communicating editor: J. Wolf
Literature Cited
- Balding D. J., Nichols R. A., 1995. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 96: 3–12. [DOI] [PubMed] [Google Scholar]
- Baldwin-Brown J. G., Long A. D., Thornton K. R., 2014. The power to detect quantitative trait loci using resequenced, experimentally evolved populations of diploid, sexual organisms. Mol. Biol. Evol. 31: 1040–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis W. D., 1994. The power and deceit of QTL experiments: lessons from comparative QTL studies, pp. 250–266 in Proceedings of the Forty-Ninth Annual Corn & Sorghum Industry Research Conference American Seed Trade Association, Washington, DC. [Google Scholar]
- Brookfield J. F. Y., 2001. Population genetics: the signature of selection. Curr. Biol. 11: R388–R390. [DOI] [PubMed] [Google Scholar]
- Careau V., Wolak M. E., Carter P. A., Garland T., Jr, 2013. Limits to behavioral evolution: the quantitative genetics of a complex trait under directional selection. Evolution 67: 3102–3119. [DOI] [PubMed] [Google Scholar]
- Carter P. A., Garland T., Jr., Dohm M. R., Hayes J. P., 1999. Genetic variation and correlations between genotype and locomotor physiology in outbred laboratory house mice (Mus domesticus). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 123: 155–162. [DOI] [PubMed] [Google Scholar]
- Chan Y. F., Jones F. C., McConnell E., Bryk J., Bunger L., et al. , 2012. Parallel selection mapping using artificially selected mice reveals body weight control loci. Curr. Biol. 22: 794–800. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham C. C., 1969. Variance of gene frequencies. Evolution 23: 72–84. [DOI] [PubMed] [Google Scholar]
- Cui Y., Zhang F., Xu J., Li Z., Xu S., 2015. Mapping quantitative trait loci in selected breeding populations: a segregation distortion approach. Heredity 115: 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion J. P., Morgan A. P., Yadgary L., Bell T. A., McMullan R. C., et al. , 2016. R2d2 drives selfish sweeps in the house mouse. Mol. Biol. Evol. 33: 1381–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Pearson, Harlow, England. [Google Scholar]
- Franssen S. U., Kofler R., Schlötterer C., 2017. Uncovering the genetic signature of quantitative trait evolution with replicated time series data. Heredity 118: 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Becker L. C., Becker D. M., Starmer J. D., Province M. A., 2010. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet. Epidemiol. 34: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T., Rose M. R., 2009. Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments. University of California Press; Berkeley, CA. [Google Scholar]
- Garland T., Jr., Kelly S. A., Malisch J. L., Kolb E. M., Hannon R. M., et al. , 2011a How to run far: multiple solutions and sex-specific responses to selective breeding for high voluntary activity levels. Proc. Biol. Sci. 278: 574–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T., Jr, Schutz H., Chappell M. A., Keeney B. K., Meek T. H., et al. , 2011b The biological control of voluntary exercise, spontaneous physical activity and daily energy expenditure in relation to obesity: human and rodent perspectives. J. Exp. Biol. 214: 206–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschka T. S., Mirand E. A., 1973. The “Breeder: HA(ICR)” Swiss mouse, a multipurpose stock selected for fecundity, pp. 319–331 in Perspectives in Cancer Research and Treatment, edited by Murphy G. P., Pressman D., Mirand E. A. Alan R. Riss, New York, NY. [Google Scholar]
- Hirschhorn J. N., Daly M. J., 2005. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6: 95–108. [DOI] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konczal M., Koteja P., Orlowska-Feuer P., Radwan J., Sadowska E. T., et al. , 2016. Genomic response to selection for predatory behavior in a mammalian model of adaptive radiation. Mol. Biol. Evol. 33: 2429–2440. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C. L., Lossie A. C., Liang T., Liu Y., Xuei X., et al. , 2016. High resolution genomic scans reveal genetic architecture controlling alcohol preference in bidirectionally selected rat model. PLoS Genet. 12: e1006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Xu S., 2003. Mapping viability loci using molecular markers. Heredity 90: 459–467. [DOI] [PubMed] [Google Scholar]
- Luo L., Zhang Y. M., Xu S., 2005. A quantitative genetics model for viability selection. Heredity 94: 347–355. [DOI] [PubMed] [Google Scholar]
- Mantel N., 1963. Chi-square tests with one degree of freedom, extensions of the Mantel–Haenszel procedure. J. Am. Stat. Assoc. 58: 690–700. [Google Scholar]
- Morgan A. P., Fu C. P., Kao C. Y., Welsh C. E., Didion J. P., et al. , 2016. The mouse universal genotyping array: from substrains to subspecies. G3 (Bethesda) 6: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir W. M., 1986. Estimation of response to selection and utilization of control populations for additional information and accuracy. Biometrics 42: 381–391. [PubMed] [Google Scholar]
- Rao C. R., 1971a Estimation of variance and covariance components-MINQUE theory. J. Multivariate Anal. 1: 257–275. [Google Scholar]
- Rao C. R., 1971b Minimum variance quadratic unbiased estimation of variance components. J. Multivariate Anal. 1: 445–456. [Google Scholar]
- Ren Y., Overmyer K. A., Qi N. R., Treutelaar M. K., Heckenkamp L., et al. , 2013. Genetic analysis of a rat model of aerobic capacity and metabolic fitness. PLoS One 8: e77588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J. S., Kawecki T. J., 2009. Behavior and neurobiology, pp. 263–300 in Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments, edited by Garland T., Jr, Rose M. R. University of California Press, Berkeley, CA. [Google Scholar]
- Rogers S. M., Bernatchez L., 2005. Integrating QTL mapping and genome scans towards the characterization of candidate loci under parallel selection in the lake whitefish (Coregonus clupeaformis). Mol. Ecol. 14: 351–361. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc. , 2009. SAS/STAT: Users’ Guide, Version 9.3. SAS Institute Inc., Cary, NC. [Google Scholar]
- Schlotterer C., Kofler R., Versace E., Tobler R., Franssen S. U., 2015. Combining experimental evolution with next-generation sequencing: a powerful tool to study adaptation from standing genetic variation. Heredity 114: 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow J. G., Carter P. A., Garland T., Jr, 1998. Artificial selection for increased wheel-running behavior in housemice. Behav. Genet. 28: 227–237. [DOI] [PubMed] [Google Scholar]
- Swallow J. G., Hayes J. P., Koteja P., Garland T., Jr, 2009. Selection experiments and experimental evolution of performance and physiology, pp. 301–351 in Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments, edited by Garland Jr. T., Rose M. R. University of California Press, Berkeley, CA. [Google Scholar]
- Turner T. L., Stewart A. D., Fields A. T., Rice W. R., Tarone A. M., 2011. Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genet. 7: e1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl C., Xu S. Z., 2000. Multipoint mapping of viability and segregation distorting loci using molecular markers. Genetics 155: 1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace I. J., Garland T., Jr, 2016. Mobility as an emergent property of biological organization: insights from experimental evolution. Evol. Anthropol. 25: 98–104. [DOI] [PubMed] [Google Scholar]
- Weir B. S., 1996. Genetic Data Analysis II - Methods for Discrete Population Genetic Data. Sinauer Associates, Inc. Publishers, Synderland, MA. [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wright S., 1950. Genetical structure of populations. Nature 166: 247–249. [DOI] [PubMed] [Google Scholar]
- Wright S., 1951. The genetic structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Wurschum T., 2012. Mapping QTL for agronomic traits in breeding populations. Theor. Appl. Genet. 125: 201–210. [DOI] [PubMed] [Google Scholar]
- Xu S., 1996. Mapping quantitative trait loci using four-way crosses. Genet. Res. 68: 175–181. [Google Scholar]
- Xu S., 2003. Theoretical basis of the Beavis effect. Genetics 165: 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R.-C., 1998. Estimating hierarchical F-statistics. Evolution 52: 950–956. [DOI] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Vroh Bi I., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included as supplementary files. File S2 shows a marker map of the 25,318 SNPs used in the mouse data analysis. File S3 outlines mouse population information including treatments (0 and 1), lines (1, 2, 4, 5, 3, 6, 7, and 8), mouse ID (1, 2, …, 72), and allele (1 and 2). File S4 presents allelic data of 144 alleles from 72 mice for 25,318 SNP loci, where 1 and 0 represent the presence and absence of the reference allele. File S5 shows genotypic data of 72 mice for 25,318 SNP loci, where each genotypic value takes one of the three values: 0, 0.5, and 1. File S6 gives gene frequencies of eight lines (p1, p2, …, p8) of the mouse population for 25,318 SNP loci, where yi is the count of the reference alleles and ni is the total number of alleles for the ith line. File S7 presents the significant loci and their test statistics, where the column with header “Mixed” shows the 152 significant loci identified by the permutation test of the mixed model procedure. File S8 shows allelic information for SNP UNC2173488 used in the working example. File S9 provides genotypic information for SNP UNC2173488 used in the working example. File S10 outlines gene frequencies of the eight lines for SNP UNC2173488 used in the working example.