Abstract
Risk stratification of acute myeloid leukemia (AML) patients needs improvement. Several AML risk classification models based on somatic mutations or gene-expression profiling have been proposed. However, systematic and independent validation of these models is required for future clinical implementation. We performed whole-transcriptome RNA-sequencing and panel-based deep DNA sequencing of 23 genes in 274 intensively treated AML patients (Clinseq-AML). We also utilized the The Cancer Genome Atlas (TCGA)-AML study (N=142) as a second validation cohort. We evaluated six previously proposed molecular-based models for AML risk stratification and two revised risk classification systems combining molecular- and clinical data. Risk groups stratified by five out of six models showed different overall survival in cytogenetic normal-AML patients in the Clinseq-AML cohort (P-value<0.05; concordance index >0.5). Risk classification systems integrating mutational or gene-expression data were found to add prognostic value to the current European Leukemia Net (ELN) risk classification. The prognostic value varied between models and across cohorts, highlighting the importance of independent validation to establish evidence of efficacy and general applicability. All but one model replicated in the Clinseq-AML cohort, indicating the potential for molecular-based AML risk models. Risk classification based on a combination of molecular and clinical data holds promise for improved AML patient stratification in the future.
Introduction
The cytogenetic karyotype is the most important prognostic factor in acute myeloid leukemia (AML) and forms the basis of the current risk classification in the disease.1 However, 40–45% of all AML patients are cytogenetic normal (CN)2, 3 and belong in the intermediate cytogenetic risk group, where prognostic stratification needs to be improved for clinical decision-making. Improved risk stratification will provide information that has the potential to lead to improved therapy decisions and outcomes.
In the last decade, many individual genes as well as biomarker panels based on multiple somatic mutations or expression levels of multiple genes have been proposed to facilitate improved prognostic risk classification. Individual somatic alterations with prognostic significance that have been reported include NPM1, CEBPA, FLT3, IDH1, IDH2, KIT, WT1 and RUNX1.4 However, in current clinical routine only aberrations in NPM1, CEBPA and FLT3-ITD are currently broadly utilized, these aberrations are also part of the European Leukemia Net (ELN) risk classification system.5 Multiple studies have proposed novel biomarker panels and predication models aimed at improving AML risk stratification. Patel et al.6 proposed a panel of somatic mutations for risk stratification based on sequencing of a set of 18 genes. Recently, Papaemmanuil et al.7 suggested a new molecular-based classification system based on mutation panel of 76 genes in a large cohort (N=1540). Several studies have characterized gene expression for stratification of intermediate risk group of AML or CN-AML. Bullinger et al.8 developed a model for stratification of CN-AML patients based on clustering of gene-expression profiles. Metzeler’s 86-probe-set,9 Li’s 24-gene10 and Marcucci’s 7-gene11 signatures were derived by applying supervised modeling methodologies. Eppert et al.12 proposed leukemia stem cells related (LSCR) and hematopoietic stem cells related (HSCR) gene signatures based on a stem cell model. These models were proposed to have prognostic value in their respective studies (Table 1).
Table 1. Molecular markers associated with AML prognosis.
| Gene set | Methods | Significance | Clinical subtype | No. of patients |
|---|---|---|---|---|
| Papaemmanuil-mutation7 | DNA sequencing of 76 genes or regions | Propose molecular-based classification | All | 1540 |
| Marcucci-7-gene11 | Microarray-based GEP; genes with promoter DMRs and expression levels significantly associated with OS | Define subgroups in CN-AML that are associated with disease prognosis | CN-AML | Training: 126 VS1 (validation): 72 VS2 (validation): 134 VS3 (validation): 65 VS4 (validation): 84 |
| Li-24-gene10 | Microarray-based GEP; top genes associated with OS from meta-analysis of four cohorts | Define subgroups that are associated with disease prognosis, and improve ELN risk classification | All | USA-Set-1 (training): 65 USA-Set-2 (training): 87 Netherlands-Set-1 (training): 241 Germany-Set-1 (training): 106 Netherlands-Set-2 (validation): 277 Germany-Set-2 (validation): 548 |
| Eppert-LSCR and HSCR12 | Microarray-based GEP; LSCR and HSCR gene profile generated from cancer stem cells model | Define subgroups in CN-AML that are associated with disease prognosis | CN-AML | AMLCG-1999: 160 |
| Patel-mutation6 | DNA sequencing of 18 genes | Define subgroups that are associated with disease prognosis | All | Test cohort: 398 Validation cohort: 104 |
| Metzeler-86-probe9 | Microarray-based GEP; supervised principal components by OS | Define subgroups in CN-AML that are associated with disease prognosis | CN-AML | Training cohort: 163 Test cohort: 79 Validation cohort: 64 |
| Bullinger-133-gene8 | Microarray-based GEP; unsupervised clustering | Define subgroups in CN-AML that are associated with disease prognosis. It was validated in an independent study23 | CN-AML | Training set: 59 Test set: 57 Test set: 22 Radmacher et al. 2006: 64 |
Abbreviations: CN-AML, cytogenetic normal-acute myeloid leukemi; DMR, DNA methylation region; ELN, European Leukemia Net; GEP, gene expression profiling; HSCR, hematopoietic stem cells related; LSCR, leukemia stem cells related; OS, overall survival.
Clinical implementation of an improved AML risk classification model has the potential to provide a significant advancement in prognostication, and to improve outcomes for AML patients through refined patient stratification and more relevant information in clinical decision making. Although previously reported studies have indicated promising prognostic results, surprisingly, neither of these biomarker panels and models have undergone systematic and independent validation to provide further evidence of their effectiveness.
In this study, we evaluate and attempt to replicate six of the most promising molecular AML risk stratification models that have been proposed to date (Table 1) in two independent cohorts, in-house Clinseq-AML study (N=274) and The Cancer Genome Atlas (TCGA) AML (N=142) study. All of the six models evaluated are based on molecular biomarkers and some of the models also integrate cytogenetic information. These studies share the common aim of improving AML prognostication and risk stratification. Our primary objective is to investigate to what extent these previously proposed models can be replicated and thereby establish independent evidence of their effectiveness and potential for future clinical implementation.
Patients and methods
Patients
Bone marrow or peripheral blood samples were obtained at the time of diagnosis from 274 AML patients of the Clinseq-AML cohort treated in Sweden between February 1997 and August 2014. Samples were separated for mononuclear cells and stored in isothermal liquid nitrogen freezers at −180 °C. All patients were treated with intensive induction regimens, including anthracyclines and cytosine arabinoside, according to national guidelines.13 Clinical data was retrieved from the Swedish Adult Acute Leukemia Registry3 or from patient records. The regional ethical review board in Stockholm, Sweden approved the study.
The TCGA study includes 142 AML patients with intensive induction treatment. Clinical and mutational data were retrieved from the data portal of TCGA (https://gdc.cancer.gov) and Supplementary Table S1 of the publication of the TCGA-AML study.14
Cytogenetic aberrations
Karyotype was available on 261 out of 274 patients in the Clinseq-AML cohort and 140 out of 142 patients in the TCGA-AML cohort. Cytogenetic risk classification was assigned following the Medical Research Council criteria.2 The ELN risk classification was also assigned by the ELN classification system using cytogenetics and mutational status of NPM1, FLT3-ITD and CEBPA.5 The Intermediate-I and –II groups were combined since the difference is minor and the number of patients in the intermediate-II groups is limited.
Sample and library preparation, sequencing and bioinformatics processing
Transcriptomic RNA and a somatic mutation panel of genes were sequenced. Details about sample preparation and bioinformatics processing were described in Supplementary Methods. The average depth of DNA sequencing was 360X. The median reads of RNA-sequence was 33 million. For RNA-sequencing data, after processing, genes with zero count in either the Clinseq-AML or the TCGA-AML data set were excluded. Thus, 18 744 genes remained for further statistical analyses. For DNA sequencing, 23 genes were manually curated for the identification of somatic variants relevant for prognostication (Supplementary Tables S14 and S15). Normalized RNA sequencing data is open assessable (https://doi.org/10.5281/zenodo.292986).
Assessment of prognostic model performance
In this study, we applied six models developed for prognostic risk stratification in patients with CN-AML in the Clinseq and TCGA cohorts. To assess the performance of the six prediction models, we used the concordance index (C-index)15, 16 of the prognostic score and the hazard ratio of overall survival between dichotomized risk groups in Cox proportional hazards regression model.17 Overall survival was measured as the date of diagnosis to the date of death from any cause, and patients alive at last follow-up were censored.
The prognostic score for each model was derived following the methods described in their respective original study (Supplementary Methods). The C-index of the prognostics score in each model was calculated to assess the ability of discrimination on predicting the overall survival. C-index is the most commonly used measurement of discrimination, which is the probability that a randomly selected person with the event will have a higher predicted risk than a randomly selected person without the event.18 A C-index of 0.5 means no discrimination, 1 means perfect discrimination. The C-index and confidence interval (CI) were calculated using R package survcomp.19
Under each model, the patients were further stratified to high- and low-risk groups based on the prognostic score according to the original study. Kaplan–Meier curves were used to visualize the probability of survival outcomes over time in each group. Univariable and multivariable Cox proportional hazards regression models were fitted at time-on scale. Variables included in the multivariable cox-regression model were age (dichotomized at 60 years), sex, etiology (de novo, secondary or therapy-related AML), and mutational status of NPM1, FLT3-ITD and CEBPA. Proportional hazards assumptions were checked using Schoenfeld residuals. The survival analysis was conducted by R package survival.20
Results
Previously published prognostic biomarker panels for AML
To review published studies on gene signatures developed to predict prognosis in AML, a systematic search was conducted (details are described in Supplementary Methods). Seven studies, including five studies reporting gene-expression-based biomarker panels and associated models, and two studies using mutational panel, were identified and selected for replication (Table 1). Marcucci-7-gene, Eppert-LSCR and HSCR, Metzeler-86-probe and Bullinger-133-gene were aimed at stratification of CN-AML, while Li-24-gene and Patel-mutation panel were proposed to improve upon the current cytogenetic-based risk classification system. Each model was implemented as described in the original study (for details, see Supplementary Methods). The gene IDs of the gene-expression-based signatures are listed in Supplementary Tables S7–S12.
The AML cohorts
The clinical and molecular characteristics of the patients in the Clinseq-AML and TCGA-AML cohorts are described in Supplementary Table S1. Among the 274 patients with intensive induction treatment in the Clinseq-AML cohort, 222 were de novo AML, 24 secondary AML and 26 therapy-related AML. The distribution of cytogenetic aberrations was similar to previously published population-based data in Sweden.3 We performed mutational analyzes of 23 genes (Supplementary Table S1). Somatic alterations were identified in 94.5% of the patients. The frequencies of somatic mutations are summarized in Supplementary Table S1 and plotted in Supplementary Figure 2. The characteristics of the patients in TCGA-AML cohort were described in their original report.14 In order to compare the cohorts used in the prognostic models, only the 142 patients with intensive induction treatment were selected from TCGA-AML. The frequencies of mutations were compared between the Clinseq-AML and the TCGA-AML in Supplementary Figure 1. To note, all patients in the TCGA-AML cohort were de novo AML, which contrasts to the Clinseq-AML cohort that represents unselected consecutive AML patients. The median follow-up time was 346.5 days in the Clinseq-AML cohort and 455.5 days in the TCGA-AML cohort (Supplementary Table S1).
Replication results across risk stratification models in CN-AML
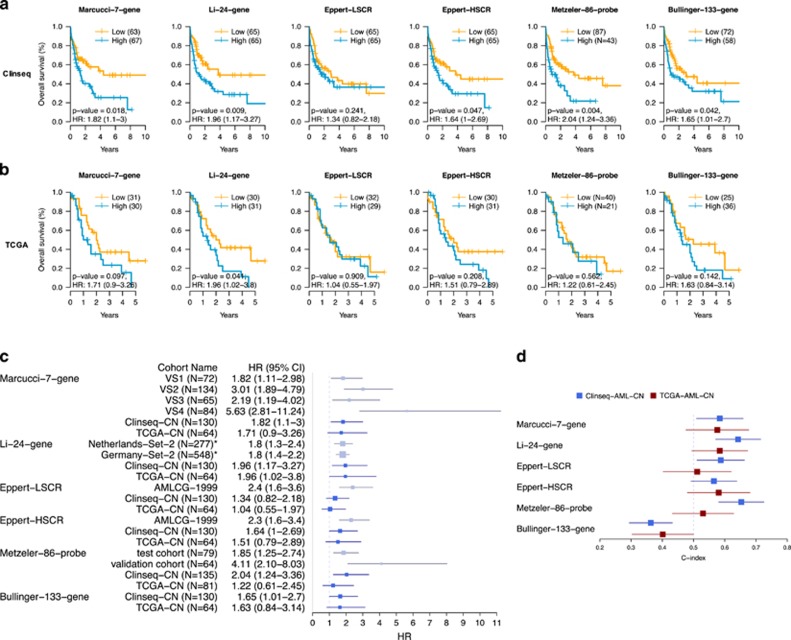
In order to evaluate the prognostic ability of each gene signature (Marcucci-7-gene, Li-24-gene, Eppert-LSCR, Eppert-HSCR, Metzeler-86-probe and Bullinger-133-gene), we applied each model for stratification of CN-AML patients in both the Clinseq-AML and the TCGA-AML cohorts (Figures 1a and b, Supplementary Table S2). There were 130 and 64 CN-AML patients in the Clinseq-AML and TCGA-AML cohorts, respectively. In the Clinseq CN-AML cohort, patients could be stratified into a high-risk and a low-risk group, with significantly different overall survival (P-value<0.05), using five of the six models and only Eppert-LSCR failed to stratify patients into low and high-risk with significantly different overall survival (Figure 1a and Supplementary Table S2). After adjusting for other risk factors, only the Bullinger-133-gene stratification remained statistically significant (Supplementary Table S3). The effect size estimates from the multivariable models had similar magnitudes and directions as the univariable results although not meeting the significance level threshold in the Marcucci-7-gene and Li-24-gene models This may be an effect of limited statistical power rather than a lack of independent prognostic power of these models. However, the prognostic value of these molecular risk stratification models beyond the traditional clinical parameters would have to be tested in larger cohorts with better statistical power before any conclusions could be drawn. The C-index of the prognostic score assess the ability of discrimination of each model. The Patel-mutation model and Li-24-gene model showed relatively good discrimination (C-index=0.65 and 0.64, respectively), while Bullinger-133-gene had no discrimination (Figure 1d and Supplementary Table S4). In the TCGA CN-AML cohort, only the Li-24-gene model provided a significant difference in overall survival between the subgroups. The C-index of models indicated no discrimination (CI of C-index included 0.5). Estimated HRs of the six models evaluated in the Clinseq-AML and the TCGA-AML cohorts were compared with HRs of the validation cohorts reported in the original studies (Figure 1c).
Figure 1.
Overall survival of CN-AML patients stratified by gene signature, Marcucci-7-gene, Li-24-gene, Eppert-LSCR, Eppert-HSCR, Metzeler-86-probe and Bullinger-133-gene in the Clinseq-AML (a) and the TCGA-AML (b). P-value is the P-value of log-rank test comparing two groups. HR is the HR and 95% CI comparing high-risk group to low-risk group. (c) HRs (95% CI) in the Clinseq-AML, the TCGA-AML cohort (blue) and HRs reported in the validation cohorts from the original studies (grey). *Not CN-AML only. (d) C-index (95% CI) in the Clinseq and the TCGA AML-CN patients.
Although different gene sets were developed based on distinct theories, there are some degrees of overlap between Li-24-gene, Eppert-HSCR, Metzeler-86-probe and Bullinger-133-gene. The gene lists were compared in pairs (Supplementary Table S6). The gene DAPK1, a tumor suppressor, appears in the gene sets of Li-24-gene, Eppert-HSCR, Metzeler-86-probe and Bullinger-133-gene. It has been reported that the DAPK1 is repressed via a pathway promoted by FLT3-ITD.21 The consistent selection of this gene from different gene sets also indicates that it may play an important role in AML.
Comparison of novel risk classification systems to cytogenetic and ELN risk classification
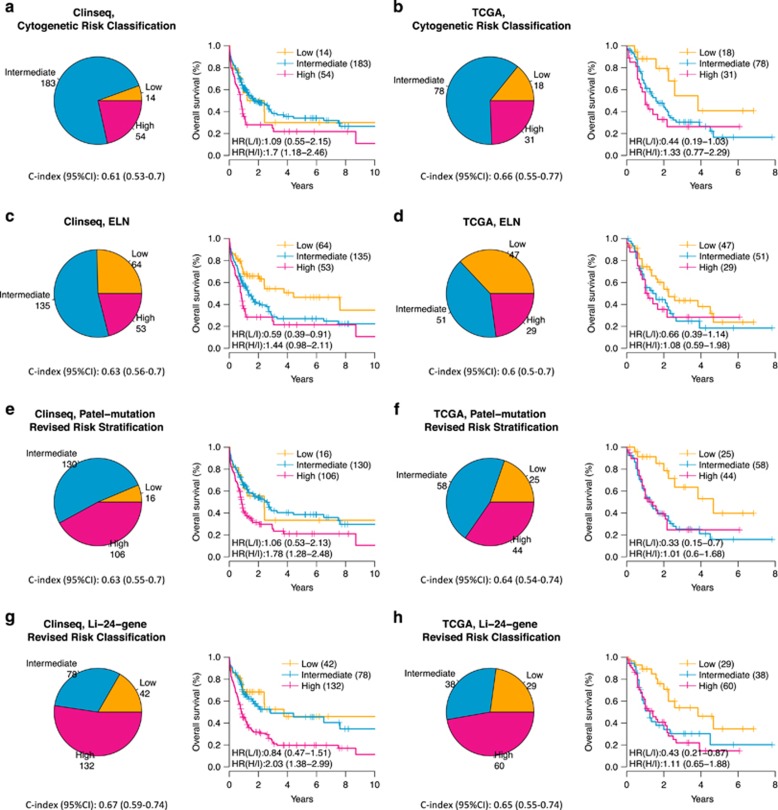
We then compared four risk classification systems (Figure 2), including conventional cytogenetic risk, ELN, Patel-mutation and Li-24 for the whole-AML cohorts, also including patients with cytogenetic low and high-risk. Patients with acute promyelocytic leukemia (translocation (15;17) were excluded (N=252 patients remaining in the Clinseq cohort after exclusion, N=127 remaining in the TCGA cohort). Stratified by cytogenetic information only, the intermediate risk group dominated and accounted for 73% of the patients in the Clinseq-AML cohort and 61% of the patients in the TCGA-AML cohort, further underlining the need to improve cytogenetic stratification. In the Clinseq-AML cohort, the three risk groups showed different overall survival rates (Figure 2). Based on mutation status of FLT3-ITD, NPM1 and CEBPA, the ELN classification system5 redistributed a subset of the CN-AML into the low-risk group (Figure 3). In Clinseq-AML, the survival outcome of this subset of patients is good. However, in TCGA-AML, the survival outcome of this subgroup was found to be similar to the intermediate risk group (Supplementary Figures S4A and B). The 3-year survival rate in this larger low-risk group increased from 30 to 54% in the Clinseq-AML cohort whereas survival decreased from 54 to 44% in the TCGA-AML cohort (Table 2).
Figure 2.
Risk group distribution and overall survival of AML risk classifications in the Clinseq-AML cohort (a, c, e and g) and the TCGA-AML cohort (b, d, f and h). Patients with acute promyelocytic leukemia were excluded. (a) and (b) are based on cytogenetic risk classification, (c) and (d) are based on the ELN classification system, (e) and (f) are based on the Patel-mutation panel revised risk classification, and (g) and (h) are based on the LI-24-gene revised ELN risk classification.
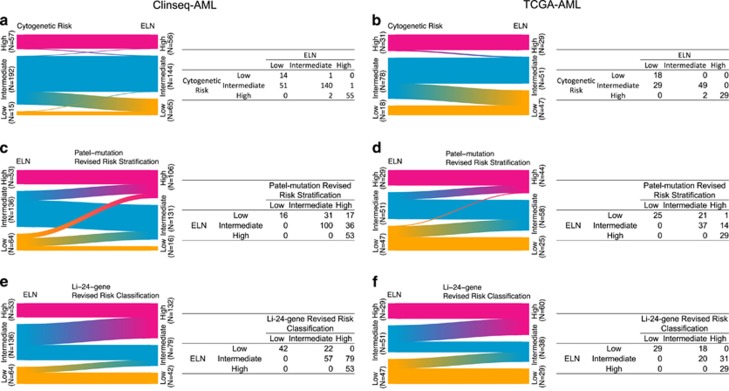
Figure 3.
Reclassification from cytogenetic risk to the ELN (a and b), from the ELN to the Patel’s revised risk stratification (c and d), and from the ELN to the Li’s revised risk classification (e and f) in the Clinseq and the TCGA cohorts.
Table 2. 3-year survival rate (%) with 95% CI stratified by risk classification systems.
|
Cytogenetic risk classification |
ELN |
Patel-mutation revised risk stratification |
Li-24-gene revised risk classification |
|||||
|---|---|---|---|---|---|---|---|---|
| N | 3-year survival rate (%) with 95% CI | N | 3-year survival rate (%) with 95% CI | N | 3-year survival rate (%) with 95% CI | N | 3-year survival rate (%) with 95% CI | |
| Clinseq | ||||||||
| Low | 14 | 30 (12.4–72.8) | 64 | 54.3 (41.7–70.6) | 16 | 33.3 (14.4–77.4) | 42 | 52.5 (36.5–75.6) |
| Intermediate | 183 | 38.3 (30.4–48.1) | 135 | 28.7 (20.5–40.1) | 130 | 42 (32.9–53.7) | 78 | 49 (36.9–65.1) |
| High | 54 | 24.8 (14.8–41.5) | 53 | 24.9 (14.7–42.2) | 106 | 25.5 (17.2–37.7) | 132 | 22.3 (15.5–32) |
| TCGA | ||||||||
| Low | 18 | 54.4 (29.9–98.9) | 47 | 43.5 (29.6–64) | 25 | 63.5 (43.3–93.3) | 29 | 53.9 (35.9–81) |
| Intermediate | 78 | 30.3 (20.4–45) | 51 | 24.7 (13.6–44.7) | 58 | 25.3 (15.3–41.8) | 38 | 30.3 (17.5–52.3) |
| High | 31 | 26.2 (12.8–53.9) | 29 | 28.4 (13.9–57.9) | 44 | 24.6 (12.4–48.9) | 60 | 21.9 (11.9–40.6) |
Abbreviations: CI, confidence interval; ELN, European Leukemia Net.
Patel et al.6 proposed to improve the cytogenetic risk classification system by including mutation status of additional genes (IDH1, IDH2, ASX1, MLL-PTD, PHF6, TET2 and DNMT3A). We performed mutational analysis on 23 genes, including the 18 genes reported in Patel et al.’s study and found that mutations in ASXL1, CEBPA, EZH2, KRAS, SF3B1 and TP53 in Clinseq-AML and EZH2, FLT3-ITD, RUNX1, SF3B1 and TP53 in the TCGA-AML studies to be associated with overall survival (P-value<0.05, log-rank test, Supplementary Table S5). We found that the Patel et al. classification system significantly redistributed patients between risk groups, resulting in a substantial reduction of the low-risk group and a doubling of the high-risk group in both AML cohorts compared to the ELN classification (Figure 3). Patients reclassified from ELN-Low and ELN-Intermediate groups to the Patel’s-High group show poor survival in the Clinseq-AML cohort (Supplementary Figure S4C). However, patients in the ELN-Low group that were reclassified to Patel’s-Intermediate group displayed a relatively good survival outcome the Clinseq-AML cohort. The number of patients in the intermediate risk group was slightly reduced. Compared to the cytogenetically determined risk stratification, there was a major redistribution of intermediate risk patients to the high-risk group. Compared with the cytogenetic risk classification, the 3-year survival rate of the high-risk group remained at a similar level (from 25 to 25% in the Clinseq-AML cohort, from 25 to 26% in the TCGA-AML cohort, Table 2).
We then evaluated the Li-24-gene model, which is based on the ELN and a 24-gene-expression panel. With the Li classification, the number of patients in the intermediate group was substantially reduced both compared to the ELN classification, from 135 to 78 in the Clinseq-AML cohort and from 51 to 38 in the TCGA-AML cohort (Figure 3). Patients reclassified from the ELN-Intermediate to the Li-high-risk group show similar survival outcomes as the patients classified as high risk by both ELN and Li’s classifications (Supplementary Figures S4E and F). Instead, the proportion of high-risk patients increased to ~50% which was paralleled by a reduction in survival in the high-risk patients. For the low-risk patients, the 3-year survival rate remained similar to the ELN classification with the Clinseq-AML data set whereas it was increased (from 44 to 54%) in the TCGA-AML data set (Table 2). Moreover, the Li-24-gene risk model shows a statistically significant effect after adjusting for age, gender and ELN, which indicates that the Li-24-gene provides extra prognostic value beyond the current ELN system. In summary, the integrated risk classification by Patel-mutation and Li-24-gene reduced the number of patients with intermediate risk, and provided improved prognostic value although the result differed somewhat between the two AML cohorts considered here.
Replication of Papaemmanuil et al.’s genomic classification of AML
Recently, Papaemmanuil et al. proposed a genomic-based classification with 11 subgroups in a large cohort combining driver mutations in 76 genes with cytogenetic information to improve the current WHO classification of AML.7 Their genomic-based classification is aimed at defining new subtypes of AML, not risk classification, hence their model is not directly comparable with other risk prediction models above. Nonetheless, new subgroups defined by mutational pattern in chromatin and RNA-spliceosome, and TP53 with chromosomal aneuploidy showed prognostic differences in that study. The 11 subgroups are defined based on genomic aberrations to reflect the biological characteristics of AML. We applied this mutational-based classification to Clinseq (N=274) and TCGA (N=142) cohort. The frequencies of subgroup membership in the three cohorts were found to be similar (Supplementary Table S16). A biologically focused approach, in contrast to approaches focused on prognosis, has the potential to also provide information that may provide target leads in future efforts to develop targeted AML therapies. In the Clinseq cohort, the survival rate in the different subgroups revealed similar pattern to the original study cohort (Supplementary Figure 3), although the difference between groups were not statistically significant due to limited number of sample. We did not conduct survival analysis in the TCGA cohort because of the small sample size.
Discussion
Despite several proposed AML reclassification models based on somatic mutations or gene-expression profiling, no systematic evaluation of these models has been carried out to date. In this study, we validated the prognostic value of six of the most important and cited AML molecular-based risk stratification models. Validation was performed by applying targeted exome sequencing and full RNA-sequencing to profile 274 intensively treated AML patients (Clinseq-AML) and as a second validation cohort we used the TCGA-AML data set.
Revised risk classification models proposed by Patel et al.6 and Li et al.10 were found to add prognostic value to cytogenetic-based classification. These two models represent two major directions of improving risk classification, (1) combining the somatic mutation profile or (2) the gene-expression profile with cytogenetic information. By combining information from the 18-gene mutation profile with conventional cytogenetic risk classification, the Patel-mutation model reclassified ~30% of patients in the intermediate risk group to the high-risk group. Considering the genes in the 18-gene mutational profile in Patel et al.’s study in three cohorts (Clinseq-AML, TCGA-AML and ECOG-E1900,6 Supplementary Table S5), only CEBPA and TP53 showed association with overall survival in more than one cohort. The frequencies of mutations are also varied in the three cohorts (Supplementary Figure S1).
The second direction for improving current risk classification is integrating gene-expression signature with cytogenetic and genetic information. Gene expression is a dynamic molecular phenotype in many respects, particularly compared with genetic mutations, therefore making it potentially more challenging to apply for diagnostics or risk classification in a clinical setting. Technological advancements, particularly sequencing, are, however, making it easier to consistently measure RNA abundances reliably and with high-technical reproducibility. Gene-expression-based biomarkers additionally provide larger effects sizes compared with genetic variants, which is of central importance in prediction modeling, including diagnostic and prognostic applications. An example of this is a study in which a survival prediction model was developed based on the TCGA-AML cohort.22 Authors showed that the transcriptomic information provided the highest prognostic power among genomic, transcriptomic and clinical variables. We also find it interesting to highlight that although the gene-expression-based models evaluated in this study were developed from microarray-based gene-expression measurements, we were able to validate their prognostic value in data acquired by RNA-sequencing, indicating that some of these signatures were not only robust across cohorts, but also across the measurement technology applied (microarrays or RNA-sequencing). Among the four risk classification systems we compared, the Li-24-gene score provided the greatest reduction of patients classified in the intermediate risk group (from ~70% to ~30%). Although the optimal set of variables (genes) may not have been established yet, and would most likely require even larger studies, the strategy of combining cytogenetic and gene-expression information to develop a better risk classification system appears promising.
In the Clinseq-AML cohort, five out of six tested expression-based signatures could separate CN-AML patients into subgroups with statistically significant distinct prognosis. However, only Li-24-gene could be validated in the TCGA-AML cohort. The discrepancy between Clinseq-AML and TCGA-AML results can at least partly be understood in terms of inclusion criteria and sampling. Whereas the TCGA-AML cohort only is comprised by de novo AML, the Clinseq-AML cohort consists of consecutive AML patients including also secondary and therapy-related AML. There was a larger proportion of cytogenetically intermediate and smaller proportion of low-risk patients in the Clinseq-AML data set compared to TCGA-AML. Mutations that have higher frequencies in the Clinseq-AML cohort are ASXL1, IDH2, TET2, cohesion and spliceosome mutations. The results presented here should also be interpreted in the context of the available sample sizes in the Clinseq-AML and TCGA-AML cohorts, where the smaller sample size in the TCGA-AML cohort may account for the lack of statistical significance in some cases. Although the Clinseq-CN cohort is larger than the other CN validation cohorts used in the original studies, failure to confirm previously reported different prognosis in the different risk groups could be an effect of statistical power and should therefore not necessarily be interpreted as evidence against the model. We also encourage interpretation of the results in terms of estimated effect sizes (Figure 1c), which agree in respect to the direction of the effects (HRs) from the original studies and findings in the Clinseq and TCGA cohorts and in many cases also have estimates that are of similar size. The C-index (Figure 1d) also offers another complementary statistic that provide means for interpreting the different risk scores in terms of their ability to correctly rank survival times.
In this study, we performed whole-transcriptomic RNA-sequencing and panel mutational sequencing on a large cohort comprised of 274 AML patients (Clinseq-AML). The TCGA-AML study is also comprised of multi-level sequencing information. The multi-omics data allows us to implement and compare different types of models. We succeeded to replicate and validate several models. We note that not all of the genes/probes from Eppert-LSCR, HSCR, Metzeler-86-probe and Bullinger-133-gene could be matched in the Clinseq-AML and the TCGA-AML studies, which may lead to a potential reduction in their prognostic values (Eppert-LSCR: 42 out of 43 genes matched, HSCR: 91 out of 122 genes matched, Metzeler-86-probe: 59 out of 63 genes matched and Bullinger-133-gene: 108 out of 112 genes matched). We also note that some models were easy to implement, while others less so. For example, Marcucci-7-gene model’s inputs are dichotomized at the median of expression value, which is easy to implement in data from diverse platforms irrespective of the original distribution of the data. The weights of predictors to generate a summarized prognostic score in Metzeler-86-probe model were given at probe level and the cutoff to dichotomize the prognosis score was a given value, leading to potential challenges in implementation. Although they may all have prognostic value, to outperform other models in respect to application, a standardized and ‘user-friendly’ procedure could pave the way for clinical practice.
Improvement in AML patient stratification is important in clinical practice. Next generation sequencing technologies are rapidly becoming cost and time efficient, and also gradually implemented in the clinical setting to facilitate diagnosis and treatment decisions in a variety of diseases, including cancer and hematological malignancies. This validation study further confirms and adds evidence that some of the gene-expression as well as mutation–based AML risk classification models are reproducible and provide prognostic information. Nonetheless, it will be necessary to evaluate these prognostic models further in larger cohorts and clinical trials before implementation in routine clinical care.
Acknowledgments
We acknowledge funding from Swedish Cancer Society (Cancerfonden), Swedish e-Science Research Centre (SERC)–‘e-Science for Cancer Prevention and Control (ecpc)’, the Swedish Research Council (Vetenskapsrådet), the Strategic Research Programme in Cancer (StratCan) in Karolinska Institutet and the Stockholm County Council.
Author contributions
MW, JL and MR developed the concept and designed the study, analyzed data and wrote the paper. DK analyzed the data. CN collected and assembled the data. ASM analyzed the data. SL developed the concept and designed the research and wrote the paper. HG developed the concept and designed the research.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu).
The authors declare no conflict of interest.
Supplementary Material
References
- Grimwade D, Mrozek K. Diagnostic and prognostic value of cytogenetics in acute myeloid leukemia. Hematol Oncol Clin North Am 2011; 25: 1135–1161, vii. [DOI] [PubMed] [Google Scholar]
- Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010; 116: 354–365. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Horstedt AS, Johansson B, Antunovic P, Billstrom R, Derolf A et al. Incidence and prognostic significance of karyotypic subgroups in older patients with acute myeloid leukemia: the Swedish population-based experience. Blood Cancer J 2014; 4: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol 2011; 29: 475–486. [DOI] [PubMed] [Google Scholar]
- Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010; 115: 453–474. [DOI] [PubMed] [Google Scholar]
- Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012; 366: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016; 374: 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med 2004; 350: 1605–1616. [DOI] [PubMed] [Google Scholar]
- Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008; 112: 4193–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Herold T, He C, Valk PJ, Chen P, Jurinovic V et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol 2013; 31: 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol 2014; 32: 548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011; 17: 1086–1093. [DOI] [PubMed] [Google Scholar]
- Wahlin A, Billstrom R, Bjor O, Ahlgren T, Hedenus M, Hoglund M et al. Results of risk-adapted therapy in acute myeloid leukaemia. A long-term population-based follow-up study. Eur J Haematol 2009; 83: 99–107. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996; 15: 361–387. [DOI] [PubMed] [Google Scholar]
- Pencina MJ, D'Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med 2004; 23: 2109–2123. [DOI] [PubMed] [Google Scholar]
- Cox DR. Regression models and life-tables. Breakthroughs in statistics. Springer: New York, NY, USA, 1992; pp 527–541. [Google Scholar]
- McGeechan K, Macaskill P, Irwig L, Liew G, Wong TY. Assessing new biomarkers and predictive models for use in clinical practice: a clinician's guide. Arch Intern Med 2008; 168: 2304–2310. [DOI] [PubMed] [Google Scholar]
- Schroder MS, Culhane AC, Quackenbush J, Haibe-Kains B. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics 2011; 27: 3206–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM, Grambsch PM Modeling survival data: extending the Cox model: Springer Science & Business Media 2000.
- Shanmugam R, Gade P, Wilson-Weekes A, Sayar H, Suvannasankha A, Goswami C et al. A noncanonical Flt3ITD/NF-kappaB signaling pathway represses DAPK1 in acute myeloid leukemia. Clin Cancer Res 2012; 18: 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung M, Pellagatti A, Malcovati L, Giagounidis A, Porta MG, Jadersten M et al. Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nat Commun 2015; 6: 5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmacher MD, Marcucci G, Ruppert AS, Mrozek K, Whitman SP, Vardiman JW et al. Independent confirmation of a prognostic gene-expression signature in adult acute myeloid leukemia with a normal karyotype: a Cancer and Leukemia Group B study. Blood 2006; 108: 1677–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.