Summary
T‐helper cell type 17 (Th17) mediated inflammation is associated with various diseases including autoimmune encephalitis, inflammatory bowel disease and lung diseases such as chronic obstructive pulmonary disease and asthma. Differentiation into distinct T helper subtypes needs to be tightly regulated to ensure an immunological balance. As microRNAs (miRNAs) are critical regulators of signalling pathways, we aimed to identify specific miRNAs implicated in controlling Th17 differentiation. We were able to create a regulatory network model of murine T helper cell differentiation by combining Affymetrix mRNA and miRNA arrays and in silico analysis. In this model, the miR‐212~132 and miR‐182~183 clusters were significantly up‐regulated upon Th17 differentiation, whereas the entire miR‐106~363 cluster was down‐regulated and predicted to target well‐known Th17 cell differentiation pathways. In vitro transfection of miR‐18b, miR‐106a and miR‐363‐3p into primary murine Cd4+ lymphocytes decreased expression of retinoid‐related orphan receptor c (Rorc), Rora, Il17a and Il17f, and abolished secretion of Th17‐mediated interleukin‐17a (Il17a). Moreover, we demonstrated target site‐specific regulation of the Th17 transcription factors Rora and nuclear factor of activated T cells (Nfat) 5 by miR‐18b, miR‐106a and miR‐363‐3p through luciferase reporter assays. Here, we provide evidence that miRNAs are involved in controlling the differentiation and function of T helper cells, offering useful tools to study and modify Th17‐mediated inflammation.
Keywords: microRNA, miR‐106a, miR‐18b, miR‐363a‐3p, nuclear factor of activated T cells 5, retinoid‐related orphan receptor α, retinoid‐related orphan receptor γt, T helper cell differentiation, T helper type 17
Abbreviations
- 3′ UTR
3′ untranslated region
- AFC
array fold change
- EAE
experimental autoimmune encephalitis
- FC
fold change
- Foxp3
forkhead box protein 3
- Gatm
glycine aminotransferase
- Gimap1
GTPase of the immunity‐associated protein family 1
- IL
interleukin
- IPA
ingenuity pathway analysis
- miRNA/miR
microRNA
- Nfat
nuclear factor of activated T cells
- qRT‐PCR
quantitative real‐time PCR
- ROR
retinoid‐related orphan receptor
- Tanc2
tetratricopeptide repeat, ankyrin repeat and coiled‐coil containing 2
- TGF
transforming growth factor
Introduction
Interleukin‐17 (IL‐17) ‐secreting T helper cells (Th17) are essential for protection from bacterial and fungal infections.1, 2 Recent literature suggests that Th17 cells are also involved in chronic, non‐infectious diseases, such as experimental autoimmune encephalitis (EAE)3 and inflammatory bowel disease,4 as well as chronic lung diseases such as chronic obstructive pulmonary disease and severe asthma.5 The differentiation from naive Th0 cells to Th17 cells is regulated by Il‐6, Il‐23 and transforming growth factor β (Tgf‐β). These initiate downstream signalling cascades including activation of T‐cell receptor, signal transducer and activator of transcription 3 (Stat3), and retinoid‐related orphan receptor γt (Rorγt).6 However, the precise regulatory networks of Th17 differentiation in complex diseases are still unknown. Identification thereof may potentially enable to develop novel therapies for Th17‐related diseases.
The expression of Rorγt and additional host proteins, such as the aryl hydrocarbon receptor (Ahr) for Th17 cells,7 needs to be carefully controlled to ensure an immunological balance. Such regulatory fine‐tuning is often modulated by small, single‐stranded non‐coding RNAs, the so‐called microRNAs (miRNAs),8, 9 the binding of which to the 3′ untranslated region (3′ UTR) of their target mRNAs either leads to degradation of the respective mRNA or a translational repression leading to a decreased expression of the target gene.8, 9 Hence, miRNAs are able to control gene or protein expression at the post‐transcriptional level. Single miRNAs have been shown to play a pivotal role in many biological systems, and in regulating the immune system.10 Several studies have proposed a role for miRNAs in the regulation of T helper cell differentiation,11, 12, 13, 14, 15 although information on the specific role of single miRNAs during differentiation into distinct T helper subtypes is scarce. Therefore, we aimed to decipher the regulatory interactions of miRNAs during Th17 differentiation to better understand the underlying molecular mechanisms of Th17‐driven diseases. We performed mRNA and miRNA microarray analyses of primary, in‐vitro‐differentiated Th17, Th2 and Th0 cells and subsequently validated the candidates on a functional level.
Materials and methods
Mice
Female wild‐type BALB/c mice were purchased from Taconic (Silkeborg, Denmark). Mice were maintained under specific pathogen‐free conditions in individually ventilated cages according to the federal guidelines for the use and care of laboratory animals. At the age of 10–14 weeks, mice were killed for spleen collection.
Isolation and in vitro differentiation of Cd4+ T helper cells
Splenic Cd4+ cells were isolated by using the mouse Cd4+ Isolation Kit II (Miltenyi Biotech, Teterow, Germany). Isolation was performed according to the manufacturer's guidelines, and yielded at least 96% purity of Cd4+ Cd3+ Cd8− T lymphocytes. A total of 200 000 Cd4+ cells per well were seeded in a 96‐well plate, and were cultured with RPMI‐1640 media containing 10% fetal calf serum, 1% penicillin/streptomycin and antibodies against Cd3 (4 µg/ml) and Cd28 (30 ng/ml) (both BioLegend, San Diego, CA). Th17 cells were also cultured with Il‐6 (20 ng/ml), Tgf‐β (5 ng/ml), Il‐23 (10 ng/ml) (all R&D Systems, Wiesbaden‐Nordenstadt, Germany) and an antibody against Interferon‐γ (Ifn‐γ) (10 µg/ml) (BioLegend). Th2 cells were differentiated with Il‐4 (100 ng/ml) (R&D Systems) and an antibody against Ifn‐γ (10 µg/ml) (BioLegend). Th0 cells served as a control and were cultured with antibodies against Cd3 and Cd28 (BioLegend). Primary Th cells were cultivated at 37°C in 5% CO2 in a humidified incubator. After 72 hr the medium was replaced with fresh differentiation media. After 120 hr of culture, cells were stimulated with 50 ng/ml PMA and 1 µg/ml ionomycin (both Sigma‐Aldrich, St Louis, MO) for 4 hr.
Intracellular cytokine staining
Before intracellular staining, secretion of cytokines during the 4‐hr stimulation was blocked by Monensin GolgiStop™ (BD, Franklin Lakes, NY) according to the manufacturer's recommendations. Cells were washed twice with PBS (containing 2% fetal calf serum and 0·01 m EDTA) and stained with surface antibodies against Cd3 (Pacific Blue), Cd8 (FITC) and Cd4 (allophycocyanin‐H7) (all 1 : 100, all BioLegend). Intracellular Il‐17a (FITC) and Il‐4 (phycoerythrin) (both Becton Dickinson, Franklin Lakes, NY) were stained by using the Cytofix/Cytoperm Plus Fixation/Permeabilization Kit (BD) according to the manufacturer's instructions. After additional washing, cells were analysed on an LSRII flow cytometer (BD).
Quantitative real‐time PCR
RNAs containing small RNAs were isolated with the miRNeasy micro kit (Qiagen, Venlo, The Netherlands) according to the manufacturer's recommendations. The quality and quantity of the isolated RNA samples were validated with the Nanodrop ND‐1000 Spectrometer (peq Lab Bioscience, Erlangen, Germany) and the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA). Only RNAs with an RNA integrity number > 7.25 were used for further analyses Quantitative real‐time PCR (qRT‐PCR) was performed using an LC480 (Roche, Basel, Switzerland) and the recommended ‘Light Cycler DNA master SYBR Green I’ Kit (Roche). Primer sequences were self‐designed and are listed in the Supplementary material (Table S1). Analysis of all qRT‐PCRs was done with the ‘LC 480 SW1.5’ software (Roche) using the second derivative maximum method and fold changes between groups were calculated by the ∆∆Ct method.16
Messenger RNA profiling
For mRNA arrays, total RNA (30 ng) of four independent differentiations was amplified using the Ovation PicoSL WTA System V2 (Nugen, San Carlos, CA) in combination with the Encore Biotin Module (Nugen). Amplified cDNA was hybridized on an Affymetrix Mouse Gene 2.0 ST array (Affymetrix, Santa Clara, CA). Staining and scanning were performed according to the Affymetrix expression protocol, except for minor modifications as suggested in the Encore Biotion protocol (Nugen). Expression console (v.1.3.0.187, Affymetrix) was used for quality control and to obtain annotated normalized Robust Multichip Average (RMA) data (standard settings including median polish and sketch‐quantile normalization). Statistical analyses were performed by using the statistical programming environment R17 implemented in CARMaweb.18 Genewise testing for differential expression was carried out employing the (limma) t‐test (P < 0·05). Heat maps were generated with CARMaweb. Array data have been submitted to GEO (GSE55013).
In silico analysis and Ingenuity pathway analysis
Pathway analyses and expression pairing were generated through the use of QIAGEN's Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) using Fisher's exact test P‐values.
MicroRNA profiling
For miRNA arrays, total RNA (600 ng) was labelled with the FlashTag Biotin HSR kit (Genisphere, Sterling, VA) and hybridized on miRNA 3.0 arrays (Affymetrix). Staining and detection of the array was performed as described for mRNA arrays. Expression console (v.1.3.0.187, Affymetrix) was used for quality control and to obtain annotated normalized RMA data [RMA + DABG (Robust Multichip Average + Detected Above BackGround)]. Statistical analyses were performed by using the statistical programming environment R17 implemented in CARMAweb.18 For miRNA analysis a filter for detection in at least n−1 samples in at least one group was used. Heatmaps were generated with CARMAweb. Array data have been submitted to GEO (GSE55013).
MicroRNA qRT‐PCR
For the miRNA analysis, all chemicals were purchased from Exiqon (Vedbaek, Denmark). The cDNA of miRNAs were synthesized with the Universal cDNA Synthesis Kit II followed by qPCR, which was done with ExiLENT SYBR®Green Master Mix. Pre‐designed primer sets for miRNA qPCR (LNA™ Primer sets, Exiqon) were used for the analysis of the following miRNAs: miR‐183‐5p (204652), miR‐212‐3p (205589), miR‐301a‐3p, miR‐363‐3p (204726), miR‐18b‐5p (205076) and miR‐106a‐5p (205061). The small nuclear RNA U6 (203907) was used as a reference miRNA for normalization.
Dual‐luciferase reporter assays
Potential miRNA binding sites within the genes Rorc, Rora and Nfat5 were identified with the Whitehead Institute for Biomedical Research target prediction tool ‘targetscan mouse’ (http://www.targetscan.org/mmu_71/). Due to the large size of the 3′ UTRs (> 7 kb) with the presence of multiple miRNA binding sites, we decided to clone smaller fragments, containing miRNA binding sites of the respective 3′ UTR into reporter plasmids (primer sequences used for cloning are listed in the Supplementary material, Table S1). For a more detailed analysis, we used synthetic DNA duplexes spanning ~80‐nucleotide regions of the respective 3′ UTR each with a single miRNA binding site (or a mutation thereof comprising seven or eight sequential T or A nucleotides, all sequences are listed in the Supplementary material, Table S5) in the reporter assays as described previously.19 Synthesized 80‐bp DNA fragments within the 3′ UTRs of Rora, Rorc and Nfat5 with binding sites for either miR‐18b, miR‐106a or miR‐363‐3p were ordered as DNA duplexes from Metabion (Planegg, Germany). To ease cloning into vectors, the DNA oligos carried the overhang recognition sites for the restriction enzymes XhoI (TCGAG on the 5′ end, C on the 3′ end) and NotI (GGCCGC on the 5′ end and GC on the 3′ end) and are phosphorylated (p) on the 5′ ends (sequences are listed in the Supplementary material, Table S5). All fragments were cloned into individual psiCheck™‐2 vectors (Promega, Madison, WI) using XhoI and NotI restriction sites. Reporter assays were performed by transfecting 100 ng of the transgene psiCheck™‐2 vectors and 5 nm of each miRNA precursor or a scrambled miRNA into A549 human alveolar basal epithelial cells by using peqFECT siRNA transfection reagent (PeqLab Biotech GmbH, Erlangen, Germany). Cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum in 96‐well plates (Nunc, Roskilde, Denmark ). After 48 hr, luciferase activity was measured in a luminescence plate reader (Berthold, Bad Wildbad, Germany) after the addition of the respective substrates by using the DualGlo Luciferase Assay System (Promega) according to the manufacturer's recommendations. Renilla luciferase (fused to 3′ UTR) activity was normalized to firefly luciferase activity (transfection control).’
Transfection of miRNAs
Primary Cd4+ T cells were transfected with miRNA mimics for miR‐106a‐5p, miR‐18b‐5p, miR‐363‐3p or a scrambled miRNA as control (Ambion, Life Technologies, Carlsbad, CA). Freshly isolated cells were cultivated in 75% normal differentiation medium and 25% transfection medium. The latter contained serum‐free medium, 0·12% transfection reagent (PeqFECT siRNA, PeqLab Biotech GmbH) and the miRNA precursors (6·5 nm final concentration). After 4 hr, the media was exchanged for fresh differentiation medium. Cells were harvested after 72 hr and analysed for gene expression, cytokine secretion and viability.
Analysis of cell viability
Viability of primary cells after transfection was measured by an MTT‐assay (Thermo Fischer Scientific, Waltham, MA). MTT dye (3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide) was added to the cells and incubated for 4 hr. Purple formazan produced by living cells was then analysed by measuring the absorbance at 500–600 nm with a plate reader (Tecan, Männedorf, Switzerland).
Statistical analysis
Statistical validation of arrays was performed using limma t‐test and Benjamini–Hochberg multiple testing correction. For all other experiments, statistical differences between groups were calculated using either one‐way analysis of variance with Tukey post‐test or unpaired Student's t‐test. The calculations and graphs were made with graphpad prism 5.01.
Results
Validation of in vitro Th cell differentiation
To study mRNA and miRNA expression in mature Th17 cells, we isolated Cd4+ cells from murine spleens, and differentiated them in vitro into Th17, Th2 and Th0 cells. A successful differentiation into distinct T‐helper subtypes was verified and quantified by intracellular cytokine staining for Il‐4 for Th2 cells, and Il‐17a for Th17 after 120 hr of culture. Both cytokines were significantly elevated in the respective T‐helper subtype. Compared to Th0 control cells, Th2 cells produced double the amount of Il‐4 (Fig. 1a) and Th17 cells showed a threefold increase of Il‐17a production (Fig. 1b).
Figure 1.
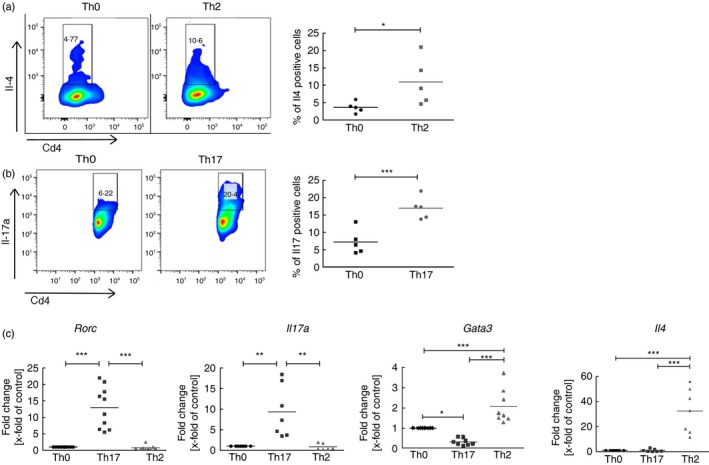
Characterization of primary, in‐vitro‐differentiated T helper cells. (a) Protein expression of the Th2 effector cytokine interleukin‐4 (Il‐4). Depicted are representative flow cytometry density blots for Il4 and Cd4. Numbers are positive counted cells in %. n = 5 independent experiments are summarized in the dot blot on the right column. (b) Protein expression of the Th17 effector cytokine Il‐17a. Depicted are density blots for Il17a and Cd4. n = 5 independent experiments are summarized in the dot blot on the right column. (c) Gene expression of major transcription factors and effector cytokines of Th17 cells and Th2 cells. Depicted are dot blots of fold changes compared with the control (Th0) and normalized to Hprt. Medians in dot blots are represented by a black line. For (a) and (b) significant differences between Th0 control cells and Th17 or Th2 cells were calculated with Student's t‐test. For (c) statistical differences were calculated with one‐way analysis of variance and Tukey post‐test. *P < 0·05, **P < 0·01,***P < 0·001.
Next, we analysed the mRNA expression of distinct Th17 and Th2 markers by qRT‐PCR. The gene encoding the Th17 transcription factor Rorγt, Rorc, and Il17a were significantly up‐regulated in Th17 cells compared with Th0 and Th2 cells. The two specific Th2 cell markers, Gata3 and Il4, were significantly increased in Th2 cells and unchanged or lower in expression in Th0 and Th17 cells (Fig. 1c), confirming specific differentiation.
mRNA profiling of Th17, Th2 and Th0 cells
In a next step, we aimed to identify regulatory networks in the distinct in‐vitro‐differentiated T helper subtypes. We therefore performed mRNA arrays involving > 35 000 transcripts. By comparing the expression values between Th17 and Th0 cells, we identified 2052 significantly regulated genes (P < 0·05), and 1918 genes that were significantly different between Th17 and Th2 cells (see Supplementary material, Tables 1 and 2 accessible at GEO GSE55013). From these gene sets, 544 genes were similarly regulated in Th17 compared with Th0 cells or compared with Th2 cells (see Supplementary material, Fig. S1). All genes that were increased in expression in Th17 cells compared with both Th0 and Th2 cells were then defined as Th17‐specific genes (see Supplementary material, Table 3, accessible at GEO GSE55013).
Next, we used qRT‐PCR to confirm the regulation of three Th17‐specific and three Th2‐specific genes. As Th17 cell validation markers we chose the aryl hydrocarbon receptor (Ahr) with an array fold change (AFC) of 5·9x, Il17f, the second main cytokine of Th17 cells with an AFC of 29·8, and the highly increased glycine aminotransferase (Gatm) with an AFC of 15·0. All three specific Th17 genes were significantly increased (Fig. 2a). For Th2 cells we chose the Th2 cytokines Il5 (AFC: 1·7), Il24 (AFC: 2·0) and tetratricopeptide repeat, ankyrin repeat and coiled‐coil containing 2 (Tanc2) (AFC: 3·0). Il5 and Il24 were significantly increased in Th2 cells compared with Th17 and Th0 cells (Fig. 2b). We observed a trend for Tanc2 up‐regulation in Th2 compared with Th0 and Th17 cells, although this did not reach statistical significance.
Figure 2.
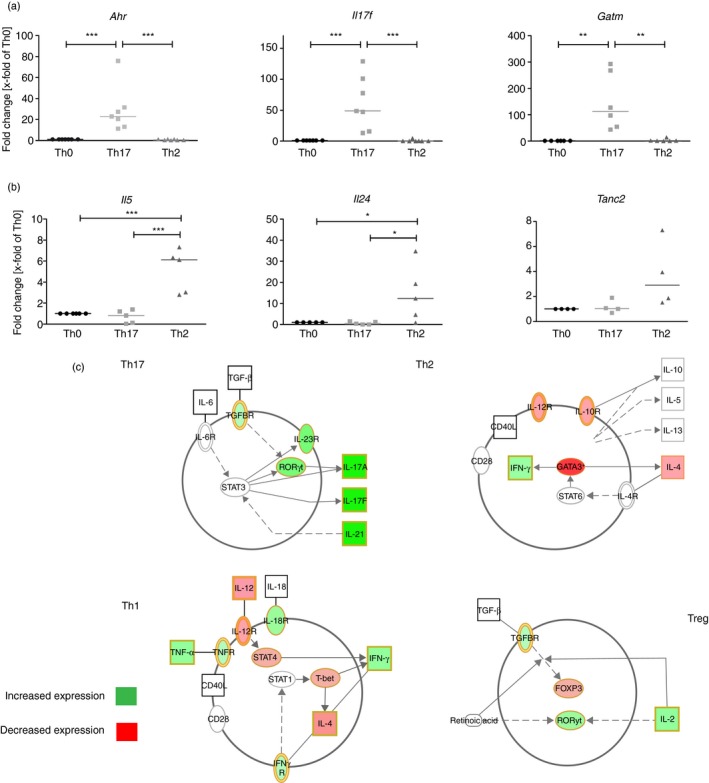
Messenger RNA (mRNA) profiling in T helper cell subtypes. (a) and (b) Quantitative RT‐PCR validation of T helper type 17 (Th17) and Th2 up‐regulated genes. Depicted are dot blots of fold changes compared with the control (Th0) and normalized to Hprt. Medians in dot blots are represented by a black line. For (b) and (c) significant differences between Th0 and Th17 or Th2 cells were calculated of n = 4 independent differentiations with one‐way analysis of variance and Tukey post‐test. *P < 0·05, **P < 0·01, ***P < 0·001. (d) Regulation of Th17 genes in the context of T helper cell differentiation. Depicted are up‐ and down‐regulated genes of Th17 mRNA arrays, which are responsible for the differentiation of Th17, Th2, Th1 and regulatory T (Treg) cells. In silico analysis and schematic figures were conducted with IPA software.
To determine which genes are involved in the regulation of Th17 cell differentiation and function, we then conducted an in silico analysis with the 2052 genes differentially regulated in Th17 cells versus. Th0. Ingenuity Pathway Analysis revealed ‘differentiation of Th17 cells’ as the most significantly enriched canonical pathway (see Supplementary material, Table S2). Of note, nearly all Th17‐specific genes described in the literature were found to be increased in our Th17 cells (Fig. 2c). In contrast, the Th2‐specific genes Gata3 and Il4 and the Th1‐specific T‐box transcription factor Tbx21 (Tbet) and Stat4 were all decreased in Th17 cells. Many genes that are connected to T regulatory (Treg) cells were also increased in Th17 cells, including TGF‐β receptor (Tgfbr), Rorc and Il2; however, the main transcription factor of Treg, forkhead‐box protein 3 (Foxp3), was decreased in expression (Fig. 2c).
Additional prediction of functional properties of Th17‐regulated genes revealed a significant enrichment of genes associated with activated lymphocytes and specific Th17 functions with the top four being ‘cell movement of T lymphocytes’, ‘T‐cell migration’, ‘recruitment of neutrophils’ and ‘phosphorylation of protein’, and a decrease in ‘bacterial infections’ and fungal infections’ (see Supplementary material, Table S2). Hence, the microarray analysis indicated correct differentiation into Th2 and Th17 cells in our in‐vitro system.
microRNA profiling in Th17, Th2 and Th0 cells
To identify regulatory miRNAs involved in Th17 differentiation, we performed Affymetrix miRNA 3.0 arrays with the same RNA samples that were used for the mRNA array. Comparing Th17 and Th0 cells, 92 miRNAs were significantly regulated in Th17 cells (P < 0·05). In comparison to Th2 cells, 162 miRNAs (P < 0·05) were significantly regulated in Th17 cells (see Supplementary material, Fig. S2, Tables 4 and 5 accessible at GEO GSE55013). We identified 60 miRNAs that were differentially expressed compared with both Th0 and Th2 cells (see Table 6 accessible at GEO GSE55013), and might therefore influence Th17 differentiation or cytokine secretion.
We further compared the significantly regulated mRNAs and miRNAs of all three T helper cell subtypes to identify Th17‐specific regulatory miRNA and mRNA interactions (Fig. 3a). This expression pairing with IPA predicted a number of miRNAs that are potentially involved in differentiation and induction of Th17 cells, with the top hits listed in Fig. 3(b,c). The miRNAs showing the highest expression were the miR‐212~132 cluster, the miR‐182~183 cluster and miR‐338‐5p (Fig. 3c). The most decreased miRNAs were miR‐301a and almost all of the miR‐106~363 cluster, including miR‐18b, miR‐20b and miR‐363‐3p (Fig. 3d).
Figure 3.
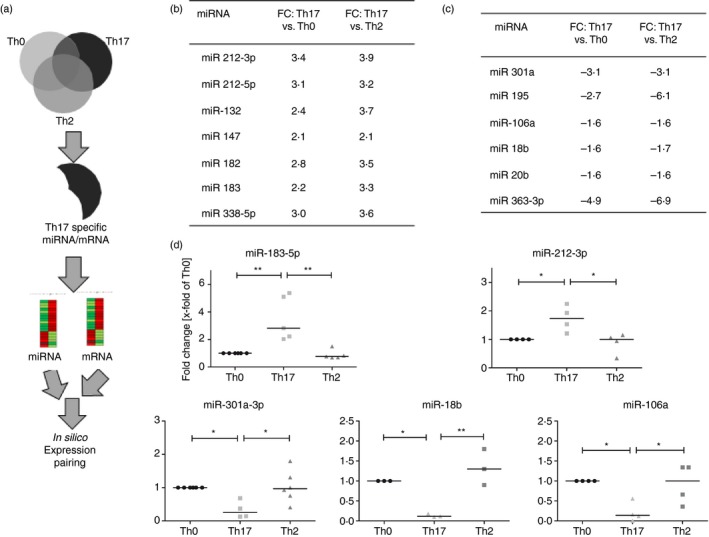
MicroRNA (miRNA) profiling in T helper cell subtypes. (a) Schematic depiction of the comparison strategy used for the identification of a T helper type 17 (Th17) miRNAs or mRNAs signature. (b) and (c) Array data of Th17‐cell‐specific increased (b) or decreased (c) miRNAs. (d) Validation of chosen miRNAs. Quantification of miRNAs was performed by quantitative RT‐PCR. Shown are the fold changes compared with the Th0 control cells. The miRNA expression levels are normalized to the expression of U6 snRNA. Statistical differences were calculated for n = 4 to n = 6 independent differentiations with one‐way analysis of variance and Tukey post‐test. *P < 0·05, **P < 0·01, *** P < 0·001. FC = Fold change.
The expression of the most prominent up‐ and down‐regulated miRNA candidates was further validated by qRT‐PCR. miR‐183‐5p and miR‐212‐3p were significantly increased in Th17 cells compared with Th0 and Th2 cells; miR‐301‐3p, miR‐18b and miR‐106a were decreased, confirming the array data (Fig. 3d).
Th17 cell differentiation involves miRNA regulation
After identifying distinct Th17‐specific miRNAs, we were interested in whether these could play a functional role in the Th17 differentiation process. Hence, we created an in silico model of T helper cell differentiation by using the miRNA and mRNA expression pairing data (Fig. 4). According to this in silico model the increased miRNA clusters 182~183 and 212~132 in Th17 cells might regulate essential genes of Th1 and Treg differentiation. The miRNAs 182 and 183 were predicted to inhibit the expression of GTPase of the immunity‐associated protein family 1 (Gimap1), which was decreased in Th17 cells, and which is important for Th1 differentiation. Moreover, the 182~183 cluster may also interfere with Foxo1/3 and Foxp3 expression, and so Treg differentiation. The Ahr‐induced miRNA cluster 212~132 may inhibit Th1 differentiation by reducing Stat4 expression. In contrast the down‐regulated miR‐363‐3p, miR‐106a and miR‐18b are predicted to bind to the Th17 key transcription factors Rorc, Rora and Nfat5. All predicted miRNA‐mRNA pairings are listed in the Supplementary material (Tables S3 and S4). Changes in expression of these transcription factors could therefore directly affect Il‐17a/f secretion (Fig. 4).
Figure 4.
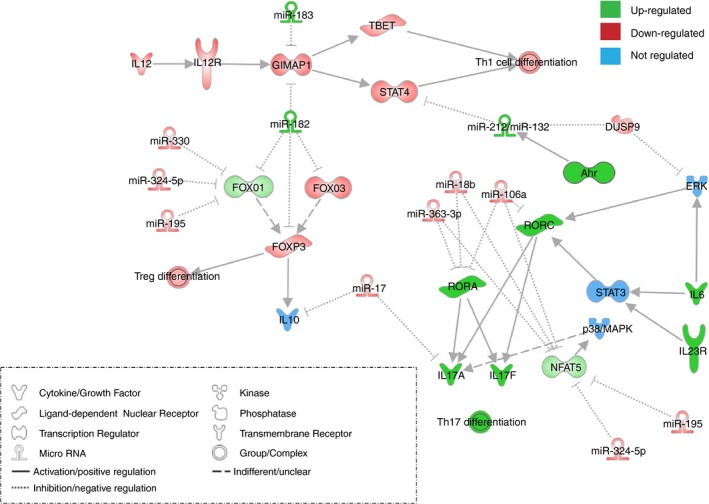
In silico pathway prediction of T helper type 17 (Th17) ‐specific miRNA/mRNA pairs. The depicted pathway was identified with IPA software using Th17‐specific regulated miRNA/mRNA pairs. Green coloured genes are up‐regulated, red coloured genes are down‐regulated; blue coloured genes are not regulated.
Over‐expression of miR‐106a, miR‐18b and miR‐363‐3p decreases Th17 differentiation and Il‐17 secretion
According to TargetScan S (mouse) (http://www.targetscan.org/mmu_71/), the 3′ UTRs of Rora and Nfat5 contain three binding sites for miR‐18b, one for miR‐106a and two for miR‐363‐3p, while the Rorc 3′ UTR only features one binding site for miR‐106a. To address this, we performed dual‐reporter luciferase‐reporter assays, with one specific region of the 3′ UTR of the three transcription factors each containing a predicted binding site for the respective miRNAs.
We therefore demonstrated a specific binding of miR‐18b and miR‐106a to the 3′ UTR fragment of Rora and Nfat5 via a diminished signal of the reporter luciferase renilla compared with the control luciferase firefly (Figs 5a and see Supplementary material, Fig. S3) and to the scrambled miRNA transfection. miR‐363‐3p also significantly bound to the 3′ UTR of Rora and trendwise also to Nfat5. Mutating the binding sites for the respective miRNAs abolished the reduction of the renilla luciferase (Fig. 5a), so indicating a specific regulation by the respective miRNA. The regulation of Rorc by miR‐106a could not be verified by this assay.
Figure 5.
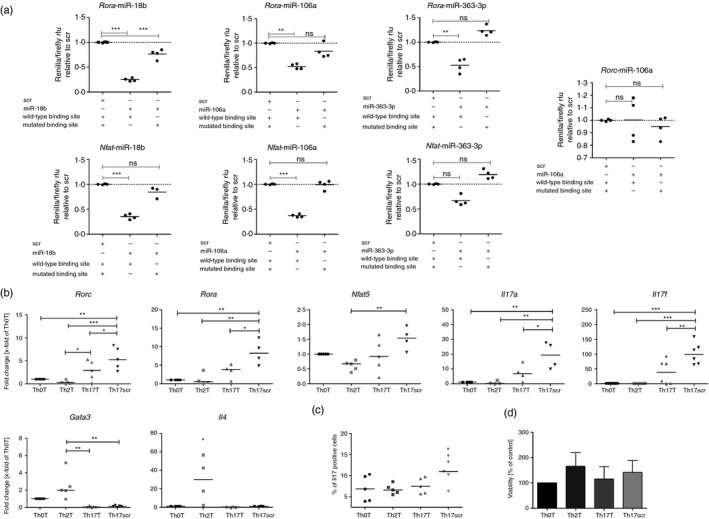
Target gene validation for microRNA (miRNA) 106a‐5p, 18b‐5p and 363‐3p and function in T helper cells. (a) Luciferase reporter assay with plasmids containing either wild‐type or mutated miRNA binding sites. Depicted are the luciferase values (Renilla/Firefly) relative to the scrambled negative control. (b) Quantitative RT‐PCR analysis of transfected T helper subsets. Cells were simultaneously transfected with 6·5 nM miRNA 106, 18b and 363 (Th0T, Th2T and Th17T) or the same concentration of a scrambled miRNA (Th17scr). Depicted are fold changes compared with the transfected Th0T subset and normalized to the expression of Hprt. (c) Flow cytometry analysis of interleukin‐17 (Il‐17) protein production of transfected T helper cells. (a–c) Medians are depicted of four to six independent experiments. (d) Viability (MTT assay) of T helper cells after transfection. Shown are the mean (± SD) relative amounts of viable cells compared with the control (n = 4). Differences are not significant. Statistical differences were calculated with one‐way analysis of varaince and Tukey post test. *P < 0·05, **P < 0·01, ***P < 0·001, ns = non‐significant.
To investigate the functional relevance of this miRNA cluster in the differentiation of Th0 to Th17 cells, we transiently over‐expressed miR‐363‐3p, miR‐106a and miR‐18b in Th0 cells at the beginning of the in vitro differentiation towards Th17. Compared with Th17 cells transfected with the same concentration of scrambled miRNA, simultaneous transfection of miR‐363‐3p, miR‐106a and miR‐18b decreased the mRNA expression of Rorc, Rora, Il17a and Il17f (Fig. 5b). Furthermore, down‐regulation of these genes by miRNA over‐expression resulted in a significant decrease of Il‐17a protein production in Th17 cells compared with the scrambled transfected Th17 cells (Fig. 5c). The viability of transfected cells did not vary between the different conditions; in particular not between miRNA‐transfected Th17 cells and scrambled transfected Th17 cells (Fig. 5d).
Discussion
This study aimed to identify regulatory miRNA/mRNA networks that control the differentiation and function of Th17 cells. Hence, we combined mRNA and miRNA microarray data of in‐vitro‐differentiated murine primary Th2 and Th17 cells in an in silico IPA. The identified Th17‐specific miRNA clusters were predicted to be functionally involved in Th17 differentiation and cytokine production. For example, the miR‐106a~363 cluster was decreased in expression upon the differentiation of Th17 cells, and we confirmed its direct interference with the specific Th17 transcription factors Rorγt, Rora and Nfat5. Further, in vitro transfections of this cluster into Th17 cells reduced the secretion of Il‐17 cytokines. Hence, we suggest that the miR‐106a~363 cluster plays an important role in fine‐tuning the T helper differentiation towards Th17.
A correct in vitro differentiation of naive Cd4+ T cells into Th17 was confirmed by (i) expression of Th17‐ and Th2‐specific genes and production of respective cytokines; (ii) a unique gene expression signature in the microarray analysis of previously described genes involved in Th17 differentiation and effector function, including Rorc, Il17a, Il17f, Ahr, Il23r, Rora and Il22;6, 20 and (iii) strong enrichment of Th17‐associated functions such as defending against bacterial or fungal infections, and preventing/hindering recruitment of neutrophilic cells in IPA.
To identify the complex regulatory networks of Th cell differentiation, we combined our mRNA array data with a miRNA microarray of our Th17, Th2 and Th0 cells, creating in silico a hypothetical T helper cell differentiation model. Although this model is based on our own expression data of miRNA or mRNA and published data, it has to be emphasized that IPA only creates an in silico prediction of potential biological models but does not provide evidentiary facts. Nonetheless, in this theoretical model miR‐182 had a central role in Th17 cell function by potentially inhibiting the differentiation pathways of Th1 and Treg cells. Additionally, miR‐182 would affect the expression of Foxo1 and Foxo3, indirectly suppressing Foxp3 expression and consequently Treg cell differentiation. Binding of miR‐182 to Foxo1 and Foxo3 mRNA has been confirmed,21, 22 and the entire miRNA cluster 183~182 has just been shown by Ichiyama et al. to promote Th17 pathogenicity through direct repression of Foxo1.23 Further, Foxo1 and Foxo3 conditional knockout mice are depleted of Treg cells and have higher Cd4+ lymphocyte populations and inflammation.24, 25 Hence, Foxo1 regulation seems to be fine‐tuned by miRNAs in Th17 cells, influencing their pathogenicity and function. Taken together, these findings not only support our in vitro Th17 differentiation, but also the IPA‐driven target prediction approach of this study.
In our study, the miR‐212~132 cluster was highly increased in Th17 cells. Ahr signalling has been shown to induce the expression of this miRNA cluster and the production of Il‐17 in vitro and in vivo,11 which is similar to our data where both Ahr and the miR‐212~132 cluster were increased during Th17 differentiation. Additionally, we speculate that this might prevent Th1 differentiation as the miR‐212~132 cluster has been shown to inhibit Stat4, indirectly repressing Ifng.26 miR‐10b,27 miR‐210,28 miR‐15529 and miR‐30a30 have been previously described to be involved in Th17 regulation and function, but are not significantly altered in our arrays.
In our study, the expression of the entire cluster of miR‐106~363 was decreased. This cluster is located on the X chromosome,31 but little is known about its role in immune function. miR‐17, miR‐106a and miR‐20a up‐regulation has been shown to lead to macrophage activation after lipopolyaccharide stimulation32 and miR‐106a, miR‐20a, miR‐18b were found to be increased in Th1 cells compared with Th2 cells in a gene array study.33 We observed a decreased expression of these miRNAs in Th17 cells when compared with Th2 cells. Hence, this miRNA cluster seems to be expressed highest in Th1 cells, on intermediate levels in Th2 cells, and lowest in Th17 cells. We speculate that this differential, hierarchical regulation might be a hint for a functional relevance in the differentiation of Th cells into distinct subtypes. In our in silico model, luciferase reporter assays and in vitro functional analyses, we demonstrated that miR‐106a, miR‐18b and miR‐363‐3p can bind to the 3′ UTR of Nfat5 and Rora, leading to a consequent decrease of Il17a/f gene expression and reduction of Il‐17a protein production. This might be further amplified by additional binding of these miRNAs to the transcription factor Stat3, as has been shown for miR‐106a.34 Stat3 is, next to Rorα and Rorγt, essential for Th17 differentiation and Il17 gene expression.35 As the miR‐106~363 cluster is decreased in Th17 cells, Stat3 might remain at baseline levels, inducing Th17 differentiation and Il‐17 production.
As our data suggest a role for miRNA cluster 106a~363 in Th17 differentiation, it will be crucial to analyse its expression in animal models with ongoing Th17‐mediated inflammation, such as autoimmune encephalomyelitis, Crohn's disease or chronic respiratory diseases such as severe asthma and chronic obstructive pulmonary disease.3, 4, 5, 36 Given our findings, we speculate that therapy approaches using miR‐106a, miR‐18b and miR‐363‐3p might potentially ameliorate or prevent Th17‐cell‐mediated inflammation. Along this line, another member of the miR‐106a~363 cluster, miR‐20b, has been shown to be down‐regulated in the blood of patients with multiple sclerosis37 and in a Th17‐driven experimental model for EAE.37 A genetic depletion of the miR‐106a~363 cluster resulted in a more severe EAE course and up‐regulation of the miR‐20b target genes Rorgt, and Stat3,37 whereas lentiviral over‐expression of miR‐20b led to decreased Th17 cells and reduced EAE severity.38 Hence, these studies confirm the disease relevance of our in silico model, strengthening the suggestion that the miR‐106a~363 cluster might be an interesting target for therapeutic interventions.
In summary, by using microarray profiling we were able to create an in silico miRNA/mRNA regulatory network of Th17 cell differentiation. We observed that the most abundant up‐ and down‐regulated miRNAs are organized in distinct clusters, and the expression of the entire cluster of miR‐106a~363 was decreased in Th17 cells. Target prediction and a luciferase reporter assay revealed that this cluster interferes directly with key transcription factors of Th17 differentiation. Over‐expression of these miRNAs reduced Th17 differentiation and the secretion of Il‐17a. Hence, the present work is a first step towards the identification and understanding of the underlying molecular mechanisms involved in Th17 cell differentiation, which is crucial to develop novel and effective treatment strategies for Th17‐mediated inflammatory diseases.
Funding sources
This study was partially supported by grants from the Helmholtz Portfolio Theme ‘Metabolic Dysfunction and Common Disease’ (J.B.) and the Helmholtz Alliance ‘Imaging and Curing Environmental Metabolic Diseases, ICEMED’ (J.B.) and INSERM (S.K‐E.).
Authors contributions
S.K‐E. conceived the manuscript, designed and supervised the study and had the primary responsibility for writing; M.K. and S.B. performed and analysed the experiments and wrote the manuscript. K.G‐K. performed cloning experiments of the 3′ UTRs. M.I. and J.B. performed the mRNA and miRNA arrays and performed initial bioinformatics analysis. B.R. and O.E. participated in critical data interpretation and design of the study. All authors contributed to the writing of the manuscript.
Disclosures
The authors declare that there are no conflicts of interests regarding this study.
Supporting information
Figure S1. Messenger RNA heatmaps of all significantly regulated transcripts.
Figure S2. MicroRNA heatmaps of all significantly regulated transcripts.
Figure S3. Luciferase reporter assays with fragments of the 3′ untranslated region (UTR).
Table S1. Primer sequences used for quantitative RT‐PCRs and cloning.
Table S2. Function of T helper type 17 (Th17) enriched genes (Th17 versus Th0).
Table S3. Predicted miRNA–mRNA expression pairings (down‐regulated miRNAs, up‐regulated mRNAs) of in‐vitro‐differentiated T helper type 17 (Th17) versus Th2 versus Th0 cells as analysed by Ingenuity Pathway Analysis and TargetScan®.
Table S4. Predicted miRNA–mRNA expression pairings (up‐regulated miRNAs, down‐regulated mRNAs) of in‐vitro‐differentiated Th17 versus Th2 versus Th0 cells as analysed by Ingenuity Pathway Analysis and TargetScan®.
Table S5. DNA duplexes used for luciferase reporter assays.
Acknowledgements
We would like to thank Anke Bettenbrock and Rabea Imker for excellent technical support. S.B., O.E. and S.K.E. are members of COST Action BM1201.
Marc Kästle and Sabine Bartel contributed equally to this study.
References
- 1. Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel‐Moreno J, Cilley GE et al IL‐23 and IL‐17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol 2007; 8:369–77. [DOI] [PubMed] [Google Scholar]
- 2. Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of interleukin‐17A for systemic anti‐Candida albicans host defense in mice. J Infect Dis 2004; 190:624–31. [DOI] [PubMed] [Google Scholar]
- 3. Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD et al IL‐23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 2005; 201:233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feng T, Qin H, Wang L, Benveniste EN, Elson CO, Cong Y. Th17 cells induce colitis and promote Th1 cell responses through IL‐17 induction of innate IL‐12 and IL‐23 production. J Immunol 2011; 186:6313–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnes PJ. Immunology of asthma and chronic obstructive pulmonary disease. Nat Rev Immunol 2008; 8:183–92. [DOI] [PubMed] [Google Scholar]
- 6. Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y et al T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR α and ROR γ . Immunity 2008; 28:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld J‐C et al The aryl hydrocarbon receptor links TH17‐cell‐mediated autoimmunity to environmental toxins. Nature 2008; 453:106–9. [DOI] [PubMed] [Google Scholar]
- 8. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466:835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denli AM, Tops BBJ, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature 2004; 432:231–5. [DOI] [PubMed] [Google Scholar]
- 10. O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol 2010; 10:111–22. [DOI] [PubMed] [Google Scholar]
- 11. Nakahama T, Hanieh H, Nguyen NT, Chinen I, Ripley B, Millrine D et al Aryl hydrocarbon receptor‐mediated induction of the microRNA‐132/212 cluster promotes interleukin‐17‐producing T‐helper cell differentiation. Proc Natl Acad Sci USA 2013; 110:11964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J et al Molecular dissection of the miR‐17‐92 cluster's critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood 2011; 118:5487–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA‐21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol Immunol 2013; 4:435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D et al The microRNA cluster miR‐17~92 promotes TFH cell differentiation and represses subset‐inappropriate gene expression. Nat Immunol 2013; 14:840–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med 2005; 202:261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfaffl MW. A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. R Core Team (2011). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3‐900051‐07‐0 [Google Scholar]
- 18. Rainer J, Sanchez‐Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R‐ and bioconductor‐based web service for microarray data analysis. Nucleic Acids Res 2006; 34:W498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin Y, Chen Z, Liu X, Zhou X. Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol Biol 2013; 936:117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glader P, Smith ME, Malmhäll C, Balder B, Sjöstrand M, Qvarfordt I et al Interleukin‐17‐producing T‐helper cells and related cytokines in human airways exposed to endotoxin. Eur Respir J 2010; 36:1155–64. [DOI] [PubMed] [Google Scholar]
- 21. Stittrich A‐B, Haftmann C, Sgouroudis E, Kühl AA, Hegazy AN, Panse I et al The microRNA miR‐182 is induced by IL‐2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol 2010; 11:1057–62. [DOI] [PubMed] [Google Scholar]
- 22. Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez‐Diaz S et al Aberrant miR‐182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia‐associated transcription factor. Proc Natl Acad Sci USA 2009; 106:1814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ichiyama K, Gonzalez‐Martin A, Kim B‐S, Jin HY, Jin W, Xu W et al The microRNA‐183‐96‐182 cluster promotes T helper 17 cell pathogenicity by negatively regulating transcription factor foxo1 expression. Immunity 2016; 44:1284–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerdiles YM, Stone EL, Beisner DR, Beisner DL, McGargill M a, Ch'en IL et al Foxo transcription factors control regulatory T cell development and function. Immunity 2010; 33:890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ouyang W, Beckett O, Ma Q, Paik J, DePinho R a, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol 2010; 11:618–27. [DOI] [PubMed] [Google Scholar]
- 26. Huang Y, Lei Y, Zhang H, Hou L, Zhang M, Dayton AI. MicroRNA regulation of STAT4 protein expression: rapid and sensitive modulation of IL‐12 signaling in human natural killer cells. Blood 2011; 118:6793–802. [DOI] [PubMed] [Google Scholar]
- 27. Chen L, Al‐Mossawi MH, Ridley A, Sekine T, Hammitzsch A, de Wit J et al miR‐10b‐5p is a novel Th17 regulator present in Th17 cells from ankylosing spondylitis. Ann Rheum Dis 2016; doi: 10.1136/annrheumdis‐2016‐210175. [DOI] [PubMed] [Google Scholar]
- 28. Wang H, Flach H, Onizawa M, Wei L, McManus MT, Weiss A. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia‐regulated microRNA miR‐210. Nat Immunol 2014; 15:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang A, Wang K, Zhou C, Gan Z, Ma D, Ye P et al Knockout of microRNA‐155 ameliorates the Th1/Th17 immune response and tissue injury in chronic rejection. J Hear Lung Transplant 2016; 36:175–84. [DOI] [PubMed] [Google Scholar]
- 30. Zhao M, Sun D, Guan Y, Wang Z, Sang D, Liu M et al Disulfiram and diphenhydramine hydrochloride upregulate miR‐30a to suppress IL‐17‐associated autoimmune inflammation. J Neurosci 2016; 36:9253–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindsay MA. microRNAs and the immune response. Trends Immunol 2008; 29:343–51. [DOI] [PubMed] [Google Scholar]
- 32. Zhu D, Pan C, Li L, Bian Z, Lv Z, Shi L et al MicroRNA‐17/20a/106a modulate macrophage inflammatory responses through targeting signal‐regulatory protein α . J Allergy Clin Immunol 2013; 132:426–36.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sasaki K, Kohanbash G, Hoji A, Ueda R, McDonald HA, Reinhart TA et al miR‐17‐92 expression in differentiated T cells – implications for cancer immunotherapy. J Transl Med 2010; 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang M, Ye Y, Cong J, Pu D, Liu J, Hu G et al Regulation of STAT3 by miR‐106a is linked to cognitive impairment in ovariectomized mice. Brain Res 2013; 1503:43–52. [DOI] [PubMed] [Google Scholar]
- 35. Harris TJ, Grosso JF, Yen H, Xin H, Kortylewski M, Albesiano E et al Cutting Edge: An In Vivo Requirement for STAT3 Signaling in TH17 Development and TH17‐Dependent Autoimmunity. J Immuno 2014; 179:4313–17. [DOI] [PubMed] [Google Scholar]
- 36. Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang T‐H et al MicroRNA‐19a‐3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra‐1 proto‐oncogene. Oncogene 2014; 33:3014–23. [DOI] [PubMed] [Google Scholar]
- 37. Ingwersen J, Menge T, Wingerath B, Kaya D, Graf J, Prozorovski T et al Natalizumab restores aberrant miRNA expression profile in multiple sclerosis and reveals a critical role for miR‐20b. Ann Clin Transl Neurol 2015; 2:43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu E, Wang X, Zheng B, Wang Q, Hao J, Chen S et al miR‐20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORγt and STAT3. J Immunol 2014; 192:5599–609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Messenger RNA heatmaps of all significantly regulated transcripts.
Figure S2. MicroRNA heatmaps of all significantly regulated transcripts.
Figure S3. Luciferase reporter assays with fragments of the 3′ untranslated region (UTR).
Table S1. Primer sequences used for quantitative RT‐PCRs and cloning.
Table S2. Function of T helper type 17 (Th17) enriched genes (Th17 versus Th0).
Table S3. Predicted miRNA–mRNA expression pairings (down‐regulated miRNAs, up‐regulated mRNAs) of in‐vitro‐differentiated T helper type 17 (Th17) versus Th2 versus Th0 cells as analysed by Ingenuity Pathway Analysis and TargetScan®.
Table S4. Predicted miRNA–mRNA expression pairings (up‐regulated miRNAs, down‐regulated mRNAs) of in‐vitro‐differentiated Th17 versus Th2 versus Th0 cells as analysed by Ingenuity Pathway Analysis and TargetScan®.
Table S5. DNA duplexes used for luciferase reporter assays.


