Abstract
Background
Attention deficit hyperactivity disorder (ADHD) is an etiologically complex childhood onset neurobehavioral disorder characterized by age-inappropriate inattention, hyperactivity, and impulsivity. Symptom severity varies widely and boys are diagnosed more frequently than girls. ADHD probands were reported to have abnormal transmissions of dopamine, serotonin, and/or noradrenaline. Monoamine oxidase A (MAOA) and B (MAOB), mitochondrial outer membrane bound two isoenzymes, mediate degradation of these neurotransmitters and thus regulating their circulating levels. Case-control analyses in different populations, including Indians, suggested involvement of MAOA and MAOB genes in the etiology of ADHD. Due to high heritability rate of ADHD, we tested familial transmission of MAOA and MAOB variants to ADHD probands in 190 nuclear families having ADHD probands from Indo-Caucasoid ethnicity.
Methods
Subjects were recruited following the Diagnostic and Statistical Manual of Mental Disorders-4th edition (DSM-IV). Appropriate scales were used for measuring the behavioral traits in probands. Genotyping was performed through PCR-based amplification of target sites followed by DNA-sequencing and/or gel-electrophoresis. Data obtained were analyzed by family based statistical methods.
Results
Out of 58 variants present in the analyzed sites only 15 were found to be polymorphic (30 bp-uVNTR, rs5906883, rs1465107, rs1465108, rs5905809, rs5906957, rs6323, rs1137070 from MAOA and rs4824562, rs56220155, rs2283728, rs2283727, rs3027441, rs6324, rs3027440 from MAOB). Statistically significant maternal transmission of alleles to male probands was observed for MAOA rs5905809 ‘G’ (p = 0.04), rs5906957 ‘A’ (p = 0.04), rs6323 ‘G’ (p = 0.0001) and MAOB rs56220155 ‘A’ (p = 0.002), rs2283728 ‘C’ (p = 0.0008), rs2283727 ‘C’ (p = 0.0008), rs3027441 ‘T’ (p = 0.003), rs6324 ‘C’ (p = 0.003), rs3027440 ‘T’ (p = 0.0002). Significantly preferential maternal transmissions of different haplotype combinations to male probands were also noticed (p < 0.05), while female probands did not reveal such transmission bias. Behavioral traits of male probands exhibited significant association with gene variants. Age of the mother at pregnancy also revealed association with risk variants of male probands.
Conclusions
It may be inferred that the MAOA and MAOB variants may contribute to the etiology of ADHD in the Indo-Caucasoid population and could be responsible for higher occurrence of ADHD in the boys.
Electronic supplementary material
The online version of this article (10.1186/s12881-017-0469-5) contains supplementary material, which is available to authorized users.
Keywords: ADHD, MAOA, MAOB, Genotyping, Maternal transmission, Behavioral trait, Maternal age, Linkage disequilibrium, Indo-Caucasoid population
Background
Attention deficit hyperactivity disorder (ADHD) is an etiologically complex neurobehavioral disorder, diagnosed mostly during early childhood [1]. The disorder is highly prevalent throughout the world, including India, and boys are more frequently diagnosed with ADHD as compared to girls [2–6]. Age-inappropriate persistent and pervasive symptoms of inattention, hyperactivity, and impulsivity [1] often lead to impairments in academic performances as well as social life [7, 8]. Other psychiatric conditions, frequently detected as co-morbidity in subjects with ADHD, may make the situation worse [9, 10]. Thus an early diagnosis, leading to early intervention, becomes crucial for successful management.
Being a multi-factorial disorder with almost 76% heritability [11, 12], ADHD is believed to have significant influence of multiple gene variants [13–16]. Candidate genes involved in the regulation of dopamine, serotonin, and noradrenalin were widely studied in ADHD subjects since behavioral traits are regulated by these neurotransmitters [13, 15, 17, 18] and both dopaminergic [15, 19–21] and serotonergic [22] transmissions revealed significant impact on behavioral as well as cognitive features.
Two flavin containing isoenzymes, monoamine oxidase A (MAOA) and B (MAOB), help in the deamination of biogenic amines from both endogenous and exogenous sources [23, 24]. Both the enzymes are localized in the mitochondrial outer membrane and metabolize dopamine, tyramine, and tryptamine with equal efficiency [25–27]. Comparative analysis also revealed that while MAOA preferentially deaminates serotonin, noradrenalin, adrenaline, and melatonin, preferred substrates for MAOB are phenylethylamine (PEA) and benzylamine [25–27].
In men of a Dutch family, MAOA deficiency showed association with aggressive behavior [28] and deletion of the MAOA gene showed association with aggressive phenotypes across species [29–31]. MAOA also exhibited association with vulnerability to disorders of attention and impulsivity [32] and a possible link between predisposition to novelty seeking [33], making the MAOA gene, encoding for the MAOA enzyme, a prime candidate for ADHD [31, 32, 34, 35].
MAOB was also postulated to regulate impulsivity, attention and vulnerability to ADHD through metabolism of dopamine, although to a lesser extent as compared to MAOA [20, 32]. Correlation of platelet MAOB activity with sensation seeking and other behavioral abnormalities have also been reported [36, 37]. Negative emotionality of healthy volunteers showed association with MAOB polymorphisms [38]. However, platelet MAOB activity failed to correlate with brain MAOB activity, thus questioning the usefulness of MAOB as a marker for psychiatric behavior [39].
Proportional analysis in patients revealed that the absence of MAOA leads to greater change in neurotransmitter metabolism than absence of MAOB [40]. While MAOA knockout mice showed increased levels of serotonin, norepinephrine and dopamine in the brain [41], only level of PEA was increased in MAOB knockout mice [42]. MAOA and MAOB double knockout mice showed an increased reactivity to stress [30] and increased levels of serotonin, norepinephrine, dopamine, and PEA in the brain to a much greater degree than in either MAOA or MAOB single knockout mice [43].
In this backdrop of information, both MAOA and MAOB genes, located on the X chromosome [26], were considered to contribute to the etiology of ADHD [31, 32, 34, 44–46]. However, worldwide only a few MAOA and MAOB gene variants were studied and the data obtained were neither consistent nor conclusive [31, 35, 37, 44–47]. Our population-based analysis on 58 MAO gene variants revealed association of a number of variants with ADHD [48, 49]. Due to high heritability of ADHD traits, in the present study all these variants were explored to identify familial transmission pattern. Additionally, based on the X-chromosomal location of MAOA and MAOB [46], we have performed gender based stratified analysis to identify whether any variant is preferentially transmitted to the probands and thus may have a role in the gender bias often reported in ADHD.
Methods
Subject recruitment
A total of 190 ADHD probands (166 males and 24 females) and their biological parents, of Indo-Caucasoid ethnicity from the eastern India, were recruited from the out-patient department of Manovikas Kendra Rehabilitation and Research Institute for the Handicapped, Kolkata, India. Diagnosis was performed by child psychiatrist and clinical psychologist following the Diagnostic and Statistical Manual of Mental Disorders-4th edition (DSM-IV) criteria [50]. 74.74% of the recruited ADHD probands were of the combined subtype, while inattentive and hyperactive-impulsive subtypes were of 13.68% and 11.58% respectively. Mean age of the ADHD probands was 8.01 ± 0.22 years (Mean ± SE). Psychological evaluation was done through – the revised Conners’ Parents Rating Scale (CPRS-R) [51] and Wechsler Intelligence Scale for Children >5 yrs. [52] / Developmental Screening Test [53] for children <5 yrs. for the inattention-hyperactivity level and intelligent quotient (IQ)/ developmental quotient (DQ) status respectively. Oppositional defiant disorder (ODD) and conduct problems of ADHD probands were assessed using the DSM-IV score and Parental Account of Children’s Symptoms (PACS) score respectively. Probands with any other neuropsychiatric disorders, mental retardation (IQ ≤ 70) including Down syndrome and Fragile-X syndrome, pervasive developmental disorder were excluded from the study. Among 166 male ADHD probands 133 were trios, 25 were duos (6 excluding mother and 19 excluding father) and 8 were single probands. Among the 24 female probands, 15 were trios, 6 were duos (having mother only) and 3 were single probands. Informed written consent was obtained from guardians / biological parents of the probands participating in the study and the protocol was approved by the Institutional Human Ethical Committee.
Genotyping
Peripheral blood of the study participants was collected by a trained phlebotomist and used for genomic DNA preparation following the standard protocol [54]. The target regions were amplified via polymerase chain reaction (PCR) using oligonucleotides designed in the lab using the Primer3 software [55] and the amplicons were utilized for genotyping the samples either through gel electrophoresis or using Sanger sequencing by capillary electrophoresis method [56]. For sequence analysis of the amplicons, Applied Biosystems 3130 Genetic Analyzer was used [48, 49]. Chromatograms were also analyzed manually and for the identification of heterozygous SNPs, >25% base calling was accepted. Detailed analytic protocols for PCR amplification and genotyping were published earlier [48, 49].
Statistical analyses of data
Genotypic counts of only the female subjects (i.e. female ADHD probands and mother of the probands) were used for analyzing the Hardy-Weinberg equilibrium [57], since the MAO genes are located on the X-chromosome. For the same reason, family-based analysis on male ADHD probands was carried out considering only the maternal transmission. Paternal transmission was considered for female ADHD probands only. Haplotype-based haplotype relative risk (HHRR) analysis [58] was performed using UNPHASED v 3.1.5 [59] to identify allelic and haplotypic transmission patterns. Correction for multiple testing was done while running the UNPHASED at 1000-fold iteration. Relative risk or Risk ratio for variants showing significant association was calculated online [60]. Pair-wise linkage disequilibrium (LD) between the variants was calculated using the Haploview program version 4.2 [61], considering male and female probands separately. Since MAO genes are X-linked, for female probands the parental genotype data and for male probands only the maternal genotype data were used for comparative analysis.
Stratified analysis on behavioral traits, age-of-onset, and maternal age at pregnancy
Based on the CPRS-R, respective ‘T scores’ for oppositional behavior, cognitive problems / inattention, hyperactivity, and ADHD index were obtained for ADHD probands (N = 166). DSM-IV score for ODD trait and PACS for conduct problems of ADHD probands were also obtained. CPRS-R ‘T scores’ ranged between 38 to 90 while DSM-IV scores and PACS scores ranged between 0 to 36 and 0 to 90 respectively. Behavioral traits/scores of ADHD probands were utilized for genetic association analysis. Male probands were divided into two sub-groups based on the presence/absence of the derived allele of each variant. Allelic association with behavioral scores was analyzed using Student’s t-test [62] in the presence of normal distribution of data and equal variances in the two comparing groups. Age of the male ADHD probands at the time of onset of the disorder were used for stratified analysis; probands with detectable symptoms at an age ≤ 7 years (N = 109) were considered under ‘early onset’, while those with detectable symptoms after 7 years (N = 57) were classified under the ‘late onset’. Association of alleles with age-of-onset of ADHD in the male probands was calculated using the chi-square test [62]. To calculate the impact of maternal age at pregnancy, allelic frequencies of male probands born to mothers ≤26 years (N = 71) were compared to that of male probands born to mothers at >26 years (N = 81) of age using the chi-square test [62]. As the number of female probands was limited, association of variants with behavioral scores, age-of-onset of ADHD, and maternal age at pregnancy were not analyzed.
Results
Out of 58 variants present in the analyzed sites, only 15 (30 bp-uVNTR, rs5906883, rs1465107, rs1465108, rs5905809, rs5906957, rs6323, rs1137070 from MAOA and rs4824562, rs56220155, rs2283728, rs2283727, rs3027441, rs6324, rs3027440 from MAOB) were found to be polymorphic. All the studied polymorphic variants followed the Hardy-Weinberg equilibrium in the female subjects, i.e. female ADHD probands and mother of the ADHD probands (Additional file 1). Family-based analysis showed statistically significant maternal over-transmission of MAOA rs5905809 ‘G’ (p = 0.04), rs5906957 ‘A’ (p = 0.04) and rs6323 ‘G’ (p = 0.0001) alleles and MAOB rs56220155 ‘A’ (p = 0.002), rs2283728 ‘C’ (p = 0.0008), rs2283727 ‘C’ (p = 0.0008), rs3027441 ‘T’ (p = 0.003), rs6324 ‘C’ (p = 0.003) and rs3027440 ‘T’ (p = 0.0002) alleles to the male probands (Table 1). The relative risk was also statistically significant for MAOA rs5905809 ‘G’, rs5906957 ‘A’ and rs6323 ‘G’ and all the MAOB variants excepting for rs4824562 (Table 1). No such parental bias in transmissions was observed in the female probands (Additional file 2). Several intra-genetic as well as inter-genetic haplotypic combinations also showed statistically significant maternal over-transmission to the male probands (p ≤ 0.05) (Table 2 & Additional file 3). Most significantly over-transmitted (p = 0.002) intra-genetic haplotypes of MAOA were rs5906883-rs6323 ‘C-G’, rs5905809-rs6323 ‘G-G’, and rs5906957-rs6323 ‘A-G’ (Table 2). Most significantly over-transmitted (p = 1.66E-05) intra-genetic haplotypes of MAOB were rs3027440-rs2283727 ‘T-C’ and rs3027440-rs2283728 ‘T-C’ (Table 2). Inter-genetic haplotype combinations, between MAOA and MAOB, showing over-transmission (p = 2.05E-05) were rs6323-rs2283727 ‘G-C’ and rs6323-rs2283728 ‘G-C’ (Table 2). Preferential non-transmission (p ≤ 0.05) of several haplotypes were also noticed (Additional file 4) in the male probands. No such significant biased parental transmissions of haplotypes were observed in the female probands (Additional file 5).
Table 1.
Maternal allelic transmission to male ADHD probands
| Genes | Variants | Alleles | Transmitted | Non-transmitted | Chi-square (p-value) | Relative risk (95% confidence interval) |
|---|---|---|---|---|---|---|
| MAOA | 30 bp-uVNTR | 3R | 0.68 | 0.65 | 0.37 (0.54) | – |
| 4R | 0.32 | 0.35 | ||||
| rs5906883 | A | 0.67 | 0.64 | 0.36 (0.55) | – | |
| C | 0.33 | 0.36 | ||||
| rs1465107 | G | 0.31 | 0.39 | 2.44 (0.12) | – | |
| A | 0.69 | 0.61 | ||||
| rs1465108 | A | 0.69 | 0.61 | 2.44 (0.12) | – | |
| G | 0.31 | 0.39 | ||||
| rs5905809 | C | 0.33 | 0.44 | 4.03 (0.04) | 1.20 (1.00 – 1.44) | |
| G | 0.67 | 0.56 | ||||
| rs5906957 | A | 0.67 | 0.56 | 4.03 (0.04) | 1.20 (1.00 – 1.44) | |
| G | 0.33 | 0.44 | ||||
| rs6323 | T | 0.24 | 0.45 | 15.15 (0.0001) | 1.38 (1.17 – 1.63) | |
| G | 0.76 | 0.55 | ||||
| rs1137070 | C | 0.32 | 0.41 | 2.40 (0.12) | – | |
| T | 0.68 | 0.59 | ||||
| MAOB | rs4824562 | A | 0.80 | 0.73 | 2.23 (0.14) | – |
| G | 0.20 | 0.27 | ||||
| rs56220155 | G | 0.26 | 0.43 | 9.16 (0.002) | 1.29 (1.09 – 1.52) | |
| A | 0.74 | 0.57 | ||||
| rs2283728 | T | 0.19 | 0.36 | 11.26 (0.0008) | 1.27 (1.10 – 1.46) | |
| C | 0.81 | 0.64 | ||||
| rs2283727 | C | 0.81 | 0.64 | 11.26 (0.0008) | 1.27 (1.10 – 1.46) | |
| A | 0.19 | 0.36 | ||||
| rs3027441 | C | 0.20 | 0.35 | 8.86 (0.003) | 1.23 (1.07 – 1.42) | |
| T | 0.80 | 0.65 | ||||
| rs6324 | C | 0.80 | 0.65 | 8.86 (0.003) | 1.23 (1.07 – 1.42) | |
| T | 0.20 | 0.35 | ||||
| rs3027440 | T | 0.86 | 0.68 | 13.67 (0.0002) | 1.26 (1.11 – 1.43) | |
| C | 0.14 | 0.32 |
Statistically significant differences are presented in bold
Table 2.
Intra- and inter-genetic haplotypes transmitted most significantly to male ADHD probands from mothers
| Combinations | Variants | Haplotype | Transmitted | Non-transmitted | Chi-square (p-value) |
|---|---|---|---|---|---|
| Intra genetic in MAOA | rs5906883-rs6323 | C-G | 0.15 | 0.05 | 9.47 (0.002) |
| rs5905809-rs6323 | G-G | 0.62 | 0.44 | 9.63 (0.002) | |
| rs5906957-rs6323 | A-G | 0.62 | 0.44 | 9.63 (0.002) | |
| Inter genetic between MAOA and MAOB | rs6323-rs2283727 | G-C | 0.58 | 0.34 | 18.15 (2.05E-05) |
| rs6323-rs2283728 | G-C | 0.58 | 0.34 | 18.15 (2.05E-05) | |
| Intra genetic in MAOB | rs3027440-rs2283727 | T-C | 0.80 | 0.57 | 18.55 (1.66E-05) |
| rs3027440-rs2283728 | T-C | 0.80 | 0.57 | 18.55 (1.66E-05) |
Statistically significant differences are presented in bold
In the male ADHD probands, intra-genetic pair-wise LDs for MAOA and MAOB variants were found to be same as reported earlier [48, 49]. Inter-genetic pair-wise analysis revealed strong LD of MAOA rs6323 with MAOB rs3027440, rs6324, rs3027441, rs2283727, rs2283728, and rs56220155 in the male probands (Fig. 1a, Additional file 6). In mothers of the male probands, all the variants from MAOA gene were found to be in pair-wise LDs with each other (Fig. 1b, Additional file 6). Complete pair-wise LDs were found between rs1465107 and rs1465108; and rs5905809 and rs5906957 respectively (Fig. 1b, Additional file 6). In the MAOB, all the variants, except rs4824562, were found to be in strong pair-wise LDs with each other (Fig. 1b, Additional file 6). Complete pair-wise LDs were found between rs6324 and rs3027441; and rs2283727 and rs2283728 respectively (Fig. 1b, Additional file 7). Strong pair-wise LDs of rs56220155 with rs2283727 and rs2283728 respectively were also observed (Fig. 1b, Additional file 6). In the female ADHD probands, no inter-genetic pair-wise LDs were observed (Fig. 2a, Additional file 6). Intra-genetic pair-wise LDs were found to be same as reported earlier [48, 49]. In the group of parents of the female probands all the variants from MAOA gene, except 30 bp-uVNTR and rs1137070, were found to be in pair-wise LDs with each other (Fig. 2b, Additional file 6). Complete pair-wise LDs were found between rs1465107 and rs1465108; and rs5905809 and rs5906957 respectively (Fig. 2b, Additional file 6). Strong pair-wise LDs of rs5906883 with rs1465107 and rs1465108 respectively were observed (Fig. 2b, Additional file 6). Strong pair-wise LDs of rs6323 with rs5905809 and rs5906957 respectively were also observed (Fig. 2b, Additional file 6). From MAOB gene, all the variants, except rs4824562, were found to be in strong pair-wise LDs with each other (Fig. 2b, Additional file 6). Complete pair-wise LDs were found between rs6324 and rs3027441; and rs2283727 and rs2283728 respectively (Fig. 2b, Additional file 6). Strong pair-wise LDs of rs3027440 with rs6324 and rs3027441 respectively were observed (Fig. 2b, Additional file 6). Strong pair-wise LDs of rs56220155 with rs2283727 and rs2283728 respectively were also noticed (Fig. 2b, Additional file 6).
Fig. 1.
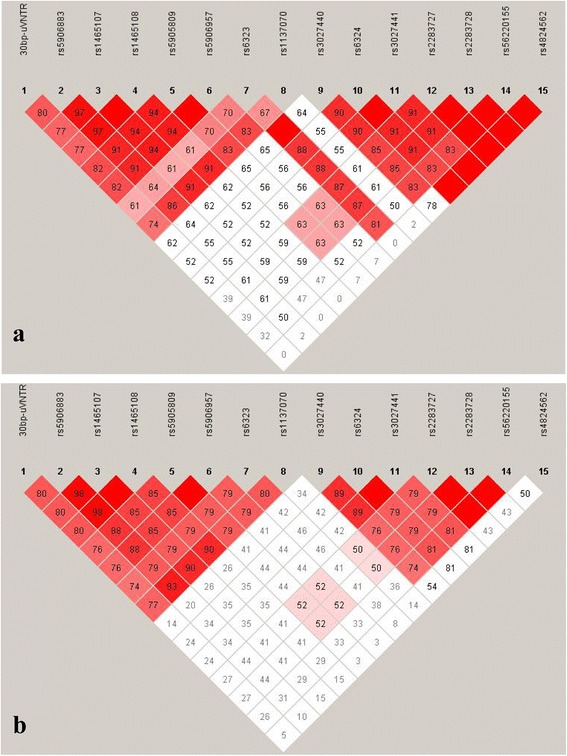
Plot of pair-wise linkage disequilibrium (LD) between the polymorphic variants from MAOA and MAOB genes in a. Male ADHD cases; b. Mothers of the male ADHD cases. Diamonds without numbers represent D’ values of 1.0; all numbers represent the D’ value expressed as a percentile. D’ is a measure of the frequency of association of alleles at 2 loci
Fig. 2.
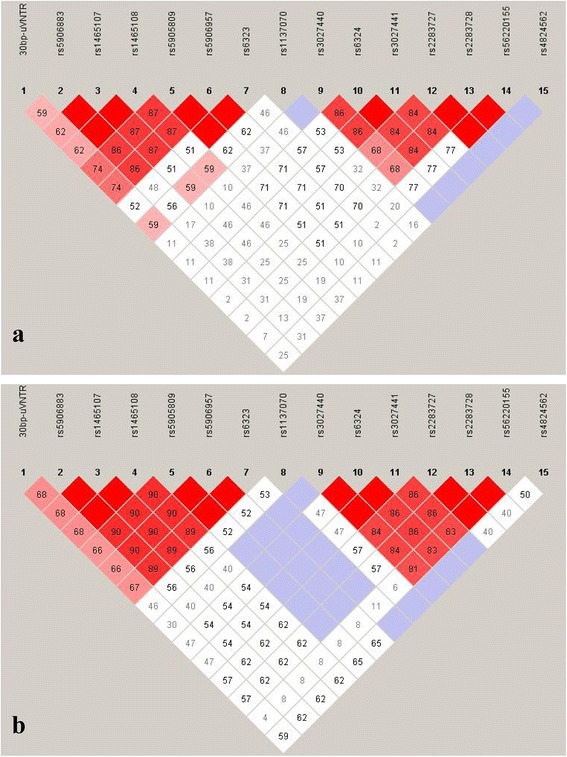
Plot of pair-wise linkage disequilibrium (LD) between the polymorphic variants from MAOA and MAOB genes in a. Female ADHD cases; b. Parents of the female ADHD cases. Diamonds without numbers represent D’ values of 1.0; all numbers represent the D’ value expressed as a percentile. D’ is a measure of the frequency of association of alleles at 2 loci
Behavioral traits were calculated by obtaining Mean ± standard error (SE). CPRS-R ‘T scores’ obtained were 62.81 ± 1.23 for oppositional behavior, 72.38 ± 0.79 for cognitive problems / inattention, 74.50 ± 0.99 for hyperactivity, and 71.74 ± 0.68 for ADHD index. DSM-IV scores for ODD trait was 15.05 ± 0.85 and PACS scores for conduct problems was 16.77 ± 1.19. Male ADHD probands having rs6323 ‘G’ allele showed statistically significant higher mean ‘T score’ for hyperactivity and ADHD index as compared to those with the ‘T’ allele (Table 3). Significantly higher mean ‘DSM-IV score’ for ODD trait was also noticed in the male ADHD probands having rs1137070 ‘C’ allele as compared to the male probands having the ‘T’ allele (Table 3). MAOB rs2283728 ‘T’ and rs2283727 ‘A’ showed statistically significant association with higher mean ‘T score’ for hyperactivity in the male ADHD probands in comparison to probands having the ‘C’ alleles of the respective variants (Table 3). Significantly higher mean ‘T score’ for cognitive problems / inattention and ADHD index were also noticed in the male ADHD probands having rs3027441 ‘C’ and rs6324 ‘T’ alleles than the male ADHD probands having ‘T’ and ‘C’ alleles of the respective variants (Table 3). No significant association between ‘T scores’ for oppositional behavior and PACS scores for conduct problems were noticed (Additional file 7). Age of onset of ADHD in male probands also failed to show any significant association (Additional file 8).
Table 3.
Analysis of association between alleles & ADHD associated trait scores of male ADHD probands
| Genes | Variants | Alleles | ‘T scores’ for cognitive problems / inattention | ‘T scores’ for hyperactivity | ‘T scores’ for ADHD index | DSM-IV scores for ODD trait | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SE | p | Mean ± SE | p | Mean ± SE | p | Mean ± SE | p | |||
| MAOA | 30 bp-uVNTR | 3R | 71.90 ± 1.03 | 0.26 | 75.16 ± 1.23 | 0.15 | 71.27 ± 0.83 | 0.29 | 14.75 ± 1.10 | 0.32 |
| 4R | 70.77 ± 1.37 | 72.93 ± 1.74 | 70.45 ± 1.19 | 15.68 ± 1.76 | ||||||
| rs5906883 | A | 72.26 ± 1.01 | 0.11 | 75.34 ± 1.27 | 0.11 | 71.48 ± 0.84 | 0.17 | 14.42 ± 1.06 | 0.17 | |
| C | 70.15 ± 1.42 | 72.70 ± 1.63 | 70.11 ± 1.16 | 16.32 ± 1.84 | ||||||
| rs1465107 | G | 71.14 ± 1.47 | 0.37 | 72.74 ± 1.71 | 0.12 | 70.51 ± 1.21 | 0.31 | 16.11 ± 1.80 | 0.21 | |
| A | 71.71 ± 1.00 | 75.22 ± 1.23 | 71.24 ± 0.83 | 14.53 ± 1.08 | ||||||
| rs1465108 | A | 71.71 ± 1.00 | 0.37 | 75.22 ± 1.23 | 0.12 | 71.24 ± 0.83 | 0.31 | 14.53 ± 1.08 | 0.21 | |
| G | 71.14 ± 1.47 | 72.74 ± 1.71 | 70.51 ± 1.21 | 16.11 ± 1.80 | ||||||
| rs5905809 | C | 70.17 ± 1.37 | 0.12 | 72.24 ± 1.60 | 0.06 | 69.83 ± 1.13 | 0.10 | 16.21 ± 1.80 | 0.19 | |
| G | 72.24 ± 1.03 | 75.58 ± 1.27 | 71.63 ± 0.85 | 14.47 ± 1.08 | ||||||
| rs5906957 | A | 72.24 ± 1.03 | 0.12 | 75.58 ± 1.27 | 0.06 | 71.63 ± 0.85 | 0.10 | 14.47 ± 1.08 | 0.19 | |
| G | 70.17 ± 1.37 | 72.24 ± 1.60 | 69.83 ± 1.13 | 16.21 ± 1.80 | ||||||
| rs6323 | T | 69.48 ± 1.62 | 0.08 | 70.88 ± 2.12 | 0.02 | 69.00 ± 1.34 | 0.04 | 15.35 ± 2.11 | 0.43 | |
| G | 72.20 ± 0.95 | 75.60 ± 1.12 | 71.67 ± 0.78 | 14.95 ± 1.02 | ||||||
| rs1137070 | C | 70.35 ± 1.46 | 0.15 | 73.00 ± 1.66 | 0.15 | 70.22 ± 1.18 | 0.20 | 17.32 ± 1.77 | 0.04 | |
| T | 72.15 ± 1.00 | 75.17 ± 1.26 | 71.42 ± 0.83 | 13.91 ± 1.07 | ||||||
| MAOB | rs4824562 | A | 71.29 ± 0.95 | 0.29 | 73.79 ± 1.10 | 0.11 | 70.59 ± 0.77 | 0.12 | 14.88 ± 1.04 | 0.35 |
| G | 72.44 ± 1.65 | 76.85 ± 2.41 | 72.59 ± 1.43 | 15.81 ± 2.20 | ||||||
| rs56220155 | G | 72.08 ± 1.36 | 0.34 | 76.86 ± 1.81 | 0.07 | 72.67 ± 1.15 | 0.07 | 13.82 ± 1.40 | 0.21 | |
| A | 71.31 ± 1.02 | 73.50 ± 1.19 | 70.38 ± 0.82 | 15.51 ± 1.17 | ||||||
| rs2283728 | T | 72.25 ± 1.42 | 0.32 | 77.79 ± 1.63 | 0.04 | 73.04 ± 1.25 | 0.06 | 13.73 ± 1.57 | 0.25 | |
| C | 71.33 ± 0.98 | 73.51 ± 1.18 | 70.45 ± 0.79 | 15.35 ± 1.09 | ||||||
| rs2283727 | C | 71.33 ± 0.98 | 0.32 | 73.51 ± 1.18 | 0.04 | 70.45 ± 0.79 | 0.06 | 15.35 ± 1.09 | 0.25 | |
| A | 72.25 ± 1.42 | 77.79 ± 1.63 | 73.04 ± 1.25 | 13.73 ± 1.57 | ||||||
| rs3027441 | C | 74.31 ± 1.60 | 0.04 | 76.83 ± 1.58 | 0.10 | 73.72 ± 1.27 | 0.02 | 14.15 ± 1.74 | 0.34 | |
| T | 70.74 ± 0.95 | 73.74 ± 1.20 | 70.23 ± 0.78 | 15.23 ± 1.07 | ||||||
| rs6324 | C | 70.74 ± 0.95 | 0.04 | 73.74 ± 1.20 | 0.10 | 70.23 ± 0.78 | 0.02 | 15.23 ± 1.07 | 0.34 | |
| T | 74.31 ± 1.60 | 76.83 ± 1.58 | 73.72 ± 1.27 | 14.15 ± 1.74 | ||||||
| rs3027440 | T | 71.08 ± 0.90 | 0.12 | 73.77 ± 1.14 | 0.08 | 70.57 ± 0.76 | 0.08 | 14.90 ± 1.06 | 0.34 | |
| C | 73.73 ± 2.00 | 77.64 ± 1.83 | 73.14 ± 1.45 | 16.00 ± 1.68 | ||||||
Statistically significant differences are presented in bold
Stratified analysis revealed higher occurrence of MAOA 30 bp-uVNTR 3-repeat (3R), rs6323 ‘G’, and rs1137070 ‘T’ variants in the male ADHD probands (Table 4) born to younger mothers (maternal age at pregnancy ≤26 years). Significant risk of association of these variants was also evident from higher relative risk (Table 4). MAOB variants failed to show any statistically significant difference (Table 4).
Table 4.
Analysis of association between maternal age at pregnancy and MAO variants of male ADHD probands
| Genes | Variants | Alleles | ≤ 26 yrs | > 26 yrs | Chi-square (p-value) | Relative risk (95% confidence interval) |
|---|---|---|---|---|---|---|
| MAOA | 30 bp-uVNTR | 3R | 0.76 | 0.61 | 4.03 (0.05) | 1.26 (1.01 – 1.57) |
| 4R | 0.24 | 0.39 | ||||
| rs5906883 | A | 0.73 | 0.61 | 2.66 (0.10) | – | |
| C | 0.27 | 0.39 | ||||
| rs1465107 | G | 0.25 | 0.36 | 2.3 (0.13) | – | |
| A | 0.75 | 0.64 | ||||
| rs1465108 | A | 0.75 | 0.64 | 2.3 (0.13) | – | |
| G | 0.25 | 0.36 | ||||
| rs5905809 | C | 0.28 | 0.39 | 2.09 (0.15) | – | |
| G | 0.72 | 0.61 | ||||
| rs5906957 | A | 0.72 | 0.61 | 2.09 (0.15) | – | |
| G | 0.28 | 0.39 | ||||
| rs6323 | T | 0.15 | 0.33 | 7.22 (0.007) | 1.27 (1.06 – 1.52) | |
| G | 0.85 | 0.67 | ||||
| rs1137070 | C | 0.25 | 0.40 | 3.87 (0.05) | 1.23 (1.00 – 1.54) | |
| T | 0.75 | 0.60 | ||||
| MAOB | rs4824562 | A | 0.83 | 0.81 | 0.06 (0.81) | – |
| G | 0.17 | 0.19 | ||||
| rs56220155 | G | 0.33 | 0.20 | 3.66 (0.06) | 1.19 (1.00 – 1.44) | |
| A | 0.67 | 0.80 | ||||
| rs2283728 | T | 0.23 | 0.16 | 0.98 (0.32) | – | |
| C | 0.77 | 0.84 | ||||
| rs2283727 | C | 0.77 | 0.84 | 0.98 (0.32) | – | |
| A | 0.23 | 0.16 | ||||
| rs3027441 | C | 0.25 | 0.15 | 2.51 (0.11) | – | |
| T | 0.75 | 0.85 | ||||
| rs6324 | C | 0.75 | 0.85 | 2.51 (0.11) | – | |
| T | 0.25 | 0.15 | ||||
| rs3027440 | T | 0.83 | 0.86 | 0.31 (0.58) | – | |
| C | 0.17 | 0.14 |
Statistically significant differences are presented in bold
Discussion
Studies on human emphasized a role of MAOA in behavioral attributes, as it is the prime enzyme degrading serotonin, a known regulator of human behavior [63]. Nevertheless, MAOA and MAOB equally degrades dopamine, another monoamine neurotransmitter which also regulates human behavior [64, 65] and interplays with the serotonergic system [66, 67], making both the enzymes crucial while studying human behavior.
Several investigators have tried to find out whether MAOA and MAOB confer risk of ADHD using family-based association studies, though the data obtained were inconclusive [31, 44–47, 68]. In the Israeli population, family based association studies revealed significant association of MAOA 30 bp-uVNTR with ADHD [47]. In the Irish ADHD probands, significantly preferential transmission of MAOA rs6323 ‘G’ allele and a haplotype was reported, while MAOB variants failed to exhibit any association [44]. In European Caucasoid subjects from eight different countries, family based study revealed positive association of MAOA with ADHD, while MAOB variants failed to do so [45]. In the Taiwanese ADHD population also, significant over-transmission of MAOA rs6323 ‘G’ allele and higher transmission of the ‘3R-G’ (30 bp-uVNTR-rs6323) haplotype was observed [35]. In Caucasian female ADHD probands from USA, a stronger association of MAOA variant was reported by family based study [69]. In the Han Chinese population also, preferential transmission of specific alleles and haplotypes to the probands was reported [46]. In the same population, MAOA polymorphisms were reported to be transmitted to only the male probands having hyperactive/impulsive subtype [70].
On the contrary, in Caucasian ADHD subjects from the United Kingdom, family based association studies found no significant association of MAOA variants with the disorder [31, 71]. A large-scale family based study, recruiting ADHD subjects residing in Ireland and Australia, also failed to identify any significant association of MAOA variants [68]. This reported discrepancy in association of MAOA and MAOB with ADHD could be due to ethnic variations in the frequency of risk variants in the population.
Our earlier investigation on limited number of Indo-Caucasoid ADHD probands (N = 73) revealed preferential maternal transmission of the 30 bp-uVNTR ‘3R’ allele to the male probands [34]. A follow up study (N = 126) revealed biased transmission of ‘3R-T’ haplotype (30 bp-uVNTR-rs6323) with high relative risk (8.06e + 007) indicating significant risk of association with ADHD [72]. A later investigation also revealed maternal transmission bias for the ‘3R-T’ haplotype to the male probands [73]. In the present family-based study, analysis was conducted on 58 variants located in the MAOA and MAOB genes. Out of the 58 variants, only 15 were polymorphic in this population. Statistical analysis was conducted on ADHD subjects (N = 190) stratified based on gender since MAO genes are located on the X-chromosome and dosage is different in the male probands as compared to the females. Significantly preferential maternal transmissions were observed for MAOA (rs5905809, rs5906957, rs6323) and MAOB (rs56220155, rs2283728, rs2283727, rs3027441, rs6324, rs3027440) variants in the male probands. Maternal transmissions to the male probands were also biased for several haplotypes. The 30 bp-uVNTR ‘3R’ allele, as part of haplotypes with other variants, showed transmission bias in the male probands. This observation provided further support to our earlier notion that the ‘30 bp-uVNTR 3R allele’ could be a risk factor for ADHD and is maternally transmitted to the male probands. This bias may, at least partly, be responsible for the male preponderance of the disorder. Further, preferential transmission of MAOA and MAOB variants from the mother to the male probands could be responsible for the higher heritability of the disorder, at least in this population. In the female ADHD probands, no such bias in parental transmissions was observed. However, the number of female probands was limited in the present investigation. Further investigation, involving large cohort of subjects from different ethnicity, would help us to elucidate the actual role of MAO in the etiology of ADHD.
In Swedish ADHD probands, the MAOA 30 bp-uVNTR ‘3R’ allele showed association with disruptive behavior in boys [74]. Our earlier population based analysis on this group of subjects revealed significant contribution of MAOA rs6323 [48] and MAOB rs56220155 [49] in ADHD associated conduct disorder as well as ODD. The present investigation revealed significant association of MAOA rs6323 and rs1137070 as well as MAOB rs2283728, rs2283727, rs3027441 and rs6324 with behavioral traits of male ADHD probands. It may be inferred on the basis of the information obtained that variants from both MAO genes may contribute to the behavioral traits of ADHD probands warranting further in depth investigation.
This first ever investigation on association between maternal age and MAO gene variants revealed statistically significant association between maternal age at pregnancy (≤ 26 years) and three MAOA variants, 30 bp-uVNTR ‘3R’, rs6323 ‘G’, and rs1137070 ‘T’ in the male ADHD probands. Out of these three variants, rs6323 also exhibited transmission bias, association with behavioral problems, formed part of haplotypes and was in strong LD with various variants. On the basis of this observation, it may be concluded that these MAOA variants, with higher occurrence in probands born to younger mothers, may be contributing to the pathophysiology of ADHD.
Limitations of the study
The major limitation of the present study was the sample size and hence, further in depth analysis on a large cohort of samples belonging to different ethnic groups would help in validation of the present observation.
Conclusions
It may be inferred from the data obtained that both MAOA and MAOB gene variants could be considered as risk factors for ADHD in the Indo-Caucasoid population from eastern India. Our study also revealed association of gene variants with behavioral problems often detected in ADHD subjects and thus could be useful for therapeutic intervention of these subjects. Probands with 30 bp-uVNTR ‘3R’ allele may not show improvement in behavioral attributes after treatment with MAOA-inhibitor, since they already possess a compensated amount of the enzyme [75].
Additional files
The Hardy-Weinberg equilibrium test performed in female subjects. Description: The table summarizes the genotypic distributions and Hardy-Weinberg equilibrium test performed for MAO polymorphic variants in female subjects. (PDF 29 kb)
Parental allelic transmission in female ADHD probands. Description: The table summarizes the parental allelic transmission of MAO alleles in female ADHD probands. (PDF 19 kb)
Maternal haplotypic transmission to male ADHD probands (only significant data presented). Description: The table summarizes the maternal haplotypic transmission of MAO haplotypes to male ADHD probands. (PDF 37 kb)
Maternal haplotypes not-transmitted to male ADHD probands (only significant data presented). Description: The table summarizes the maternal MAO haplotypes not-transmitted to male ADHD probands. (PDF 35 kb)
Parental haplotypic transmission to female ADHD probands. Description: The table summarizes the parental haplotypic transmission of MAO haplotypes to female ADHD probands. (PDF 101 kb)
Pair-wise Linkage Disequilibrium (LD) pattern of MAO polymorphic variants (analyzed using Haploview 4.2). Description: The table summarizes the D’ and r 2 values for pair-wise LD analysis in male probands, mothers of male probands, female probands and parents of female probands. (PDF 67 kb)
Analysis of allelic association with CPRS-R ‘T scores’ for oppositional behavior and PACS scores for conduct problems in male ADHD probands. Description: The table summarizes statistical comparisons between the mean scores and MAO alleles. (PDF 23 kb)
Analysis of allelic association with the status of age-of-onset of ADHD in the male probands. Description: The table summarizes statistical comparison of the early and/or late onset of disorder with MAO alleles. (PDF 25 kb)
Acknowledgements
Authors are thankful to all the volunteers for active participation.
Funding
This work was sponsored by grant # BT/PR14637/MED/30/561/2010 of the Department of Biotechnology, Government of India.
Availability of data and materials
The genotype data are presented in detail in the Tables and Additional files. Data that do not pertain to individual patients will be freely available from the corresponding author on reasonable request. Any other information will be shared based on the ethical clearance.
Abbreviations
- ADHD
Attention deficit hyperactivity disorder
- bp
Base pair
- CPRS-R
Revised conners’ parents rating scale
- DNA
Deoxyribonucleic acid
- DSM-IV
Diagnostic and statistical manual of mental disorders-4th edition
- HHRR
Haplotype-based haplotype relative risk
- IQ
Intelligent quotient
- LD
Linkage disequilibrium
- MAO
Monoamine oxidase
- MAOA
Monoamine oxidase A
- MAOB
Monoamine oxidase B
- ODD
Oppositional defiant disorder
- PACS
Parental account of children’s symptoms
- PCR
Polymerase chain reaction
- PEA
Phenylethylamine
- RR
Relative risk or risk ratio
- rs
Reference SNP
- SNP
Single-nucleotide polymorphism
- uVNTR
Upstream variable number of tandem repeats
Authors’ contributions
AK carried out genotyping, data acquisition, statistical analyses, data interpretation, and drafted the manuscript. RG participated in genotyping and initial analysis. TS collected the data on ADHD probands’ maternal age at pregnancy and assisted in analysis. SM collected the data on age-of-onset of ADHD in the probands and assisted in interpretation. ARo and CKP helped in sequence analysis of target sites. SS and ARa recruited the study subjects and provided clinical assessment. KPM, UR and KM conceptualized as well as designed the study and monitored its completion. KM supervised the work, edited and revised the manuscript. All the authors read, helped in revision and approved the final manuscript.
Ethics approval and consent to participate
Subjects were recruited as part of a research project approved by the Human Ethical Committee of the Institute (2011/04/28) which follows the guidelines of the Indian Council of Medical Research. Informed written consent was obtained from the guardians/caregivers/ adult individuals for participation in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12881-017-0469-5) contains supplementary material, which is available to authorized users.
Contributor Information
Arijit Karmakar, Email: research.arijitkarmakar@gmail.com.
Rishov Goswami, Email: rishovgoswami@gmail.com.
Tanusree Saha, Email: tanusreesaha_microbio@yahoo.in.
Subhamita Maitra, Email: msubhamita4u@yahoo.com.
Anirban Roychowdhury, Email: anirbanr91@gmail.com.
Chinmay Kumar Panda, Email: ckpanda.cnci@gmail.com.
Swagata Sinha, Email: swagata7sinha@hotmail.com.
Anirban Ray, Email: dranirbanray@gmail.com.
Kochupurackal P. Mohanakumar, Email: kpmohanakumar@yahoo.com
Usha Rajamma, Email: ushamvk@yahoo.co.in.
Kanchan Mukhopadhyay, Phone: 91-033-4001-9179, Email: kanchanmvk@yahoo.com.
References
- 1.APA . Diagnostic and statistical manual of mental disorders. 5. Washington, DC: American Psychiatric Association; 2013. [Google Scholar]
- 2.Malhi P, Singhi P. Spectrum of attention deficit hyperactivity disorders in children among referrals to psychology services. Indian Pediatr. 2000;37:1256–1260. [PubMed] [Google Scholar]
- 3.Ajinkya S, Kaur D, Gursale A, Jadhav P. Prevalence of parent-rated attention deficit hyperactivity disorder and associated parent-related factors in primary school children of Navi Mumbai--a school based study. Indian J Pediatr. 2013;80:207–210. doi: 10.1007/s12098-012-0854-1. [DOI] [PubMed] [Google Scholar]
- 4.Venkata JA, Panicker AS. Prevalence of attention deficit hyperactivity disorder in primary school children. Indian J Psychiatry. 2013;55:338–342. doi: 10.4103/0019-5545.120544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies W. Sex differences in attention deficit hyperactivity disorder: candidate genetic and endocrine mechanisms. Front Neuroendocrinol. 2014;35:331–346. doi: 10.1016/j.yfrne.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Thomas R, Sanders S, Doust J, Beller E, Glasziou P. Prevalence of attention-deficit/hyperactivity disorder: a systematic review and meta-analysis. Pediatrics. 2015;135:e994–1001. doi: 10.1542/peds.2014-3482. [DOI] [PubMed] [Google Scholar]
- 7.Daley D, Birchwood J. ADHD and academic performance: why does ADHD impact on academic performance and what can be done to support ADHD children in the classroom? Child Care Health Dev. 2010;36:455–464. doi: 10.1111/j.1365-2214.2009.01046.x. [DOI] [PubMed] [Google Scholar]
- 8.Classi P, Milton D, Ward S, Sarsour K, Johnston J. Social and emotional difficulties in children with ADHD and the impact on school attendance and healthcare utilization. Child Adolesc Psychiatry Ment Health. 2012;6:33. doi: 10.1186/1753-2000-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booster GD, Dupaul GJ, Eiraldi R, Power TJ. Functional impairments in children with ADHD: unique effects of age and comorbid status. J Atten Disord. 2012;16:179–189. doi: 10.1177/1087054710383239. [DOI] [PubMed] [Google Scholar]
- 10.Reinhardt MC, Reinhardt CA. Attention deficit-hyperactivity disorder, comorbidities, and risk situations. J Pediatr. 2013;89:124–130. doi: 10.1016/j.jped.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Fonseca DJ, Mateus HE, Galvez JM, Forero DA, Talero-Gutierrez C, Velez-van-Meerbeke A. Lack of association of polymorphisms in six candidate genes in colombian adhd patients. Ann Neurosci. 2015;22:217–221. doi: 10.5214/ans.0972.7531.220405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faraone SV, Bonvicini C, Scassellati C. Biomarkers in the diagnosis of ADHD--promising directions. Curr Psychiatry Rep. 2014;16:497. doi: 10.1007/s11920-014-0497-1. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Chang SH, Zhang LY, Gao L, Wang J. Molecular genetic studies of ADHD and its candidate genes: a review. Psychiatry Res. 2014;219:10–24. doi: 10.1016/j.psychres.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Martin J, Hamshere ML, Stergiakouli E, O'Donovan MC, Thapar A. Genetic risk for attention-deficit/hyperactivity disorder contributes to neurodevelopmental traits in the general population. Biol Psychiatry. 2014;76:664–671. doi: 10.1016/j.biopsych.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schachar R. Genetics of attention deficit hyperactivity disorder (adhd): recent updates and future prospects. Curr Dev Disord Rep. 2014;1:41–49. doi: 10.1007/s40474-013-0004-0. [DOI] [Google Scholar]
- 17.Banaschewski T, Becker K, Scherag S, Franke B, Coghill D. Molecular genetics of attention-deficit/hyperactivity disorder: an overview. Eur Child Adolesc Psychiatry. 2010;19:237–257. doi: 10.1007/s00787-010-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cortese S. The neurobiology and genetics of attention-deficit/hyperactivity disorder (ADHD): what every clinician should know. Eur J Paediatr Neurol. 2012;16:422–433. doi: 10.1016/j.ejpn.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Johansen EB, Aase H, Meyer A, Sagvolden T. Attention-deficit/hyperactivity disorder (ADHD) behaviour explained by dysfunctioning reinforcement and extinction processes. Behav Brain Res. 2002;130:37–45. doi: 10.1016/S0166-4328(01)00434-X. [DOI] [PubMed] [Google Scholar]
- 20.Solanto MV. Dopamine dysfunction in AD/HD: integrating clinical and basic neuroscience research. Behav Brain Res. 2002;130:65–71. doi: 10.1016/S0166-4328(01)00431-4. [DOI] [PubMed] [Google Scholar]
- 21.Mueller KL, Tomblin JB. Diagnosis of ADHD and its behavioral, neurologic and genetic roots. Top Lang Disord. 2012;32:207–227. doi: 10.1097/TLD.0b013e318261ffdd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oades RD, Lasky-Su J, Christiansen H, Faraone SV, Sonuga-Barke EJ, Banaschewski T, et al. The influence of serotonin- and other genes on impulsive behavioral aggression and cognitive impulsivity in children with attention-deficit/hyperactivity disorder (ADHD): Findings from a family-based association test (FBAT) analysis. Behav Brain Funct. 2008;4:48. doi: 10.1186/1744-9081-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edmondson DE, Mattevi A, Binda C, Li M, Hubalek F. Structure and mechanism of monoamine oxidase. Curr Med Chem. 2004;11:1983–1993. doi: 10.2174/0929867043364784. [DOI] [PubMed] [Google Scholar]
- 24.Tipton KF, Boyce S, O'Sullivan J, Davey GP, Healy J. Monoamine oxidases: certainties and uncertainties. Curr Med Chem. 2004;11:1965–1982. doi: 10.2174/0929867043364810. [DOI] [PubMed] [Google Scholar]
- 25.Bach AW, Lan NC, Johnson DL, Abell CW, Bembenek ME, Kwan SW, et al. cDNA cloning of human liver monoamine oxidase A and B: molecular basis of differences in enzymatic properties. Proc Natl Acad Sci U S A. 1988;85:4934–4938. doi: 10.1073/pnas.85.13.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimsby J, Chen K, Wang LJ, Lan NC, Shih JC. Human monoamine oxidase A and B genes exhibit identical exon-intron organization. Proc Natl Acad Sci U S A. 1991;88:3637–3641. doi: 10.1073/pnas.88.9.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu QS, Grimsby J, Chen K, Shih JC. Promoter organization and activity of human monoamine oxidase (MAO) A and B genes. J Neurosci. 1992;12:4437–4446. doi: 10.1523/JNEUROSCI.12-11-04437.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunner HG, Nelen M, Breakefield XO, Ropers HH, van Oost BA. Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science. 1993;262:578–580. doi: 10.1126/science.8211186. [DOI] [PubMed] [Google Scholar]
- 29.Alia-Klein N, Goldstein RZ, Kriplani A, Logan J, Tomasi D, Williams B, et al. Brain monoamine oxidase A activity predicts trait aggression. J Neurosci. 2008;28:5099–5104. doi: 10.1523/JNEUROSCI.0925-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shih JC, Chen K, Ridd MJ. Monoamine oxidase: from genes to behavior. Annu Rev Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawson DC, Turic D, Langley K, Pay HM, Govan CF, Norton N, et al. Association analysis of monoamine oxidase A and attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2003;116B:84–89. doi: 10.1002/ajmg.b.10002. [DOI] [PubMed] [Google Scholar]
- 32.Trent S, Davies W. The influence of sex-linked genetic mechanisms on attention and impulsivity. Biol Psychol. 2012;89:1–13. doi: 10.1016/j.biopsycho.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiraishi H, Suzuki A, Fukasawa T, Aoshima T, Ujiie Y, Ishii G, et al. Monoamine oxidase A gene promoter polymorphism affects novelty seeking and reward dependence in healthy study participants. Psychiatr Genet. 2006;16:55–58. doi: 10.1097/01.ypg.0000199447.62044.ef. [DOI] [PubMed] [Google Scholar]
- 34.Das M, Bhowmik AD, Sinha S, Chattopadhyay A, Chaudhuri K, Singh M, et al. MAOA promoter polymorphism and attention deficit hyperactivity disorder (ADHD) in indian children. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:637–642. doi: 10.1002/ajmg.b.30385. [DOI] [PubMed] [Google Scholar]
- 35.Xu X, Brookes K, Chen CK, Huang YS, Wu YY, Asherson P. Association study between the monoamine oxidase A gene and attention deficit hyperactivity disorder in Taiwanese samples. BMC Psychiatry. 2007;7:10. doi: 10.1186/1471-244X-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oreland L, Damberg M, Hallman J, Berggard C, Garpenstrand H. Risk factors for the neurohumoral alterations underlying personality disturbances. Neurotox Res. 2002;4:421–426. doi: 10.1080/10298420290031405. [DOI] [PubMed] [Google Scholar]
- 37.Staif R, Drtilkova I, Theiner P, Didden W, Pitelova R, Mikes V, et al. Two candidate gene polymorphisms in ADHD children: a case-control study of catechol-O-methyltransferase (COMT) and monoamine oxidase B (MAOB) genes. Arch Med Sci. 2006;2:235–239. [Google Scholar]
- 38.Dlugos AM, Palmer AA, de Wit H. Negative emotionality: monoamine oxidase B gene variants modulate personality traits in healthy humans. J Neural Transm. 2009;116:1323–1334. doi: 10.1007/s00702-009-0281-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitfield JB, Pang D, Bucholz KK, Madden PA, Heath AC, Statham DJ, et al. Monoamine oxidase: associations with alcohol dependence, smoking and other measures of psychopathology. Psychol Med. 2000;30:443–454. doi: 10.1017/S0033291799001798. [DOI] [PubMed] [Google Scholar]
- 40.Lenders JW, Eisenhofer G, Abeling NG, Berger W, Murphy DL, Konings CH, et al. Specific genetic deficiencies of the A and B isoenzymes of monoamine oxidase are characterized by distinct neurochemical and clinical phenotypes. J Clin Invest. 1996;97:1010–1019. doi: 10.1172/JCI118492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cases O, Seif I, Grimsby J, Gaspar P, Chen K, Pournin S, et al. Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science. 1995;268:1763–1766. doi: 10.1126/science.7792602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grimsby J, Toth M, Chen K, Kumazawa T, Klaidman L, Adams JD, et al. Increased stress response and beta-phenylethylamine in MAOB-deficient mice. Nat Genet. 1997;17:206–210. doi: 10.1038/ng1097-206. [DOI] [PubMed] [Google Scholar]
- 43.Chen K, Holschneider DP, Wu W, Rebrin I, Shih JC. A spontaneous point mutation produces monoamine oxidase A/B knock-out mice with greatly elevated monoamines and anxiety-like behavior. J Biol Chem. 2004;279:39645–39652. doi: 10.1074/jbc.M405550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Domschke K, Sheehan K, Lowe N, Kirley A, Mullins C, O'Sullivan R, et al. Association analysis of the monoamine oxidase A and B genes with attention deficit hyperactivity disorder (ADHD) in an Irish sample: preferential transmission of the MAO-A 941G allele to affected children. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:110–114. doi: 10.1002/ajmg.b.30158. [DOI] [PubMed] [Google Scholar]
- 45.Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N, et al. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry. 2006;11:934–953. doi: 10.1038/sj.mp.4001869. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Wang Y, Hu S, Zhou R, Yu X, Wang B, et al. The monoamine oxidase B gene exhibits significant association to ADHD. Am J Med Genet B Neuropsychiatr Genet. 2008;147:370–374. doi: 10.1002/ajmg.b.30606. [DOI] [PubMed] [Google Scholar]
- 47.Manor I, Tyano S, Mel E, Eisenberg J, Bachner-Melman R, Kotler M, et al. Family-based and association studies of monoamine oxidase A and attention deficit hyperactivity disorder (ADHD): preferential transmission of the long promoter-region repeat and its association with impaired performance on a continuous performance test (TOVA) Mol Psychiatry. 2002;7:626–632. doi: 10.1038/sj.mp.4001037. [DOI] [PubMed] [Google Scholar]
- 48.Karmakar A, Maitra S, Verma D, Chakraborti B, Goswami R, Ghosh P, et al. Potential contribution of monoamine oxidase A gene variants in ADHD and behavioral co-morbidities: scenario in eastern Indian probands. Neurochem Res. 2014;39:843–852. doi: 10.1007/s11064-014-1276-4. [DOI] [PubMed] [Google Scholar]
- 49.Karmakar A, Maitra S, Chakraborti B, Verma D, Sinha S, Mohanakumar KP, et al. Monoamine oxidase B gene variants associated with attention deficit hyperactivity disorder in the Indo-Caucasoid population from West Bengal. BMC Genet. 2016;17:92. doi: 10.1186/s12863-016-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.APA . Diagnostic and statistical manual of mental disorders. 4. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 51.Conners CK, Sitarenios G, Parker JD, Epstein JN. The revised conners’ parent rating scale (CPRS-R): factor structure, reliability, and criterion validity. J Abnorm Child Psychol. 1998;26:257–268. doi: 10.1023/A:1022602400621. [DOI] [PubMed] [Google Scholar]
- 52.Wechsler D. The Wechsler intelligence scale for children. 3. San Antonio, TX: The Psychological Corporation; 1991. [Google Scholar]
- 53.Bharat RJ. AIISH norms on SFB with Indian children. J All India Inst Speech Hear. 1971;2:34–39. [Google Scholar]
- 54.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terwilliger JD, Ott J. A haplotype-based 'haplotype relative risk’ approach to detecting allelic associations. Hum Hered. 1992;42:337–346. doi: 10.1159/000154096. [DOI] [PubMed] [Google Scholar]
- 59.Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calculator for confidence intervals of relative risk. http://www.hutchon.net/confidrr.htm. Accessed 21 Sep 2017.
- 61.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 62.VassarStats: Website for Statistical Computation. http://vassarstats.net/index.html. Accessed 09 Jan 2017.
- 63.Crockett MJ, Clark L, Hauser MD, Robbins TW. Serotonin selectively influences moral judgment and behavior through effects on harm aversion. Proc Natl Acad Sci U S A. 2010;107:17433–17438. doi: 10.1073/pnas.1009396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volkow ND, Wang GJ, Maynard L, Jayne M, Fowler JS, Zhu W, et al. Brain dopamine is associated with eating behaviors in humans. Int J Eat Disord. 2003;33:136–142. doi: 10.1002/eat.10118. [DOI] [PubMed] [Google Scholar]
- 65.Egerton A, Mehta MA, Montgomery AJ, Lappin JM, Howes OD, Reeves SJ, et al. The dopaminergic basis of human behaviors: a review of molecular imaging studies. Neurosci Biobehav Rev. 2009;33:1109–1132. doi: 10.1016/j.neubiorev.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oades RD. Dopamine-serotonin interactions in attention-deficit hyperactivity disorder (ADHD) Prog Brain Res. 2008;172:543–565. doi: 10.1016/S0079-6123(08)00926-6. [DOI] [PubMed] [Google Scholar]
- 67.Seo D, Patrick CJ, Kennealy PJ. Role of serotonin and dopamine system interactions in the neurobiology of impulsive aggression and its comorbidity with other clinical disorders. Aggress Violent Behav. 2008;13:383–395. doi: 10.1016/j.avb.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hawi Z, Matthews N, Barry E, Kirley A, Wagner J, Wallace RH, et al. A high density linkage disequilibrium mapping in 14 noradrenergic genes: evidence of association between SLC6A2, ADRA1B and ADHD. Psychopharmacology. 2013;225:895–902. doi: 10.1007/s00213-012-2875-x. [DOI] [PubMed] [Google Scholar]
- 69.Biederman J, Kim JW, Doyle AE, Mick E, Fagerness J, Smoller JW, et al. Sexually dimorphic effects of four genes (COMT, SLC6A2, MAOA, SLC6A4) in genetic associations of ADHD: a preliminary study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1511–1518. doi: 10.1002/ajmg.b.30874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu L, Guan LL, Chen Y, Ji N, Li HM, Li ZH, et al. Association analyses of MAOA in Chinese Han subjects with attention-deficit/hyperactivity disorder: family-based association test, case-control study, and quantitative traits of impulsivity. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:737–748. doi: 10.1002/ajmg.b.31217. [DOI] [PubMed] [Google Scholar]
- 71.Payton A, Holmes J, Barrett JH, Hever T, Fitzpatrick H, Trumper AL, et al. Examining for association between candidate gene polymorphisms in the dopamine pathway and attention-deficit hyperactivity disorder: a family-based study. Am J Med Genet. 2001;105:464–470. doi: 10.1002/ajmg.1407. [DOI] [PubMed] [Google Scholar]
- 72.Das M, Das Bhowmik A, Bhaduri N, Sarkar K, Ghosh P, Sinha S, et al. Role of gene-gene/gene-environment interaction in the etiology of eastern Indian ADHD probands. Prog Neuro-Psychopharmacol Biol Psychiatry. 2011;35:577–587. doi: 10.1016/j.pnpbp.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 73.Das Bhowmik A, Sarkar K, Ghosh P, Das M, Bhaduri N, Sarkar K, et al. Significance of dopaminergic gene variants in the male biasness of ADHD. J Atten Disord. 2017;21:200–208. doi: 10.1177/1087054713494004. [DOI] [PubMed] [Google Scholar]
- 74.Malmberg K, Wargelius HL, Lichtenstein P, Oreland L, Larsson JO. ADHD and disruptive behavior scores - associations with MAO-A and 5-HTT genes and with platelet MAO-B activity in adolescents. BMC Psychiatry. 2008;8:28. doi: 10.1186/1471-244X-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sabol SZ, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 1998;103:273–279. doi: 10.1007/s004390050816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Hardy-Weinberg equilibrium test performed in female subjects. Description: The table summarizes the genotypic distributions and Hardy-Weinberg equilibrium test performed for MAO polymorphic variants in female subjects. (PDF 29 kb)
Parental allelic transmission in female ADHD probands. Description: The table summarizes the parental allelic transmission of MAO alleles in female ADHD probands. (PDF 19 kb)
Maternal haplotypic transmission to male ADHD probands (only significant data presented). Description: The table summarizes the maternal haplotypic transmission of MAO haplotypes to male ADHD probands. (PDF 37 kb)
Maternal haplotypes not-transmitted to male ADHD probands (only significant data presented). Description: The table summarizes the maternal MAO haplotypes not-transmitted to male ADHD probands. (PDF 35 kb)
Parental haplotypic transmission to female ADHD probands. Description: The table summarizes the parental haplotypic transmission of MAO haplotypes to female ADHD probands. (PDF 101 kb)
Pair-wise Linkage Disequilibrium (LD) pattern of MAO polymorphic variants (analyzed using Haploview 4.2). Description: The table summarizes the D’ and r 2 values for pair-wise LD analysis in male probands, mothers of male probands, female probands and parents of female probands. (PDF 67 kb)
Analysis of allelic association with CPRS-R ‘T scores’ for oppositional behavior and PACS scores for conduct problems in male ADHD probands. Description: The table summarizes statistical comparisons between the mean scores and MAO alleles. (PDF 23 kb)
Analysis of allelic association with the status of age-of-onset of ADHD in the male probands. Description: The table summarizes statistical comparison of the early and/or late onset of disorder with MAO alleles. (PDF 25 kb)
Data Availability Statement
The genotype data are presented in detail in the Tables and Additional files. Data that do not pertain to individual patients will be freely available from the corresponding author on reasonable request. Any other information will be shared based on the ethical clearance.


