Abstract
Messenger RNA (mRNA) represents a promising class of nucleic acid-based therapeutics. While numerous nanocarriers have been developed for mRNA delivery, the inherent labile nature of mRNA results in a very low transfection efficiency and poor expression of desired protein. Here we preassemble the mRNA translation initiation structure through an inherent molecular recognition between 7-methyl guanosine (m7G) capped mRNA and eukaryotic initiation factor 4E (eIF4E) protein to form ribonucleoproteins (RNPs), thereby mimicking the first step of protein synthesis inside cells. Subsequent electrostatic stabilization of RNPs with structurally tunable cationic carriers leads to nano-sized complexes (nanoplexes), which elicit high levels of mRNA transfection in different cell types by enhancing intracellular mRNA stability and protein synthesis. By investigating a family of synthetic polypeptides bearing different side group arrangements of cationic charge, we find that the molecular structure modulates the nano-scale distance between the mRNA strand and the eIF4E protein inside the nanoplex, which directly impacts the enhancement of mRNA transfection. To demonstrate the biomedical potential of this approach, we use this approach to introduce mRNA/eIF4E nanoplexes to murine dendritic cells, resulting in increased activation of cytotoxic CD8 T cells ex vivo. More importantly, eIF4E enhances gene expression in lungs following a systemic delivery of luciferase mRNA/eIF4E in mice. Collectively, this bio-inspired molecular assembly method could lead to a new paradigm of gene delivery.
Keywords: mRNA, nucleic acid delivery, eIF4E, nanoplex
Delivery of mRNA as a therapeutic has been extensively investigated in both preclinical and clinical studies.1–4 Unlike DNA, mRNA does not need to enter the nucleus to be functional and therefore can more successfully transfect non-dividing primary cells.5 In addition, mRNA does not integrate into the genome, and hence has no risk of insertional mutagenesis.6–7 For most pharmaceutical applications, it is also advantageous that mRNA is only transiently active and completely degradable via physiological metabolic pathways.8–9 Nevertheless, the lack of efficient delivery and translation of mRNA inside cells remains a key barrier to broad applications of mRNA in the treatment of diseases.
Whereas a robust translation of endogenous mRNA involves sequentially coordinated steps of mRNA-protein interactions from the nucleus to cytoplasmic ribosomes, the state-of-the-art approach to introducing exogenous mRNA remains direct packaging of mRNA within cationic carriers. The primary role of such carriers has been to package and protect mRNA from degradation, modulate uptake by cells, and facilitate some degree of endosomal escape; however, once released, the mRNA must go through additional steps for translation. Upon delivery into the cytoplasm, mRNA is subjected to two competing processes. Eukaryotic mRNA contains a 7-methyl guanosine (m7G) cap on the 5′ end. Removal of the m7G cap leads to an RNA degradation pathway mediated by 5′-3′ RNA exonucleases and dramatically reduces the stability of delivered mRNAs.10–11 On the other hand, recognition of the m7G cap by the eukaryotic initiation factor 4E (eIF4E) is required for the assembly of protein translation initiation complex.12–13 Studies have shown that the intracellular binding of eIF4E to the m7G cap is the rate-limiting step for protein translation.14–15 In addition, a recent study indicated that certain cationic carriers for mRNA delivery could actually shield the m7G cap from the endogenous eIF4E in a rabbit reticulocyte lysate system and therefore reduced the translation of transfected mRNA.16
We devised a bio-inspired method that addresses the above challenge by first preassembling mRNA with the eIF4E protein to form ribonucleoproteins (RNPs), mimicking the rate-limiting step of protein translation initiation. These RNPs were subsequently packaged with a series of cationic carriers with an identical polyaspartamide backbone but distinct oligoalkylamine side chains (referred to as polyamines in this study). As reported here, the delivery of RNPs packaged with versions of these polyamines resulted in an up to 70-fold increase of mRNA expression observed in a range of different mammalian cell types. Since eIF4E itself has a relatively low binding constant with the m7G cap,17–18 we discovered that the number of aminoethylene repeats or spacing between two neighboring amine groups on the side chains of polyamine carriers could dramatically impact the clustering of mRNA and eIF4E protein; the result is that a slight change in the side chain structure of the polyamine can vary the degree of enhancement of mRNA transfection by more than an order of magnitude. We show that the arrangement of amines along the side chain yield differences in the association of the eIF4E protein and the mRNA strand. Furthermore, to gain an in-depth understanding of biological mechanisms, we experimentally demonstrated that transfection enhancement is attributed to increases in mRNA stability and recruitment of ribosomes to mRNA. We applied this preassembly structure to improve the efficacy of mRNA-pulsed dendritic cells in the stimulation of the cytotoxic T cell response ex vivo. Additionally, a systemic delivery of luciferase mRNA/eIF4E nanoplexes via tail veins enhanced the transgene expression over the conventional mRNA delivery method in mouse lungs.
Results and Discussion
An overview of bio-inspired assembly for enhanced mRNA delivery
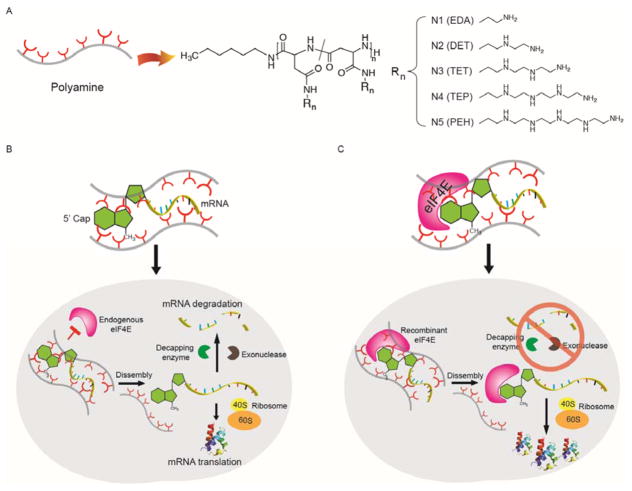
We speculated that the gene delivery approach of encapsulating mRNA alone in a cationic complex could potentially hamper exogenously administered mRNA from being actively engaged in the protein synthesis happening inside cells (Figure 1B); whereas co-encapsulation of the mRNA with the protein might allow increased transfection (Figure 1C). To overcome steric hindrance and mRNA degradation that may take place due to the limited binding of m7G capped mRNA to endogenous eIF4E, we designed a systematic series of synthetic polypeptides derived using N-carboxyanhydride polymerization of L-benzyl aspartate, followed by exhaustive amination of the side chain with various N-amine substituents with a varied number of aminoethylene repeats in the side chain to create a library of polycations for RNP encapsulation (Figure 1A and C).
Figure 1. An overview of bio-inspired assembly for enhanced mRNA delivery.
(A) Molecular structures of polycation carriers used in this study. Note that the amination of the backbone likely induces an intramolecular isomerization of the repeating unit, aspartamide, generating two isomers. (B) The state-of-the-art approach through directly complexing mRNA with cationic carriers for mRNA delivery. When associating with cationic carriers, the m7G cap on mRNA is not readily accessible to endogenous eIF4E for the mRNA translation initiation due to steric hindrance. Upon release from the complex, however, mRNA is susceptible to degradation, which results in poor expression of a desired protein. (C) Preassembly of mRNA/eIF4E nanoplexes with polyamines mimics the very first step of mRNA translation, which can improve both mRNA stability and protein translation.
mRNA/eIF4E nanoplex results in superior mRNA expression
Following purification from E.coli, recombinant human eIF4E retained its m7G cap-binding ability and spontaneously assembled with m7G capped mRNA in vitro to form RNPs as confirmed by the gel shift assay (Supplementary Fig. 1). RNPs were subsequently packaged with N-substituted polyaspartamides. The polyamines with one to four side chain aminoethylene repeat units have been previously characterized, demonstrating potent gene delivery in vitro and in vivo.19–21 Moreover, in this study, an additional polyamine with five aminoethylene repeats was synthesized and characterized (Figure 1A, Supplementary Scheme 1 and Supplementary Figs. 2A–F). At a fixed 15:1 N/P ratio (protonable amine on polyamines relative to phosphate on mRNA) in which the cell viability was confirmed to be >95%, luciferase mRNA or luciferase mRNA/eIF4E formed stable nanoplexes (~100 nm) with all five polyamines and exhibited positive zeta potential (Supplementary Figs. 3A–C).
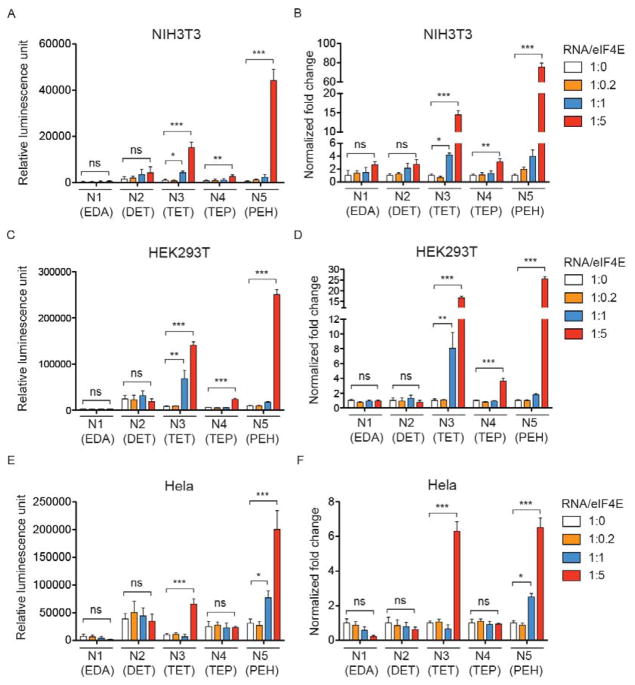
We tested whether the preassembled mRNA/eIF4E would increase luciferase expression following transfection with five different polyamines in mouse fibroblast cells (NIH3T3), human embryotic kidney cells (HEK293T), and ovarian cancer cells (HeLa). A broad range of relative compositions of the protein and mRNA was evaluated while keeping the N/P ratio of polyamine to mRNA at 15:1 (Figure 2). In NIH3T3 and HEK293T cells, co-delivery of eIF4E resulted in a maximum of 70-fold increase in luciferase expression relative to mRNA transfection alone by the same polyamine at a mass ratio of 1:5 (mRNA to eIF4E) (Figure 2A–D). Considering the molecular weights of luciferase mRNA and eIF4E, a mass ratio of 1:5 corresponds to ~ 1:10 molar ratio of mRNA to eIF4E. Because the natural stoichiometric ratio between mRNA and eIF4E is 1:1 and each mRNA only has one m7G cap at the 5′ end, the relatively high eIF4E to mRNA molar ratio required for increased mRNA expression likely reflects the fact that eIF4E itself has a low affinity to the cap structure.22 Interestingly, in NIH3T3 and HEK293T cells, only N3 (TET), N4 (TEP), and N5 (PEH) resulted in increases of luciferase expression via co-delivery of eIF4E, with N3 (TET) and N5 (PEH) having much more pronounced enhancement than N4 (TEP). In comparison, the expression enhancement was lower by about an order of magnitude for N3 (TET) and N5 (PEH) in HeLa cells, and increased luciferase expression was not detected at all with N4 (TEP) (Figure 2E and F). Furthermore, we found that the co-delivery of mRNA/eIF4E nanoplex using two additional N/P ratios (5:1 and 10:1), which resulted in minimal cytotoxicity, also significantly enhanced mRNA expression in comparison to mRNA alone (Supplementary Figs. 4A and B). In addition to luciferase, we examined the delivery of GFP mRNA/eIF4E nanoplexes. A similar trend was observed in the same cell lines with an up to 200-fold increase of GFP expression in NIH3T3 cells (Supplementary Figs. 5A–F). These data suggest that such eIF4E-mediated increase of mRNA expression is independent of mRNA species or cell type. Considering the fact that all five polyamines were derived from the same polyaspartamide backbone, these results further imply that the mRNA expression enhancement through preassembly with eIF4E is dependent on the side chains of polyamines.
Figure 2. mRNA/eIF4E nanoplex enhances luciferase expression dependent on the side chains of polyamines.
100 ng luciferase mRNA was preassembled with eIF4E at different mass ratios and transfected with polyamines at a 15:1 N/P ratio. Luciferase expression was quantified 24 hr after transfection in (A) NIH3T3, (C) HEK293T and (E) HeLa. For each type of polyamine, the increased luciferase expression via delivery of mRNA/eIF4E nanoplex was normalized to transfection with mRNA alone and presented as the fold change in (B) NIH3T3, (D) HEK293T and (F) HeLa. All experiments were performed twice in quadruplicates. Data represent the mean ± SEM (n=4). *, P<0.05; **, P<0.01; ***, P<0.001; ns, no significance.
Side chains of polyamine carriers modulate nano-scale distances and functional assembly of mRNA and eIF4E
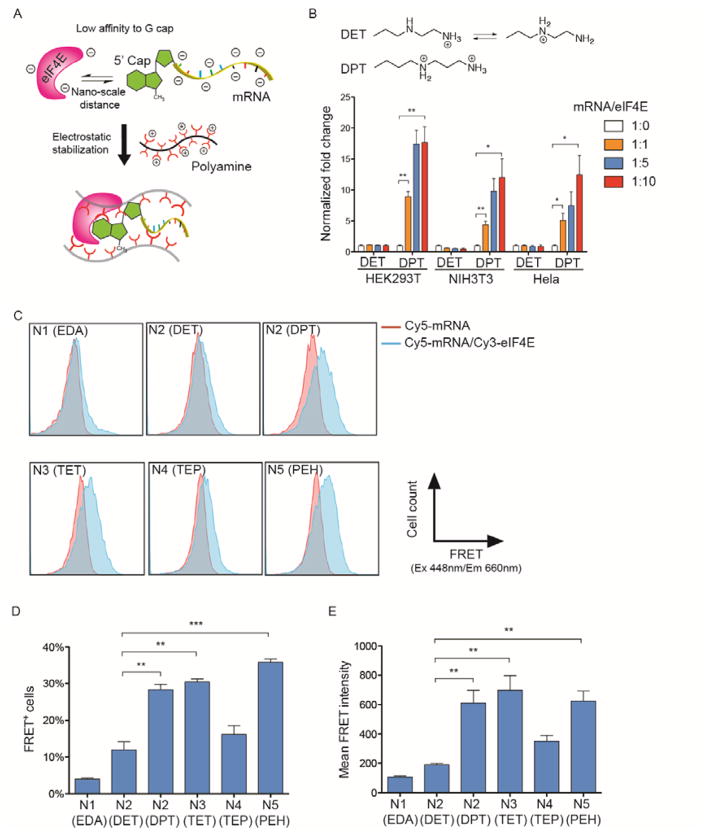
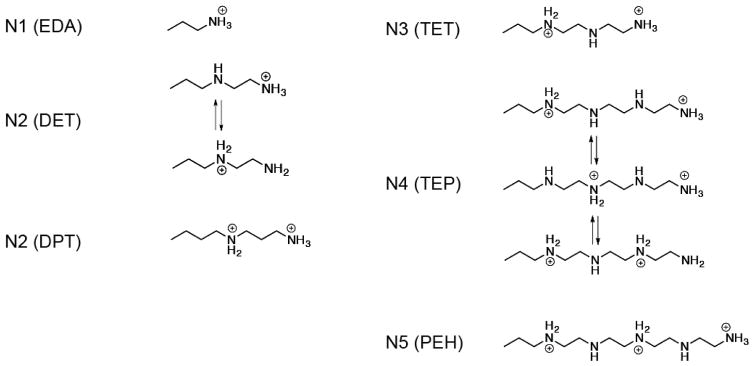
Biophysical studies revealed that eIF4E by itself has a low affinity to the m7G cap on mRNA.23–24 In a separate study, synthetic m7G cap analogs chemically modified with enhanced affinity for eIF4E were incorporated into the 5′ end of mRNA and elicited increased mRNA translation in vitro. In light of these findings, we reasoned that the physical association between mRNA and eIF4E protein in the nanoplex is critical for increased mRNA expression. The isoelectric point of eIF4E is estimated to be 5.8, and it has a weakly negative charge in the cytoplasmic environment (~pH 7.4) where mRNA translation occurs. Considering the highly anionic nature of mRNA, polyamine carriers may regulate the physical distances between mRNA and eIF4E through electrostatic stabilization (Figure 3A). Since all five polyamines were derived from the same poly(benzyl-L-aspartate) backbone, we sought to investigate the side chain structures. An earlier study indicates that a cationic charge at the end of the side chain of N1 (EDA), N2 (DET), N3 (TET) and N4 (TEP) is crucial for the electrostatic stabilization on RNA.25 Therefore, we examined the protonation structures of amine groups in different polyamines at the cytoplasmic pH 7.4 where mRNA translation occurs. Previous studies have shown that protonating neighboring amines on the 1,2-diaminoethane moiety (-NH2CH2CH2NH2-) is thermodynamically unfavorable due to electrostatic repulsion. This phenomenon, along with a potentiometric titration, has been utilized to predict the protonation structures of N1 (EDA), N2 (DET), N3 (TET) and N4 (TEP) at pH 7.4, with N2 (DET) and N4 (TEP) bearing two and three protonation structures in equilibrium, respectively (Scheme 1).25–26 Using the same approach, we found that N5 (PEH) assumes a ~56% degree of protonation at pH 7.4. This implies that nearly three out of five amine groups on the side chain are protonated. Thereby, the distribution of these three protonated amines that results in the lowest thermodynamic energy is most likely the protonation of every other amine group on the side chain as shown in Scheme 1. Further comparison of the protonation structures in different polyamines suggests half and one-third of the terminal amine groups may be in the nonprotonated form on N2 (DET) and N4 (TEP), respectively.25 Thus, we attempted to increase the fraction of protonated terminal amines on N2 (DET) by introducing an additional methylene group to create an N-substituted polyaspartamide possessing a side chain of 1,3-diaminopropane (-NHCH2CH2CH2NH2), which is referred to as N2 (DPT) in this study. According to previous studies, N2 (DPT) bears a fully protonated terminal amine at pH 7.4. Interestingly, as shown in Figure 3B, N2 (DPT) enabled an up to 15-fold increase of luciferase mRNA expression when delivering mRNA/eIF4E nanoplexes relative to mRNA alone. In contrast, N2 (DET) failed to elicit increased mRNA expression with the same nanoplexes. Therefore, N2 (DPT) improves mRNA transfection likely through increased clustering between mRNA and eIF4E as a result of enhanced protonation on the terminal amines.
Figure 3. Side chains of polyamine carriers modulate nano-scale distances and functional assembly of mRNA and eIF4E.
(A) A schematic of stabilized mRNA/eIF4E assembly via polyamines. Given the anionic charge of both eIF4E and mRNA as well as the low affinity of eIF4E to the m7G cap, polyamines may serve to stabilize the physical clustering between mRNA and eIF4E. The degree of such stabilization is associated with the fraction of charged terminal amines on the side chains of polyamines at a physiological pH. (B) “Gain-of-function” of eIF4E-mediated transfection enhancement by introducing a methylene group on the polyamine side chain. The estimated protonation structures of N2 (DET) and N2 (DPT) at pH 7.4 are shown on the top. While N2 (DPT) enabled an up to 15-fold increase of luciferase expression when delivering mRNA/eIF4E nanoplexes relative to mRNA alone, N2 (DET) failed to increase mRNA expression through eIF4E. Data represent the mean ± SEM (n=4). *, P<0.05; **, P<0.01. (C) Representative histograms of the intracellular FRET assay to measure the degree of co-localization between Cy5-mRNA and Cy3-eIF4E. At 4hr after transfection of Cy5-mRNA or Cy5-mRNA/Cy3-eIF4E into HEK293T cells with each polyamine, cells were analyzed by flow cytometer with 488 nm laser and 660 nm detector. (D) Percentage of FRET+ and (E) mean FRET intensity were calculated by subtracting the fluorescence signal of Cy5-mRNA-transfected cells from that of Cy5-mRNA/Cy3-eIF4E -transfected cells. Data represent the mean ± SEM (n=3). **, P<0.01, ***, P<0.001.
Scheme 1.
Predicted protonation structures on the side chain of polyamines at pH 7.4
To directly confirm whether polyamines differentially modulate the physical distances between mRNA and eIF4E inside the nanoplex, eIF4E and luciferase mRNA were labeled with Cyanine 3 (Cy3) and Cyanine 5 (Cy5), respectively. In principle, when these two dyes are in vicinity of each other (2–9 nm) inside cells, Förster resonance energy transfer (FRET) can be detected by flow cytometer. As a result, an intracellular FRET assay is able to quantitatively measure the proximity between eIF4E and mRNA in a cellular environment. By measuring the percentage of FRET-positive cells and mean FRET intensity, it was found that only N2 (DPT), N3 (TET) and N5 (PEH) resulted in relatively high degrees of co-localization at a 1:5 (Cy5-mRNA/Cy3-eIF4E) mass ratio 4 hr after transfection into HEK293T cells (Figure 3C–E). Consistently, across all three cell lines tested (HEK293T, NIH3T3 and HeLa), only N2 (DPT), N3 (TET) and N5 (PEH) were able to substantially enhance luciferase or GFP mRNA expression following delivery of mRNA/eIF4E nanoplexes (Figure 2, Figure 3B, Supplementary Figs. 4 and 5). Notably, an intermediate degree of co-localization between Cy3-eIF4E and Cy5-mRNA packaged with N4 (TEP) was detected in comparison to other polyamines (Figure 3D and E), which is consistent with a marginal increase (~2 fold) of reporter mRNA expression in HEK293T (Figure 2 and Supplementary Figs. 4 and 5). In comparison, N1 (EDA) and N2 (DET) gave rise to a minimal degree of co-localization based on the FRET assay (Figure 3D and E), which is in agreement with their lack of ability to increase reporter mRNA expression through delivery of mRNA/eIF4E (Figure 2 and Supplementary Figs. 4 and 5). Through tuning the side chain structure along with intracellular FRET assays, our results suggest that in addition to facilitating the internalization of nanoplexes into cells, polyamine carriers also serve to stabilize the association between mRNA and eIF4E, which is critical for the eIF4E-mediated enhancement of mRNA transfection. We deduce that such an effect is likely dictated by the protonation degree of terminal amine and cationic charge density on the side chain (Figure 3A).
Preassembly of mRNA/eIF4E is indispensable for increased mRNA expression
Given the fact that eIF4E is considered the least abundant protein among translation initiation factors,14, 27 we asked whether it is the assembly of mRNA/eIF4E nanoplex or the restoring of stoichiometric amount of eIF4E within cells that resulted in enhanced mRNA expression.27 To test the latter possibility, NIH3T3 and HEK293T were modified to stably overexpress eIF4E, and the increased level of eIF4E was confirmed by western blotting (Supplementary Figs. 6C and D). Next, luciferase mRNA was transfected into these cell lines via each of the five different polyamines. At 24 hr after transfection, it was found that raising endogenous eIF4E levels failed to enhance mRNA expression (Supplementary Figs. 6A and B). To rule out the possibility that overexpressing eIF4E by the genetic modification may not have increased eIF4E to the same levels as direct delivery of eIF4E protein, we monitored the total eIF4E levels in cells upon delivery of preassembled luciferase mRNA/eIF4E (1:5 mass ratio) packaged with N5 (PEH) over 12 hr by western blotting. The amount of eIF4E protein added into culture medium relative to the cell number in western blotting assays remained the same as the in vitro transfection studies shown in Figure 2. Unexpectedly, the amount of eIF4E introduced into cells through delivery of mRNA/eIF4E nanoplexes is nearly undetectable. In contrast, cells overexpressing eIF4E contained a substantially high level of eIF4E (Supplementary Figs. 6C and D). Therefore, simply raising the level of eIF4E inside cells may not be able to explain the enhanced expression of mRNA via delivery of mRNA/eIF4E nanoplexes. If this is true, preassembly of mRNA/eIF4E prior to transfection is likely indispensable for increased mRNA expression.
Preassembled translation initiation nanoplex increases intracellular mRNA stability
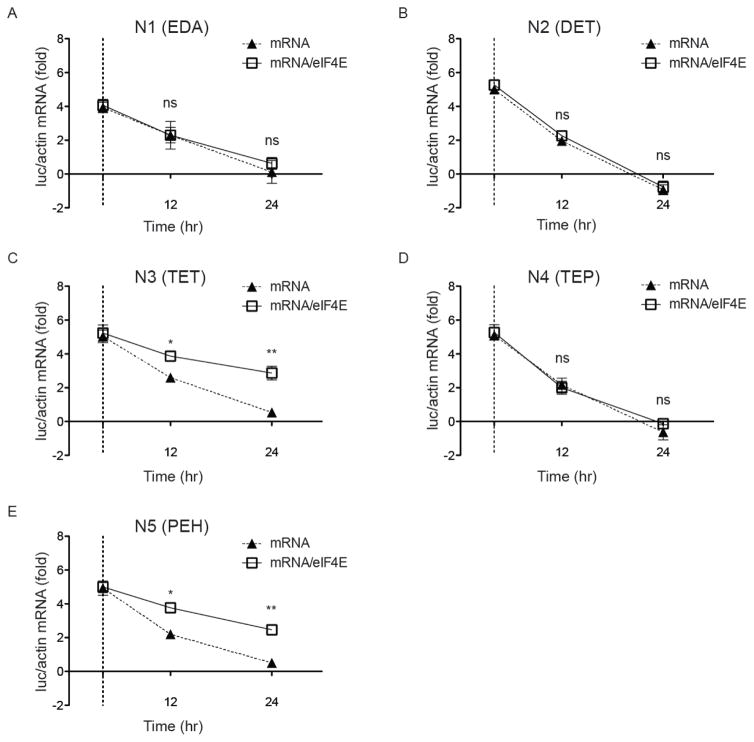
The 5′-3′ exonuclease-mediated hydrolysis represents a predominant mRNA degradation pathway inside cells. This degradation pathway is initiated through the removal of the m7G cap from mRNA with decapping enzymes, and is inhibited when eIF4E binds to the cap.10, 28 Therefore, we tested whether the mRNA/eIF4E nanoplex could protect mRNA from decapping and subsequent degradation, thereby enhancing the intracellular mRNA stability. Equal amounts of luciferase mRNA alone or together with eIF4E at 1:5 mass ratio (RNA/eIF4E) were transfected into HEK293T cells by each of the five polyamines. At 4 hr after transfection, cells were washed extensively and fresh media was added to remove nontransfected mRNA. A fraction of cells were harvested at this time point to set a baseline for quantification of intracellular luciferase mRNA.
We found no difference in the mRNA uptake between mRNA alone and mRNA/eIF4E transfected with all five polyamines in the first 4 hours (Figure 4A–E). In a separate flow cytometry study, Cy5-labeled mRNA alone and mRNA/eIF4E nanoplexes were transfected into HEK293T cells via the same polyamines. At 4 hr after transfection, >95% of cells had internalized mRNA at comparable levels in mRNA- and mRNA/eIF4E-transfected cells based on mean fluorescence intensity (MFI) (Supplementary Fig. 7). This demonstrated that eIF4E does not improve mRNA expression through increasing the mRNA uptake. However, after removing nontransfected mRNA at the 4 hr time point, eIF4E significantly enhanced intracellular mRNA stability over 24 hr in cells transfected with N3 (TET) and N5 (PEH) (Figure 4C and E). In comparison, N1 (EDA), N2 (DET) and N4 (TEP) failed to improve mRNA stability through delivering mRNA/eIF4E nanoplexes (Figure 4A, B and D). This enhanced intracellular mRNA stability via preassembly with eIF4E may partly explain the increase of mRNA expression. Moreover, the observation that such eIF4E-mediated increase of mRNA stability was only detected in N3 (TET) and N5 (PEH) polyamines is consistent with the results in which only these two polyamines support substantial enhancement of mRNA expression in different cell types (Figure 2 and Supplementary Figs. 4 and 5).
Figure 4. Preassembled translation initiation nanoplex increases intracellular mRNA stability.
Equal amount of luciferase mRNA alone or mRNA/eIF4E (1:5 mass ratio) were transfected into HEK293T cells by each of five polyamines. At 4 hr after transfection, nontransfected mRNA was removed from culture media and a fraction of cells were harvested to set a baseline for quantification of intracellular luciferase mRNA (vertical dash line). At 12 hr and 24 hr time points, it was found that eIF4E significantly enhanced intracellular mRNA stability in cells transfected with (C) N3 (TET) and (E) N5 (PEH) but not with (A) N1 (EDA), (B) N2 (DET) and (D) N4 (TEP). Data represent the mean ± SEM (n=3). *, P<0.05; **, P<0.01; ns, no significance.
Preassembled translation initiation nanoplex enhances mRNA translation
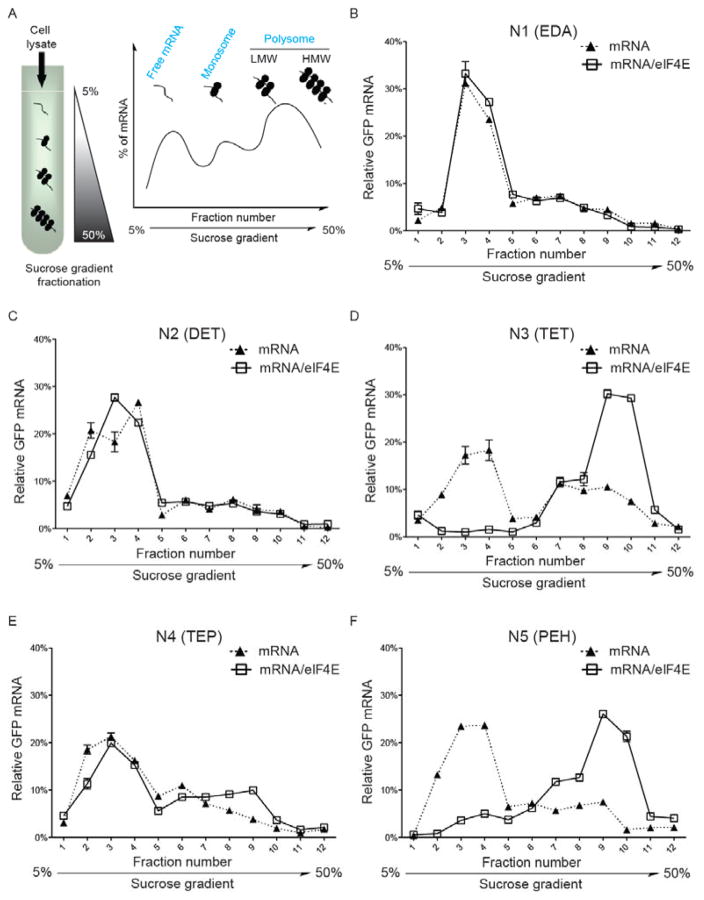
eIF4E is responsible for initiating protein synthesis from a mRNA template (i.e. mRNA translation). Inside cells, an efficient mRNA translation is characterized by multiple ribosomes (polysome) engaged in a single mRNA strand during protein synthesis and therefore the number of ribosomes bound to mRNA is commonly proportional to the degree of translational activity.29 We adapted a well-established polysome-profiling assay to answer whether this preassembled translation initiation nanoplex, mRNA/eIF4E, could increase the recruitment of ribosomes to co-transfected mRNA. At 24 hr after transfection of GFP mRNA or mRNA/eIF4E into HEK293T cells with five different polyamines, cells were treated with an eukaryote protein synthesis inhibitor, cycloheximide (CHX) to “fix” ribosomes with their associated mRNA. Then cell lysates were prepared and separated by ultracentrifugation in a linear sucrose gradient (5%–50%) with 24 fractions. The binding of ribosomes to mRNA changes the sedimentation coefficient of mRNAs. As a result, free mRNA and mRNA populations associated with a single ribosome (monosome) and multiple ribosomes (polysome) can be identified by spectral absorbance (OD254nm) (Figure 5A and Supplementary Fig. 8). Additionally, detection of a small 40S ribosomal protein (S6) by western blotting confirmed the presence of these two populations (Supplementary Fig. 9). Since the OD254nm absorbance quantifies total RNA species, a majority of which are ribosomal RNA, we combined two neighboring fractions after absorbance measurement to obtain a total of 12 fractions and quantified the global distribution of transfected GFP mRNA through qPCR. By referring to the OD254nm absorbance spectrum along with western blotting against the 40S ribosomal protein S6 (Supplementary Figs. 8 and 9), we could assign free GFP mRNA to fractions #1 through #5, monosome-associated GFP mRNA to fraction #6 and polysome-associated GFP mRNA to fractions #9 through #11 (Figure 5B–F). It was found that the GFP mRNA/eIF4E nanoplexes dramatically shifted the distribution of mRNA from ribosome-free species to polysome-associated ones (a feature of active mRNA translation) relative to transfection with mRNA alone in the presence of N3 (TET) and N5 (PEH) polyamines (Figure 5D and F). In contrast, N4 (TEP) showed a marginal increase of actively translated mRNA with eIF4E, but no such shift was detected with N1 (EDA) and N2 (DET) (Figure 5B, C and E). This trend was consistent with the distinct profile of transfection enhancement through preassembled mRNA/eIF4E nanoplexes with different polyamine carriers (Figure 2 and Supplementary Figs. 4 and 5). Therefore, in addition to improving the intracellular mRNA stability, eIF4E protein increases the translation of mRNA through active recruitment of ribosomes. Moreover, a clear difference among different polyamines was observed at the mRNA translation level, which further suggests that different side chain structures play a critical role in modulating such effect.
Figure 5. Preassembled translation initiation nanoplex enhances mRNA translation.
(A). A schematic of sucrose gradient fractionation to separate free mRNA, monosome, low molecular weight (LMW) and high molecular weight (HMW) polysomes. 24 hr after transfection of GFP mRNA or mRNA/eIF4E into HEK293T cells packaged with each of five polyamines, relative abundance of GFP mRNA in 12 fractions was measured by qPCR. Delivery of mRNA/eIF4E nanoplexes dramatically shifted the distribution of mRNA towards polysome-associated fractions relative to transfection with mRNA alone when packaged with (D) N3 (TET) and (F) N5 (PEH). In comparison, (E) N4 (TEP) enabled a marginal shift of actively translated mRNA when delivering mRNA/eIF4E and no such effects were detected with (B) N1 (EDA) and (C) N2 (DET). Results are representative data of two independent experiments.
Delivery of mRNA/eIF4E nanoplexes enhances mRNA expression ex vivo and in vivo
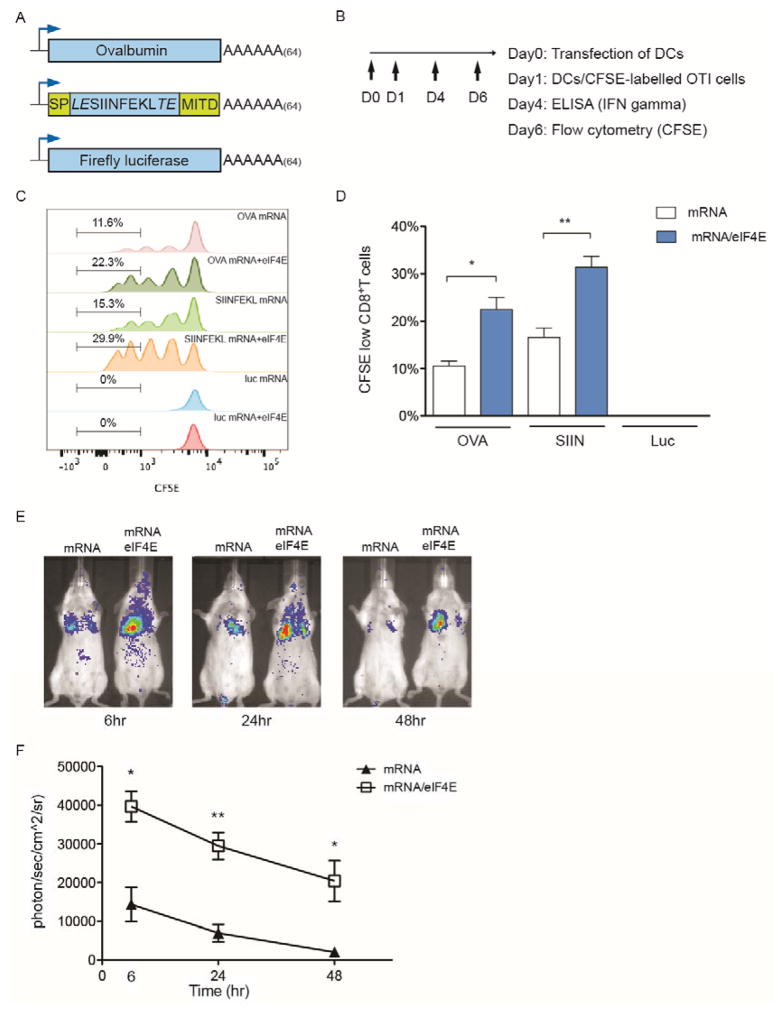
mRNA represents an increasingly important agent for gene delivery. We investigated whether delivering antigen mRNA/eIF4E nanoplexes into primary dendritic cells (DCs) can enhance an antigen-specific T cell activation ex vivo. Bone marrow-derived dendritic cells (BMDCs) from C57BL/6 mice were transfected with various combinations of mRNA/eIF4E with N5 (PEH): Two different modes of chicken ovalbumin (OVA) antigen mRNA were investigated. One includes a full-length OVA mRNA, which represents a vaccination scenario where the antigen epitopes are not defined. The second mode utilizes a chimeric mRNA construct, which includes a N-terminal leader peptide with a MHC class I trafficking signal (MITD) attached to the C terminus of a short mRNA fragment encoding an OVA-derived antigen epitope (SIINFEKL peptide) (Figure 6A). As shown in Figure 6B, 24 hr after transfection of BMDCs, immune cells were harvested from OTI mouse lymph nodes (LNs), which contain CD8 T cells transgenically expressing a T cell receptor recognizing the MHC I-restricted SIINFEKL peptide. LN-derived cells were pre-labeled with 1μM CFSE fluorescent dyes and subsequently co-cultured with the BMDCs. The activation of naïve OTI CD8 T cells and proliferation of activated OTI CD8 T cells were monitored by fluorescence intensity of CFSE gated on CD8 T cells using flow cytometer. On day 6, it was found that with full OVA and SIINFEKL mRNA, eIF4E significantly enhanced the proliferation of CD8 T cells but not other cells. In contrast, BMDCs transfected with luciferase mRNA or along with eIF4E failed to stimulate the proliferation of OTI CD8 T cells or other cells (Figure 6C and D and Supplementary Fig. 10A). In addition to stimulating the T cell proliferation, on day 4, delivery of OVA mRNA/eIF4E or SIINFEKL mRNA/eIF4E nanoplexes into BMDCs resulted in increased secretion of IFN-gamma in comparison to mRNA alone, which reflects the enhanced conversion of naïve T cells to cytotoxic T cells (Supplementary Fig. 10B). We next investigated whether eIF4E could enhance mRNA delivery in vivo. As a proof-of-concept, luciferase mRNA or luciferase mRNA/eIF4E was packaged with the polyamine and introduced into Balb/c mice via tail veins. The transgene expression peaked at 6 hr following the injection and was primarily found in lungs. The signals decayed over 48 hr in both mRNA/polyamine and mRNA/eIF4E/polyamine groups, which indicated the nature of transient gene expression in the mRNA-mediated gene delivery. However, the preassembly with eIF4E significantly increased luciferase expression over injecting mRNA/polyamine alone (Figure 6E and F). Collectively, in both ex vivo and in vivo experiments, we demonstrated that the preassembly of mRNA/eIF4E nanoplex can enhance the transfection efficiency of mRNA in primary cells and mice. Nevertheless, the degree of such enhancement is lower than the transfection studies in cell lines. This difference may reflect the increased difficulty in transfecting primary cells and the lower stability of mRNA/eIF4E nanoplexes through the systemic delivery route in mice.
Figure 6. Delivery of mRNA/eIF4E nanoplexes enhances mRNA expression ex vivo and in vivo.
(A) Schematics of different mRNA templates used in the study: Full length OVA; SIINFEKL peptide flanked by a MHC class I signal peptide fragment (78 bp) and a MHC class I trafficking signal (MITD, 168 bp); Luciferase mRNA. (B) Procedures for ex vivo antigen presentation. (C) Representative histograms of proliferation of OTI CD8 T cells on day 6 in response to antigen presentation by DCs. DCs were transfected with antigen mRNA alone or mRNA/eIF4E nanoplexes on day 0 through N5 (PEH). (D) Percentage of activated CD8 T cells through CFSE labeling based on the same gating as in (C). The activation of CD8 T cells was identified by the decrease of CFSE fluorescence intensity via flow cytometer. (E) Representative bioluminescence imaging (BLI) at 6, 24 and 48 hr after the tail vein injection of luciferase mRNA or mRNA/eIF4E packaged with the polyamine. (F) Quantification of luciferase expression in lungs of Balb/c mice via BLI. Data represent the mean ± SEM (n=3). *, P<0.05. **, P<0.01.
Safety considerations of delivering mRNA/eIF4E nanoplexes
Last but not the least, we addressed the safety concerns associated with co-delivery of mRNA/eIF4E, since constitutive overexpression of eIF4E has been found in several human cancers.30–31 As shown in Supplementary Figs 6C and D, a bolus delivery of mRNA/eIF4E into cells resulted in a negligible change of total eIF4E levels inside cells. We further tested whether repeated delivery of eIF4E into cells induces cell proliferation. HEK293 or NIH3T3 cells were transfected daily for four days with 100 ng luciferase mRNA alone or 100 ng mRNA/500 ng eIF4E in 96-well plates. Cell proliferation was measured daily by the MTT assay. Over 96 hr, no appreciable stimulation of cell proliferation was detected in the cells receiving mRNA/eIF4E compared to those transfected with reporter mRNA alone. Conversely, cells overexpressing eIF4E proliferated significantly faster, which was consistent with the role of eIF4E overexpression in certain cancers (Supplementary Figs. 11A and B). We examined whether delivery of eIF4E proteins could elevate c-Myc expression, which is known to be upregulated in cancer cells with eIF4E overexpression. 2 μg luciferase mRNA/10 μg eIF4E protein, along with N5 (PEH) were added into culture medium containing 2×105 HEK293T cells daily for four days. Consistent with the cell proliferation experiment, delivery of eIF4E protein failed to up-regulate c-Myc expression, whereas in eIF4E-overexpressing cells, c-Myc expression appreciably elevated (Supplementary Fig. 11C). Note that over a four-day transfection period, a total of 40 μg eIF4E was added into 2 ml cell culture medium. Additionally, we estimated a total of ~500 μg of proteins from cell lysates at 96 hr by the Bradford assay. Considering that no change of total eIF4E level was detected (Supplementary Fig. 11C), we reasoned that two explanations were possible. First, only a small fraction of mRNA/eIF4E may have been internalized into cells. Second, eIF4E may have a relatively short half-life. In fact, it has been found that ectopically expressed eIF4E is subjected to ubiquitination and a proteasome-dependent degradation pathway within 24 hr.32 Similar to existing protein delivery-based applications including Cas9 protein-sgRNA for precise genome editing and OSKC transcription factor proteins (Oct4, Sox2, Klf4 and c-Myc) for induced pluripotent stem cells, co-delivery of eIF4E protein may have the advantage of immediate action and relatively short half-life for mRNA-based therapeutics.33–34
Conclusions
By integrating a deep understanding of mRNA biology with material chemistry, we developed a distinct approach to engage mRNA in active protein expression inside cells. Through preloading capped mRNA with the translation initiation factor protein, eIF4E, a substantial increase of mRNA expression inside cells has been achieved. Nevertheless, our study shows that simply overexpressing eIF4E inside cells prior to mRNA transfection does not increase mRNA translation. Our result appears to contradict an earlier study in which co-delivering eIF4E mRNA with EGF mRNA was found to augment wound healing.27 Note that this earlier study did not provide evidence that transient overexpression of eIF4E directly increased the translation of EGF mRNA, which was responsible for wound healing. Instead, the collagen expression was used as an indirect readout for the effect of EGF expression on wound healing. In fact, separate studies demonstrated that the upregulation of eIF4E can induce collagen expression indirectly through activation of TGFβ signaling.35–36 Therefore, it is likely that co-delivering eIF4E mRNA with EGF mRNA does not enhance wound healing directly through increasing EGF expression. Given the promiscuous G cap binding characteristic of eIF4E and presence of abundant endogenous mRNAs, we speculate that simply elevating the intracellular level of eIF4E does not enable eIF4E to exclusively recognize the G cap of transfected mRNA.
Furthermore, our results with synthetic polyamines strongly suggest that a physical co-localization between mRNA and eIF4E protein is indispensable for eIF4E-mediated increase of mRNA expression. By comparing a series of polyamines with the same backbone and distinct side chain amines, we discovered that the ability of eIF4E to increase mRNA expression is dependent on the side chain structure. Among six polyamines examined in the study, a clear correlation between eIF4E-mediated increase of mRNA expression and stability of mRNA/eIF4E was observed. By fine-tuning the structure of the side chain, we found that such stabilization effects are likely attributed to the protonation degree of the terminal amine group. Nevertheless, considering that N1 (EDA) assumes one fully protonated terminal amine, we do not rule out other possible mechanisms as contributions, such as the overall cationic charge density. However, we reasoned that N1 (EDA) may be an exception since the expression of GFP or luciferase mRNA transfected with N1 (EDA) was nearly undetectable in different cell lines. Additionally, we also observed that poly-L-lysine with a similar degree of polymerization failed to transfect reporter mRNAs alone or in the co-delivery with eIF4E (data not shown). This is likely due to an inherent low transfection efficiency of polyamines with only one protonable side chain amine, which was found in previous studies as well,25, 37 and to the fact that only a primary amine is present, which has a higher pKa and has been thought to lower endosomal escape.
The method presented here should provide both practical and theoretical implications for the mRNA-based gene delivery. The co-delivery of antigen mRNA/eIF4E in dendritic cells demonstrated in this study may have implications on boosting the efficacy of mRNA-based vaccines. Alternatively, co-delivering eIF4E could be utilized to dramatically improve mRNA expression in the treatment of certain genetic diseases as demonstrated before with chemically modified mRNA.38–39 Additionally, such co-delivery strategies can be applied to other nucleic acid-based therapeutics including small interference RNA (siRNA) and DNA by identifying proteins that can potentially facilitate the function of these nucleic acids inside cells. With respect to the theoretical significance, many polymer- and lipid-based mRNA carriers have been developed to enable robust mRNA delivery and expression in vitro and in vivo. However, studies on the detailed mechanisms are usually focused on the material properties such as buffering effects and charge distribution, often independent of the nucleic acid vector. Our work implies that characterization of the dynamic interactions between the synthetic delivery vehicle, mRNA, and the cellular protein machinery responsible for mRNA stability and translation may offer a distinct perspective that enables examination of the mechanisms underlying efficient mRNA delivery.
Methods and Materials
Chemicals and antibodies
β-benzyl-L-aspartate and triphosgene were purchased from Chem-Impex (Wood Dale, IL, USA) and used without further purification. Diethylenetriamine, triethylenetetraamine, tetraethylenepentamine, and pentaethylenehexamine were respectively purchased from Alfa Aesar (Haverhill, MA, USA), MP Biomedicals (Santa Ana, CA, USA), TCI America (Portland, OR, USA), and Acros Organics (Pittsburgh, PA, USA) and used as received. N, N-Dimethylformamide (DMF) was dried and stored over 3Å molecular sieves under an argon atmosphere prior to use. All other reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received. Fire fly luciferase assay kit was purchased from Biotium (Fremont, CA, USA) and performed per manufacturer’s instruction. GFP, fire fly luciferase and OVA mRNA were from Trilink (San Diego, CA, USA). These capped (Cap 0) and polyadenylated mRNA have been optimized for mammalian systems and modified with pseudouridine and 5-methylcytidine to reduce immune stimulation. Cyanine 3 and Cyanine 5 succinimidyl esters (potassium salt) were purchased from AAT bioquest (Sunnyvale, CA, USA). CFSE cell proliferation kit was from Invitrogen (Grand Island, NY, USA). Primary antibodies used in this study are: β Tubulin (G-8), eIF4E (P-2) are from Santa Cruz Biotech (Dallas, TX, USA). APC anti-mouse CD8a and TruStain fcX™ (anti-mouse CD16/32) antibodies are from Biolegend (San Diego, CA, USA). Secondary antibodies are: goat anti-rabbit IgG-HRP and goat anti-mouse IgG-HRP (Santa Crutz Biotech, Santa Cruz, CA, USA).
Synthesis and characterization of β-benzyl-L-aspartate N-carboxyanhydride (NCA)
To a dispersion of β-benzyl-L-aspartate (25.0 g, 112 mmol) in THF (200 mL) in a 1 L two neck round bottom flask equipped with a magnetic stir bar and rubber septum was added triphosgene (30.0 g, 100 mmol) in one portion. The mixture was stirred and sparged with a steady stream of argon under reflux for 2 hr then cooled to RT. Hexanes (700 mL) were added and the mixture was allowed to sit for 7 hr. The resulting precipitate was collected by vacuum filtration and washed 4 times with hexanes under a blanket of argon. The resulting solid was added to a flame dried 1 L flask, dissolved in anhydrous THF (300 mL), and recrystallized via solvent diffusion under a layer of hexanes (700 mL) in an argon atmosphere at RT. The solids were collected by vacuum filtration under a blanket of argon, washed 4 times with hexanes, and dried in vacuo to give white crystals (25.7 g, 103 mmol, 92%).
1H NMR (400 MHz, CDCl3) δ 7.47 – 7.30 (m, 5H), 6.13 (s, 1H), 5.19 (s, 2H), 4.60 (dd, J = 9.5, 2.4 Hz, 1H), 3.09 (dd, J = 17.7, 3.1 Hz, 1H), 2.84 (dd, J = 17.7, 9.6 Hz, 1H)
13C{1H} NMR (101 MHz, CDCl3) δ 169.55 (s), 168.31 (s), 151.32 (s), 134.76 (s), 129.03 (s), 128.95 (s), 128.71 (s), 67.98 (s), 53.98 (s), 36.22 (s)
Synthesis and characterization of poly(β-benzyl-L-aspartate) (PBLD)
A flame dried 100 mL Schlenk flask equipped with a magnetic stir bar and rubber septum was charged with NCA (3.28 g, 13.2 mmol) and DMF (30 mL). Hexylamine (17.4 μL, 0.132 mmol) was added via micropipette with stirring and the reaction was sparged with a steady stream of argon at RT for 48 hr. The reaction was added to water and the resulting precipitate was collected by centrifugation, washed 3 times with water, and dried in vacuo to give a white powder (2.44 g, 11.9 mmol repeat units, 90%).
1H NMR (400 MHz, DMSO): δ 8.37 – 7.98 (m, 1H), 7.51 – 7.08 (m, 5H), 5.21 – 4.92 (m, 2H), 4.74 – 4.48 (m, 1H), 2.98 – 2.54 (m, 2H).
Synthesis and characterization of polyamines
Polyamines were synthesized according to a modified procedure of Uchida and coworkers.25 Briefly, to a chilled solution of PBLD in N-Methyl-2-pyrrolidone (NMP) (2 mL) was added dropwise with stirring 50 equivalents of oligoalkylamines (relative to the reactive benzyl ester on the PBLD) diluted two-fold with NMP. After stirring for 2 hr at 0°C, the pH was adjusted to 1 with dropwise addition while stirring of cold 6 N HCl. The resulting solution was dialyzed from a regenerated cellulose membrane bag (Spectrum Laboratories, 1 kDa MWCO) against 0.01 N HCl followed by mQ water, frozen, and lyophilized to give a white powder.
N1 (EDA): 1H NMR (400 MHz, D2O) MR (400 MHz, Dwith NMP. After stirring for 2 hours at 0°C,
N2 (DET): 1H NMR (400 MHz, D2O) δ 4.71 (s, 1H), 3.45 (s, 2H), 3.22 (s, 2H), 3.11 (s, 2H), 2.97 (s, 2H), 2.83 (s, 2H).
N3 (TET): 1H NMR (400 MHz, D2O) δ 3.70 – 3.50 (m, 7H), 3.49 – 3.41 (m, 2H), 3.35 (s, 2H), 3.23 – 2.62 (m, 4H).
N4 (TEP): 1H NMR (400 MHz, D2O) δ 4.72 (s, 1H), 3.64 – 3.39 (m, 9H), 3.37 – 3.05 (m, 5H), 3.00 – 2.62 (m, 4H).
N5 (PEH): 1H NMR (400 MHz, D2O) δ 3.80 – 2.57 (m, 23H).
1H NMR and 13C NMR spectra were obtained in CDCl3, dimethyl sulfoxide-d6 or deuterium oxide (Cambridge Isotope Laboratories) using a Bruker Avance 400 MHz NMR spectrometer at 25°C.
Cell lines and mice
HEK293T, NIH3T3, and HeLa cells were obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). All cells were cultured in DMEM (Invitrogen, Grand Island, NY, USA) with 10% FBS and 1% Penicillin/Streptomycin. Cells were screened for mycoplasma-free by High Throughput Screening Facility at The Koch Institute for Integrative Cancer Research at MIT (Cambridge, MA, USA). OT-1 transgenic mice were housed in the MIT Animal Facility. We performed all mouse studies in the context of an animal protocol approved by the MIT Division of Comparative Medicine following federal, state and local guidelines.
Production and purification of recombinant eIF4E
Human eIF4E cDNA was synthesized by IDT (Coralville, Iowa, USA) and cloned into pSH200 plasmid (A gift from Prof. Xiling Shen at Duke University) via NcoI and HindIII. His-tagged eIF4E was expressed in BL21 E.coli through overnight induction with 0.1 mM IPTG at 20°C for 12 hr. Bacteria were lysed in the presence of 1mg/ml lysozyme at room temperature for 20min followed by sonication at 20 watts for 5 min at 10 sec interval on ice. Cell lysate was centrifuged at 14,000×g, 4 °C for 90 min. Cell lysate was cleared through 0.45 μm filter and subjected to cobalt NTA column-based purification. After purification, proteins were buffer exchanged to 20 mM HEPES, 150 mM NaCI, 10% glycerol, 1 mM DTT and stored at −80°C. Protein function was confirmed by gel shift assay with capped and uncapped mRNA. Protein purity was verified by SDS-PAGE.
Preparation of SIINFEKL mRNA by in vitro transcription
cDNA encoding SIINFEKL peptide flanked by a MHC class I signal peptide fragment (78 bp, secretion signal (sec)) and the transmembrane and cytosolic domains including the stop-codon (MHC class I trafficking signal (MITD), 168 bp) were synthesized by IDT according to previous studies.40–41 cDNA was cloned into pGEM4Z/GFP/A64 (A gift from Dr. Smita K. Nair at Duke University) by replacing GFP fragment with XbaI and NotI. Linearization with SpeI, followed by in vitro transcription (IVT) with HiScribe™ T7 High Yield RNA Synthesis Kit (NEB), yields a transcript containing 61 nucleotides of vector-derived sequence, the coding sequence and 64 A residues. In a typical 20 μl reaction, the following nucleotides were prepared: ATP (10 mM), pseudo-UTP (10 mM), Methyl-CTP (10 mM), GTP (2 mM), anti reverse Cap analog (8 mM). RNA was purified by RNeasy purification kit (Qiagen, Hilden, Germany). RNA quality was confirmed by running in 1% Agarose gel. Concentration was determined by OD260nm.
Preparation of nanoplexes for transfection
Polyamines were dissolved in 10 mM HEPES buffer (pH 7.4). For each well of a 96-well plate, 100 ng mRNA diluted in 5 μl OptiMEM was mixed with 5 μl OptiMEM containing eIF4E at room temperature for 10 min. The formation of mRNA/eIF4E complex was confirmed by gel shift assay in native agarose gel performed at 70V, 4°C for 30 min. Afterwards, 5 μl OptiMEM containing polyamine was added and incubated at room temperature for 15 min prior to transfection. Polyamine was adjusted to achieve 15 to 1 (N/P) ratio for transfection in HEK293T, NIH3T3 and HeLa cells. 10 to 1 (N/P) ratio was used for transfection in dendritic cells.
Dynamic light scattering (DLS) and zeta potential measurements of nanoplexes
Naked luciferase mRNA or mRNA/eIF4E complexes were mixed with each polyamine at a 15 to 1 (N/P) ratio in 10mM HEPES (pH 7.4). These nanoplexes were prepared in the same way as those for cell transfection studies. Final mRNA concentration in DLS and zeta potential measurements was 10 μg/ml. Hydrodynamic size and polydispersity index were measured using dynamic light scattering (Malvern ZS90 particle analyzer, λ = 633 nm). Zeta potential measurements were made using laser Doppler electrophoresis with the Malvern ZS90. From the obtained electrophoretic mobility, zeta potential was calculated by the Smoluchowski equation: ζ = 4πηυ/ε, where η is the viscosity of the solvent, υ is the electrophoretic mobility, and ε is the dielectric constant of the solvent.
Potentiometric titration
The polyamine in a chloride salt form was dissolved in 0.1 N HCl (5 mL) containing 50 mM NaCl to obtain a solution with 100 mM amine, and then sequentially titrated with 0.1 N NaOH containing 50 mM NaCl. The pH values were acquired with a pH meter. A plot of pH against NaOH volume was made to calculate different apparent pKa associated with each estimated protonation structure and the degree of protonation at pH 7.4.
Quantification of reporter gene expression
24 hr prior to transfection, cells were seeded in 100 μl culture media in a 96-well plate. 5000 cells per well for NIH3T3 and HeLa. 10000 cells per well for HEK293T. On the day of transfection, nanoplexes containing 100 ng reporter mRNA was added into each well. For luciferase, 24 hr after transfection, cells were lysed and a luciferase substrate was added per the manufacturer’s instructions. Bioluminescence was quantified by a Tecan plate reader (Switzerland) with an integration time of 1000 ms. Control wells with nontransfected cells were included each time for subtraction of the background. For GFP detection, the FACS LSR Fortessa HTS flow cytomer was used after trypsinizing cells. 10000 live cells were measured and analyzed for percentage of GFP positive cells and mean fluorescence intensity (MFI).
Detection of intracellular RNA abundance by quantitative PCR
RNA was extracted by Trizol reagent (Invitrogen) and converted to cDNA via Ecodry cDNA synthesis kit (Clontech). cDNA was amplified in LightCycler® 480 SYBR Green I Master reagent and quantified by Roche LightCycler 480 Real-Time PCR System. Primer sequences used for detection are: Luciferase forward: gaaatgtccgttcggttggc; Luciferase reverse: tccgataaataacgcgccca; GFP forward: ggagcgcaccatcttcttca; GFP reverse: agggtgtcgccctcgaa; human actin forward:tccctggagaagagctacga; human actin reverse: agcactgtgttggcgtacag; mouse actin forward: tggcgcttttgactcaggat; mouse actin reverse: gggatgtttgctccaaccaa.
Polysome profiling
Polysome profiling was performed by following a previous protocol.29 Briefly, in one 10 cm petri dish, 106 HEK293T cells were seeded 24 hr before transfection. 4 μg GFP mRNA alone or mixed with 20 μg eIF4E protein were complexed with polyamines at a 15:1 N/P ratio in 600 μl Opti-MEM medium at room temperature for 15 min. Complexes were added into cells and incubated at 37°C, 5% CO2 for 24 hr. In the meantime, 5 and 50% sucrose were prepared in sucrose gradient buffer (20 mM HEPES, 100 mM KCl, 50 mM MgCl2, pH 7.6) with EDTA-free protease inhibitor cocktail and 100 μg/ml cycloheximide (CHX). A linear sucrose gradient was made by filling half of centrifuge tubes with 50% followed by 5% sucrose buffers. Tubes were laid horizontally in the cold room for 6 hr and were lift upright in the cold room for sample loading the next day. On the day of sucrose gradient fractionation, in each 10cm petri dish, cells were incubated with 100 μg/ml CHX at 37°C for 5 min followed by washing with 10 ml ice-cold PBS containing 100 μg/ml CHX twice. Cell pellet was collected and subjected to 500 μl hypotonic lysis (5 mM Tris-HCl, 2.5 mM MgCl2, 1.5 mM KCl, 0.5% TritonX-100, 0.5% sodium deoxycholate, 100 μg/ml CHX, 2 mM DTT, pH 7.5) supplemented with 1xEDTA-free protease inhibitor cocktail and 50U Recombinant RNasin™ Ribonuclease Inhibitor for each 10 cm petri dish. Cells were lysed on ice for 5 min and lysates were collected by centrifugation at 17000×g, 10 min. Cell lysates were loaded onto 5–50% linear sucrose gradient and were ultracentrifuged in a SW41 rotor at 36,000 rpm, 2 hr, 4°C. After ultracentrifugation was finished, a hole at the bottom of centrifuge tube was drilled with a 25 gauge needle. 0.5 ml fraction was collected sequentially starting from the tube bottom. A total of 24 fractions from a 12 ml tube were collected. The absorbance at OD254nm was measured by DU 800 spectrophotometer (Beckman Coulter, Brea, CA, USA). An OD254nm absorbance was plotted against 24 fractions from a sucrose gradient (5–50%) to identify free RNA, monosome and polysome populations. Afterwards, two neighboring fractions were combined into one tube (i.e. a total of 800 μl after removing 100 μl from each fraction for OD254nm measurement). 10 ng luciferase mRNA was spiked into 400 μl combined fraction as an internal reference and was mixed with equal volume of Trizol for RNA isolation. Total mRNAs were converted into cDNA and the percentage of GFP mRNA in each fraction was calculated by first normalizing GFP mRNA (transfected alone or with eIF4E) to luciferase mRNA (internal control) in each fraction and then comparing it to the sum of normalized GFP abundance from a total of 12 combined fractions.
To identify the location of 40S small subunit within the fractions, 400 μl sample from the rest of combined factions was precipitated by trichloroacetic acid protein precipitation followed by washing with acetone twice. Protein pellets were dried completely before re-suspension in 1xSDS PAGE loading buffer (50 mM Tris-Cl, pH 6.8), 2% SDS, 0.1% bromophenol blue, 10% glycerol, 100 mM beta mercaptoethanol, pH 6.8). Total proteins were analyzed by western blotting. S6 Ribosomal Protein (5G10) Rabbit mAb (Cell Signaling Technology) was used to detect the presence and abundance of S6 (a major compotent of 40S small subunit of ribosome) in each fraction.
Flow cytometry analysis of intracellular Förster resonance energy transfer (FRET)
eIF4E protein was labeled by Cy3-NHS (AAT bioquest, Sunnyvale, CA, USA) at a 5:1 (dye/protein) molar ratio in the protein storage buffer (20 mM HEPES, 150 mM NaCI, 10% glycerol, 1 mM DTT) at room temperature for one hour. Protein-dye conjugates were separated from free dyes through 0.5 ml Zeba™ Spin Desalting Columns, 7K MWCO (Thermofisher). Luciferase mRNAs were labeled with Label IT Cy5 Labeling Kit (Mirus Bio LLC, Madison, WI, USA) at a 10:1 (dye/mRNA) molar ratio per the provider’s instruction. Labeled mRNAs were purified by ammonia acetate/ethanol precipitation. In a 96-well plate, 104 HEK293T or NIH3T3 cells were seeded in 100 μl growth medium per well 24 hr before transfection. On the day of transfection, Cy5-mRNA (100 ng RNA) alone or Cy3-eIF4E/Cy5-mRNA (100 ng RNA/500 ng eIF4E) were transfected with polyamines at 15:1 N/P molar ratio into cells. Cells were harvested at 4 hr post transfection. Fluorescence intensity of the cells was monitored and evaluated with a FACSCelesta (BD Biosciences) equipped with Diva software (BD Biosciences) using a 488 laser for excitation and a 660/20 nm filter.
Ex vivo antigen presentation assay
In a 48-well plate, 104 BMDCs were seeded in 200 μl RPMI 1640, 10% FBS 24 hr before transfection. On day 0, each well received 200 ng OVA mRNA, SIINFEKL mRNA, luciferase mRNA or together with 1 μg eIF4E proteins transfected with N3 (TET) or N5 (PEH). On day 1, mononuclear cells (MNCs) were isolated from inguinal, mesenteric, cervical, axillary and brachial lymph nodes of OTI mice and labeled with 1 μM CFSE fluorescence dye for tracking of cell proliferation and activation. 2×105 CFSE-labeled MNCs were added into each well. Activation of CD8 T cells was detected by measuring fluorescence intensity of intracellular CFSE gated on CD8 T cells. The percentage of CFSE-low population was quantified. Only viable cells (DAPI-negative) were analyzed in flow cytometer through simultaneous labeling of DAPI. On day 4, supernatant from co-culture wells was removed for quantification of interferon gamma concentration by Murine IFN-γ Mini TMB ELISA Development Kit (Peprotech, Rocky Hill, NJ, USA).
In vivo mRNA delivery
Female Balb/c mice aged 8–10 weeks were purchased from Taconic Biosciences (Hudson, NY, USA). 5 μg luciferase mRNA or 5 μg mRNA/5 μg eIF4E packaged with polyamines at a 50:1 N/P ratio were prepared in 120 μl OptiMEM medium and were injected into animals via tail veins.
Bioluminescence imaging
Bioluminescent imaging was performed with a CCD camera mounted in a light-tight specimen box (Xenogen, Waltham, MA, USA). Imaging and quantification of signals were controlled by the acquisition and analysis software Living Image (Xenogen). Anaesthetised mice were placed in the IVIS Imaging System and imaged from ventral views 10 min after intraperitoneal injection of D-luciferin at 150 mg/kg body weight.
Cell proliferation assay
Cell proliferation was measured by detecting mitochondrial dehydrogenase activity using MTT as the substrate. Cells in a 96-well plate were incubated with MTT at a concentration of 0.5 mg/mL, at 37°C for 1 hr. The purple MTT product was solubilized with DMSO and measured at 570 nm using a Tecan plate reader.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5.0a for Mac OS X (San Diego, CA, USA). A one-way ANOVA followed by Tukey post test or Student’s t test was used to compare statistical significance in the studies.
Supplementary Material
Acknowledgments
This work was supported by the Department of Defense Ovarian Cancer Research Program Teal Innovator Award (PTH) and the Koch Institute Quinquennial Postdoctoral Fellowship (JL). We thank: Glenn Paradis at the David H. Koch Institute Flow Cytometry Core at MIT for providing assistance in the intracellular FRET experiment; Dr. Mariane Bandeira Melo and Department of Comparative Medicine at MIT for providing transgenic OTI mice; Dr. Jeerapond Leelawattanachai at the National Nanotechnology Center (NANOTEC) of Thailand for scientific discussion. Celestine Hong at the Department of Chemical Engineering of MIT for her precious critique and proofreading. DJI is an investigator of the Howard Hughes Medical Institute.
Footnotes
Contributions
J.L. designed the experiments. J.L., W.W., Y.H., E.Y., Y.L. and K.Z. carried out experiments. P.T.H. and D.J.I. supervised the study. J.L. and P.T.H. wrote the manuscript.
Supporting Information Available: Additional figures, schemes, results, and method description as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jirikowski GF, Sanna PP, Maciejewski-Lenoir D, Bloom FE. Reversal of Diabetes Insipidus in Brattleboro Rats: Intrahypothalamic Injection of Vasopressin mRNA. Science. 1992;255:996–8. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- 2.Conry RM, LoBuglio AF, Wright M, Sumerel L, Pike MJ, Johanning F, Benjamin R, Lu D, Curiel DT. Characterization of a Messenger RNA Polynucleotide Vaccine Vector. Cancer Res. 1995;55:1397–400. [PubMed] [Google Scholar]
- 3.Boczkowski D, Nair SK, Snyder D, Gilboa E. Dendritic Cells Pulsed with RNA Are Potent Antigen-Presenting Cells In Vitro and In Vivo. J Exp Med. 1996;184:465–72. doi: 10.1084/jem.184.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoerr I, Obst R, Rammensee HG, Jung G. In Vivo Application of RNA Leads to Induction of Specific Cytotoxic T Lymphocytes and Antibodies. Eur J Immunol. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Zou S, Scarfo K, Nantz MH, Hecker JG. Lipid-Mediated Delivery of RNA Is More Efficient Than Delivery of DNA in Non-Dividing Cells. Int J Pharm. 2010;389:232–243. doi: 10.1016/j.ijpharm.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahin U, Kariko K, Tureci O. mRNA-Based Therapeutics - Developing a New Class of Drugs. Nat Rev Drug Discovery. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 7.Schlake T, Thess A, Fotin-Mleczek M, Kallen KJ. Developing mRNA-Vaccine Technologies. RNA Biology. 2012;9:1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phua KK, Leong KW, Nair SK. Transfection Efficiency and Transgene Expression Kinetics of mRNA Delivered in Naked and Nanoparticle Format. J Control Release. 2013;166:227–33. doi: 10.1016/j.jconrel.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK, Madden TD, Hope MJ, Weissman D. Expression Kinetics of Nucleoside-Modified mRNA Delivered in Lipid Nanoparticles to Mice by Various Routes. J Control Release. 2015;217:345–51. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song MG, Li Y, Kiledjian M. Multiple mRNA Decapping Enzymes in Mammalian Cells. Mol Cell. 2010;40:423–32. doi: 10.1016/j.molcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piccirillo C, Khanna R, Kiledjian M. Functional Characterization of the Mammalian mRNA Decapping Enzyme HDCP2. RNA. 2003;9:1138–47. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Leary SE, Petrov A, Chen J, Puglisi JD. Dynamic Recognition of the mRNA Cap by Saccharomyces Cerevisiae eIF4E. Structure. 2013;21:2197–207. doi: 10.1016/j.str.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal Structure of the Messenger RNA 5′ Cap-Binding Protein (eIF4E) Bound to 7-Methyl-GDP. Cell. 1997;89:951–61. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 14.Duncan R, Milburn SC, Hershey JW. Regulated Phosphorylation and Low Abundance of HeLa Cell Initiation Factor eIF-4F Suggest a Role in Translational Control. Heat Shock Effects on eIF-4F. J Biol Chem. 1987;262:380–8. [PubMed] [Google Scholar]
- 15.Sonenberg N, Hinnebusch AG. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uchida H, Itaka K, Uchida S, Ishii T, Suma T, Miyata K, Oba M, Nishiyama N, Kataoka K. Synthetic Polyamines to Regulate mRNA Translation through the Preservative Binding of Eukaryotic Initiation Factor 4e to the Cap Structure. J Am Chem Soc. 2016;138:1478–81. doi: 10.1021/jacs.5b11726. [DOI] [PubMed] [Google Scholar]
- 17.Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, Proud CG. Phosphorylation of Eukaryotic Initiation Factor 4e Markedly Reduces Its Affinity for Capped mRNA. J Biol Chem. 2002;277:3303–9. doi: 10.1074/jbc.M103607200. [DOI] [PubMed] [Google Scholar]
- 18.Hagedorn CH, SpivakKroizman T, Friedland DE, Goss DJ, Xie YP. Expression of Functional eIF-4E (Human): Purification, Detailed Characterization, and Its Use in Isolating eIF-4E Binding Proteins. Protein Expression Purif. 1997;9:53–60. doi: 10.1006/prep.1996.0661. [DOI] [PubMed] [Google Scholar]
- 19.Itaka K, Ishii T, Hasegawa Y, Kataoka K. Biodegradable Polyamino Acid-Based Polycations as Safe and Effective Gene Carrier Minimizing Cumulative Toxicity. Biomaterials. 2010;31:3707–3714. doi: 10.1016/j.biomaterials.2009.11.072. [DOI] [PubMed] [Google Scholar]
- 20.Miyata K, Oba M, Nakanishi M, Fukushima S, Yamasaki Y, Koyama H, Nishiyama N, Kataoka K. Polyplexes from Poly(Aspartamide) Bearing 1,2-Diaminoethane Side Chains Induce pH-Selective, Endosomal Membrane Destabilization with Amplified Transfection and Negligible Cytotoxicity. J Am Chem Soc. 2008;130:16287–16294. doi: 10.1021/ja804561g. [DOI] [PubMed] [Google Scholar]
- 21.Aini H, Itaka K, Fujisawa A, Uchida H, Uchida S, Fukushima S, Kataoka K, Saito T, Chung UI, Ohba S. Messenger RNA Delivery of a Cartilage-Anabolic Transcription Factor as a Disease-Modifying Strategy for Osteoarthritis Treatment. Sci Rep. 2016;6:18743. doi: 10.1038/srep18743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin Suppresses eIF4E-Mediated Oncogenic Transformation by Physical Mimicry of the 7-Methyl Guanosine mRNA Cap. Proc Natl Acad Sci U S A. 2004;101:18105–10. doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, Burley SK, Stolarski R. Biophysical Studies of eIF4E Cap-Binding Protein: Recognition of mRNA 5′ Cap Structure and Synthetic Fragments of eIF4G and 4E-BP1 Proteins. J Mol Biol. 2002;319:615–35. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- 24.Haghighat A, Sonenberg N. eIF4G Dramatically Enhances the Binding of eIF4E to the mRNA 5′-Cap Structure. J Biol Chem. 1997;272:21677–80. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 25.Uchida H, Itaka K, Nomoto T, Ishii T, Suma T, Ikegami M, Miyata K, Oba M, Nishiyama N, Kataoka K. Modulated Protonation of Side Chain Aminoethylene Repeats in N-Substituted Polyaspartamides Promotes mRNA Transfection. J Am Chem Soc. 2014;136:12396–405. doi: 10.1021/ja506194z. [DOI] [PubMed] [Google Scholar]
- 26.Uchida H, Miyata K, Oba M, Ishii T, Suma T, Itaka K, Nishiyama N, Kataoka K. Odd-Even Effect of Repeating Aminoethylene Units in the Side Chain of N-Substituted Polyaspartamides on Gene Transfection Profiles. J Am Chem Soc. 2011;133:15524–15532. doi: 10.1021/ja204466y. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz KW, Murray MT, Sylora R, Sohn RL, Dulchavsky SA. Augmentation of Wound Healing with Translation Initiation Factor eIF4E mRNA. J Surg Res. 2002;103:175–82. doi: 10.1006/jsre.2002.6360. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz DC, Parker R. mRNA Decapping in Yeast Requires Dissociation of the Cap Binding Protein, Eukaryotic Translation Initiation Factor 4e. Mol Cell Biol. 2000;20:7933–42. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandin V, Sikstrom K, Alain T, Morita M, McLaughlan S, Larsson O, Topisirovic I. Polysome Fractionation and Analysis of Mammalian Translatomes on a Genome-Wide Scale. J Vis Exp. 2014 doi: 10.3791/51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E--from Translation to Transformation. Oncogene. 2004;23:3172–9. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 31.Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, Chen Y, Zhang H, Sissons S, Cox K, McNulty AM, Parsons SH, Wang T, Sams L, Geeganage S, Douglass LE, et al. Therapeutic Suppression of Translation Initiation Factor eIF4E Expression Reduces Tumor Growth without Toxicity. J Clin Invest. 2007;117:2638–2648. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murata T, Shimotohno K. Ubiquitination and Proteasome-Dependent Degradation of Human Eukaryotic Translation Initiation Factor 4e. J Biol Chem. 2006;281:20788–800. doi: 10.1074/jbc.M600563200. [DOI] [PubMed] [Google Scholar]
- 33.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cell. 2009;4:472–6. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Cationic Lipid-Mediated Delivery of Proteins Enables Efficient Protein-Based Genome Editing In Vitro and In Vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Decarlo L, Mestel C, Barcellos-Hoff MH, Schneidera RJ. Eukaryotic Translation Initiation Factor 4e Is a Feed-Forward Translational Coactivator of Transforming Growth Factor Beta Early Protransforming Events in Breast Epithelial Cells. Mol Cell Biol. 2015;35:2597–2609. doi: 10.1128/MCB.00324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng F, Fornoni A, Elliot SJ, Guan YF, Breyer MD, Striker LJ, Striker GE. Upregulation of Type I Collagen by TGF-beta in Mesangial Cells Is Blocked by PPARgamma Activation. American Journal of Physiology-Renal Physiology. 2002;282:F639–F648. doi: 10.1152/ajprenal.00189.2001. [DOI] [PubMed] [Google Scholar]
- 37.Bettinger T, Carlisle RC, Read ML, Ogris M, Seymour LW. Peptide-Mediated RNA Delivery: A Novel Approach for Enhanced Transfection of Primary and Post-Mitotic Cells. Nucleic Acids Res. 2001;29:3882–91. doi: 10.1093/nar/29.18.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turnbull IC, Eltoukhy AA, Fish KM, Nonnenmacher M, Ishikawa K, Chen J, Hajjar RJ, Anderson DG, Costa KD. Myocardial Delivery of Lipidoid Nanoparticle Carrying Modrna Induces Rapid and Transient Expression. Mol Ther. 2016;24:66–75. doi: 10.1038/mt.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Su HH, Yang Y, Hu Y, Zhang L, Blancafort P, Huang L. Systemic Delivery of Modified mRNA Encoding Herpes Simplex Virus 1 Thymidine Kinase for Targeted Cancer Gene Therapy. Mol Ther. 2013;21:358–67. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreiter S, Selmi A, Diken M, Sebastian M, Osterloh P, Schild H, Huber C, Tureci O, Sahin U. Increased Antigen Presentation Efficiency by Coupling Antigens to MHC Class I Trafficking Signals. J Immunol. 2008;180:309–18. doi: 10.4049/jimmunol.180.1.309. [DOI] [PubMed] [Google Scholar]
- 41.Kranz LM, Diken M, Haas H, Kreiter S, Loquai C, Reuter KC, Meng M, Fritz D, Vascotto F, Hefesha H, Grunwitz C, Vormehr M, Husemann Y, Selmi A, Kuhn AN, Buck J, Derhovanessian E, Rae R, Attig S, Diekmann J, et al. Systemic RNA Delivery to Dendritic Cells Exploits Antiviral Defence for Cancer Immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.