Abstract
Mitochondrial genome maintenance plays a central role in preserving brain health. We previously demonstrated accumulation of mitochondrial DNA damage and severe neurodegeneration in transgenic mice inducibly expressing a mutated mitochondrial DNA repair enzyme (mutUNG1) selectively in forebrain neurons. Here, we examine whether severe neurodegeneration in mutUNG1-expressing mice could be rescued by feeding the mice a ketogenic diet, which is known to have beneficial effects in several neurological disorders. The diet increased the levels of superoxide dismutase 2, and mitochondrial mass, enzymes, and regulators such as SIRT1 and FIS1, and appeared to downregulate N-methyl-D-aspartic acid (NMDA) receptor subunits NR2A/B and upregulate γ-aminobutyric acid A (GABAA) receptor subunits α1. However, unexpectedly, the ketogenic diet aggravated neurodegeneration and mitochondrial deterioration. Electron microscopy showed structurally impaired mitochondria accumulating in neuronal perikarya. We propose that aggravation is caused by increased mitochondrial biogenesis of generally dysfunctional mitochondria. This study thereby questions the dogma that a ketogenic diet is unambiguously beneficial in mitochondrial disorders.
Keywords: MtDNA damage, Ketogenic diet, Biogenesis, Neurodegeneration
1. Introduction
Mitochondrial dysfunction plays a major role in neurodegeneration, including in Alzheimer’s disease and Parkinson’s disease (Haun et al., 2013; Itoh et al., 2013; Lu, 2009). Neurons have a high energy demand and are mostly powered by mitochondria. Deficient energy supplies due to defective mitochondria can lead to neuronal death and disease (Attwell and Laughlin, 2001; Chen and Chan, 2006). In addition, dysfunctional mitochondria are known to generate high levels of reactive oxygen species (ROS), which are harmful to all cellular macromolecules (Adam-Vizi, 2005; Rodell et al., 2013).
A ketogenic diet, high on fat, moderate on protein, and low on carbohydrates, reproduces biochemical changes that take place during fasting (Gjedde and Crone, 1975). The diet leads to the accumulation of ketone bodies, produced in the liver from fatty acids when the insulin-to-glucagon ratio is low, and to reduced blood glucose levels (Masino and Rho, 2012; Meidenbauer et al., 2014). A ketogenic diet is known to have a positive effect in certain neurological diseases, especially in drug-resistant epilepsy, in which it can reduce the frequency of seizures leading to an improved quality of life (Freeman et al., 2007; Sirven et al., 1999). Recently, it has been proposed that a ketogenic diet would be beneficial for patients with neurodegenerative disorders, such as Alzheimer’s disease (Van der Auwera et al., 2005), Parkinson’s disease (Vanitallie et al., 2005), Huntington’s disease (Ruskin et al., 2011), amyotrophic lateral sclerosis (Zhao et al., 2006), and even glaucoma (Zarnowski et al., 2012) and that this diet could alleviate or slow the progression of these disorders.
Although the mechanisms of neuroprotective effects of a ketogenic diet have not been identified, several mechanisms have been proposed, including augmented endothelial transport of monocarboxylates, alterations in the antioxidant status, changes in neurotransmitter levels in the brain, especially GABA, and metabolic actions of the diet on the energy metabolism of the cell (Lauritzen et al., 2015; Rho and Sankar, 2008). Several studies have shown that a ketogenic diet increases the number and performance of mitochondria in neurons (Bough et al., 2006; Hughes et al., 2014; Nylen et al., 2009; Rho and Rogawski, 2007). Induction of mitochondrial biogenesis leads to increased energy reserves and improved energy metabolism in neurons and enables them to cope better with detrimental effects that occur in neurological disease (Rho and Rogawski, 2007). It is therefore plausible that the favorable effects of the ketogenic diet on neurological disorders, such as epilepsy and neurodegenerative diseases, in part stem from an increased mass of mitochondria in the neurons.
We have generated a transgenic mouse model that reproduces hallmarks of neurodegenerative disease. In this transgenic mouse, we induce damage to mitochondrial DNA (mtDNA) selectively in forebrain neurons, notably in the hippocampus (Lauritzen et al., 2010, 2011a, 2011b). This is achieved by inducibly expressing a mutated version (mutUNG1) of the mitochondrial isoform of the DNA base excision repair protein uracil-DNA glycosylase UNG1, which removes thymine (in addition to its natural substrate uracil) thereby creating high levels of abasic (apyrimidinic) sites (AP-sites) in mtDNA (Kavli et al., 1996; Kubota et al., 1996; Otterlei et al., 1998). AP-sites are highly cytotoxic as they stall polymerases and enhance the risk of DNA strand breaks. They may be particularly harmful to mtDNA because mitochondria are deficient in translesion synthesis (Avkin et al., 2002; Pages et al., 2008; Zhang et al., 2006). By placing mutUNG1 under the control of the Tet-on system, we temporally and spatially control its expression (Sun et al., 2007). The control by the CaMKIIa-promoter restricted the mutUNG1 expression to forebrain neurons (Michalon et al., 2005). The expression of mutUNG1 is initiated in young adult mice to avoid developmental defects that could be caused by mutUNG1. The mutUNG1-expressing mice display neurodegeneration and concomitant behavioral impairments with reduced cognitive abilities. The affected hippocampal tissue showed mitochondrial dysfunction with reduced mtDNA copy number and impaired expression and function of mtDNA-encoded proteins (reduced complex I respiration, reduced expression of complexes I, III, IV, and V), as well as severely altered mitochondrial dynamics. In addition, there was a strong induction of endogenous antioxidant defense mechanisms indicative of oxidative stress (Lauritzen et al., 2010, 2011a). These features mimic features of many neurodegenerative conditions including Alzheimer’s disease.
Given that a ketogenic diet is beneficial in several neurodegenerative disorders, we tested the effects of this diet in neuronal tissues with high levels of mtDNA damage and determined whether it would alleviate or slow neurodegeneration in the mutUNG1-expressing mice. The main conclusion of this study is that in spite of signs of upregulation of antioxidant defenses (superoxide dismutase 2 [SOD2]), mitochondrial mass, and regulators (SIRT1, FIS1), a ketogenic diet is not beneficial for mutUNG1-expressing mice. On the contrary, the diet aggravates neurodegenerative changes, comprising severe atrophy of the hippocampus, neuronal loss, astrogliosis, and breakdown of mitochondrial dynamics. Specifically, there is a more severe reduction in NMDA receptor subunits NR2A/B at excitatory synapses and a slight increase in GABAA receptor subunit α1, compared with mice fed a standard diet, indicating synaptic imbalance in receptor composition.
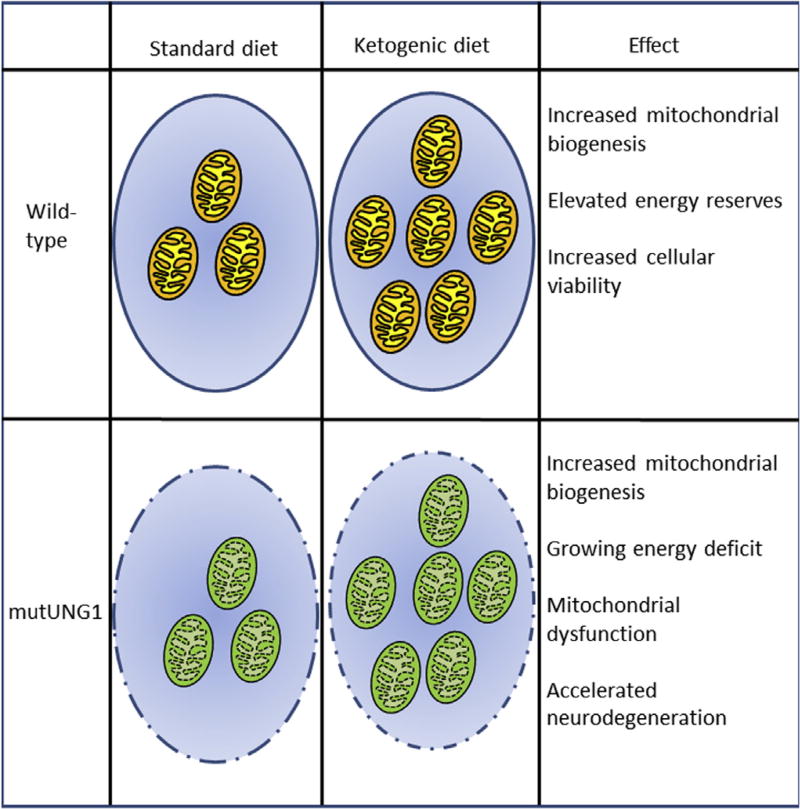
We propose that the energetic cost of generating new mitochondria is higher than the bioenergetic gain because the mitochondria are all dysfunctional. The mitochondria have inefficient electron transport chains and are thereby unable to provide the necessary adenosine triphosphate-levels despite their increased number. The mitochondrial biogenesis therefore turns into an energy drain rather than a solution to the imminent energy crisis (Fig. 1).
Fig. 1.
Proposed model for how a ketogenic diet aggravates neurodegenerative effects of mtDNA toxicity in mutUNG1-expressing mice. A ketogenic diet induces mitochondrial biogenesis, which enhances the energy metabolism and energy reserves in neural tissues. It thereby makes the neurons better equipped to deal with cellular stress and increases cellular viability. In mutUNG1-expressing mice, the mutUNG1 protein is targeted to all mitochondria, generating high levels of mtDNA damage, which causes dysfunctional mitochondria. We propose that in this scenario, an increase in mitochondrial biogenesis will lead to an energetic cost that is higher than the gain and that this ultimately leads to an energy deficit. This together with the harmful effects of increased ROS levels, resulting from a greater number of dysfunctional mitochondria, leads to increased neurodegeneration rather than an alleviating effect. Abbreviation: mtDNA, mitochondrial DNA.
To our knowledge, this is the first study to find severe detrimental effects of a ketogenic diet postdevelopmentally.
2. Materials and methods
2.1. Transgenic mice and treatment
Transgenic mice expressing a mutated version of the human mtDNA repair enzyme UNG1 (mutUNG1) were generated in the C57BL/6 strain and treated with doxycycline to induce transgene expression as previously described (Lauritzen et al., 2010). The expression of mutUNG1 was induced by adding doxycycline to the chow at 6 mg/g. Food characterized as a standard diet in this study is the “Rat and Mouse No. 3 Breeding” from Special Diets Services, the routine diet used for small rodents at the Section for Comparative Medicine at the Oslo University Hospital where the animals were housed. The energy content (Atwater fuel energy) of this diet is specified as 12% fats, 27% proteins, and 61% carbohydrates. As a ketogenic diet in this study, we used the “High-Fat Ketogenic Diet (Rodent)” (5TJQ) from Test Diet, the energy content of which is specified as 84% fats, 16% proteins, and 0% carbohydrates. The mice were fed a standard diet until they were 8 weeks old, at which age they were fed either a standard diet or a ketogenic diet, both mixed with doxycycline, administered ad libitum, and maintained until the time of death at 4 to 6 months of age. As the animal welfare regulation required mutUNG1 mice to be euthanized if losing more than 15% of weight, the weight was monitored weekly (Supplementary Fig. S1). There was no loss of weight in the mutUNG1 mice; although weight was stable on ketogenic diet, it increased on standard diet. In our previous studies on mutUNG1 expression in brain (Lauritzen et al., 2010, 2011a, 2011b), we found that CaMKIIa-rtTa mice have no phenotype, as is true also of wild-type mice fed doxycycline. Importantly, mutUNG1 mice (CaMKIIa-rtTa-Luciferase × TetOn-mutUNG1 mice) not induced by doxycycline showed no phenotype. We therefore used wild-type mice of the same litter as controls.
All experiments were approved by the National Animal Research Authority in Norway and conducted in accordance with the laws and regulations controlling experimental procedures in live animals in Norway and the European Union’s Directive 86/609/EEC.
2.2. Light microscopy and biochemistry
Whole brains were fixed in 10% (v/v) neutral-buffered formalin (Richard-Allan Scientific) for at least 48 hours and paraffin embedded. Brains were cut into 4-µm-thick coronal sections using a rotary microtome (Microm 355 S), and the sections were dried on glass slides at 37 °C overnight.
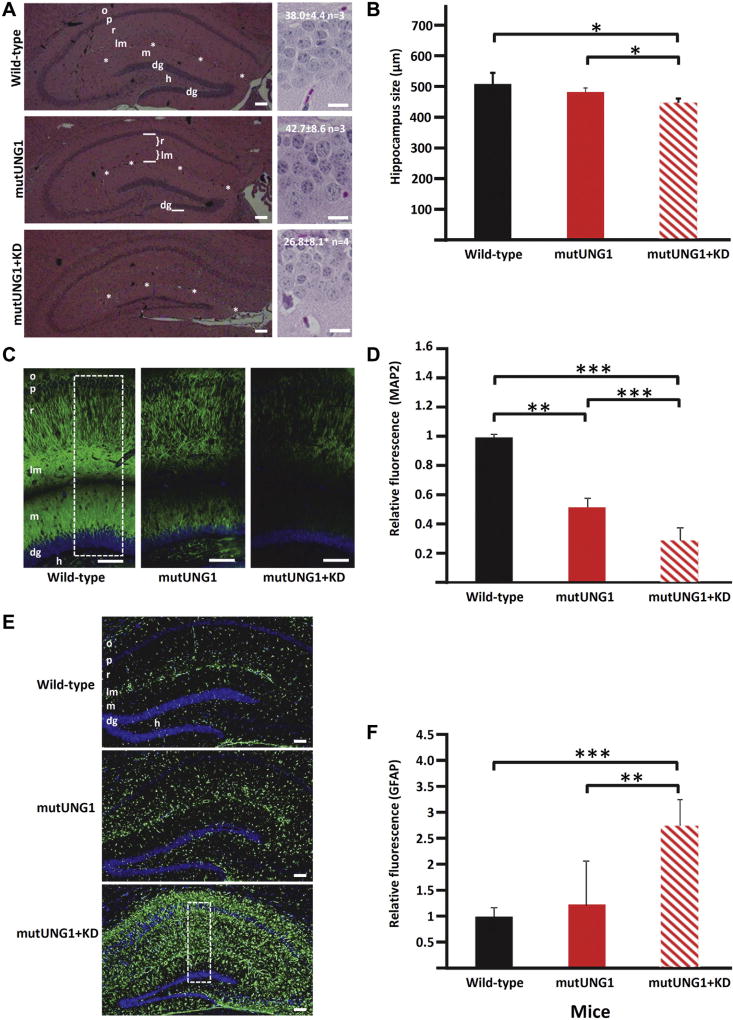
For hematoxylin and eosin (HE) staining, sections were treated with hematoxylin and eosin (HE; Richard-Allan Scientific). HE-stained sections were studied with a Zeiss Axioplan 2 microscope, and pictures were taken and analyzed with an AxioCamHRc and AxioVision Rel. 4.6 software. For cell counting, high-resolution images of histological sections were acquired with an ×20 objective lens using an automated slide scanner system (Axio Scan Z1, Carl Zeiss Microscopy, Munich, Germany). Images were inspected using the Zen Lite Blue software (Carl Zeiss Microscopy) and analyzed via the Navigator Data Management Interface (http://cmbn-navigator.uio.no/navigator). Numbers of cells were counted in a 100-µm long stretch of the infrapyramidal part of the granular cell layer of the dentate gyrus (Fig. 2A) on pictures of the dorsal hippocampus scanned at high magnification. The “size” of the hippocampal formation was quantified as the combined thicknesses of the layers radiatum (r) and lacunosummoleculare (lm) (Fig. 2A, middle panel).
Fig. 2.
A ketogenic diet accelerates atrophy, neurodegeneration, and reactive astrogliosis in the hippocampus in mutUNG1-expressing mice. (A) Representative HE-stained coronal paraffin sections of hippocampus illustrate increased atrophy and neuronal degeneration in mutUNG1-expressing mice fed a ketogenic diet (KD, bottom panels) compared with wild-type (top panels) and mutUNG1-expressing mice fed a standard diet (middle panel). Numbers of cells were counted in a 100-µm long stretch of the infrapyramidal part of the granular cell layer (dg, marked in the middle left panel) on pictures scanned at high magnification. Samples pictures are shown in right panels together with results (mean ± standard deviation, n mice, p = 0.05 for difference between the lower 2). (B) Quantification of the thickness of hippocampal layers r + lm (in coronal sections, A, marked in the middle frame) shows a significant decrease in hippocampus size in mutUNG1-expressing mice fed a ketogenic diet compared with wild-type and mutUNG1-expressing mice fed a standard diet, respectively, (n = 3 mice per group, p = 0.02 and p = 0.03). (C) Coronal paraffin-embedded sections of hippocampus immunolabeled for the neuronal marker MAP2 in wild-type and mutUNG1-expressing mice fed a standard diet, and mutUNG1-expressing mice fed a ketogenic diet, the latter showing loss of labeled pyramidal and granule cell dendrites. (D) Quantification of immunostaining shows a significant decrease of MAP2 in mutUNG1-expressing mice fed a ketogenic diet compared with wild-type and mutUNG1-expressing mice fed a standard diet (n = 3 mice per group, p < 0.0001 and p < 0.0001); mutUNG1-expressing mice fed a standard diet also differed from wild-type mice (p = 0.01). (E) Coronal paraffin-embedded sections of hippocampus immunostained for the astrocyte marker GFAP in wild-type and mutUNG1-expressing mice fed a standard diet, and mutUNG1-expressing mice fed a ketogenic diet. (F) Quantification of immunofluorescence shows a significant increase of GFAP in mutUNG1-expressing mice fed a ketogenic diet compared with wild-type and mutUNG1-expressing mice fed a standard diet (n = 3 mice per group, p = 0.003, p < 0.0001). Stippled rectangles in (C) and (E) indicate regions of interest used for quantification of immunoreactivity. Scale bars = 100 µm (C and E) and 20 µm (right panels in A). Asterisk series marks fissura hippocampi between lm and m. In this and subsequent figures, levels of statistical significance are indicated by asterisks (*p≤ 0.05; **p≤ 0.01; ***p≤ 0.001). Abbreviations: dg, granule cell layer of dentate gyrus; h, hilus of the dentate gyrus; HE, hematoxylin and eosin; lm, stratum lacunosum-moleculare of hippocampus CA1; m, stratum moleculare of dentate gyrus; o, stratum oriens; p, pyramidal cell layer; r, stratum radiatum.
For immunohistochemical studies, sections were deparaffinized by heating at 57 °C for 10 minutes and incubated twice in Clear Rite 3-solution (Richard-Allan Scientific) for 5 minutes each. Thereafter, sections were rehydrated by incubating for 1 minute each in a series of descending concentrations of ethanol (100%, 96%, and 70%), ending in distilled H2O. Sections were then subjected to antigen retrieval by heating for 15 minutes at 95 °C in a Tris-EDTA buffer (10-mM Tris base, 3.8-mM EDTA, pH adjusted to 9.0), cooled at room temperature for 10 minutes, incubated for 5 minutes in distilled H2O, and then 5 minutes in phosphate buffered saline (PBS)/0.1% Tween-20. Sections were then permeabilized with 0.1% Triton X-100 (Sigma) in PBS for 15 minutes, washed twice with 0.1% Tween-20 in PBS, and blocked by incubating with a solution containing 5% bovine serum albumin (BSA; Sigma), 5% goat serum (Sigma), and 0.1% Triton X-100 (Sigma). Sections were incubated overnight at 4 °C with primary antibodies diluted in 0.5% BSA/0.5% goat serum/0.1% Tween-20 in PBS. After washing, sections were incubated with Alexa Fluor 594-conjugated goat anti-mouse IgG (H + L) (Invitrogen, A11005) or Alexa Fluor 488-conjugated goat anti-rabbit IgG (H + L) (Invitrogen, A11008) secondary antibodies, as appropriate. Nuclei in sections were stained by incubating with 1 µg/mL 4′,6-diamidino-2-phenylindole (Sigma). Sections were coverslip-mounted using Mowiol 4–88 Reagent (Merck Biosciences Ltd) and examined with a Zeiss Axiovert 200 M microscope. Images were acquired and analyzed using AxioCamMRm and AxioVision Rel. 4.6 software. All microscope and software settings were equal for images being compared. 4′,6-diamidino-2-phenylindole-staining was used to visualize cell-nuclei. Quantification of immunoreactivities was performed by measuring green and red fluorescence intensity, using ImageJ software (http://rsb.info.nih.gov/ij/) with the Analyze:Measure RGB plugin. The numbers of green or red pixels were expressed relative to the measured area (“area of interest”), a rectangle containing all layers between the pyramidal layer in CA1 and the hilus of the dentate area (Figs. 2C, E and 3A).
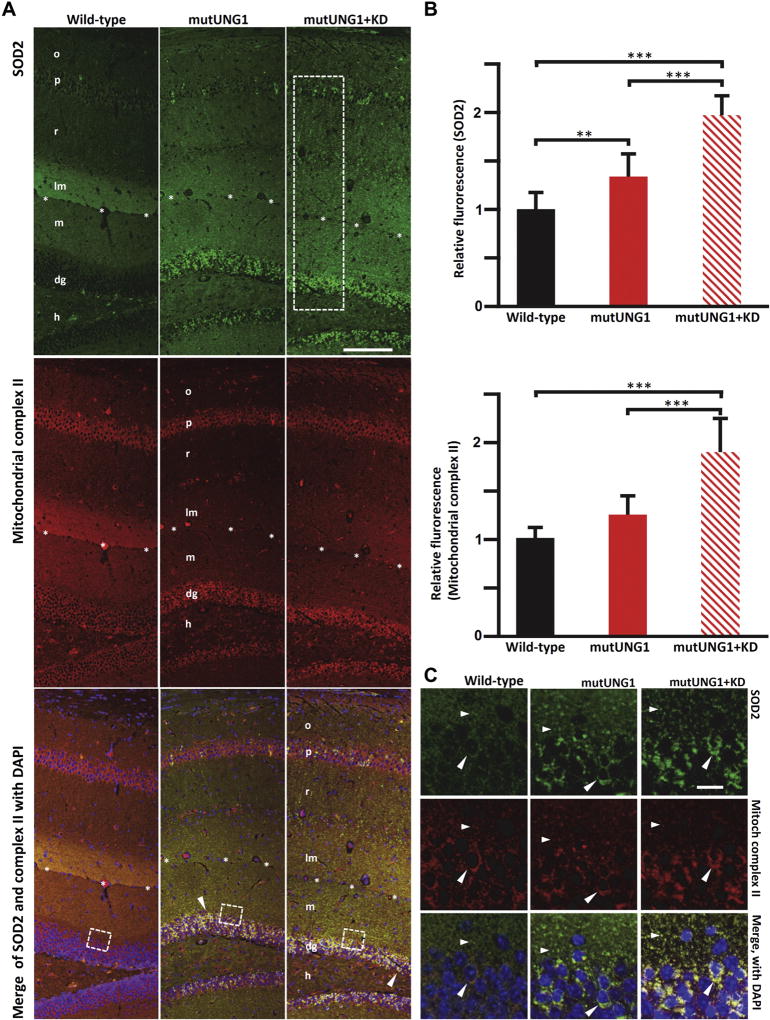
Fig. 3.
mutUNG1-expressing mice fed a ketogenic diet have upregulated mitochondrial antioxidant defenses. (A) Coronal sections of paraffin-embedded hippocampus immunolabeled for SOD2 (green) indicate induction in mitochondrial antioxidant defenses in mutUNG1-expressing mice fed a standard diet, and a further strong increase in mutUNG1-expressing mice fed a ketogenic diet (KD). The nuclear DNA encoded electron transport complex II (red, middle row) shows a similar distribution as SOD2 in all 3 conditions. In the merged frames (bottom row), yellow-orange color signifies colocalization of immunoreactivities in the same structures. Strong mitochondrial labeling of perikarya (arrowhead) in mutUNG1 mice with and without ketogenic diet but not in wild-type mice. Both red and green are brighter for mutUNG1 + KD than for only mutUNG1. Stippled rectangle indicates region of interest used for quantification of immunoreactivity (B), stippled squares indicate approximate positions of close-up pictures (C). Scale bar = 100 µm (objective lens 10 ×). (B) Quantification of SOD2 and mitochondrial complex II immunofluorescence (average per area in rectangular regions of interest as indicated in A (top). SOD2 increases in mutUNG1 mice compared to wild-type (p = 0.003) and in mutUNG1 mice fed a ketogenic diet (p < 0.0001). Complex II increases in mutUNG1 mice when fed a ketogenic diet (p = 0.001). (C) High magnification of areas equivalent to the stippled squares in A demonstrates colocalization of SOD2 and complex II in mitochondria-like structures in neuronal perikarya (dg cells, large arrowheads) as well as in the neuropil (small arrowheads). Clusters of such structures accumulate in neuronal perikarya in mutUNG1-expressing mice, particularly when on ketogenic diet (lower right frame). Scale bar = 10 µm (objective lens 63× oil immersion). Abbreviations: dg, granule cell layer; h, hilus of the dentate gyrus; lm, stratum lacunosum-moleculare of hippocampus CA1; m, stratum moleculare of dentate gyrus; o, stratum oriens; p, pyramidal cell layer; r, stratum radiatum; SOD2, superoxide dismutase 2; asterisk series marks fissura hippocampi between lm and m. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Antibodies against the following proteins were used for immunocytochemistry at the indicated concentrations: mitochondrial complex II, 70 kDA subunit (Invitrogen, Cat-number: 459200; dilution 1:1000), voltage-dependent anion channel 1 (VDAC1; Abcam, ab15895; dilution 1:100); microtubule-associated protein 2 (MAP2; Chemicon, AB5622; dilution 1:500); glial fibrillary acidic protein (GFAP; Sigma, G3893 [monoclonal]; dilution 1:1000); superoxide dismutase 2 (SOD2; Abcam, ab13533; dilution 1:1000); cytochrome c (Cyt C; Invitrogen, 456100 [monoclonal]; dilution 1:500).
Hippocampus of wild-type, mutUNG1, and mutUNG1-keto mice was homogenized in lysis buffer containing 0.5% Igepal CA-630 (NP-40), 2-mM dithiothreitol, 1% protease inhibitor cocktail, and 0.5% Triton X-100 in PBS, and total protein concentration was measured using Direct DetectTM (Millipore). Western blotting was performed according to our previously published article (Lauritzen et al., 2010). In brief, the proteins in SDS-PAGE gel were transferred onto the nitrocellulose membrane and incubated with primary antibody against SOD2 (Abcam ab13533; 1:5000), VDAC1 (Abam ab15895; 1:1000), sirtuin 1 (SIRT1; Sigma AV32386; 1:2500), mitochondrial fission 1 protein (FIS1; Sigma HPA017430; 1:1000), and β-actin (ActB; Milipore MAB1501R; 1:5000). The nitrocellulose membrane was incubated for 1 hour with secondary antibody conjugated with horseradish peroxidase (HRP) (BIO-RAD 170–6515 goat anti-rabbit IgG [H + L] HRP conjugate 1:20,000, or Sigma–Aldrich A9044 rabbit anti-mouse IgG [whole molecule] HRP conjugate 1:20,000). Notably, all protein targets were developed on the same membrane followed by consecutive stripping of the membrane for 30 minutes using stripping buffer (Thermo Scientific).
The protein lysates were also used for Oxyblot (OxyBlot Protein Oxidation Detection Kit, Millipore), and OxiSelect (OxiSelect Protein Carbonyl Fluorometric Assay, Cell Biolabs). Oxyblots were performed according to the manufacturer’s instructions with 15-µg proteins. Briefly, 30 µg of protein lysate from each sample was reduced adding dithiothreitol to 50 mM, denatured in SDS at a final concentration of 6%, and then, the sample was split into 2 tubes, 15 µg protein in each. Next, 2,4-dinitrophenylhydrazine was added to 1 tube, to derivatize the carbonyl groups. Derivatization control solution was added to the other tube of each sample, as a negative control. Following derivatization and neutralization, the proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane (0.45 µm), blocked with 1% BSA in PBS-T for 1 hour, incubated with primary antibody rabbit anti-DNP at 1:150 dilution in blocking buffer for 1–2 hours at room temperature, washed in PBS-T, incubated with goat anti-rabbit IgG HRP secondary antibody at 1:300 in blocking buffer, and washed in PBS-T before ECL (Super Signal West Dura Extended Duration Substrate, Thermo Scientific) was added, and the Oxyblot was scanned in a LAS 3000 Image Analyzer to visualize the oxidized protein carbonyl groups of the samples. In the oxyblot anti-ActB, antibody-HRP (ab20272, Abcam) was used as a loading control. The oxyblot was washed in PBS-T and stripped (30 minutes in stripping solution, Thermo Scientific), blocked for 1 hour in 5% BSA in PBS-T, and incubated for 1.5 hours at room temperature with anti-ActB-HRP diluted 1:2000.
For the OxiSelect Protein Carbonyl Fluorometric Assay (Cell Biolabs Inc), 75 µg of the indicated protein lysates from wild-type, mutUNG1 and mutUNG1+KD mice were used and treated according to the product manual of the kit. Briefly, protein carbonyls in the protein samples were first derivatized with the Protein Carbonyl Fluorophore. Proteins were then TCA precipitated and free fluorophore was removed by washing the protein pellet with acetone. The protein pellet was dissolved in guanidine hydrochloride, the absorbance of protein-fluorophore product was measured fluoro-metrically with a 485/535-nm filter set in a Wallac plate reader, and the protein carbonyl was calculated.
2.3. Ultrastructure and postembedding immunogold electron microscopy
The procedure was adapted from Bergersen et al. (Bergersen et al., 2008) with tissues from 9 mice. Three mice from each group were used for the immunogold postembedding procedures. The mice were anesthetized by an intraperitoneal injection of pentobarbital (100 mg/mL) (0.2 mL/100 g of body weight) and perfused transcardially at 5 mL/min for 10 minutes with 4% paraformaldehyde and 0.1% glutaraldehyde in 0.1-M sodium phosphate buffer (PB; pH 7.4, 4 °C) for electron microscopy. The brains were removed and stored in fixative at 4 °C until further processing. Small rectangular sections (approximately 0.5 mm × 0.5 mm × 1 mm) of the CA1 region of the hippocampus were dissected out of the fixed brains and cryoprotected by incrementally permeating the tissue in 10% (30 minutes), 20% (30 minutes), and 30% (overnight) glycerol in 0.1-M PB. The tissue sections were then submerged in liquid propane cooled to −170 °C by liquid nitrogen in a Universal Cryofixation System KF80 (Reichert-Jung, Austria). The tissue sections were transferred into Reichert capsules within a flow through chamber using precooled forceps. The tissue was freeze-substituted by immersion in a solution of anhydrous methanol and 1.5% uranyl acetate for 30 hours at a temperature of −90 °C. After 30 hours, the temperature was increased from −90 °C to −45 °C in 4 °C increments per hour, and kept at −45 °C for the following steps. The tissue was then washed 3 times in anhydrous methanol before being immersed in a mixture of methanol and Lowicryl HM20 (Lowi, Germany) to allow resin infiltration. The mixture of methanol and Lowicryl was decreased stepwise from 2:1 (1 hour), 1:1 (1 hour), and 1:2 (1 hour) until the solution was pure Lowicryl (overnight). The tissue was then transferred into a precooled chamber filled with ethanol and polymerized under ultraviolet light (360-nm wavelength) for 24 hours at −45 °C. The polymerization was completed by increasing the temperature from −45 °C to 0 °C in 5 °C increments per hour followed by 35 hours at 0 °C. Ultrathin sections (90 nm) were cut using a Reichert-Jung ultramicrotome with a diamond knife and mounted onto nickel mesh grids (Electron Microscopy Sciences, USA).
The grids were processed in a moisture chamber at room temperature in solutions containing TBS and 0.1% Triton X-100 (TBST), with additions as stated. The grids were etched in TBS containing H2O2 (10 minutes), and rinsed in 0.1-M PB. The grids were then incubated in sodiumborohydride and glycine solution (10-mg sodiumborohydride and 37.5-mg glycine in 10 mLTBST; 10 minutes), TBST (10 minutes), blocking solution (2% human serum albumin in TBST; 10 minutes), and then overnight with primary antibodies against NR2AB (1:100), GluR1 (1:100), GluR2/3 (1:50), GABAAα1 (1:100), and GABAAγ2 (1:150) in blocking solution. The grids were rinsed in TBST followed by 10 minutes of incubation in TBST twice before being incubated in blocking solution (10 minutes). The grids were incubated (2 hours) with goat F(ab′) anti-rabbit immunoglobulin conjugated with 10-nm colloidal gold (1:20) in blocking solution to visualize the bound antibodies. The secondary antibody in blocking solution was spun at 1000 rpm (10 minutes) before use to sediment any aggregated gold particles. The grids were then rinsed in purified water and dried before being contrasted in 5% uranyl acetate and 30% lead citrate. For ultrastructural analysis, ultrathin sections were contrasted without immunogold processing.
The sections were studied using a Tecnai 12 electron microscope, and electron micrographs were recorded at magnifications of ×26,000 and ×43,000. Electron micrographs of clearly visible synapses were recorded randomly in the selected subregions of the hippocampal formation. Excitatory synapses were identified as axon terminals forming asymmetric synapses onto dendritic spines within the stratum radiatum in the CA1 region, whereas inhibitory synapses were identified as axon terminals forming symmetrical synapses onto the cellular soma of the CA1 pyramidal cells. Between 30 and 55 electron micrographs were recorded for the excitatory synapses, and between 15 and 25 micrographs were recorded for the inhibitory synapses, yielding 30–80 individual excitatory synapses and 15–30 individual inhibitory synapses.
The lengths (nm) of the excitatory postsynaptic membrane (identified as overlying the postsynaptic density) and the inhibitory postsynaptic membrane (identified as the part of the perikaryal membrane closely apposed to the inhibitory type nerve terminal membrane) were measured, and the densities of gold particles associated with the postsynaptic membranes were calculated (Bergersen et al., 2008). The densities of the gold particles were calculated by counting the number of gold particles located ±25 nm from the midline of the postsynaptic membranes, that is, within the lateral resolution of the immunogold method (about ±30 nm) (Bergersen et al., 2003, 2008) and approximating the area to that of a rectangle with the length of the postsynaptic membrane and 50-nm wide.
Antibodies used for immunogold labeling: The primary antibodies used were rabbit anti-NR2A/B (Chemicon, USA, AB1548), anti-GluR1 (Chemicon, USA, AB1504), anti-GluR2/3 (Millipore, USA, AB1506), anti-GABAA α1 (Alomone Labs, Israel, AGA-001), and anti-GABAAγ2 (Alpha Diagnostic International, USA, GAG21-A). The secondary antibody used was goat F(ab′) anti-rabbit immunoglobulin conjugated with 10-nm colloidal gold (British Biocell International, UK, EM.GFAR10).
For morphological analysis of ultrastructure, electron micrographs were taken randomly in the pyramidal cell layer and the stratum oriens of the CA1, about 20 micrographs per mouse. The area of mitochondria in the cytoplasm of pyramidal cells and in the cytoplasm of excitatory type nerve terminals (i.e., forming a synaptic bouton opposite a dendritic spines) in stratum radiatum was recorded along with the area of cytoplasm in which they were observed, and the percent of mitochondrial area was calculated (according to the Delesse principle, the percent of the area in a section is equal to the percent of the tissue volume). All identifiable pyramidal cells and excitatory type terminals in the electron micrographs were included in the analysis. The data were pooled for each animal and presented as mean ± standard deviation (SD), or mean ± standard error of the mean (SEM) when stated, of n = 3 mice per group.
2.4. Statistical analysis
Quantitative data are presented as mean ± SD (or ±SEM when stated), and statistical significance was determined using unpaired 2-tailed Student’s t-tests unless otherwise stated. Electron microscopic observations on individual synapses varied more within than between experimental animals; therefore, when stated, the total number of synapses observed was alternatively taken as n and statistical significance analyzed by the Mann–Whitney U test (equivalent to the Wilcoxon rank sum test). The null hypothesis was rejected at the 0.05 level. In the figures, levels of significance are indicated by stars (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001).
3. Results
3.1. A ketogenic diet increases the mitochondrial mass in the hippocampus of mice
A high fat and low carbohydrate ketogenic diet previously was shown to increase the mitochondrial mass in neuronal tissue (Bough et al., 2006; Hughes et al., 2014; Nylen et al., 2009; Rho and Rogawski, 2007), which may account for the beneficial effects of the diet in certain neurological pathologies. To validate our experimental setup, we first investigated if a ketogenic diet increased mitochondrial mass in the mouse strain (C57BL/6) used to generate the transgene mutUNG1 mice. Wild-type controls were fed a ketogenic diet for 2 months and compared with wild-type littermates fed a standard diet. Immunohistochemical studies indicated that there was an increased amount of mitochondria in mice fed a ketogenic diet, as seen in sections of hippocampus immunostained for the mitochondrial markers mitochondrial complex II and VDAC1 (immunofluorescence intensity in wild-type mice on ketogenic diet increased to 133 ± 25% and 152 ± 19%, respectively, of the intensity in mice on standard diet, mean ± SD, n = 3 mice, p < 0.05 and p = 0.01), whereas other cell proteins (SOD2, MAP2, GFAP, and GAD67) did not change (Supplementary Figs. S2 and S3). We therefore tested the effect of this diet in mice with mtDNA damage induced by mutUNG1.
3.2. A ketogenic diet accelerates atrophy, neurodegeneration, and reactive astrogliosis in the hippocampus in mutUNG1-expressing mice
HE-stained coronal brain sections revealed atrophy of the hippocampus of mutUNG1-expressing mice after just 2 months of induction with the mtDNA-damaging enzyme (Fig. 2A). Atrophy of the hippocampus is a characteristic of neurodegenerative disorders such as Alzheimer’s disease and aging (Dhikav and Anand, 2007). The thickness of hippocampal cortex neuropil, quantified as the distance from the surface of the dentate gyrus in the hippocampal fissure to the CA1 pyramidal cell-soma layer (i.e., the combined thickness of the stratum radiatum and stratum lacunosummoleculare, Fig. 2A), showed a significant shrinkage in mutUNG1-expressing mice fed a ketogenic diet compared with wild-type and mutUNG1-expressing mice fed a standard diet (Fig. 2B). The hilus, a region particularly vulnerable to neuropathologic damage (Hsieh, 1999), was profoundly shrunken in mutUNG1-expressing mice on the ketogenic diet (Fig. 2A, bottom frame). In addition, we observed a reduced number of neurons in mutUNG1 mice fed a ketogenic diet by counting the number of granule cells in 100 µm stretches of the lower limb of the granular layer (Fig. 2A, right part). The reduced tissue volume therefore likely represents a combined reduction of the number of neurons and in the volume of each neuron.
We next immunolabeled coronal sections of the hippocampus with an antibody against the neuronal marker MAP2 in mice with mutUNG1 expression induced for 4 months. This revealed a decrease in MAP2-staining in mutUNG1-expressing mice, which was further aggravated by a ketogenic diet (Fig. 2C). Quantification showed a significant reduction in MAP2-staining in mutUNG1-expressing mice fed a standard diet compared with wild-type controls. A ketogenic diet further reduced MAP2-staining in mutUNG1-expressing mice (Fig. 2D). These data imply severe neurodegeneration in the hippocampus of mutUNG1-expressing mice, intensified in mice fed a ketogenic diet. Astrocytes play an important part in various neurodegenerative disorders including Alzheimer’s disease, amyotrophic lateral sclerosis, Parkinson’s disease, and various forms of dementia. Through reactive astrogliosis, astrocytes contribute to the neuroinflammatory component of neurodegeneration (Verkhratsky et al., 2010). We therefore investigated the state of the astrocytes in the different groups of mice using immunolabeling with the astrocyte marker GFAP, which is known to be upregulated in reactive astrocytes (Pekny et al., 2014). Astrocyte-staining increased greatly in mutUNG1-expressing mice on a ketogenic diet for 2 months compared with mice on a standard diet (Fig. 2E). Quantification of the GFAP-immunostaining showed a significant increase in mutUNG1-expressing mice on a ketogenic diet compared with mice fed a standard diet (Fig. 2F). This demonstrates reactive astrogliosis in mutUNG1-expressing mice fed a ketogenic diet.
Despite these strong signs of changes in the brain, there was no loss of weight in the mutUNG1 mice during 17 weeks: the weight was stable on ketogenic diet and increased moderately on standard diet (Supplementary Fig. S1). This indicates that there was no gross deterioration of the condition of the mutUNG1-expressing mice, but also that the energy they were able to extract from the ketogenic diet was insufficient to sustain a normal gain of weight. In contrast to the weight developments (from age 8 weeks) shown here for mutUNG1 expressing C57BL/6 mice (Supplementary Fig. S1), normal C57BL/6J mice gain weight (from age 17 weeks) on a ketogenic diet but not on a standard diet (Meidenbauer et al., 2014). These observations are consistent with the assumption that mutUNG1-generated mitochondrial damage hampers lipid metabolism more than it hampers carbohydrate metabolism. It also calls for examination of whether the high lipid load of ketogenic diet could contribute to mitochondrial deterioration in the mutUNG1-expressing mice.
3.3. mutUNG1-expressing mice fed a ketogenic diet have upregulated mitochondrial antioxidant defenses and show impaired mitochondrial dynamics and function
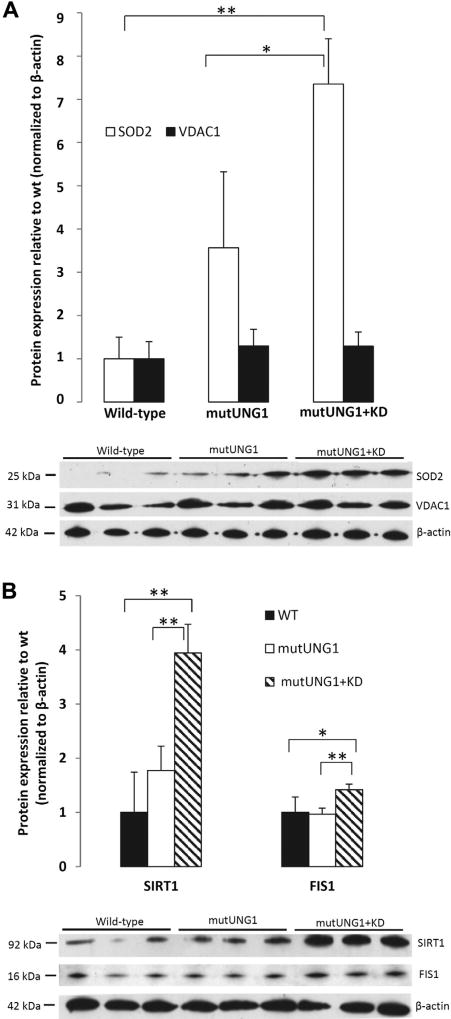
Superoxide anion is the main form of reactive oxidative species generated by the electron transport chain in the mitochondria, and SOD2 (also known as Mn-superoxide dismutase) is important for detoxifying this form of ROS in the mitochondria (Jung et al., 2009). SOD2 may be considered the key member of the defenses against potentially damaging radicals from the electron transport chain (Flynn and Melov, 2013). By immunostaining, we investigated the presence of SOD2 in coronal sections of hippocampus and detected an upregulation in mutUNG1-expressing mice compared with wild-type controls, with the strongest upregulation in mutUNG1-expressing mice fed a ketogenic diet (Fig. 3A and B). Double immunolabeling showed that the SOD2 was present in mitochondria as it colocalized with mitochondrial complex II in neuronal cell bodies as well as in the neuropil (Fig. 3C). In mutUNG1 mice, particularly when on ketogenic diet, but not in wild-type mice, mitochondria-like structures were seen to accumulate in principal neuron perikarya (Fig. 3C). Upregulation of SOD2 was also demonstrated by quantitative Western blotting (Fig. 4A). This upregulation of the mitochondrial antioxidant defenses is consistent with increased production of ROS in the mitochondria of the hippocampus of mutUNG1-expressing mice fed a ketogenic diet. Interestingly, the mitochondrial fission protein, FIS1, was also upregulated (Fig. 4B). This protein separates healthy from defective parts of mitochondria and couples stressinduced mitochondrial fission with reparative mitochondrial degradation processes (Shen et al., 2014); it is increased in mild cognitive impairment and Alzheimer’s disease (Wang et al., 2012).
Fig. 4.
A ketogenic diet upregulates antioxidative defense (SOD2), mitochondrial fission activity (FIS1), and a longevity-associated deacetylase (SIRT1) in mutUNG1-expressing mice. (A) SOD2 and the mitochondrial marker VDAC1 were quantified in Western blots, normalized to ActB as loading control. In mutUNG1 mice, SOD2 increases several fold compared with wild-type (p < 0.001) and in mutUNG1 mice it is doubled by a ketogenic diet (p = 0.03). VDAC1 shows only minor changes in the same conditions. (B) Similarly, FIS1 and SIRT1 are strongly upregulated by ketogenic diet in mutUNG1 mice versus mutUNG1 mice on standard diet (SIRT1 p = 0.006, FIS1 p = 0.007), as well as versus wild-type mice (SIRT1 p = 0.005, FIS1 p = 0.03); *p≤ 0.05; **p≤ 0.01. Abbreviations: SOD2, superoxide dismutase 2; VDAC1, voltage-dependent anion channel 1.
In an effort to evaluate the degree of ROS influence in the tissue, protein carbonylation was recorded by a qualitative protein oxidation detection kit (Oxyblot, Millipore) (Supplementary Fig. S4) as well as a quantitative (OxiSelect, Cell Biolabs) protein oxidation detection kit. The latter gave the following carbonylation values (% of wild-type, mean ± SD, n = 3 mice): wild-type 100 ± 68, mutUNG1 32 ± 8, mutUNG1 on ketogenic diet 40 ± 35. Thus the degree of protein carbonylation varied considerably among the samples (although duplicates of each sample agreed well), and there was no clear effect of the ketogenic diet. Therefore, no firm conclusion can be drawn regarding ROS action. The apparent lack of a consistently increased protein carbonylation in mutUNG1-expressing mice, whether on a ketogenic or a normal diet, could perhaps mean that antioxidant defense mechanisms compensate for the presumably increased generation of ROS caused by mutUNG1.
Mitochondria were then examined more closely in electron micrographs, quantifying the mitochondrial area (corresponding to the mitochondrial volume and thus mitochondrial mass in the tissue) in different compartments of neuronal cells in the hippocampus. The mitochondrial area was significantly larger in the soma of mutUNG1-expressing mice fed a ketogenic diet compared with mice (mutUNG1 or wild-type or both) fed a standard diet (Fig. 5A). However, in the presynaptic terminals, the mitochondrial area was significantly smaller in mutUNG1-expressing mice fed a ketogenic diet than in mice fed a standard diet (Fig. 5B). These data demonstrate a perturbed distribution of mitochondria in the affected neurons of these mice, and indicate disturbed intracellular transport of mitochondria. Because neuronal cells have high local energy demands (especially in synaptic areas), deficient transport and distribution of mitochondria can disturb neuronal function and lead to neurodegeneration due to lack of sufficient energy. Also, mitochondria accumulate in the nerve cell soma in Alzheimer’s disease (Lu, 2009; Wang et al., 2009).
Fig. 5.
mutUNG1-expressing mice fed a ketogenic diet. show a deranged distribution of mitochondria. (A) Quantification of the mitochondrial area in CA1 pyramidal cell somas from electron micrographs shows a significant increase in mitochondrial mass in mutUNG1-expressing mice fed a ketogenic diet (p = 0.02) and in mutUNG1-expressing mice fed a standard diet (p = 0.01), compared with wild-type mice fed a standard diet. (B) Quantification of the mitochondrial area in presynaptic terminals in the CA1 stratum radiatum from electron micrographs shows a significant decrease in mitochondrial mass in mutUNG1-expressing mice fed a ketogenic diet (p = 0.05), and in mutUNG1-expressing mice fed a standard diet (p = 0.03), compared with wild-type mice fed a standard diet. The data are presented as mean ± standard deviation; *p≤ 0.05; **p≤ 0.01. Abbreviation: KD, ketogenic diet.
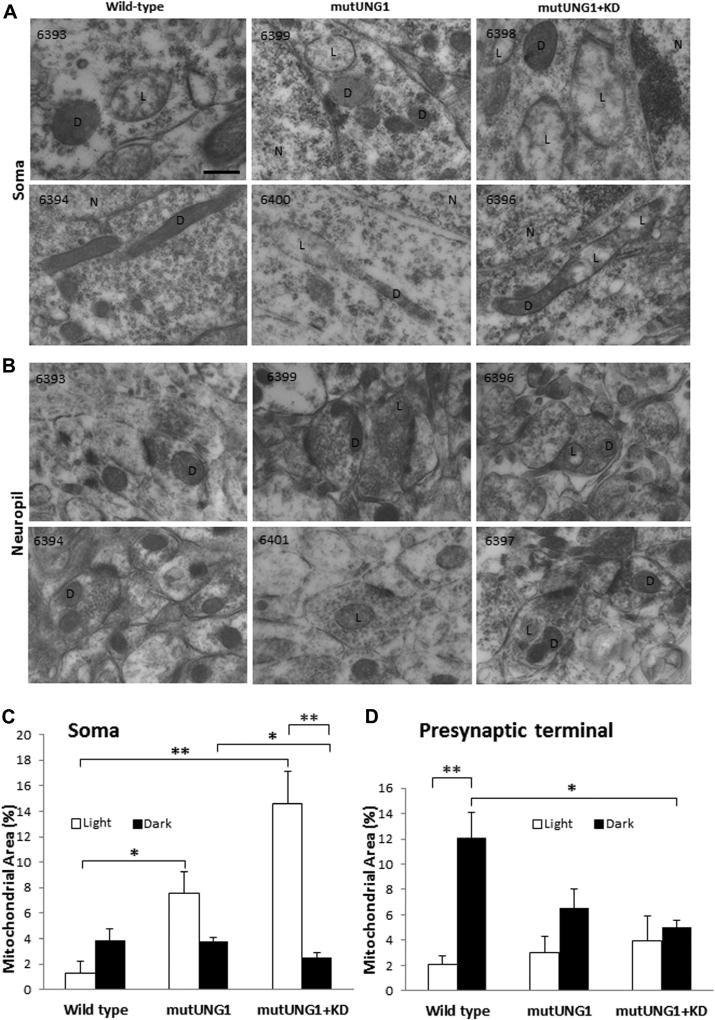
A proportion of the mitochondria appeared swollen and less electron dense than normal. This proportion was small in wild-type but increased on mutUNG1 expression, and further rose nearly 3-fold with a ketogenic diet (Fig. 6A and B). The appearance of the light mitochondria is similar to that described for dysfunctional mitochondria induced by a diabetogenic diet (wild-type mice fed 20% sucrose-sweetened water for 7 months) and in triple transgenic Alzheimer’s disease mice (Carvalho et al., 2012), asserting that the light appearance indicates deteriorated function. When mitochondria were categorized as “light” or “dark”, it turned out that the increased mitochondrial mass in pyramidal cell somas induced by mutUNG1 expression and ketogenic diet was due to an increase in light mitochondria (Fig. 6C). In contrast, the reduced mitochondrial mass in excitatory nerve terminals was due to the loss of dark mitochondria (Fig. 6D). These observations indicate lack of supply of normal mitochondria from the cell body to the excitatory nerve terminals, and an accumulation of abnormal, swollen mitochondria in the soma. The swollen mitochondria could be very large in the perikarya (Fig. 6A, 6398), and at places, light and dark regions occurred along the same mitochondrion (Fig. 6A, 6396 and 6400), possibly a sign of fission-fusion dynamics. In the nerve terminals, the mitochondria differed less in appearance (Fig. 6B), consistent with retrograde transport of dysfunctional mitochondria to the cell body.
Fig. 6.
A ketogenic diet aggravates perikaryal accumulation and swelling of mitochondria in mutUNG1-expressing mice. (A and B) Representative electron micrographs from pyramidal cell somas (A) and stratum radiatum neuropil (B) in hippocampus CA1 showing abnormal mitochondrial morphology in mutUNG1-expressing mice, on control and ketogenic diet (KD) indicating mitochondrial malfunction and breakdown of mitochondrial dynamics. Two electron micrographs were selected among the 3 mice analyzed for each condition (identification numbers in upper left corner) to illustrate typical morphology. Mitochondria were classified as showing the normal dark (D) appearance, and a swollen, light (L) appearance, respectively. (N = nucleus.) Scale bar = 200 nm. (C and D) Quantification of the percent mitochondrial mass (determined as the percent area of mitochondrial profiles in the tissue sections) in the cytoplasm of CA1 pyramidal cell somas (C) and of nerve terminals in excitatory synapses (D). The mitochondrial and cytoplasm areas, respectively, were pooled for each type of structure in each animal (n = 3 mice per group), and the percentage calculated and presented as mean ± standard error of the mean (SEM) for each condition. In somas of mutUNG1 mice, a ketogenic diet (KD) significantly increased the number of light mitochondria compared with dark mitochondria (p = 0.01). Wild-type mice had significantly less light mitochondria than mutUNG1-expressing mice fed a standard diet (p = 0.03), and than mutUNG1-expressing mice fed a ketogenic diet (p = 0.009). In presynaptic excitatory terminals, wild-type mice had significantly more dark than light mitochondria (p = 0.01), and more dark mitochondria compared with mutUNG1-expressing mice fed a ketogenic diet (p = 0.03). Note that a ketogenic diet significantly increased the accumulation of light mitochondria in the soma: the proportion of light to dark mitochondria was 2.1 ± 0.6 in mutUNG1 mice on a normal diet, and 5.7 ± 0.4 (mean ± SEM) in mutUNG1 mice fed a ketogenic diet (p = 0.008); *p≤ 0.05; **p ≤ 0.01.
The oppositely directed changes in the mass of presumably functional and dysfunctional mitochondria revealed by electron microscopy (Fig. 6C and D) are consistent with the minor changes in the mitochondrial marker VDAC1 (Fig. 4A), indicating small net changes in the amount of functional mitochondria.
Mechanistically, the ability of a ketogenic diet to rescue mitochondrial deficits in a model of early aging (Cockayne Syndrome mice) may depend on the longevity-associated histone deacetylase, SIRT1, a central transcription factor that enhances mitochondrial biogenesis (Scheibye-Knudsen et al., 2014). We therefore examined the expression of SIRT1 protein and found it to be strongly upregulated by ketogenic diet in the mutUNG1-expressing mice (Fig. 4B).
3.4. A ketogenic diet accelerates mutUNG1-induced changes in the glutamate receptor distribution at excitatory synapses
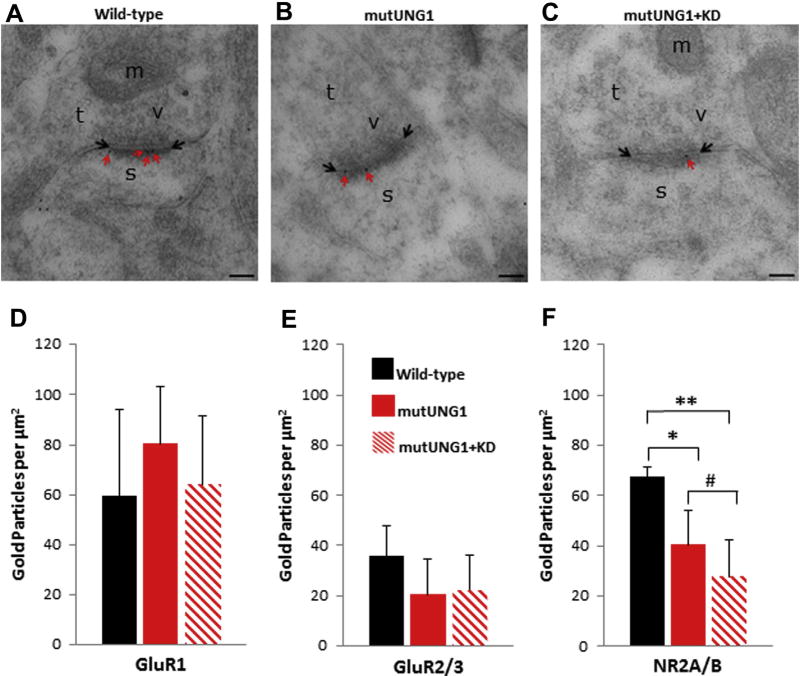
Since previous studies have shown changes in excitatory α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and NMDA receptor densities in the hippocampus of mutUNG1-expressing mice (Lauritzen et al., 2011a), we performed immunogold postembedding labeling of ultrathin sections from the stratum radiatum of the CA1 region of the hippocampus with antibodies against the AMPA receptor subunits GluR1 and GluR2/3 and against the NMDA receptor subunit NR2AB. The density of gold particles within 25 nm of either side of the postsynaptic membrane overlying the postsynaptic density of asymmetric, that is, excitatory type, synapses was quantified. Representative electron micrographs showing NR2A/B labeling in excitatory synapses of the stratum radiatum are shown in Fig. 7A – C. The AMPA and NMDA receptor densities were quantified (Fig. 7D – F). The GluR1 labeling in the stratum radiatum appears to showan increase from wild type to mutUNG1 and then a decrease in labeling density in mutUNG1 on a ketogenic diet (Fig. 7D), whereas the GluR2/3 labeling showed a decrease in labeling densities from the wild-type to the mutUNG1-expressing mice fed a standard diet that was not altered or further aggravated in response to the ketogenic diet (Fig. 7E), but none of these changes were statistically significant. However, the NR2A/B labeling showed a clear decrease in the labeling densities among the 3 groups, with the decreases between the wild-type and mutUNG1-expressing mice fed a ketogenic diet (p = 0.01), and between the wild-type and mutUNG1-expressing mice fed a standard diet (p = 0.03) being significantly different (Fig. 7F). As the results varied more between synapse than between animals, statistical significance was also evaluated by means of the Mann–Whitney U Test using the total number of synapses as n (n = 140 and n = 152 for mutUNG1 with and without ketogenic diet, respectively). This revealed that a ketogenic diet significantly reduced NR2A/B labeling in mutUNG1-expressing mice (#, p = 0.04). These data indicate that treatment of the mutUNG1-expressing mice with a ketogenic diet accelerates the loss of NMDA receptors in the stratum radiatum of the CA1 region of the hippocampus of mutUNG1-expressing mice.
Fig. 7.
A ketogenic diet accelerates changes in the glutamate receptor distribution at excitatory synapses. (A–C) Micrographs illustrating representative excitatory synapses in the stratum radiatum of the CA1 region are shown for (A) wild-type (B) mutUNG1-expressing mice fed a standard diet, and (C) mutUNG1-expressing mice fed a ketogenic diet. Red arrows indicate gold particles (black dots) representing labeled NR2A/B receptor subunits within the postsynaptic membranes (limited by black arrows). Scale bar = 100 nm. (D–F) Quantification of excitatory AMPA receptor subunits GluR1 (D), GluR2/3 (E), and NMDA receptor subunits NR2A/B (F) in the stratum radiatum of the hippocampus CA1 region. Gold particles within 25 nm of either side of the postsynaptic membranes were counted. The GluR1 and GluR2/3 labeling showed no statistically significant differences between the 3 groups of mice. The NR2A/B labeling showed a significant decrease between the wild-type and mutUNG1-expressing mice fed a standard diet (p = 0.03) and between the wild-type and mutUNG1-expressing mice fed a ketogenic diet (p = 0.01). The Mann–Whitney U test (#) revealed a significant reduction (p = 0.04) of NR2A/B labeling of synapses in mutUNG1-expressing mice on a ketogenic diet (n = 140 synapses) compared with a standard diet (n = 152 synapses). Data are shown as mean subunit-representing particle number per µm2± standard deviation, n = 3 mice per group; *p≤ 0.05; **p ≤ 0.01. Abbreviations: KD, ketogenic diet; m, mitochondria; s, postsynaptic spine; t, presynaptic terminal; v, synaptic vesicles. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. A ketogenic diet results in alterations of the GABAergic receptor distribution in the CA1 region of the hippocampus
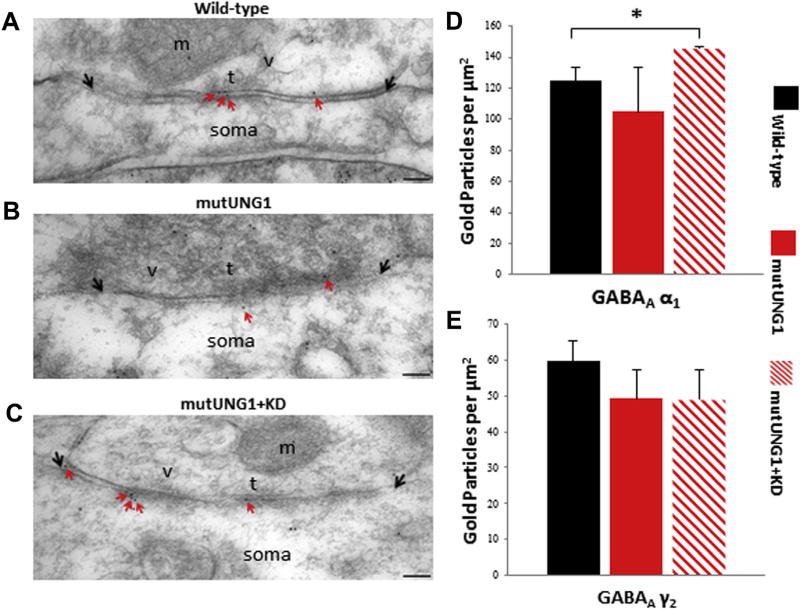
In addition to examining the asymmetric excitatory synapses, we also quantified the labeling densities of the GABAA α1 and GABAA γ2 receptor subunits at symmetric, that is, inhibitory type, synapses on the CA1 pyramidal cell somas in the hippocampus. Representative electron micrographs showing GABAA γ2 labeling in inhibitory synapses on the CA1 pyramidal cell somas in the hippocampus are shown in Fig. 8A – C. The GABAA receptor–labeling densities were quantified as previously described for the AMPA and NMDA receptor labeling densities. The GABAA α1 labeling showed a significant increase between the wild-type and mutUNG1-expressing mice fed a ketogenic diet (p = 0.01, Fig. 8D). The GABAA γ2 labeling displayed a decrease between the wild-type and mutUNG1-expressing groups, but these alterations were not statistically significant (Fig. 8E). No changes were detected in the GABA-synthesizing enzyme GAD67 (Supplementary Figs. S2 and S3). These data indicate that modest alterations of inhibitory synapses occur in mutUNG1-expressing mice fed a ketogenic diet.
Fig. 8.
GABAA receptor distribution in the CA1 region of the hippocampus. (A–C) Micrographs illustrating representative inhibitory synapses on the soma of CA1 pyramidal cells are shown for (A) wild-type, (B) mutUNG1-expressing mice fed a standard diet, and (C) mutUNG1-expressing mice fed a ketogenic diet Red arrows indicate gold particles (black dots) representing labeled GABAAγ2 receptor subunits at the synaptic membranes (limited by black arrows). Scale bar = 100 nm (D and E) Quantification of inhibitory GABAAα1 (D) and GABAAγ2(E) receptor subunits. Gold particles within 25nmof the postsynaptic membranes were counted. The GABAAα1 labeling showed a significant increase in mutUNG1-expressing mice fed a ketogenic diet (p = 0.02) compared with wild-type, whereas there were no significant differences in GABAAγ2 labeling between the 3 groups of mice. Data are shown as mean subunit-representing particle number per µm2± standard deviation, n = 3 mice per group; *p ≤ 0.05. Abbreviations: KD, ketogenic diet; m, mitochondria; soma, cell soma; t, presynaptic terminal; v, synaptic vesicles. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
We previously showed that high levels of mtDNA damage in the form of AP-sites in forebrain neurons in mice cause mitochondrial dysfunction leading to neurodegeneration, abnormal synaptic structures, and ultimately impaired behavior (Lauritzen et al., 2010, 2011a). Because a ketogenic diet is well known for its therapeutic effect in drug-resistant epilepsy, a disorder where impaired mitochondrial function and lower energy metabolism is believed to play an important role (Folbergrova and Kunz, 2012; Yuen and Sander, 2011), we investigated if a ketogenic diet had a positive effect on the phenotype of the mutUNG1-expressing mice. Interestingly, a ketogenic diet at moderate calorie restriction (90% of recommended dietary allowance) caused mitochondrial proliferation, upregulation of genes involved in oxidative metabolism, and increased energy reserves alternative to glucose and glycogen, in parallel with enhanced seizure resistance and robustness of synaptic activity in hypoglycemia (Bough et al., 2006). This is consistent with our observations that the ketogenic diet induced increase of mitochondrial mass, as well as of SOD2, mitochondrial complex II, FIS1, and SIRT1. In a recent study, we reported that a high-fat ketogenic diet is able to rescue premature aging through activation of SIRT1, a central transcription factor that promotes mitochondrial biogenesis (Scheibye-Knudsen et al., 2014), and in this current work, we show that SIRT1 is increased by ketogenic diet in the mutUNG1-expressing mice. However, in contrast to the beneficial effects reported in studies on other disease conditions, and although we do observe the same increase in SIRT1 proteins levels as previously reported, we found that the ketogenic diet aggravated neurodegeneration in mutUNG1-expressing mice.
We ascribe this observation to a “catch-22” situation: in neurons with normal mitochondria, a ketogenic diet is beneficial as it causes increased mitochondrial biogenesis affording an energetic advantage in dealing with cellular challenges (Fig. 1, top). In a situation where all the mitochondria are dysfunctional, there is an energy deficit (in addition to increased harmful ROS production). Through increased mitochondrial biogenesis, this energy deficit increases further since the production of more mitochondria is energetically costly in itself, and the original, as well as the newly generated mitochondria, will be dysfunctional due to high levels of mtDNA damage. Instead of increasing the energy levels in the neurons, the increased mitochondrial biogenesis therefore drains the energy reserves further, accelerating the neurodegeneration (Fig. 1, bottom). A ketogenic diet has been suggested to result in limited glucose availability for the brain (Cunnane and Likhodii, 2004), and this would aggravate such energy shortage. However, enhanced (Meidenbauer et al., 2014; Seyfried et al., 2003) or normal (Nandivada et al., 2016) blood glucose levels have been reported in mice on a ketogenic diet administered ad libitum. These results suggest that hypoglycemia may not be a contributing factor in our study, although this cannot be completely excluded. It should be noted that our diet contained no carbohydrate (calculated 83.9%, 16.1%, and 0.0% of energy from fat, protein, and carbohydrate, respectively), whereas the diet used by the Seyfried group (Meidenbauer et al., 2014) contained some carbohydrate, more fat, and less protein (calculated 91.8%, 7.6%, and 0.6% of energy from fat, protein, and carbohydrate). Adaptive changes in gene expression normally induced by ketogenic diet and leading to enhanced energy reserves (Bough et al., 2006) would presumably not work because they are dependent on biogenesis of healthy mitochondria rather than of mitochondria carrying mutUNG1. For the same reason, selection of healthy versus unhealthy mitochondria, which occurs as part of adaptive changes in response to stressors (such as exercise and hypoxic or toxic preconditioning) (Rodell et al., 2013), would not work. In addition, increased amounts of mitochondria that are all dysfunctional, presumably with increased ROS production, will further harm the neuronal population. The mutUNG1 is targeted to all mitochondria in the affected tissues, causing mtDNA damage. Quality control through mitochondrial dynamics, whereby fusion and fission events dilute damaged components and mitophagy removes nonrepairable organelles (Berman and Hollenbeck, 2013; Youle and van der Bliek, 2012), is unable to correct this situation of pervasive mitochondrial damage.
A recent article (Selfridge et al., 2015) reports that a ketogenic diet, and to a lesser extent a nonketogenic high-fat diet, reduces brain levels of mtDNA, but interestingly, the transcription efficiency appears to be concomitantly enhanced to the extent that mtDNA encoded mRNA and protein synthesis are kept normal. The mechanism may be that heteroplasmic cells with mutated mtDNA undergo a “heteroplasmic shift” when exposed to ketone bodies, whereby the cells select normal mtDNA versus abnormal mtDNA, resulting in a reduced proportion of damaged mtDNA, normalized mitochondrial protein synthesis, and recovered respiratory competence (Santra et al., 2004). Rather than increasing mtDNA damage, the ketogenic diet therefore helps rid the cells of damaged mtDNA to bring the proportion of normal wild-type mtDNA above a functional threshold. However, this mechanism presumes heteroplasmic expression of mutated mtDNA; therefore it may not work in mutUNG1 mice, in which cells presumably are “homoplasmic” in the sense that mtDNA damage is caused continuously in all mitochondria. In agreement, we found that a ketogenic diet provides no protection against the mutUNG1 phenotype.
We observed a strongly increased reactive astrocytosis in mutUNG1 mice fed a ketogenic diet. Reactive astrogliosis is indicative of a neuroinflammatory response and involves both protective and potentially harmful components (Pekny et al., 2014). A recent article shows that neuropathological signs of inflammation correlate with mtDNA damage and neuronal loss in mesial temporal lobe epilepsy (Volmering et al., 2016). Another recent article (Singh et al., 2016) describes a regulatory network controlling lipid metabolism in response to high-fat intake in which anti-inflammatory arachidonic acid derivatives cause enhanced mitochondrial proliferation and function through the activation of peroxisome proliferator-activated receptor gamma coactivator 1-alpha and also involves activation of SOD2 and SIRT1. These results support our findings of mitochondriogenesis and enhanced SOD2 and SIRT1 in mutUNG1-expressing mice fed a ketogenic diet.
The results presented here suggest that in some neurological disorders with mitochondrial dysfunction, patients should avoid a ketogenic diet because it may accelerate rather than reverse the neurodegenerative process. Future studies on pathologies with a mitochondrial basis should therefore focus on correcting the malfunctioning mechanism, before mitochondrial biogenesis is stimulated. Conversely, certain brain tumors such as glioblastoma have extensive tumor-specific mutations in mtDNA with respiratory chain malfunction (Lloyd et al., 2015), pointing to the possibility that the known therapeutic effect of a ketogenic diet in such tumors (Meidenbauer et al., 2015) could work in part through a mechanism similar to that proposed here. However, mutUNG1-expressing mice, as used by us, represent an extreme situation, with mitochondrial damage that is propagated to the new mitochondria formed in mitochondrial biogenesis. A modified murine model producing moderate mtDNA damage might better represent neurological disease conditions. In such a model, a ketogenic diet might have beneficial effects, including the stimulation of mitochondrial biogenesis. One example of such a situation is the mouse model carrying a homozygous mutation in the Cockayne Syndrome B locus with neurodegeneration and premature aging, where a beneficial effect of a ketogenic diet was reported (Scheibye-Knudsen et al., 2014). Stressors can promote natural selection of healthy mitochondria, a selection that may partly underlie the beneficial effects of preconditioning and exercise, as recently pointed out (Rodell et al., 2013). We propose that a ketogenic diet may represent a stressor with similar potentially beneficial effects on mitochondria but that this mechanism is obstructed by the effect of mutUNG1 expression being too pervasive to leave healthy traits that can be selected.
It is important to test this hypothesis in a modified mutUNG1-expressing mouse in which the induction step is titrated with respect to timing and doxycycline dosage. Although the ketogenic diet did not ameliorate the mutUNG1 phenotype in the present study, the findings do point to effects that partly may underlie its recognized beneficial action in epilepsy and may help counteract cytotoxicity and neurodegeneration: a reduced expression of NMDA receptor subunits at excitatory synapses and a slight increase of GABA receptor subunits at inhibitory synapses were indicated in hippocampus CA1. Interestingly, these changes may be synergistic with the energetically based enhancement of GABAergic inhibition and reduced excitability induced by a ketogenic diet (Bough et al., 2006).
Finally, this study highlights the important role of mitochondria in sustaining healthy neurons and the critical need to keep the mitochondria functional with intact mtDNA to avoid neurodegenerative disease, especially in conditions when mitochondrial biogenesis is required to meet increasing neuronal energy needs during cellular stress.
Supplementary Material
Acknowledgments
This study was supported by research grants from The Research Council of Norway, the Norwegian Health Association, The Lundbeck Foundation (Denmark), and The South-Eastern Norway Regional Health Authority. Images for cell counting were acquired at the Norbrain Slidescanning Facility at the Institute of Basic Medical Sciences, University of Oslo, a resource funded by the Research Council of Norway. The authors gratefully acknowledge this help as well as help with protein carbonyl assay from the Regional Core Facility for Mitochondria and Metabolism at the Oslo University Hospital (http://ous-research.no/mito/).
Footnotes
Disclosure statement
The authors have no actual or potential conflicts of interest. All aspects of the research described here were carried out in accordance with the laws of Norway concerning animal research.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.neurobiolaging.2016.08.005.
References
- Adam-Vizi V. Production of reactive oxygen species in brain mitochondria: contribution by electron transport chain and non-electron transport chain sources. Antioxid. Redox Signal. 2005;7:1140–1149. doi: 10.1089/ars.2005.7.1140. [DOI] [PubMed] [Google Scholar]
- Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J. Cereb. Blood Flow Metab. 2001;21:1133–1145. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergersen L, Ruiz A, Bjaalie JG, Kullmann DM, Gundersen V. GABA and GABAA receptors at hippocampal mossy fibre synapses. Eur. J. Neurosci. 2003;18:931–941. doi: 10.1046/j.1460-9568.2003.02828.x. [DOI] [PubMed] [Google Scholar]
- Bergersen LH, Storm-Mathisen J, Gundersen V. Immunogold quantification of amino acids and proteins in complex subcellular compartments. Nat. Protoc. 2008;3:144–152. doi: 10.1038/nprot.2007.525. [DOI] [PubMed] [Google Scholar]
- Berman SB, Hollenbeck PJ. Exploring the life cycle of mitochondria in neuropsychiatric diseases: mitochondrial dynamics and quality control. Neurobiol. Dis. 2013;51:1–2. doi: 10.1016/j.nbd.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Bough KJ, Wetherington J, Hassel B, Pare JF, Gawryluk JW, Greene JG, Shaw R, Smith Y, Geiger JD, Dingledine RJ. Mitochondrial biogenesis in the anticonvulsant mechanism of the ketogenic diet. Ann. Neurol. 2006;60:223–235. doi: 10.1002/ana.20899. [DOI] [PubMed] [Google Scholar]
- Carvalho C, Cardoso S, Correia SC, Santos RX, Santos MS, Baldeiras I, Oliveira CR, Moreira PI. Metabolic alterations induced by sucrose intake and Alzheimer’s disease promote similar brain mitochondrial abnormalities. Diabetes. 2012;61:1234–1242. doi: 10.2337/db11-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Critical dependence of neurons on mitochondrial dynamics. Curr. Opin. Cell Biol. 2006;18:453–459. doi: 10.1016/j.ceb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Cunnane SC, Likhodii SS. Claims to identify detrimental effects of the ketogenic diet (KD) on cognitive function in rats. Pediatr. Res. 2004;56:663–664. doi: 10.1203/01.PDR.0000142215.95720.72. author reply 664. [DOI] [PubMed] [Google Scholar]
- Dhikav V, Anand KS. Is hippocampal atrophy a future drug target? Med. Hypotheses. 2007;68:1300–1306. doi: 10.1016/j.mehy.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Melov S. SOD2 in mitochondrial dysfunction and neurodegeneration. Free Radic. Biol. Med. 2013;62:4–12. doi: 10.1016/j.freeradbiomed.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folbergrova J, Kunz WS. Mitochondrial dysfunction in epilepsy. Mitochondrion. 2012;12:35–40. doi: 10.1016/j.mito.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Freeman JM, Kossoff EH, Hartman AL. The ketogenic diet: one decade later. Pediatrics. 2007;119:535–543. doi: 10.1542/peds.2006-2447. [DOI] [PubMed] [Google Scholar]
- Gjedde A, Crone C. Induction processes in blood-brain transfer of ketone bodies during starvation. Am. J. Physiol. 1975;229:1165–1169. doi: 10.1152/ajplegacy.1975.229.5.1165. [DOI] [PubMed] [Google Scholar]
- Haun F, Nakamura T, Lipton SA. Dysfunctional mitochondrial dynamics in the pathophysiology of neurodegenerative diseases. J. Cell Death. 2013;6:27–35. doi: 10.4137/JCD.S10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh PF. Neuropathology of limbic status epilepticus induced by electrical stimulation of naive rats. Neurol. Res. 1999;21:399–403. doi: 10.1080/01616412.1999.11740950. [DOI] [PubMed] [Google Scholar]
- Hughes SD, Kanabus M, Anderson G, Hargreaves IP, Rutherford T, O’Donnell M, Cross JH, Rahman S, Eaton S, Heales SJ. The ketogenic diet component decanoic acid increases mitochondrial citrate synthase and complex I activity in neuronal cells. J. Neurochem. 2014;129:426–433. doi: 10.1111/jnc.12646. [DOI] [PubMed] [Google Scholar]
- Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JE, Kim GS, Narasimhan P, Song YS, Chan PH. Regulation of Mnsuperoxide dismutase activity and neuroprotection by STAT3 in mice after cerebral ischemia. J. Neurosci. 2009;29:7003–7014. doi: 10.1523/JNEUROSCI.1110-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavli B, Slupphaug G, Mol CD, Arvai AS, Peterson SB, Tainer JA, Krokan HE. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- Lauritzen F, Eid T, Bergersen LH. Monocarboxylate transporters in temporal lobe epilepsy: roles of lactate and ketogenic diet. Brain Struct. Funct. 2015;220:1–12. doi: 10.1007/s00429-013-0672-x. [DOI] [PubMed] [Google Scholar]
- Lauritzen KH, Cheng C, Wiksen H, Bergersen LH, Klungland A. Mitochondrial DNA toxicity compromises mitochondrial dynamics and induces hippocampal antioxidant defenses. DNA Repair (Amst) 2011a;10:639–653. doi: 10.1016/j.dnarep.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Lauritzen KH, Dalhus B, Storm JF, Bjoras M, Klungland A. Modeling the impact of mitochondrial DNA damage in forebrain neurons and beyond. Mech. Ageing Dev. 2011b;132:424–428. doi: 10.1016/j.mad.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Lauritzen KH, Moldestad O, Eide L, Carlsen H, Nesse G, Storm JF, Mansuy IM, Bergersen LH, Klungland A. Mitochondrial DNA toxicity in forebrain neurons causes apoptosis, neurodegeneration, and impaired behavior. Mol. Cell. Biol. 2010;30:1357–1367. doi: 10.1128/MCB.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RE, Keatley K, Littlewood DT, Meunier B, Holt WV, An Q, Higgins SC, Polyzoidis S, Stephenson KF, Ashkan K, Fillmore HL, Pilkington GJ, McGeehan JE. Identification and functional prediction of mitochondrial complex III and IV mutations associated with glioblastoma. Neuro Oncol. 2015;17:942–952. doi: 10.1093/neuonc/nov020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B. Mitochondrial dynamics and neurodegeneration. Curr. Neurol. Neurosci. Rep. 2009;9:212–219. doi: 10.1007/s11910-009-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masino SA, Rho JM. Mechanisms of ketogenic diet action. In: Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Jasper’s Basic Mechanisms of the Epilepsies, [Internet] 4. National Center for Biotechnology Information (US); Bethesda, MD: 2012. [PubMed] [Google Scholar]
- Meidenbauer JJ, Mukherjee P, Seyfried TN. The glucose ketone index calculator: a simple tool to monitor therapeutic efficacy for metabolic management of brain cancer. Nutr. Metab. (Lond.) 2015;12:12. doi: 10.1186/s12986-015-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meidenbauer JJ, Ta N, Seyfried TN. Influence of a ketogenic diet, fish-oil, and calorie restriction on plasma metabolites and lipids in C57BL/6J mice. Nutr. Metab. (Lond.) 2014;11:23. doi: 10.1186/1743-7075-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalon A, Koshibu K, Baumgartel K, Spirig DH, Mansuy IM. Inducible and neuron-specific gene expression in the adult mouse brain with the rtTA2S–M2 system. Genesis. 2005;43:205–212. doi: 10.1002/gene.20175. [DOI] [PubMed] [Google Scholar]
- Nandivada P, Fell GL, Pan AH, Nose V, Ling PR, Bistrian BR, Puder M. Eucaloric ketogenic diet reduces hypoglycemia and inflammation in mice with endotoxemia. Lipids. 2016;51:703–714. doi: 10.1007/s11745-016-4156-7. [DOI] [PubMed] [Google Scholar]
- Nylen K, Velazquez JL, Sayed V, Gibson KM, Burnham WM, Snead OC., 3rd The effects of a ketogenic dieton ATP concentrations and the number of hippocampal mitochondria in Aldh5a1(−/−) mice. Biochim. Biophys. Acta. 2009;1790:208–212. doi: 10.1016/j.bbagen.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei M, Haug T, Nagelhus TA, Slupphaug G, Lindmo T, Krokan HE. Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, Wilhelmsson U, Pekna M. The dual role of astrocyte activation and reactive gliosis. Neurosci. Lett. 2014;565:30–38. doi: 10.1016/j.neulet.2013.12.071. [DOI] [PubMed] [Google Scholar]
- Rho JM, Rogawski MA. The ketogenic diet: stoking the powerhouse of the cell. Epilepsy Curr. 2007;7:58–60. doi: 10.1111/j.1535-7511.2007.00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho JM, Sankar R. The ketogenic diet in a pill: is this possible? Epilepsia. 2008;49(Suppl 8):127–133. doi: 10.1111/j.1528-1167.2008.01857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodell A, Rasmussen LJ, Bergersen LH, Singh KK, Gjedde A. Natural selection of mitochondria during somatic lifetime promotes healthy aging. Front. Neuroenergetics. 2013;5:7. doi: 10.3389/fnene.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin DN, Ross JL, Kawamura M, Jr, Ruiz TL, Geiger JD, Masino SA. A ketogenic diet delays weight loss and does not impair working memory or motor function in the R6/2 1J mouse model of Huntington’s disease. Physiol. Behav. 2011;103:501–507. doi: 10.1016/j.physbeh.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra S, Gilkerson RW, Davidson M, Schon EA. Ketogenic treatment reduces deleted mitochondrial DNAs in cultured human cells. Ann. Neurol. 2004;56:662–669. doi: 10.1002/ana.20240. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Mitchell SJ, Fang EF, Iyama T, Ward T, Wang J, Dunn CA, Singh N, Veith S, Hasan-Olive MM, Mangerich A, Wilson MA, Mattson MP, Bergersen LH, Cogger VC, Warren A, Le Couteur DG, Moaddel R, Wilson DM, 3rd, Croteau DL, de Cabo R, Bohr VA. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab. 2014;20:840–855. doi: 10.1016/j.cmet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selfridge JE, Wilkins HM, E L, Carl SM, Koppel S, Funk E, Fields T, Lu J, Tang EP, Slawson C, Wang W, Zhu H, Swerdlow RH. Effect of one month duration ketogenic and non-ketogenic high fat diets on mouse brain bioenergetic infrastructure. J. Bioenerg. Biomembr. 2015;47:1–11. doi: 10.1007/s10863-014-9570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried TN, Sanderson TM, El-Abbadi MM, McGowan R, Mukherjee P. Role of glucose and ketone bodies in the metabolic control of experimental brain cancer. Br. J. Cancer. 2003;89:1375–1382. doi: 10.1038/sj.bjc.6601269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, Cho JH, Hattori N, Youle RJ, van der Bliek AM. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol. Biol. Cell. 2014;25:145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Schragenheim J, Cao J, Abraham NG, Bellner L. PGC-1 alpha regulates HO-1 expression, mitochondrial dynamics and biogenesis: role of epoxyeicosatrienoic acid [Epub ahead of print] Prostaglandins Other Lipid Mediat. 2016 doi: 10.1016/j.prostaglandins.2016.07.004. http://dx.doi.org/10.1016/j.prostaglandins.2016.07.004. [DOI] [PMC free article] [PubMed]
- Sirven J, Whedon B, Caplan D, Liporace J, Glosser D, O’Dwyer J, Sperling MR. The ketogenic diet for intractable epilepsy in adults: preliminary results. Epilepsia. 1999;40:1721–1726. doi: 10.1111/j.1528-1157.1999.tb01589.x. [DOI] [PubMed] [Google Scholar]
- Sun Y, Chen X, Xiao D. Tetracycline-inducible expression systems: new strategies and practices in the transgenic mouse modeling. Acta Biochim. Biophys. Sin (Shanghai) 2007;39:235–246. doi: 10.1111/j.1745-7270.2007.00258.x. [DOI] [PubMed] [Google Scholar]
- Van der Auwera I, Wera S, Van Leuven F, Henderson ST. A ketogenic diet reduces amyloid beta 40 and 42 in a mouse model of Alzheimer’s disease. Nutr. Metab. (Lond.) 2005;2:28. doi: 10.1186/1743-7075-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitallie TB, Nonas C, Di Rocco A, Boyar K, Hyams K, Heymsfield SB. Treatment of Parkinson disease with diet-induced hyperketonemia: a feasibility study. Neurology. 2005;64:728–730. doi: 10.1212/01.WNL.0000152046.11390.45. [DOI] [PubMed] [Google Scholar]
- Verkhratsky A, Olabarria M, Noristani HN, Yeh CY, Rodriguez JJ. As-trocytes in Alzheimer’s disease. Neurotherapeutics. 2010;7:399–412. doi: 10.1016/j.nurt.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmering E, Niehusmann P, Peeva V, Grote A, Zsurka G, Altmuller J, Nurnberg P, Becker AJ, Schoch S, Elger CE, Kunz WS. Neuropatho-logical signs of inflammation correlate with mitochondrial DNA deletions in mesial temporal lobe epilepsy. Acta Neuropathol. 2016;132:277–288. doi: 10.1007/s00401-016-1561-1. [DOI] [PubMed] [Google Scholar]
- Wang S, Song J, Tan M, Albers KM, Jia J. Mitochondrial fission proteins in peripheral blood lymphocytes are potential biomarkers for Alzheimer’s disease. Eur. J. Neurol. 2012;19:1015–1022. doi: 10.1111/j.1468-1331.2012.03670.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Su B, Zheng L, Perry G, Smith MA, Zhu X. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J. Neurochem. 2009;109(Suppl 1):153–159. doi: 10.1111/j.1471-4159.2009.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen AW, Sander JW. Impaired mitochondrial energy production: the basis of pharmacoresistance in epilepsy. Med. Hypotheses. 2011;77:536–540. doi: 10.1016/j.mehy.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Zarnowski T, Tulidowicz-Bielak M, Kosior-Jarecka E, Zarnowska I, Turski WA, Gasior M. A ketogenic diet may offer neuroprotection in glaucoma and mitochondrial diseases of the optic nerve. Med. Hypothesis Discov. Innov. Ophthalmol. 2012;1:45–49. [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Chatterjee A, Singh KK. Saccharomyces cerevisiae polymerase zeta functions in mitochondria. Genetics. 2006;172:2683–2688. doi: 10.1534/genetics.105.051029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Lange DJ, Voustianiouk A, MacGrogan D, Ho L, Suh J, Humala N, Thiyagarajan M, Wang J, Pasinetti GM. A ketogenic diet as a potential novel therapeutic intervention in amyotrophic lateral sclerosis. BMC Neurosci. 2006;7:29. doi: 10.1186/1471-2202-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.