Abstract
RlmN is a dual-specificity RNA methylase that modifies C2 of adenosine 2503 (A2503) in 23S rRNA and C2 of adenosine 37 (A37) in several Escherichia coli tRNAs. A related methylase, Cfr, modifies C8 of A2503 by a similar mechanism, conferring resistance to multiple classes of antibiotics. Herein, we report the x-ray structure of a key intermediate in the RlmN reaction, in which a Cys118→Ala variant of the protein is cross-linked to a tRNAGlu substrate through the terminal methylene carbon of a formerly methylcysteinyl residue and C2 of A37. RlmN contacts the entire length of tRNAGlu, accessing A37 using an induced-fit strategy that completely unfolds the tRNA anticodon stem loop, which is likely critical for recognition of both tRNA and rRNA substrates.
RlmN is a radical S-adenosylmethionine (SAM) enzyme that is best known for catalyzing the methylation of C2 of adenosine 2503 (A2503) (1–3) in domain V of 23S rRNA. A2503, a conserved nucleotide, resides in the peptidyltransferase (PTC) center of the ribosome near the entrance to the exit channel for the nascent polypeptide (4–7). C2 modification is not essential, but has been reported to enhance translational fidelity (8, 9). A2503 is also methylated at C8 by Cfr, which is both evolutionarily and mechanistically related to RlmN (10, 11). The C8 modification, however, confers resistance to over five classes of antibiotics that target the bacterial ribosome (12–15). While methylation of C8 of A2503 is the only known in vivo activity of Cfr, RlmN also installs a C2 methyl group at adenosine 37 (A37) in six Escherichia coli tRNAs (tRNAArgICG, tRNAAspQUC, tRNAGlncmnm5sUUG, tRNAGlnCUG, tRNAGlumnm5s2UUC, and tRNAHisQUG) (16). RlmN thus joins a pseudouridine synthase, RluA, as the only known dual-specificity RNA modification enzymes capable of acting both on ribosomal and on transfer RNA (17, 18).
Although SAM is the source of the appended methyl carbon in the reactions catalyzed by RlmN and Cfr, these enzymes operate by a mechanism that is distinctly different from that of typical SAM-dependent methyltransferases (fig. S1), as demanded by the inertness of the C2 and C8 carbons of adenosine to electrophilic attack and the low acidities of their associated protons (3, 19–23). As radical SAM (RS) enzymes, RlmN and Cfr employ very similar radical-based mechanisms of catalysis, initiated by the abstraction of a hydrogen atom (H•) from a Cys-appended methyl group via a 5′-deoxyadenosyl 5′-radical (5′-dA•) (20). Subsequent attack of the resulting methylene radical upon the carbon atom undergoing methylation affords a protein/RNA cross-linked intermediate whose resolution requires prior proton abstraction from C2 (RlmN) or C8 (Cfr) of the substrate by an unidentified base. This intermediate has been trapped via mutagenesis of a key Cys residue (C118 in RlmN) in the Cfr and RlmN reactions and characterized by spectroscopic (21, 24), proteomic (25), and biochemical methods (20, 21, 24, 25). Conversion of the intermediate to the methylated product has also been demonstrated in the Cfr reaction (21). Although x-ray structures exist for RlmN in the presence and absence of SAM (24, 26), the large and conformationally flexible nature of its rRNA target has precluded successful structure determination of an enzyme-substrate complex. Here we take advantage of our current mechanistic understanding of the system to obtain an x-ray crystal structure of a C118A variant of RlmN cross-linked to an RNA substrate.
Studies by Grove et al. (20) and McCusker et al. (25) showed that the C118A or C118S variants of RlmN are unable to resolve a covalent protein/RNA intermediate during catalysis and become cross-linked to the nucleic acid when overproduced in E. coli. Because those studies predated the recognition that RlmN is responsible for methylation of tRNA in E. coli (16), it was assumed that the protein was cross-linked to a fragment of 23S rRNA. However, when the cross-linked enzyme was isolated by ion-exchange chromatography (fig. S2), observation of a distinct RNA-containing protein fraction suggested that the enzyme was cross-linked to a homogeneous RNA. Diffraction-quality crystals of the in vivo cross-linked species yielded x-ray datasets to 2.9 Å resolution, and the structure was solved by molecular replacement using a previously published RlmN structure (PDB accession code 3RFA) (Table S1, Supplementary Materials). Surprisingly, the electron density most closely resembled tRNAGlu rather than the expected rRNA fragment (fig. S3A) (16).
Each of the tRNAs expected to be modified by RlmN (16) were generated by in vitro transcription, and tRNAGlu and tRNAArg support the greatest turnover rate in activity assays (fig. S4). The structure of transcribed tRNAGlu cross-linked to purified RlmN C118A (termed the in vitro cross-link) was solved to 2.4 Å resolution (Fig. 1). Significant positive difference density was observed between the terminal methylene unit of mCys355 and C2 of A37 (Fig. 2A), but modeling the interaction as a carbon-carbon bond fully accounts for the electron density, confirming the presence of a covalent adduct. Remarkably, methionine (Met) and 5′-deoxyadenosine (5′-dA) remain visible in the active site, even in the in vivo structure, suggesting that they dissociate after the methylated RNA is released (Fig. 2B). In the absence of the RNA substrate, mCys355 resides near the SAM methyl group, cited as ideal for methyl acquisition from the cosubstrate (26) but relatively far (~6 Å) from the site of 5′-dA• formation (Fig. 2C). Interaction with tRNA, however, brings the Cys-appended methyl group within 4.1 Å of C-5′ of the SAM cleavage product, appropriate for H• abstraction (Fig. 2B). An extended water-mediated hydrogen-bonding network (fig. S5) between the backbone of the loop and A37 of the substrate facilitates the 5 Å shift in the mCys355 loop. Structural features unique to adenine, such as N3 and the exocyclic amine (Fig. 2D), provide a basis for selective recognition of the target base. Because these interactions would ensure specificity for reaction with C2, Cfr may use a different strategy to configure the analogous Cys for C8 methylation.
Figure 1.
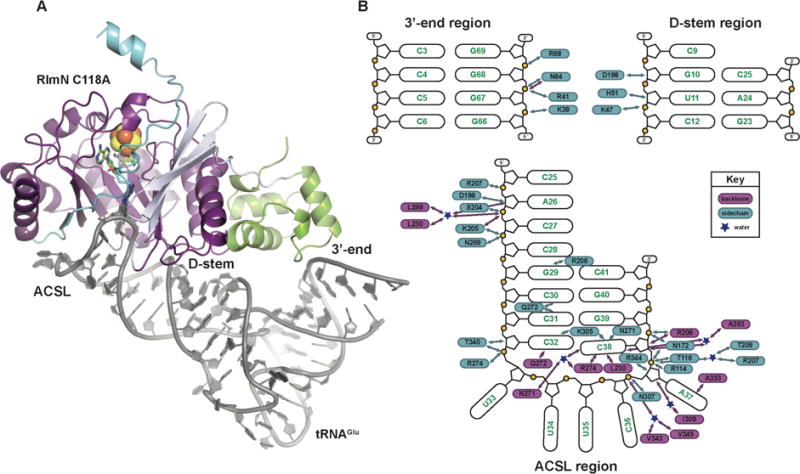
The x-ray crystal structure of the RlmN C118A-tRNAGlu in vitro cross-link. (A) The C118A RlmN protein is shown as a ribbon diagram, colored by domain, and the tRNAGlu is illustrated as a cartoon in grey. The mC355-A37 cross-link and SAM cleavage products, 5′dA and methionine, are shown in stick format and colored by atom type. The [4Fe-4S]2+ cluster is shown as a space-filling model. (B) A schematic diagram of the interactions between RlmN and the tRNA substrate. Protein residues shown in blue interact via the side chain and those shown in purple interact through the peptide backbone. Blue stars represent water molecules. The RNA is divided into three parts corresponding to the three regions of interaction observed in the complex.
Figure 2.
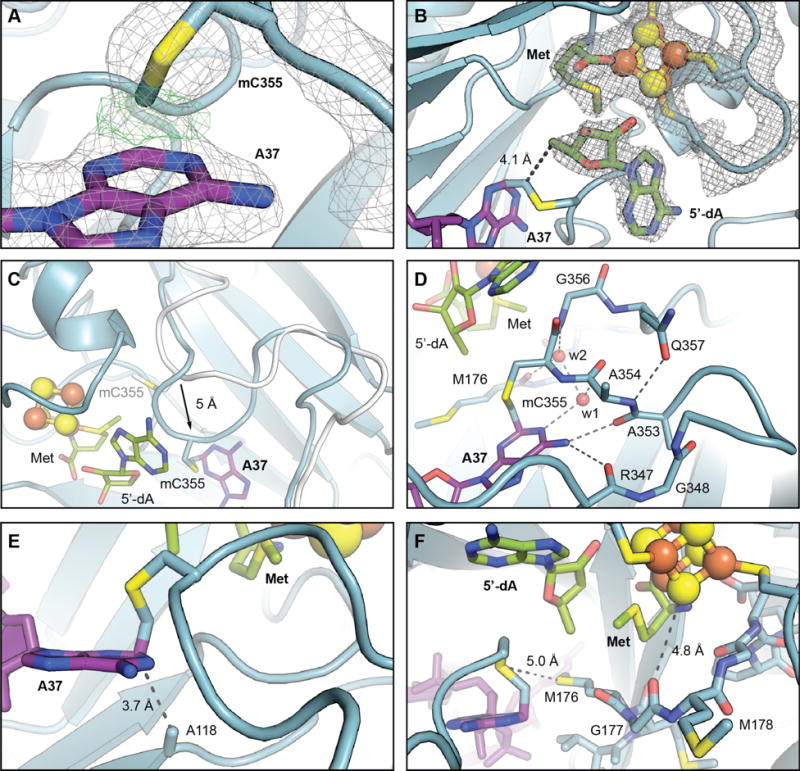
Views of the active site in the structure of the RlmN C118A-tRNAGlu in vitro cross-link. (A) A zoomed-in view of mC355 and A37 with selected residues and cofactors shown in stick format and colored by atom type. A 2Fo-Fc electron density map (gray mesh, contoured at 2.0 σ) and an omit map (green mesh, contoured at 4.0 σ) for the covalent bond between A37 C2 and the mCys355 Cδ are shown in overlay. (B) 2Fo-Fc electron density map for RlmN residues 125–132 (CX3CX2C motif, which binds [4Fe-4S] cluster), the [4Fe-4S] cluster, methionine, and 5′dA. The distance between the 5′-carbon of 5′-dA to the mC355 Cδ, consistent with H• abstraction via a 5′-dA• necessary to initiate cross-link formation, is indicated by a dashed line. (C) Overlay of loop from the wt RlmN x-ray structure with SAM (white, PDB accession code 3RFA) with the RlmN C118A in vitro cross-link (blue), illustrating the 5Å shift in position of the loop backbone upon interaction with the tRNA substrate. Selected amino acid side chains, nucleic acid bases, 5′-dA, and methionine are shown in stick format. The [4Fe-4S] cluster is illustrated as a ball-and-stick model. (D) A detailed view of the interactions involved in loop repositioning. Hydrogen bonding interactions are illustrated with dashed lines and ordered water molecules are shown as red spheres. (E) A zoomed-in view of the loop containing mC355 with the covalent cross-link to C2 of A37 shown in sticks. The distance between C2 of A37 and A118, consistent with assignment of C118 as a proton acceptor in the wt RlmN reaction, is indicated by a dashed line. (F) An alternate view of the active site with the conserved MGMGE motif and other selected amino acids displayed in stick format. Interactions potentially important in thiyl radical stabilization or in electron transfer mediated by the [4Fe-4S]1+ are shown as dashed lines.
Recent mechanistic studies suggest that resolution of the covalent cross-link requires prior deprotonation at C2, possibly by C118 (fig. S1) (24). The cross-linked structure shows A118 pointed directly at C2 of A37 3.7 Å away, supporting assignment of C118 as the general base (Fig. 2E). Upon deprotonation of C2, radical fragmentation of the C-S bond would generate a thiyl radical and an enamine, which can tautomerize to the methylated product upon return of the C2 proton initially abstracted by C118. Catalysis is then completed by reduction of the thiyl radical by one electron. Based on the structure of the cross-linked species, the thiyl radical would be ~9.9 Å away from the nearest iron ion in the cluster, consistent with direct electron transfer between the two species. The proximity (5.0 Å) of the Cys 355 side chain (the proposed site of thiyl radical formation) to the sulfur atom of Met176 (Fig. 2F), a strictly conserved residue in RlmN and Cfr, might allow formation of a transient thiosulfuranyl radical (27). In E. coli class III ribonucleotide reductase (RNR), a similar species is proposed to protect a reactive thiyl radical intermediate when reducing equivalents necessary to complete the reaction cycle are scarce (27). A similar interaction could be advantageous in RlmN if electron transfer to resolve the cross-linked intermediate is rate-limiting in vivo.
The protein component of the in vitro cross-link changes very little relative to that of the in vivo cross-link, as well as when compared to the previously solved structures of RlmN (Fig. 1A and figs. S3, S6) (26). The protein-nucleic acid interface spans the entire face of tRNAGlu using three separate regions or domains (Fig. 1B and figs S7, S8). The N-terminal domain, hypothesized to bind RNA because of its structural similarity with known nucleic acid binding proteins (26), uses only four points of contact to interact with the backbone at the 3′-end of tRNAGlu (fig. S8). The small number of substrate binding determinants is surprising, although the domain may interact more extensively with the larger rRNA substrate. Additional residues in the N-terminal domain (K47, H51) and a side chain (D198) contributed by a longer peripheral loop in the radical SAM core bind to the RNA backbone in the major groove of the D-stem of the tRNA (nt 10–12) (fig. S8). RlmN binds the concave surface of the substrate, suggesting that it requires full-length L-shaped tRNA for efficient catalysis. Other structurally characterized tRNA modifying enzymes share the same requirement (28), but the overall binding mode and type of RNA recognition elements found in RlmN more closely resemble those of tRNA synthetases (29) (fig. S9).
The most extensive RlmN-tRNA interactions involve the anti-codon stem loop (ACSL) of tRNAGlu near A37. The protein binds in the minor groove of the ACSL (Fig. 1A) and interacts more intimately with the nucleobases (Fig. 1B). Only a few sequence-specific contacts are observed, typical of ruler or position-specific RNA recognition strategies found in other tRNA modification systems (28,30), which would enable RlmN to accommodate the substantial sequence diversity of its tRNA substrates (16) in addition to its rRNA substrate (3). A strictly conserved arginine (R206) residue and G29 of tRNAGlu (Fig. 3C), found in all six E. coli tRNAs containing the m2A modification (16), form sequence-specific bidentate hydrogen bonds (H-bonds) between the exocyclic amine and N7 of G29, similar to an interaction found in the discriminating glutamyl-tRNA synthetase, wherein a single arginine-cytosine interaction allows the tRNA synthetase to distinguish between tRNAGlu and tRNAGln (31). In R206A RlmN, the ability to methylate tRNAGlu is completely abolished while methylation of a 155-mer rRNA substrate remains unaffected (Fig. 3C, inset). R206 may serve as an anchor for the tRNA substrate (Fig. 3D) to initiate base-flipping of A37 into the active site by an unusual mechanism that involves dramatic deformation of the ACSL backbone.
Figure 3.
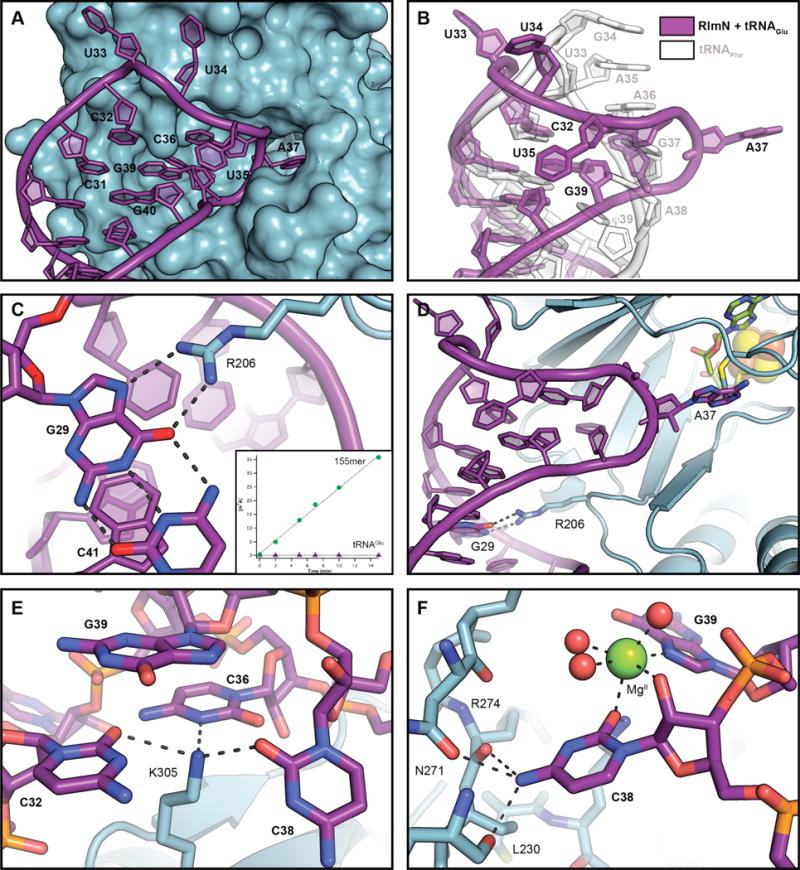
Selected views of the interactions between RlmN and tRNAGlu. (A) A surface representation of RlmN showing its interaction with the tRNA anti-codon stem loop shown in cartoon representation (purple). (B) A comparison of the structure of the anticodon stem loop in the C118A RlmN tRNAGlu cross-linked intermediate structure (purple) with the yeast tRNAPhe structure (white, PDB accession code 4TNA). (C) A zoomed-in view of R206 with G29 and C41. Selected amino acid residues and nucleic acid bases are shown in stick format. Hydrogen bonding interactions are displayed as dashed lines. In inset, the rate of formation of m2adenosine by RlmN R206A using an in vitro-transcribed 155-mer rRNA (green circles) or tRNAGlu (purple triangles) as substrates. (D) A zoomed-out view of the position of R206 in relation to the mC355/A37 cross-link and other active site components. The [4Fe-4S] cluster is illustrated as a space-filling model. (E) A view of the interactions of K305 with C32, C36, and C38. (F) The internal binding pocket of C38. The Mg2+ ion is shown as a green sphere and ordered water molecules as red spheres. Hydrogen bonding and metal coordination interactions are shown as dashed lines.
The ACSL conformational change is induced by an RlmN-specific secondary structure element (26), a seventh β-strand that forces an outward splay of all of the non Watson-Crick base-paired residues in the ACSL (Fig. 3A, B). As a result, A37 is sequestered into an active site pocket using a distinctive base-flipping mechanism that completely disrupts the single-strand aromatic base stack in the ACSL. A similar tactic is observed in other tRNA modifying enzymes, including bacterial adenosine deaminase (TadA) (32) and A37 N6-isopentenyltransferase (MiaA) (33) (fig. S10). Apart from A37 (fig. S13), C38 makes the most extensive contacts in the active site (Fig. 3E–F). The use of distinct complementary binding pockets for multiple bases in the ACSL resembles the substrate recognition strategy employed by bacterial Gln tRNA synthetase to read out the sequence in the anticodon and discriminate among potential targets (35). However, in RlmN, the C38 binding pocket is not base-specific. The observation is consistent with the sequences of the known targets for A37 C2 methylation, which would require accommodation of any nucleobase at position 38 except G (16). In RlmN, the additional C38 pocket may simply stabilize the new tRNA conformation in this region and promote extrusion of A37.
The structure of RlmN cross-linked to a tRNAGlu substrate reveals that the protein recognizes the overall shape of tRNAGlu via interaction with the sugar-phosphate backbone in the D-stem loop and the 3′-end of the tRNA. The observed analogies to tRNA synthetase structures, coupled with the known dual specificity of the enzyme towards A2503 in the central domain of 23S rRNA, provide a link between RlmN and three distinct components of the protein synthesis machinery, suggesting an early origin for A37/A2503 C2 methylation among known RNA modifications. In the ACSL region, the enzyme pries open the tRNA structure to gain access to its A37 nucleobase target. The recognition strategy is fully distinct from that used by RluA, the only other known dual specificity RNA modification enzyme (17). In that system, sequence elements conserved among rRNA and tRNA substrates dictate specificity, although the protein also relies on local RNA refolding for readout of the key motifs. The ability of RlmN to significantly remodel tRNA in the anticodon region may represent a second mechanism to confer dual specificity. The observations here highlight an intriguing structural connection between tRNA and the core of the bacterial ribosome (36), a theme that RlmN likely exploits in targeting two distinct substrates that exhibit little sequence similarity but share tertiary features common to evolutionarily ancient components of the protein synthesis machinery.
Supplementary Material
One sentence summary.
Substrate recognition in a dual specificity RNA methylase is revealed by x-ray crystallography of a cross-linked protein/RNA intermediate in the RlmN reaction.
Acknowledgments
This work has been supported by NIH grants GM100011 (A.K.B.) and GM101957 (S.J.B.), the Searle Scholars Program (A.K.B.), and Tobacco Settlement Funds (TSF13/14 SAP_4100062216) to S.J.B. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes HHR6 (C118A RlmN/tRNA in vivo crosslink) and 5HR7 (C118A RlmN/tRNA in vitro crosslink).
Footnotes
REFERENCES AND NOTES
- 1.Toh SM, Xiong L, Bae T, Mankin AS. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA. 2008;14:98–106. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kowalak JA, Bruenger E, McCloskey JA. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23S ribosomal RNA. J Biol Chem. 1995;270:17758–17764. doi: 10.1074/jbc.270.30.17758. [DOI] [PubMed] [Google Scholar]
- 3.Yan F, et al. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ban N, et al. A 9 Å resolution x-ray crystallographic map of the large ribosomal subunit. Cell. 1998;93:1105–1115. doi: 10.1016/s0092-86740081455-5. [DOI] [PubMed] [Google Scholar]
- 5.Harms J, et al. High-resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-86740100546-3. [DOI] [PubMed] [Google Scholar]
- 6.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 7.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 8.Vázquez-Laslop N, Ramu H, Klepacki D, Mankin AS. The key role of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramu H, et al. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell. 2011;41:321–330. doi: 10.1016/j.molcel.2010.12.031. DOI:j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Atkinson GC, et al. Distinction between the Cfr Methyltransferase Conferring Antibiotic Resistance and the Housekeeping RlmN Methyltransferase. Antimicrob Agents Chemother. 2013;57:4019–4026. doi: 10.1128/aac.00448-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giessing AMB, et al. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009;15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother. 2006;50:2500–2505. doi: 10.1128/aac.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith LK, Mankin AS. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob Agents Chemother. 2008;52:1703–1712. doi: 10.1128/aac.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kehrenberg C, Schwarz S, Jacobsen NE, Hansen LH, Vester B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 15.Morales G, et al. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin Infect Dis. 2010;50:821–825. doi: 10.1086/650574. [DOI] [PubMed] [Google Scholar]
- 16.Benítez-Páez A, Villarroya M, Armengod M-E. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA. 2012;18:1–13. doi: 10.1261/rna.033266.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoang C, et al. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Mol Cell. 2006;24:535–545. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Raychaudhuri S, Niu L, Conrad J, Lane BG, Ofengand J. Functional effect of deletion and mutation of the Escherichia coli ribosomal RNA and tRNA pseudouridine synthase RluA. J Biol Chem. 1999;274:18880–18886. doi: 10.1074/jbc.274.27.18880. [DOI] [PubMed] [Google Scholar]
- 19.Bauerle MR, Schwalm EL, Booker SJ. Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J Biol Chem. 2015;290:3995–4002. doi: 10.1074/jbc.r114.607044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grove TL, et al. A radically different mechanism for S-adenosylmethionine-dependent methyltansferases. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 21.Grove TL, et al. A substrate radical intermediate in catalysis by the antibiotic resistance protein Cfr. Nat Chem Biol. 2013;9:422–427. doi: 10.1038/nchembio.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grove TL, Radle MI, Krebs C, Booker SJ. Cfr and RlmN contain a single [4Fe-4S] cluster, which directs two distinct reactivities for S-adenosylmethionine: methyl transfer by SN2 displacement and radical generation. J Am Chem Soc. 2011;133:19586–19589. doi: 10.1021/ja207327v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan F, Fujimori DG. RNA methylation by radical SAM enzyme RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc Natl Acad Sci U S A. 2011;108:3930–3934. doi: 10.1073/pnas.1017781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silakov A, et al. Characterization of a cross-linked protein-nucleic acid substrate radical in the reaction catalyzed by RlmN. J Am Chem Soc. 2014;136:8221–8228. doi: 10.1021/ja410560p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCusker KP, et al. Covalent intermediate in the catalytic mechanism of the radical S-adenosyl-L-methionine methyl synthase RlmN trapped by mutagenesis. J Am Chem Soc. 2012;134:18074–18081. doi: 10.1021/ja307855d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boal AK, et al. Structural basis for methyl transfer by a radical SAM enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei Y, et al. A chemically competent thiosulfuranyl radical in the Escherichia coli class III ribonucleotide reductase. J Am Chem Soc. 2014;136:9001–9013. doi: 10.1021/ja5030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 28.Ishitani R, Yokoyama S, Nureki O. Structure, dynamics, and function of RNA modification enzymes. Curr Opin Struct Biol. 2008;18:330–339. doi: 10.1016/j.sbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 30.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 31.Sekine S, Nureki O, Shimada A, Vassylyev DG, Yokoyama S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat Struct Biol. 2001;8:203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 32.Losey HC, Ruthenburg AJ, Verdine GL. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat Struct Mol Biol. 2006;13:153–159. doi: 10.1038/nsmb1047. [DOI] [PubMed] [Google Scholar]
- 33.Chimnaronk S, et al. Snapshots of dynamics in synthesizing N(6)-isopentenyladenosine at the tRNA anticodon. Biochemistry. 2009;48:5057–5065. doi: 10.1021/bi900337d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts RJ, Cheng X. Base flipping. Annu Rev Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 35.Steitz TA, Rould MA, Perona JJ. Structural basis of tRNA discrimination as derived from the high resolution crystal structure of glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP. Mol Biol Rep. 1990;14:213–214. doi: 10.1007/bf00360479. [DOI] [PubMed] [Google Scholar]
- 36.de Farias ST, do Rêgo TG, José MV. Evolution of transfer RNA and the origin of the translation system. Front Genet. 2014;5:303. doi: 10.3389/fgene.2014.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-68799776066-X. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Keating KS, Pyle AM. RCrane: semi-automated RNA model building. Acta crystallogr D. 2012;68:985–995. doi: 10.1107/S0907444912018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lebedev AA, et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta crystallogr D. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shisler KA, Broderick JB. Glycyl radical activating enzymes: structure, mechanism, and substrate interactions. Arch Biochem Biophys. 2014;546:64–71. doi: 10.1016/j.abb.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


