Abstract
This article summarizes the contributions of high throughput genomic, proteomic, metabolomic, and gene expression investigations to our understanding of inherited or acquired risk for acute respiratory distress syndrome (ARDS). While not yet widely applied to a complex trait like ARDS, these techniques are now routinely employed to study a variety of disease states. Omic applications hold great promise for identifying novel factors that may contribute to ARDS pathophysiology or may be appropriate for further development as biomarkers or surrogates in clinical studies. Opportunities and challenges of different techniques are discussed, and examples of successful applications in non-ARDS fields are used to illustrate the potential utility of each technique.
Keywords: Acute respiratory distress syndrome, Genomic, Proteomic, Metabolomic, Gene expression, Complex trait
Acute respiratory distress syndrome (ARDS) inflicts considerable morbidity and mortality among critically ill patients and lacks any specific pharmacologic therapy.1,2 Because clinical factors alone fail to explain which patients with ARDS risk factors will develop the syndrome, or to accurately predict which patients will die as a result of ARDS, there is great interest to understand whether one could leverage new biologic techniques to better characterize risk and prognosis. With major advances in the fields of genomics, mass spectroscopy, and bioinformatics, there are numerous approaches that can now be applied to a complex trait like ARDS, yet the benefit of these is uncertain (Table 1). The goal of this paper is to review the state of knowledge of genetic contributions to ARDS risk and mortality, to briefly review broader applications of genomics to ARDS pathogenesis, and to consider examples from non-ARDS fields whereby genomic approaches have yielded major advances. Applying similar approaches to ARDS may deepen our understanding and offer new therapeutic paradigms for patients with ARDS.
Table 1. Potential applications of different ‘omic technologies to a complex trait like ARDS.
For each application, the tested analyte is named and the most likely potential applications are highlighted with an X.
| Analyte Field |
Infer Mechanism |
Candidate Marker Validation |
Candidate Marker Discovery |
Identify Subclasses |
Risk Stratify |
Improve Diagnosis |
Identify Therapeutic Targets |
|---|---|---|---|---|---|---|---|
|
| |||||||
| DNA | x | x | x | x | x | ||
| Genomics | |||||||
|
| |||||||
| DNA | x | x | x | x | |||
| methylation or acetylation | |||||||
| Epigenomics | |||||||
|
| |||||||
| mRNA | x | x | x | x | x | x | |
| - miRNA | |||||||
| - ncRNA | |||||||
| Transcriptomics | |||||||
|
| |||||||
| Proteins | x | x | x | x | x | x | x |
| Proteomics | |||||||
|
| |||||||
| Metabolites | x | x | x | x | x | x | x |
| Metabolomics | |||||||
|
| |||||||
| Systems | x | x | x | x | |||
| Interactome | |||||||
|
| |||||||
| Microbiota | x | x | x | x | x | x | x |
| Microbiome | |||||||
Shifting the Paradigm: What can genomics teach us about a trait like ARDS?
Many investigators associate the word genomic with inherited conditions that obviously cluster among families. Because there are no reported families in whom ARDS has affected multiple members, one might conclude that genomics would offer little to our understanding of ARDS. However, an alternative perspective is to consider ARDS as a pattern of response to injury, be it pneumonia, sepsis, or trauma. As a trait, the response to injury has significant heritability.3–7 In fact, death from infection was the most heritable condition when studied in a large Danish registry of adoptees, with a much stronger heritability than vascular disease or cancer.8 Rather than acting as monogenetic, or Mendelian, traits, whereby a single genetic variation explains the bulk of the observed phenotype, ARDS risk and severity are likely influenced by multiple genetic variants that each contribute to a modest degree. Small effect sizes of each contributing gene variant mandate large study populations for detection. In addition, ARDS is ripe for the utilization of intermediate traits and the identification of endotypes, or subtypes, which may demonstrate a more homogeneous genetic background.
Surveying the Landscape of ARDS Genomics
Numerous candidate gene association studies have been reported in ARDS, and several reproducible associations have emerged. While a complete review of the genetic associations with ARDS risk or ARDS mortality is beyond the scope of the present review, comprehensive reviews recently have been published.9–11 The best replicated genetic variants for ARDS risk represent our present understanding of ARDS pathophysiology; pro-and anti-inflammatory cytokine gene polymorphisms are well represented (IL6, IL10, IL1RN, MBL2),12–18 as are vascular injury markers (VEGFA, ANGPT2, ACE, MYLK),19–24 innate immunity pathway members (IRAK3, TLR1, NFKB1, NFKBIA, FAS, PI3),5,25–28 and markers of respiratory epithelial injury (SFTPB).29,30 While each of these associations has been associated with ARDS risk or outcome in at least 2 populations, none influences ARDS risk or severity to a degree that warrants genetic testing of at-risk populations. Instead, the main contribution of ARDS genetic associations to date has been to focus attention on molecular pathways at play in causing or perpetuating the syndrome.
Further, it is tempting to speculate that genetic associations may highlight potential therapeutic targets to either prevent ARDS or to improve outcomes once it has developed. For example, the association of variants in the angiopoietin-2 gene (ANGPT2) with ARDS risk in mixed ICU population and trauma populations,22,23 coupled with strong animal evidence that antagonizing angiopoietin-2 protein (ANG2) or augmenting its counterpart, angiopoietin-1 (ANG1), results in decreased mortality31–34 suggests that it may be helpful to block this protein in humans at risk for ARDS. Given numerous failed trials to prevent or treat ARDS, however, it may be that investigators should seek a molecular marker, such high ANG2 plasma protein level or carriage of an ANGPT2 genetic variant, to enrich a clinical trial population for subjects likely to respond. A similar case could be made for the use of human neutrophil elastase (hNE) inhibitors, already used in Japan to treat ARDS albeit with scant evidence of efficacy,35–37 since the gene pre-elafin (PI3) has evidence for 1) dysregulation among patients who fail to resolve ARDS,38 2) functional promoter variants that associate with ARDS,27,28 and since 3) plasma levels of its gene product elafin are reduced in ARDS patients.39 Perhaps patients with low elafin levels or high hNE activity would be more likely to respond to hNE inhibitors. The angiotensin converting enzyme (ACE) gene also may suggest a therapeutic strategy, as ACE gene variants that increase ACE1 levels have been associated with ARDS mortality,24,40–43 and the counterregulatory ACE2 enzyme seems to decrease lung injury.44,45
ARDS enters the Genomic Age
The earliest contributions of genomics to our understanding of ARDS successfully leveraged animal models, quantitative traits, and bioinformatics. Grigoryev and colleagues used bioinformatic approaches to find overlap in the gene expression, or messenger RNA (mRNA) levels from human cells or animal lung tissue subjected to repeated mechanical stress or models of ventilator-induced injury. Orthologues, or genes with common structure and function across different species, that behaved in a reproducible manner across numerous models of injury were subsequently investigated through mouse knockout models or gene silencing. In this manner, Ye and colleagues identified pre-B cell colony enhancing factor (PBEF), also known by the gene name NAMPT, as a regulator of inflammatory cytokines which in turn cause epithelial and endothelial permeability.46–48 In addition, independent groups have now replicated the association between promoter variants in NAMPT and the development of ARDS.49 New candidate genes can also arise from screening multiple rodent species for differential susceptibility to lung injury, as Leikauf and colleagues performed using inhalational injury models.50–52 When translating animal and in vitro work to human populations, however, the choice of experimental model is highly relevant. Dos Santos and colleagues demonstrated that the gene expression response to ventilator-induced lung injury is distinct from endotoxin models of sepsis,53 which is consonant with findings in human populations that gene variants associated with ARDS may be specific to ARDS risk factor.54
In the first published human genome-wide association study (GWAS) for ARDS susceptibility, Christie and colleagues identified a novel locus, PPFIA1 or liprin alpha, as a replicable risk factor for ARDS following trauma.55 While little was known about how liprin alpha would contribute to lung injury at the time of discovery, subsequent work has implicated liprin alpha as a binding partner and downregulator of Mammalian homolog of Diaphanous (mDia), a Rho effector that mediates stress fiber formation.56 Just as nonmuscle myosin light chain kinase is a critical regulator of the endothelial cytoskeleton with implications for ARDS,57,58 liprin alpha may also prove to have relevance for barrier enhancement.
As future GWAS are published, new candidates will undoubtedly emerge and meta-analysis will be possible to harness the power of multiple populations. At the same time, however, GWAS may prove more fruitful if the heterogeneity of ARDS is acknowledged a priori, and analyses are conducted within more homogenous subtypes. For instance, Tejera and Christiani demonstrated that the genetic risk factors for extrapulmonary ARDS and pulmonary ARDS were distinct,54 a finding echoed by most reviews.9,41 In addition to ARDS precipitant, factors such as ancestry,12 gender,59 age or comorbidity17 may be important. Furthermore, given the notable heterogeneity of ARDS, there are almost certaintly additional subtypes that may become apparent with additional data, as elegantly proven by Calfee and colleagues applying latent class analysis to 2 populations from the NHLBI ARDS network.60 Since the unbiased model suggested 2 classes of patients with ARDS, one characterized by higher inflammatory cytokine plasma levels, lower plasma bicarbonate, and a tendency for shock, and as the 2 subclasses had dramatically different outcomes and response to a high positive end-expiratory pressure, it may be that leveraging GWAS within each subclass would yield important insights about processes driving each type of ARDS.
Advances from Gene Expression
In addition to the animal studies mentioned above, several human investigations have explored the utility of gene expression to identify novel candidate genes in ARDS. Gene expression analysis can be very advantageous, since the output of the analysis is a quantitative trait which offers strong statistical power relative to a dichotomous trait like the presence or absence of ARDS. In addition, because mRNA reflects the mature transcript that will be translated into protein, changes in transcript abundance are likely to be functional and thus offer strong evidence for involvement in a trait. Finally, multidimensional analysis of global gene expression changes are possible using microarray technology, whereby a small amount of starting mRNA can be assessed for roughly 20,000 transcripts simultaneously. This technique is not only a very efficient way to assess gene expression level, but also allows multidimensional analytic strategies to define patterns of expression. Two general types of analyses can be performed: a supervised, or hierarchical analysis, in which samples are assigned to their class by the investigator, or an unbiased or machine learning analysis, whereby the analysis seeks to detect unobserved patterns from unlabeled data. An early example of the hierarchical analysis involved the demonstration that gene expression changes between acute myelogenous leukemia (AML) bone marrow mononuclear cells were distinct from acute lymphogenous leukemia (ALL) cells.61 A subsequent group performed a similar genome wide expression analysis of ALL blast cells in an unsupervised manner, identifying 6 major molecular subtypes that reflected different biological mechanisms and suggested differential response to treatment.62 Expression patterns of specific factors in tumor tissue are now routinely assayed in both hematolgic and solid organ malignancies in an effort to personalize therapy.63,64
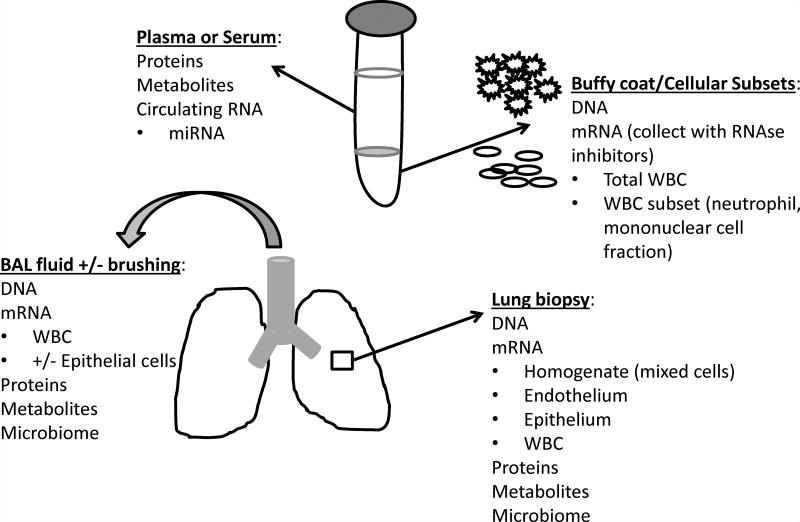
Research in ARDS transcriptomics is hampered by the lack of available lung tissue upon which to perform microarray analysis, since only the minority of patients – less than 10% – progress to lung biopsy.65 Without lung tissue, most investigations have relied upon gene expression of either specific blood cell populations or white blood cells collected from whole blood. While whole blood is a relatively convenient way to sample RNA, this collection method may introduce variability and will never reflect the expression pattern of lung endothelium or epithelium, since gene expression is cell-specific (Figure 1).66 In one of the first descriptions of applying microarray technology to human ARDS, Howrylak and colleagues reported the global whole blood gene expression of 13 patients with sepsis and ARDS compared to 20 with sepsis alone, and reported that differential expression of 8 transcripts distinguished patients with ARDS.67 The most dysregulated transcript encoded for the heavy subunit of ferritin, which is interesting given the role of iron in catalyzing reactive oxygen species (ROS) and the potential contribution of ROS towards lung injury.68–71 A novel approach was applied by Wang and colleagues to compare the whole blood gene expression profile of patients during acute ARDS and convalescence, which allowed for each patient to be her own control and to filter much of the background variability between expression across individuals. Using this approach, the investigators identified the peptidase inhibitor 3 gene PI3, encoding elafin, as downregulated in ARDS.38 Functional promoter variants in PI3 associate with lower cytokine-induced transcriptional activity and greater sepsis-associated ARDS, potentially acting through more durable binding of pre-elafin to extracellular matrix proteins.27
Figure.
Biospecimens available for ‘omic applications in ARDS
To date, there has been no large systematic application of gene expression in human samples with ARDS. Furthermore, it remains unclear whether the signature obtained from whole blood or from circulating leukocytes will provide relevant answers for a condition of alveolar epithelial and endothelial dysfunction. While questions remain about the suitability of studying whole blood to gain insights into a lung-centric condition, advances have been made by applying this approach in sepsis, generally considered a systemic vascular disorder. Wong and colleagues performed unbiased clustering analysis to whole blood gene expression of approximately 100 septic children and identified 3 subclasses that correlated with differential mortality.72 The highest mortality subclass was characterized by significant downregulation of genes annotated to glucocorticoid signaling, the adaptive immune system, and zinc-related biology. The investigators were able to extend their findings by analyzing the secreted protein products of some of the most dysregulated genes in the high-mortality group, resulting a 5-biomarker decision tool that reliably identifies a higher risk group for death when applied to both children and adults with septic shock.73,74 Thus, despite limitations of whole blood or even leukocyte gene expression to inform about lung tissue expression, large scale peripheral blood gene expression may yield advances in ARDS.
Untapped potential: transcriptomics of the future
While the field of transcriptomics to date has been dominated by microarray studies quantifying mRNA, the transcriptome encompasses all forms of RNA, including transfer RNA, ribosomal RNA, and many forms of noncoding RNA. The science of noncoding RNA molecules has exploded in the past 10 years, with the identification of multiple new classes of non-protein coding entities75 and emerging understanding of their complex roles in regulating gene expression.76 Advances in next generation sequencing capabilities, coupled with improved bioinformatic support and efficiencies of scale have fueled high throughput sequencing of RNA in both targeted and genome-wide approaches.77 Because the technology of RNA sequencing is so nascent, there are few published reports applying these methods to a complex trait like ARDS. However, applications of RNA-seq to cancer and cardiovascular traits may exemplify useful approaches for the future. MicroRNA, or miRNA, only described in 1993,78 are now understood to be small, roughly 22-nucleotide RNA sequences that typically bind the 3’ untranslated region of target mRNA and inhibit translation or promote mRNA degradation.79,80 Candidate genes influencing ARDS, including MYLK and PBEF/NAMPT, are regulated by miRNA81,82 which raises the possibility that engineered miRNA or their antagonists may be a therapeutic strategy in the future. In cancer, circulating miRNA profiles in plasma are being investigated as biomarkers with diagnostic or prognostic utility,83 approaches that may prove fruitful in ARDS.
Another as yet underexplored aspect of genomic regulation of ARDS is whether epigenetic modifications influence disease susceptibility or outcome. Epigenetic changes are classically construed as heritable changes in gene expression, function, or activity that occur without a change in DNA sequence, such as might occur due to changes in DNA methylation, histone modification, gene silencing, or imprinting.84,85 Epigenetic mechanisms are attractive explanations for severe gene by environment interactions, and thus may be relevant to a complex trait like ARDS which only manifests in the setting of a severe environmental insult like mechanical ventilation, systemic infection, or severe trauma. Much of our understanding surrounding DNA methylation and histone modification comes from cancer biology. Tumor-suppressor gene hypermethylation leads to transcriptional silencing, promoting tumorigenesis.86,87 Histones are dynamically regulated by processes including acetylation, methylation, phosphorylation, and ubiquitinylation, resulting in dynamic effects on gene expression and chromatin structure.87,88 Differential epigenetic regulation can be assessed in either targeted or unbiased, genome-wide approaches by leveraging bisulfite sequencing (for methylation) and/or mass spectrometry to detect post-translational histone modification.87 Given evidence from the Encyclopedia of DNA Elements (ENCODE) project highlighting vast complexity of genome regulation,76,89,90 future studies should explore the role of epigenomic variation in the context of ARDS and potentially for distinct ARDS precipitants.
Proteomics and Metabolomics: Searching for the key players
While it was once assumed that gene expression would adequately describe the state of expressed proteins in a body site or cellular compartment, the correlation between mRNA and protein profiles is surprisingly poor.91–93 Understanding the differential regulation of proteins relative to gene expression has enabled a better understanding of alternative splicing and post-translational regulation, while focusing on the expressed proteins has aided the identification of biomarkers for diagnosis, prognosis, and potentially, mechanistic importance. Proteomic approaches are generally conceived as large-scale analyses, going beyond individual or even multiplex protein quantification by enzyme-linked immunosorbent assay (ELISA) or bead-based immunoassays. Parallel to the explosion in next generation sequencing technology to analyze the genome have been advances in matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry to analyze high dimensional protein populations, including the identification of unknown proteins. Over 10 years ago, Bowler and colleagues performed 2 dimensional electrophoresis (2DE) followed by mass spectrometry upon edema fluid from 16 subjects with ARDS compared to 12 healthy controls.94 Compared to healthy subjects, alveolar fluid albumin, transferrin, IgG, and clusterin were increased, and surfactant protein A and alpha-1-antitrypsin were decreased in ARDS patients.94 In addition, several proteins detectable in the alveolar fluid of ARDS subjects demonstrated significant posttranslational modifications.94 Schnapp and a group from the University of Washington applied liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomics to 3 bronchoalveolar lavage (BAL) samples from ARDS subjects, and analyzed almost 900 resulting unique proteins.95 Focusing on secreted proteins, the authors identified increased insulin-like growth factor-binding protein-3 (IGFBP-3) and its ligand insulin growth factor 1 (IGF-1) early in ARDS and suggested that IGF-1 contributes to fibroblast survival in ARDS.95 Several investigators have since reported the identification of increased BAL apolipoproteins,96 S100 calcium-binding proteins,96–98 and inflammatory proteins including acute phase proteins and cytokines (TNF-α, interleukin-1β).95,98 Investigating the plasma compartment, Chen and colleagues identified 16 proteins as differentially expressed in ARDS patients compared to healthy controls, with ARDS subjects showing significantly downregulated plasma apolipoproteins (apo A-I, A-IV, B-100, C-II, and CIII) and complement factor H, with upregulated complement C9, serum amyloid A, and Creactive protein.99 Consonant with the individual protein findings, the most dysregulated pathways were annotated as acute phase signaling, complement system, interleukin-12 signaling, and production of nitric oxide and reactive oxygen species.99
A complementary approach to proteomics is to examine the small molecule metabolite profile of either the blood or lung compartment, in order to gain more information about the physiologic processes occurring in that compartment. Metabolites, the intermediate products of metabolism, can be endogenous or exogenous, and can be peptides but also lipids, carbohydrates, amino acids, nucleotides, hormones, vitamins, or foreign chemical substrates as from a drug, diet, or other exposure. Metabolites are first separated with gas and/or liquid chromatography and then quantified with mass spectrometry or isotope-labeled nuclear magnetic resonance spectroscopy (1H-NMR or 13C-NMR).100,101 Stringer and colleagues at the University of Michigan first reported on 1H-NMR-identified metabolites in the plasma of 13 sepsis-associated ARDS subjects compared to 6 healthy controls.102 Forty metabolites were identified, and ARDS plasma was characterized by higher adenosine, glutathione, sphingomyelin, and phoshatidylserine, and pathway annotation analysis suggested that each metabolite participated in a unique metabolic network.102 Because sepsis-associated ARDS samples were compared to healthy control subjects’ plasma, it remained unclear whether the analytes identified were specific to ARDS or whether they might reflect alterations from sepsis. To study ARDS-specific metabolites would require a study with non-ARDS septic subjects as controls.
Metabolomic profiling of plasma has been applied more commonly to sepsis. Seymour identified numerous differences in the initial emergency department plasma sample of sepsis survivors versus non-survivors after matching subjects on clinical characteristics and procalcitonin level.103 Metabolism of bile acids, sterols, amino acids, nucleotides, and energy were among the most dysregulated pathways in non-survivors.103 Langley and colleagues performed multiple analyses, comparing sepsis survivors to noninfected subjects presenting with non-infectious systemic inflammatory response syndrome (SIRS), and also sepsis survivors to non-survivors.104 There was a progressive decline in glycerophosphocholine and glycerophosphoethanolamine esters among septic subjects that was more pronounced among non-survivors, as well as increased lactate and increased carnitine esters, products of fatty acid metabolism, in nonsurvivors.104 One of the limitations of metabolomic investigations is the lack of consensus for analytic strategy using multiple layers of large data. This fact is highlighted by a second metabolomic investigation in the same populations as the Langley study, but done in reverse order and with a different analytic strategy. Rogers and colleagues first identified and then validated individual metabolites associated with death during critical illness, and then developed an iterative Bayesian network to risk stratify for mortality.105 Compared to Langley study which identified 4 critical metabolites, 2 clinical variables (age, hematocrit), and lactate in a predictive model, the Rogers model identified no clinical data but 7 metabolites, all of them distinct from those chosen by the Langley model.104,105 In a pediatric septic population, Mickiewicz applied 1H-NMR spectroscopy to serum samples and demonstrated with principal components analysis that the metabolic profile of septic shock differed from that in healthy children or those with SIRS, and septic shock was characterized by increased levels of metabolites associated with muscle turnover, amino acid oxidation, and decreased energy supply.106 The field of metabolomics seems ripe for the development of consensus recommendations governing analytic approach, need for independent replication, and to prompt both meta-analysis and application of unsupervised learning methodologies for class prediction.
Bronchoalveolar lavage fluid has posed a challenge for metabolomics due to the fluid’s high protein and salt content, and relatively low concentration of most metabolites.107 However, Evans and colleagues recently overcame these limitations by testing BAL fluid on multiple high-performance liquid chromatography platforms, and determining that the optimal performance was with reversed phase and hydrophilic interaction chromatography prior to mass spectrometry.102 Consonant with proteomic studies of ARDS BAL fluid that demonstrated reduced surfactant proteins, Evans reported measuring reduced phosphatidylcholine, the major phospholipid of pulmonary surfactant, in BAL fluid among ARDS subjects.108 Alveolar fluid from ARDS subjects also demonstrated higher levels of products associated with energy metabolism – lactate, citrate, creatine, and creatinine – all of which were similarly increased in the plasma of ARDS patients.102,108 As novel findings, several guanosine network metabolites were increased in ARDS BAL fluid, prompting re-examination of xanthine oxidase activity in lung injury models.108
Introducing the Interactome: a systems biology approach
The common thread through this review of various ‘omic approaches have been the exponential growth in complex data fueled by major technological advances. The explosion of data has created new challenges for computing power and analysis, but similarly new opportunities to describe how multiple aspects of human data fit together. Groundbreaking examples of integrative thinking paired whole genome genotyping to tissue-specific gene expression and identified not only the genetic determinants of specific traits, but used annotation and enrichment analysis to determine which transcription factors were critical to obesity109 or coronary artery disease (CAD).110 This approach, sometimes termed network biology, is particularly successful when it helps to prioritize further mechanistic study, translating observations in human populations back to the laboratory for testing of causality. In one notable example, Rader and colleagues built on the demonstration that one genetic variant strongly associated with CAD, low density lipoprotein (LDL) cholesterol levels, and hepatic expression of the sortilin gene SORT1.110 The investigators then performed fine mapping of the SNP’s linkage disequilibrium block to identify the functional variant, replicated the genetic association in populations of different ancestral background, and used overexpression and knockdown experiments in mice to prove that SORT1 expression modifies plasma LDL and a novel pathway of LDL regulation was identified.111,112 Thus the network analysis integrating multiple threads of data not only suggested putative candidates for mechanistic follow up, but helped to prioritize the functional candidate and suggested the causal pathway through which it acted. Network science is most effective with multiple platforms of systematic data acquisition, thus it has been leveraged successfully with several cancer networks113 and common traits like CAD.114 As investigators begin gathering ARDS-specific systematic data, a network based approach may prove feasible in the future.
Further complexity may be added to the system by considering not only the human interactome, but also the interaction between the human host and resident microbial flora. While not yet widely applied to lung injury models, careful examination of gut microbiota in animal models of metabolic syndrome has demonstrated the critical interaction between inflammasome signaling and gut microflora.115 Not only have microbes shaped genetic architecture of global populations through natural selection,116 but they may be dynamic partners in the evolution of a complex phenotype like ARDS.117 Next generation sequencing technology makes it possible to characterize the lung microbiome,118 which may yield important insights in the future.
Summary
Acute respiratory distress syndrome is a complex trait poised to benefit from the application of high throughput technologies to assay DNA, mRNA, proteins, metabolites, microbiomes, and systems. Though hindered by infrequent access to lung tissue, researchers have made important advances in the early application of multiple ‘omic applications. As we apply lessons from non-ARDS phenotypes highlighting the potential for these techniques to expand our pathophysiologic understanding, new discoveries in ARDS await.
Key points.
Numerous candidate gene association studies and protein investigations have been employed in ARDS to moderate success, but large scale genomic, proteomic, or metabolomic studies have not yet been undertaken. There exists significant opportunity to apply ‘omic platforms to advance our understanding of ARDS pathophysiology.
While small proteomic and metabolomics investigations in ARDS have proven feasibility, to date there have been limited mechanistic follow up of compelling candidates and questions remain regarding the optimal target tissue and the best analytic strategy for ARDS.
Future studies could leverage ‘omic experience gained evaluating other non-ARDS complex traits, and could explore unbiased analytic strategies for class distinction or network analysis.
The success of high throughput discovery ‘omic investigations derives from tracing observations in human populations back to their mechanistic underpinnings.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Dr. Meyer has no disclosures relevant to this publication.
References
- 1.Rubenfeld GD, Caldwell E, Peabody E, et al. Incidence and outcomes of acute lung injury. N Engl J Med. 2005 Oct 20;353(16):1685–1693. doi: 10.1056/NEJMoa050333. [DOI] [PubMed] [Google Scholar]
- 2.Ranieri VM, Rubenfeld GD, Thompson BT, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012 Jun 20;307(23):2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson J, Patel P, Shah R, et al. Race and gender variation in response to evoked inflammation. Journal of Translational Medicine. 2013;11(1):63. doi: 10.1186/1479-5876-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehta NN, Heffron SP, Patel PN, et al. A human model of inflammatory cardio-metabolic dysfunction; a double blind placebo-controlled crossover trial. J Transl Med. 2012;10:124. doi: 10.1186/1479-5876-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wurfel MM, Gordon AC, Holden TD, et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med. 2008 Oct 1;178(7):710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zabaleta J, Schneider B, Ryckman K, et al. Ethnic differences in cytokine gene polymorphisms: potential implications for cancer development. Cancer Immunology, Immunotherapy. 2008;57(1):107–114. doi: 10.1007/s00262-007-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillay V, Gaillard MC, Halkas A, Song E, Dewar JB. Differences in the genotypes and plasma concentrations of the INTERLEUKIN-1 receptor antagonist in black and white South African asthmatics and control subjects. Cytokine. 2000 Jun;12(6):819–821. doi: 10.1006/cyto.1999.0637. [DOI] [PubMed] [Google Scholar]
- 8.Sorensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988 Mar 24;318(12):727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 9.Meyer NJ, Christie JD. Genetic heterogeneity and risk for ARDS. Semin Respir Crit Care Med. 2013;34(4) doi: 10.1055/s-0033-1351121. in press. [DOI] [PubMed] [Google Scholar]
- 10.Acosta-Herrera M, Pino-Yanes M, Perez-Mendez L, Villar J, Flores C. Assessing the quality of studies supporting genetic susceptibility and outcomes of ARDS. Frontiers in genetics. 2014;5:20. doi: 10.3389/fgene.2014.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao L, Barnes KC. Recent advances in genetic predisposition to clinical acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2009 May 1;296(5):L713–725. doi: 10.1152/ajplung.90269.2008. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer NJ, Daye ZJ, Rushefski M, et al. SNP-set analysis replicates acute lung injury genetic risk factors. BMC Med Genet. 2012 Jun 28;13(1):52. doi: 10.1186/1471-2350-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Mahony DS, Glavan BJ, Holden TD, et al. Inflammation and Immune-Related Candidate Gene Associations with Acute Lung Injury Susceptibility and Severity: A Validation Study. PLoS ONE. 2012;7(12):e51104. doi: 10.1371/journal.pone.0051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores C, Ma SF, Maresso K, Wade MS, Villar J, Garcia JG. IL6 gene-wide haplotype is associated with susceptibility to acute lung injury. Transl Res. 2008 Jul;152(1):11–17. doi: 10.1016/j.trsl.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Marshall RP, Webb S, Hill MR, Humphries SE, Laurent GJ. Genetic polymorphisms associated with susceptibility and outcome in ARDS. Chest. 2002 Mar;121(3 Suppl):68S–69S. doi: 10.1378/chest.121.3_suppl.68s. [DOI] [PubMed] [Google Scholar]
- 16.Gong MN, Zhou W, Williams PL, Thompson BT, Pothier L, Christiani DC. Polymorphisms in the mannose binding lectin-2 gene and acute respiratory distress syndrome. Crit Care Med. 2007 Jan;35(1):48–56. doi: 10.1097/01.CCM.0000251132.10689.F3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong MN, Thompson BT, Williams PL, et al. Interleukin-10 polymorphism in position-1082 and acute respiratory distress syndrome. Eur Respir J. 2006 Apr;27(4):674–681. doi: 10.1183/09031936.06.00046405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer NJ, Feng R, Li M, et al. IL1RN Coding Variant Is Associated with Lower Risk of Acute Respiratory Distress Syndrome and Increased Plasma IL-1 Receptor Antagonist. Am J Respir Crit Care Med. 2013 Feb 28;187(9):950–959. doi: 10.1164/rccm.201208-1501OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medford AR, Godinho SI, Keen LJ, Bidwell JL, Millar AB. Relationship between vascular endothelial growth factor + 936 genotype and plasma/epithelial lining fluid vascular endothelial growth factor protein levels in patients with and at risk for ARDS. Chest. 2009 Aug;136(2):457–464. doi: 10.1378/chest.09-0383. [DOI] [PubMed] [Google Scholar]
- 20.Medford ARL, Millar AB. Vascular endothelial growth factor (VEGF) in acute lung injury (ALI) and acute respiratory distress syndrome (ARDS): paradox or paradigm? Thorax. 2006 Jul 1;61(7):621–626. doi: 10.1136/thx.2005.040204. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medford AR, Keen LJ, Bidwell JL, Millar AB. Vascular endothelial growth factor gene polymorphism and acute respiratory distress syndrome. Thorax. 2005 Mar;60(3):244–248. doi: 10.1136/thx.2004.034785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer NJ, Li M, Feng R, et al. ANGPT2 Genetic Variant is Associated with Trauma-Associated Acute Lung Injury and Altered Plasma Angiopoietin-2 Isoform Ratio. Am J Respir Crit Care Med. 2011 Jan 21;183:1344–1353. doi: 10.1164/rccm.201005-0701OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su L, Zhai R, Sheu CC, et al. Genetic variants in the angiopoietin-2 gene are associated with increased risk of ARDS. Intensive Care Med. 2009 Mar 7;35:1024–1030. doi: 10.1007/s00134-009-1413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall RP, Webb S, Bellingan GJ, et al. Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am J Respir Crit Care Med. 2002 Sep 1;166(5):646–650. doi: 10.1164/rccm.2108086. [DOI] [PubMed] [Google Scholar]
- 25.Pino-Yanes M, Ma SF, Sun X, et al. Interleukin-1 Receptor-associated Kinase 3 Gene Associates with Susceptibility to Acute Lung Injury. Am J Respir Cell Mol Biol. 2011 Feb 4;45(4):740–745. doi: 10.1165/rcmb.2010-0292OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pino-Yanes M, Corrales A, Casula M, et al. Common Variants of TLR1 Associate with Organ Dysfunction and Sustained Pro-Inflammatory Responses during Sepsis. PLoS ONE. 2010;5(10):e13759. doi: 10.1371/journal.pone.0013759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tejera P, O’Mahony DS, Owen CA, et al. Functional characterization of polymorphisms in the PI3 (elafin) gene and validation of their contribution to risk of ARDS. American Journal of Respiratory Cell and Molecular Biology. 2014 doi: 10.1165/rcmb.2013-0238OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tejera P, Wang Z, Zhai R, et al. Genetic Polymorphisms of Peptidase Inhibitor 3 (Elafin) Are Associated with Acute Respiratory Distress Syndrome. American Journal of Respiratory Cell and Molecular Biology. 2009 Dec 1;41(6):696–704. doi: 10.1165/rcmb.2008-0410OC. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Z, Thomas NJ, Wang Y, et al. Deletions within a CA-repeat-rich region of intron 4 of the human SP-B gene affect mRNA splicing. Biochem J. 2005 Jul 15;389(Pt 2):403–412. doi: 10.1042/BJ20042032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Z, Pearson C, Chinchilli V, et al. Polymorphisms of human SP-A, SP-B, and SP-D genes: association of SP-B Thr131Ile with ARDS. Clin Genet. 2000 Sep;58(3):181–191. doi: 10.1034/j.1399-0004.2000.580305.x. [DOI] [PubMed] [Google Scholar]
- 31.David S, Mukherjee A, Ghosh CC, et al. Angiopoietin-2 may contribute to multiple organ dysfunction and death in sepsis*. Critical Care Medicine. 2012;40(11):3034–3041. doi: 10.1097/CCM.0b013e31825fdc31. 3010.1097/CCM.3030b3013e31825fdc31831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumpers P, Gueler F, David S, et al. The synthetic Tie2 agonist peptide vasculotide protects against vascular leakage and reduces mortality in murine abdominal sepsis. Crit Care. 2011 Oct 31;15(5):R261. doi: 10.1186/cc10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.David S, Park JK, Meurs M, et al. Acute administration of recombinant Angiopoietin-1 ameliorates multiple-organ dysfunction syndrome and improves survival in murine sepsis. Cytokine. 2011 Aug;55(2):251–259. doi: 10.1016/j.cyto.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Alfieri A, Watson JJ, Kammerer RA, et al. Angiopoietin-1 variant reduces LPS-induced microvascular dysfunction in a murine model of sepsis. Crit Care. 2012 Oct 4;16(5):R182. doi: 10.1186/cc11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aikawa N, Ishizaka A, Hirasawa H, et al. Reevaluation of the efficacy and safety of the neutrophil elastase inhibitor, Sivelestat, for the treatment of acute lung injury associated with systemic inflammatory response syndrome; a phase IV study. Pulm Pharmacol Ther. 2011 Oct;24(5):549–554. doi: 10.1016/j.pupt.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Iwata K, Doi A, Ohji G, et al. Effect of neutrophil elastase inhibitor (sivelestat sodium) in the treatment of acute lung injury (ALI) and acute respiratory distress syndrome (ARDS): a systematic review and meta-analysis. Intern Med. 2010;49(22):2423–2432. doi: 10.2169/internalmedicine.49.4010. [DOI] [PubMed] [Google Scholar]
- 37.Zeiher BG, Artigas A, Vincent JL, et al. Neutrophil elastase inhibition in acute lung injury: results of the STRIVE study. Crit Care Med. 2004 Aug;32(8):1695–1702. doi: 10.1097/01.ccm.0000133332.48386.85. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Beach D, Su L, Zhai R, Christiani DC. A genome-wide expression analysis in blood identifies pre-elafin as a biomarker in ARDS. Am J Respir Cell Mol Biol. 2008 Jun;38(6):724–732. doi: 10.1165/rcmb.2007-0354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Chen F, Zhai R, et al. Plasma Neutrophil Elastase and Elafin Imbalance Is Associated with Acute Respiratory Distress Syndrome (ARDS) Development. PLoS ONE. 2009;4(2):e4380. doi: 10.1371/journal.pone.0004380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakada TA, Russell JA, Boyd JH, et al. Association of angiotensin II type 1 receptor-associated protein gene polymorphism with increased mortality in septic shock. Crit Care Med. 2011 Jul;39(7):1641–1648. doi: 10.1097/CCM.0b013e318218665a. [DOI] [PubMed] [Google Scholar]
- 41.Flores C, Pino-Yanes Mdel M, Villar J. A quality assessment of genetic association studies supporting susceptibility and outcome in acute lung injury. Crit Care. 2008;12(5):R130. doi: 10.1186/cc7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adamzik M, Frey U, Sixt S, et al. ACE I/D but not AGT (-6)A/G polymorphism is a risk factor for mortality in ARDS. Eur Respir J. 2007 Mar;29(3):482–488. doi: 10.1183/09031936.00046106. [DOI] [PubMed] [Google Scholar]
- 43.Jerng JS, Yu CJ, Wang HC, Chen KY, Cheng SL, Yang PC. Polymorphism of the angiotensinconverting enzyme gene affects the outcome of acute respiratory distress syndrome. Crit Care Med. 2006 Apr;34(4):1001–1006. doi: 10.1097/01.CCM.0000206107.92476.39. [DOI] [PubMed] [Google Scholar]
- 44.Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005 Aug;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicholls J, Peiris M. Good ACE, bad ACE do battle in lung injury, SARS. Nat Med. 2005 Aug;11(8):821–822. doi: 10.1038/nm0805-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P, Li H, Cepeda J, et al. Regulation of Inflammatory Cytokine Expression in Pulmonary Epithelial Cells by Pre-B-cell Colony-enhancing Factor via a Nonenzymatic and AP-1-dependent Mechanism. Journal of Biological Chemistry. 2009 Oct 2;284(40):27344–27351. doi: 10.1074/jbc.M109.002519. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye SQ, Simon BA, Maloney JP, et al. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med. 2005 Feb 15;171(4):361–370. doi: 10.1164/rccm.200404-563OC. [DOI] [PubMed] [Google Scholar]
- 48.Ye SQ, Zhang LQ, Adyshev D, et al. Pre-B-cell-colony-enhancing factor is critically involved in thrombin-induced lung endothelial cell barrier dysregulation. Microvasc Res. 2005 Nov;70(3):142–151. doi: 10.1016/j.mvr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Bajwa EK, Yu CJ, Gong MN, Thompson BT, Christiani DC. PBEF gene polymorphisms influence the risk of developing ARDS. Proc Am Thorac Soc. 2006;3:A272. [Google Scholar]
- 50.Leikauf GD, Concel VJ, Liu P, et al. Haplotype association mapping of acute lung injury in mice implicates activin a receptor, type 1. Am J Respir Crit Care Med. 2011 Jun 1;183(11):1499–1509. doi: 10.1164/rccm.201006-0912OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leikauf GD, Pope-Varsalona H, Concel VJ, et al. Functional genomics of chlorine-induced acute lung injury in mice. Proc Am Thorac Soc. 2010 Jul;7(4):294–296. doi: 10.1513/pats.201001-005SM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leikauf GD, Concel VJ, Bein K, et al. Functional Genomic Assessment of Phosgene-Induced Acute Lung Injury in Mice. American Journal of Respiratory Cell and Molecular Biology. 2013;49(3):368–383. doi: 10.1165/rcmb.2012-0337OC. 2013/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.dos Santos CC, Okutani D, Hu P, et al. Differential gene profiling in acute lung injury identifies injury-specific gene expression. Crit Care Med. 2008 Mar;36(3):855–865. doi: 10.1097/CCM.0B013E3181659333. [DOI] [PubMed] [Google Scholar]
- 54.Tejera P, Meyer NJ, Chen F, et al. Distinct and replicable genetic risk factors for acute respiratory distress syndrome of pulmonary or extrapulmonary origin. Journal of Medical Genetics. 2012 Oct 9; doi: 10.1136/jmedgenet-2012-100972. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christie JD, Wurfel MM, Feng R, et al. Genome Wide Association Identifies PPFIA1 as a Candidate Gene for Acute Lung Injury Risk Following Major Trauma. PLoS ONE. 2012;7(1):e28268. doi: 10.1371/journal.pone.0028268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakamoto S, Ishizaki T, Okawa K, et al. Liprin-α controls stress fiber formation by binding to mDia and regulating its membrane localization. Journal of Cell Science. 2012 Jan 1;125(1):108–120. doi: 10.1242/jcs.087411. 2012. [DOI] [PubMed] [Google Scholar]
- 57.Christie JD, Ma SF, Aplenc R, et al. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med. 2008 Oct;36(10):2794–2800. doi: 10.1097/ccm.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- 58.Gao L, Grant A, Halder I, et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol. 2006 Apr;34(4):487–495. doi: 10.1165/rcmb.2005-0404OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheu CC, Zhai R, Su L, et al. Sex-specific association of epidermal growth factor gene polymorphisms with acute respiratory distress syndrome. Eur Respir J. 2009 Mar;33(3):543–550. doi: 10.1183/09031936.00091308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calfee CS, Delucci K, Parsons PE, et al. Latent class analysis of ARDS subphenoypes: analysis of data from two randomized controlled trials. Lancet Respir Med. 2014 doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999 Oct 15;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 62.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002 Mar;1(2):133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 63.Kris MG, Johnson BE, Berry LD, et al. USing multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolff AC, Hammond MEH, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. Journal of Clinical Oncology. 2006 Jan 20;25(1):118–145. doi: 10.1200/JCO.2006.09.2775. 2006. [DOI] [PubMed] [Google Scholar]
- 65.Patel SR, Karmpaliotis D, Ayas NT, et al. The role of open-lung biopsy in ARDS. Chest. 2004 Jan;125(1):197–202. doi: 10.1378/chest.125.1.197. [DOI] [PubMed] [Google Scholar]
- 66.Feezor RJ, Baker HV, Mindrinos MN, et al. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol. Genomics. 2004;19(3):247–254. doi: 10.1152/physiolgenomics.00020.2004. 2004-01-01 00:00:00. [DOI] [PubMed] [Google Scholar]
- 67.Howrylak JA, Dolinay T, Lucht L, et al. Discovery of the gene signature for acute lung injury in patients with sepsis. Physiol. Genomics. 2009 Apr 10;37(2):133–139. doi: 10.1152/physiolgenomics.90275.2008. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lagan AL, Quinlan GJ, Mumby S, et al. Variation in iron homeostasis genes between patients with ARDS and healthy control subjects. Chest. 2008 Jun;133(6):1302–1311. doi: 10.1378/chest.07-1117. [DOI] [PubMed] [Google Scholar]
- 69.Quinlan GJ, Chen Y, Evans TW, Gutteridge JM. Iron signalling regulated directly and through oxygen: implications for sepsis and the acute respiratory distress syndrome. Clin. Sci. 2001 Feb;100(2):169–182. 2001. [PubMed] [Google Scholar]
- 70.Connelly KG, Moss M, Parsons PE, et al. Serum ferritin as a predictor of the acute respiratory distress syndrome. American Journal of Respiratory and Critical Care Medicine. 1997;155(1):21–25. doi: 10.1164/ajrccm.155.1.9001283. 1997/01/01. [DOI] [PubMed] [Google Scholar]
- 71.Marzec JM, Christie JD, Reddy SP, et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 2007 Jul;21(9):2237–2246. doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- 72.Wong HR, Cvijanovich N, Lin R, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong HR, Lindsell CJ, Pettila V, et al. A multi-biomarker-based outcome risk stratification model for adult septic shock. Crit Care Med. 2014;42(4):781–789. doi: 10.1097/CCM.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wong H, Salisbury S, Xiao Q, et al. The pediatric sepsis biomarker risk model. Critical Care. 2012;16(5):R174. doi: 10.1186/cc11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mattick JS, Makunin IV. Non-coding RNA. Human Molecular Genetics. 2006 Apr 15;15(suppl 1):R17–R29. doi: 10.1093/hmg/ddl046. 2006. [DOI] [PubMed] [Google Scholar]
- 76.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–437. doi: 10.1038/nrg3722. 06//print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mercer TR, Gerhardt DJ, Dinger ME, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotech. 2012;30(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. 12/3/ [DOI] [PubMed] [Google Scholar]
- 79.Lau NC, Lim LP, Weinstein EG, Bartel DP. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis elegans. Science. 2001 Oct 26;294(5543):858–862. doi: 10.1126/science.1065062. 2001. [DOI] [PubMed] [Google Scholar]
- 80.Lee RC, Ambros V. An Extensive Class of Small RNAs in Caenorhabditis elegans. Science. 2001 Oct 26;294(5543):862–864. doi: 10.1126/science.1065329. 2001. [DOI] [PubMed] [Google Scholar]
- 81.Adyshev DM, Moldobaeva N, Mapes B, Elangovan V, Garcia JGN. MicroRNA Regulation of Nonmuscle Myosin Light Chain Kinase Expression in Human Lung Endothelium. American Journal of Respiratory Cell and Molecular Biology. 2013;49(1):58–66. doi: 10.1165/rcmb.2012-0397OC. 2013/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adyshev DM, Elangovan VR, Moldobaeva N, Mapes B, Sun X, Garcia JGN. Mechanical Stress Induces Pre–B-cell Colony-Enhancing Factor/NAMPT Expression via Epigenetic Regulation by miR-374a and miR-568 in Human Lung Endothelium. American Journal of Respiratory Cell and Molecular Biology. 2013;50(2):409–418. doi: 10.1165/rcmb.2013-0292OC. 2014/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schrauder MG, Strick R, Schulz-Wendtland R, et al. Circulating Micro-RNAs as Potential Blood-Based Markers for Early Stage Breast Cancer Detection. PLoS ONE. 2012;7(1):e29770. doi: 10.1371/journal.pone.0029770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. 05/24/print. [DOI] [PubMed] [Google Scholar]
- 85.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. 05/24/print. [DOI] [PubMed] [Google Scholar]
- 86.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. 1/7/ [DOI] [PubMed] [Google Scholar]
- 87.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. doi: 10.1038/nrg2005. 04//print. [DOI] [PubMed] [Google Scholar]
- 88.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–1266. doi: 10.1038/nature03672. 06/30/print. [DOI] [PubMed] [Google Scholar]
- 89.ENCODE_project_consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ideker T, Thorsson V, Ranish JA, et al. Integrated Genomic and Proteomic Analyses of a Systematically Perturbed Metabolic Network. Science. 2001 May;4292(5518):929–934. doi: 10.1126/science.292.5518.929. 2001. [DOI] [PubMed] [Google Scholar]
- 92.Chen G, Gharib TG, Huang C-C, et al. Discordant Protein and mRNA Expression in Lung Adenocarcinomas. Molecular & Cellular Proteomics. 2002 Apr 1;1(4):304–313. doi: 10.1074/mcp.m200008-mcp200. 2002. [DOI] [PubMed] [Google Scholar]
- 93.Rogers S, Girolami M, Kolch W, et al. Investigating the correspondence between transcriptomic and proteomic expression profiles using coupled cluster models. Bioinformatics. 2008 Dec 15;24(24):2894–2900. doi: 10.1093/bioinformatics/btn553. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bowler RP, Duda B, Chan ED, et al. Proteomic analysis of pulmonary edema fluid and plasma in patients with acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2004 Jun;286(6):L1095–1104. doi: 10.1152/ajplung.00304.2003. [DOI] [PubMed] [Google Scholar]
- 95.Schnapp LM, Donohoe S, Chen J, et al. Mining the Acute Respiratory Distress Syndrome Proteome: Identification of the Insulin-Like Growth Factor (IGF)/IGF-Binding Protein-3 Pathway in Acute Lung Injury. The American Journal of Pathology. 2006;169(1):86–95. doi: 10.2353/ajpath.2006.050612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Torre C, Ying SX, Munson PJ, Meduri GU, Suffredini AF. Proteomic analysis of inflammatory biomarkers in bronchoalveolar lavage. Proteomics. 2006 Jul;6(13):3949–3957. doi: 10.1002/pmic.200500693. [DOI] [PubMed] [Google Scholar]
- 97.Nguyen EV, Gharib SA, Palazzo SJ, Chow Y-h, Goodlett DR, Schnapp LM. Proteomic Profiling of Bronchoalveolar Lavage Fluid in Critically Ill Patients with Ventilator-Associated Pneumonia. PLoS ONE. 2013;8(3):e58782. doi: 10.1371/journal.pone.0058782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang DW, Hayashi S, Gharib SA, et al. Proteomic and Computational Analysis of Bronchoalveolar Proteins during the Course of the Acute Respiratory Distress Syndrome. American Journal of Respiratory and Critical Care Medicine. 2008 Oct 1;178(7):701–709. doi: 10.1164/rccm.200712-1895OC. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen X, Shan Q, Jiang L, Zhu B, Xi X. Quantitative proteomic analysis by iTRAQ for identification of candidate biomarkers in plasma from acute respiratory distress syndrome patients. Biochemical and biophysical research communications. 2013 Nov 8;441(1):1–6. doi: 10.1016/j.bbrc.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 100.Nicholson JK, Foxall PJD, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H-13C NMR Spectroscopy of Human Blood Plasma. Analytical Chemistry. 1995;67(5):793–811. doi: 10.1021/ac00101a004. 1995/03/01. [DOI] [PubMed] [Google Scholar]
- 101.Dunn WB, Bailey NJC, Johnson HE. Measuring the metabolome: current analytical technologies. Analyst. 2005;130(5):606–625. doi: 10.1039/b418288j. [DOI] [PubMed] [Google Scholar]
- 102.Stringer KA, Serkova NJ, Karnovsky A, Guire K, Paine R, Standiford TJ. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma 1H-nuclear magnetic resonance quantitative metabolomics and computational analysis. American Journal of Physiology - Lung Cellular and Molecular Physiology. 2011 Jan 1;300(1):L4–L11. doi: 10.1152/ajplung.00231.2010. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seymour C, Yende S, Scott M, et al. Metabolomics in pneumonia and sepsis: an analysis of the GenIMS cohort study. Intensive Care Medicine. 2013;39(8):1423–1434. doi: 10.1007/s00134-013-2935-7. 2013/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Langley RJ, Tsalik EL, Velkinburgh JCv, et al. An Integrated Clinico-Metabolomic Model Improves Prediction of Death in Sepsis. Science Translational Medicine. 2013 Jul 24;5(195):195ra195. doi: 10.1126/scitranslmed.3005893. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rogers AJ, McGeachie M, Baron RM, et al. Metabolomic derangements are associated with mortality in critically ill adult patients. PLoS One. 2014 Jan 3;9(1):e87538. doi: 10.1371/journal.pone.0087538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mickiewicz B, Vogel HJ, Wong HR, Winston BW. Metabolomics as a Novel Approach for Early Diagnosis of Pediatric Septic Shock and its Mortality. American Journal of Respiratory and Critical Care Medicine. 2013 Mar 7; doi: 10.1164/rccm.201209-1726OC. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Serkova NJ, Standiford TJ, Stringer KA. The Emerging Field of Quantitative Blood Metabolomics for Biomarker Discovery in Critical Illnesses. American Journal of Respiratory and Critical Care Medicine. 2011 Sep 15;184(6):647–655. doi: 10.1164/rccm.201103-0474CI. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Evans CR, Karnovsky A, Kovach MA, Standiford TJ, Burant CF, Stringer KA. Untargeted LC–MS Metabolomics of Bronchoalveolar Lavage Fluid Differentiates Acute Respiratory Distress Syndrome from Health. Journal of Proteome Research. 2013;13(2):640–649. doi: 10.1021/pr4007624. 2014/02/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–428. doi: 10.1038/nature06758. 03/27/print. [DOI] [PubMed] [Google Scholar]
- 110.Schadt EE, Molony C, Chudin E, et al. Mapping the Genetic Architecture of Gene Expression in Human Liver. PLoS Biol. 2008;6(5):e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Musunuru K, Strong A, Frank-Kamenetsky M, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466(7307):714–719. doi: 10.1038/nature09266. 08/05/print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Strong A, Ding Q, Edmondson AC, et al. Hepatic sortilin regulates both apolipoprotein B secretion and LDL catabolism. The Journal of Clinical Investigation. 2012;122(8):2807–2816. doi: 10.1172/JCI63563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006 Sep 15;22(18):2291–2297. doi: 10.1093/bioinformatics/btl390. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chan SY, White K, Loscalzo J. Deciphering the molecular basis of human cardiovascular disease through network biology. Current opinion in cardiology. 2012 May;27(3):202–209. doi: 10.1097/HCO.0b013e3283515b31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482(7384):179–185. doi: 10.1038/nature10809. 02/09/print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nat Rev Genet. 2014;15(6):379–393. doi: 10.1038/nrg3734. 06//print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. The Role of the Lung Microbiome in Health and Disease. A National Heart, Lung, and Blood Institute Workshop Report. American Journal of Respiratory and Critical Care Medicine. 2013;187(12):1382–1387. doi: 10.1164/rccm.201303-0488WS. 2013/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Charlson ES, Diamond JM, Bittinger K, et al. Lung-enriched Organisms and Aberrant Bacterial and Fungal Respiratory Microbiota after Lung Transplant. American Journal of Respiratory and Critical Care Medicine. 2012;186(6):536–545. doi: 10.1164/rccm.201204-0693OC. 2012/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]