Abstract
Aim
Since whole-genome sequencing (WGS) information can have positive and negative personal utility for individuals, we examined predictors of willingness to pay (WTP) for WGS.
Patients & methods
We surveyed two independent populations: adult patients (n = 203) and college seniors (n = 980). Ordinal logistic regression models were used to characterize the relationship between predictors and WTP.
Results
Sex, age, education, income, genomic knowledge and knowing someone who had genetic testing or having had genetic testing done personally were associated with significantly higher WTP for WGS. After controlling for income and education, males were willing to pay more for WGS than females.
Conclusion
Differences in WTP may impact equity, coverage, affordability and access, and should be anticipated by public dialog about related health policy.
Keywords: access to healthcare, attitude to health, healthcare costs, human genome, personalized medicine, whole genome sequencing, willingness-to-pay
Whole-genome sequencing (WGS) involves sequencing nearly all of an individual’s genome and has multiple applications, ranging from disease screening to treatment planning and pharmacogenomic uses [1–5], ultimately aimed at improving patient outcomes. As WGS techniques continue to improve and the associated costs decline [6], it is possible that the use of WGS will become more widely adopted and integrated into routine clinical practice. WGS results can include: clinically actionable findings (treatable or preventable), findings that are not currently clinically actionable (with unclear treatment implications) and findings of uncertain significance [7]. Further, the findings can be divided into primary findings (variants in genes relevant to the diagnostic indication for which sequencing was ordered) and secondary findings (also termed incidental findings; variants in genes not relevant to a diagnostic indication for which sequencing was ordered) [8]. The volume and complexity of WGS information has both positive and negative consequences depending on individual preferences and values. WGS can also reveal pharmacogenomic findings that may be of clinical value, depending on the genes included in the panel and corresponding drug and clinical indication.
One approach to assessing the value of personalized medicine technologies such as WGS is to determine how much people are willing to give up monetarily for both health and nonhealth benefits associated with testing and treatment, estimated as ‘willingness to pay (WTP)’ [9]. WTP reflects the personal utility, or disutility where there are negative consequences. A recent nationally representative US survey found that more than half of those queried would not pay more than US$500 for actionable WGS information and one third would not pay more than US$200 for nonactionable information [10]. Marshall et al. also found that WGS information had a positive value for some but negative value for others, suggesting the importance of preferences to inform access and funding policies about WGS testing and reporting as an example of personalized medicine [10].
In addition to assessing the value of personalized medicine technologies, it is important to consider how the younger generation values these technologies as they are the future generation who are likely to be using them. In this paper, we report on the WTP for WGS from independent surveys of two different adult populations (middle-aged individuals and college seniors). We compare these two different populations to understand how differences in age, life stage and life experience impact value and access.
Patients & methods
The MedSeq Project: primary care & cardiology participants
The MedSeq Project (Brigham and Women’s Hospital at Harvard Medical School and Baylor College of Medicine), protocol published elsewhere [11], surveyed two adult populations: patients deemed generally healthy by their primary care physicians from primary care practices (40–65 years old) and patients with hypertrophic or dilated cardiomyopathy from cardiology practices (18 years and older). Participants were recruited by their physicians, who were also study participants, and randomized to standard of care, including a detailed review of their family history, or standard of care plus family history and an interpreted WGS examining over 4600 disease-associated genes. Participants completed surveys (electronically or pen and paper) after study enrollment (baseline) and after disclosure of study results, and were followed for 6 months to explore the impact of WGS on their health, healthcare utilization, emotional reactions and attitudes. This paper reports on the WTP questions for the 203 participants who completed the baseline survey prior to learning their randomization status and who responded to any of the WTP items.
Participants were directly asked about their WTP for WGS as follows:
-
If you could receive this WGS test outside of this study, how likely would you be to ask for this test if:
Your health insurance covered the cost of testing (Likert scale response options: definitely not to definitely would have testing).
You had to pay for the testing yourself (Likert scale response options: definitely not to definitely would have testing).
How much would you be willing to pay for this WGS test if you were not in this study and it was not covered by your insurance? (Free text: US dollars, measured as a continuous variable).
The baseline survey also assessed genomic knowledge using an existing scale (modified with permission from Likert to true/false response options) [12]. Demographic information, including age, sex, education level, ethnicity, annual household income and health status were collected.
College seniors
In a separate effort, senior college students (18 years or older) from University of California (UC) Berkeley and San Francisco State University, approximately 5500 and 2000, respectively, were invited to participate in an anonymous survey to investigate issues related to genetic testing. The main difference between the surveyed populations was that UC Berkeley students had previously participated in a program which explored the theme of genetics and personalized medicine which is described elsewhere [13]. A total of n = 980 students from both universities completed the survey and were included in the final analyses.
Surveys were administered electronically and participants provided informed consent before answering any questions. Students who completed the survey were entered into an optional random drawing for an iPad and/or no-cost dinner with UC Berkeley professors at a local restaurant.
Participants were asked about their willingness to have their whole genome sequenced, and their WTP (in US dollars) based on the following scenario and questions:
-
In the very near future, an individual will be able to have his/her whole-genome (DNA) sequenced in order to understand his/her entire genetic makeup. The test results each individual receives when his/her whole genome is sequenced will be a MIXTURE of:
Results for which important medical or other health related decisions CAN be made;
Results for which medical or other health related decisions CANNOT be made because they are unlikely to be useful for preventing or treating a condition BUT the results may still be of PERSONAL INTEREST;
Results of UNKNOWN importance.
Experts such as clinicians and scientists determine what findings from WGS can be used to help treat or prevent a disease.
Based on the information provided, how likely are you to have your whole genome sequenced? (Likert scale response options: very unlikely to very likely to have whole genome sequenced).
Based on the information provided, how much would you be willing to pay out of pocket for WGS? (response options: US$0–200, US$200–500, US$500–1000, US$1000–3000, >US$3000).
The survey also included a component that assessed genomic knowledge using an existing survey instrument [14]. Demographic information, including age, sex, parent’s education level, race/ethnicity, annual family household income and health status were collected.
Statistical methods
Descriptive statistics were calculated for the demographic variables. Although race/ethnicity were specified by respondents using multiple categories, in order to compare the two populations, we dichotomized race/ethnicity to non-Hispanic white or not. Categorical distributions of WTP and willingness to have WGS were calculated for the college seniors study participants. Mean WTP was calculated for the MedSeq Project participants.
Predictors of WTP identified using ordinal logistic regression for both populations. We identified a priori common variables to each dataset that we anticipated might be predictors of WTP for WGS. Categories used for the WTP outcome variable in the ordinal logistic regression analysis were: ≤US$199, US$200–499 and ≥US$500. Regression parameter estimates, standard errors (SEs) and p-values were reported for each predictor in all models. Analyses were conducted using IBM SPSS Statistics (version 24.0) and STATA (version 13.1). Results with p-values < 0.05 were considered statistically significant.
Results
The MedSeq Project: primary care & cardiology patients
Overall, mean age of participants (n = 203) was 55 years (standard deviation [SD]: 11.3) (Table 1). Participants were predominantly non-Hispanic white (88%, n = 177), the majority reported having a college degree (81%, n = 165) and an annual household income of US$100,000 or greater (61%, n = 124). In addition to reporting high levels of education, participants demonstrated strong genomic knowledge. The mean genomic knowledge score for participants at baseline was 10 out of 11 (91%) correct items and there were no significant differences in knowledge between primary care and cardiology participants. Some participants (n = 30) did not answer the WTP questions, but there were no significant demographic differences between those who responded and those who did not (p > 0.05).
Table 1.
Demographics for the MedSeq Project and college seniors.
| Characteristic | The MedSeq Project (n = 203) | College seniors (n = 980)† |
|---|---|---|
| Age (years): | ||
| Mean (SD) | 55 (11.3) | – |
| ≤21 | – | 475 (48) |
| 22–29 | n/a | 471 (48) |
| ≥30 | n/a | 23 (2) |
| ≤50 | 64 (32) | n/a |
| 51–65 | 106 (52) | n/a |
| ≥66 | 33 (16) | n/a |
|
| ||
| Sex, n (%): | ||
| Male | 101 (50) | 314 (32) |
| Female | 102 (50) | 647 (66) |
|
| ||
| Personal education level (MedSeq) and parent’s education level (college seniors), n (%): | ||
| No college degree | 38 (19) | 342 (35) |
| College degree | 165 (81) | 606 (62) |
|
| ||
| Annual household income, n (%)‡: | ||
| <US$100,000 | 71 (35) | 475 (48) |
| ≥US$100,000 | 124 (61) | 385 (39) |
|
| ||
| Race/ethnicity, n (%): | ||
| Non-Hispanic white | 177 (88) | 372 (38) |
|
| ||
| Reported previous genetic testing, n (%): | ||
| No | 144 (71) | n/a |
| Yes | 59 (29) | n/a |
|
| ||
| Know someone who had genetic testing/had genetic testing done personally, n (%): | ||
| No | n/a | 265 (27) |
| Yes | n/a | 453 (46) |
|
| ||
| Mean genomic knowledge score (SD)§ | 10 (1.2) | n/a |
|
| ||
| Mean proportion of correct genomic knowledge questions, % (SD)¶ | n/a | 85 (9.7) |
May not total 100% due to missing data.
The MedSeq Project reports annual household income and college seniors reports annual family household income.
Mean genomic knowledge score ranged from 0 to 11, with 11 representing high genomic knowledge.
Mean proportion of correct genomic knowledge questions ranged from 0 to 100%, with 100% representing 16 out of 16 genomic knowledge questions answered correctly.
n/a: Not applicable and did not include this variable due to reporting differences between the two studies; SD: Standard deviation.
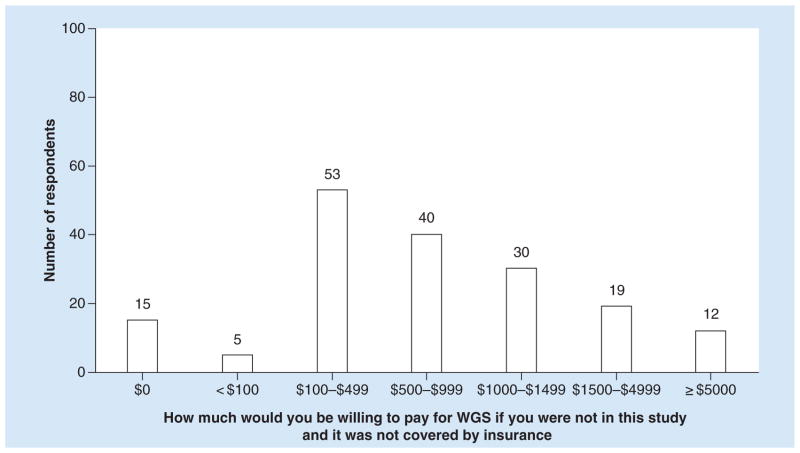
The majority of MedSeq Project participants (63%, n = 127 out of n = 201 respondents that answered the question) responded that they would definitely have testing if the cost were covered by their insurance. Participants were split on whether they would have testing if they had to self-pay, with 48% (n = 95 out of n = 200 respondents that answered the question) responding that they would probably not have testing and 31% (n = 62 out of n = 200 respondents that answered the question) responding that they probably would have testing. On average MedSeq Project participants were willing to pay US$1035 (SD: US$1707). Figure 1 summarizes the distribution of amounts that MedSeq Project respondents were willing to pay for WGS (n = 174).
Figure 1. Distribution of willingness to pay for the MedSeq Project (n = 174).
WGS: Whole-genome sequencing.
We found that sex, education, age and genomic knowledge were associated with significantly higher WTP (Table 2). Males were willing to pay more than females (p = 0.022), college graduates were willing to pay more than noncollege graduates (p = 0.045), and individuals 66 years of age and older were willing to pay more than those between 51–65 years and those 50 years of age and younger (p = 0.042 and p = 0.013, respectively). Those with better genomic knowledge were also willing to pay more (p = 0.035).
Table 2.
Ordinal logistic regression parameter estimates for predictors of willingness to pay for the MedSeq Project (n = 168) and college seniors (n = 782).
| Variables | MedSeq Project, parameter estimate (SE) | College seniors, parameter estimate (SE) |
|---|---|---|
| Age (years): | ||
| ≤21 | n/a† | – ‡ |
| 22–29 | n/a† | 0.09 (0.15) |
| ≥30 | n/a† | 0.10 (0.53) |
| ≤50 | −1.54 (0.62)* | n/a† |
| 51–65 | −1.25 (0.61)* | n/a† |
| ≥66 | –‡ | n/a† |
|
| ||
| Female | −0.76 (0.33)* | −0.32 (0.15)* |
|
| ||
| College degree | 0.87 (0.44)* | 0.04 (0.18) |
|
| ||
| Annual household income ≥US$100,000§ | 0.44 (0.37) | 0.62 (0.17)** |
|
| ||
| No previous genetic testing | −0.06 (0.37) | n/a† |
|
| ||
| Know someone who had genetic testing/had genetic testing done personally | n/a† | 0.40 (0.17)* |
|
| ||
| Genomic knowledge score (MedSeq) and mean proportion of correct genomic knowledge questions (college seniors) | 0.31 (0.15)* | −0.01 (0.01) |
Parameter estimate corresponds to an increase or decrease (depending on direction of effect) in the log odds of being in a higher level of willingness to pay. For example, a one unit increase in female (i.e., going from 0 to 1, or male to female) corresponds to a 0.76 decrease in the log odds of being in a higher level of willingness to pay for MedSeq, and 0.32 decrease in the log odds of being in a higher level of willingness to pay for college seniors, given all the other variables in the model are accounted for (held constant). Willingness to pay categories used in the ordinal logistic regression analyses were ≤US$199, US$200–499 and ≥US$500.
This variable was not included in model due to reporting differences between the two studies.
Reference group
The MedSeq Project explored annual household income, college seniors explored annual family household income.
p < 0.05.
p < 0.01.
n/a: Not applicable; SE: Standard error.
College seniors
Overall, most respondents were younger than 29 years (n = 946, 96%), with nearly half being 21 years and younger (Table 1). 38% (n = 372) of respondents reported being non-Hispanic white, having parents who are college graduates or have more than a college degree/diploma (62%, n = 606) and 39% (n = 385) reported annual family household income of US$100,000 or greater. The mean proportion of correct genomic knowledge questions was 85% (Table 1).
With regard to genetic awareness, 47% (n = 459) indicated they strongly agreed or agreed they were aware of genetic testing options for health and personalized medicine. When it came to interest in having genetic testing, 33% (n = 320) indicated they were neutral and 49% (n = 481) indicated they strongly agreed or agreed. When asked about pursuing genetic testing for themselves, 14% (n = 132) indicated they were not interested. When asked about likelihood of having their whole genome sequenced based on the scenario provided, more than half of respondents (59%, n = 578) indicated they were neutral to likely (data not shown).
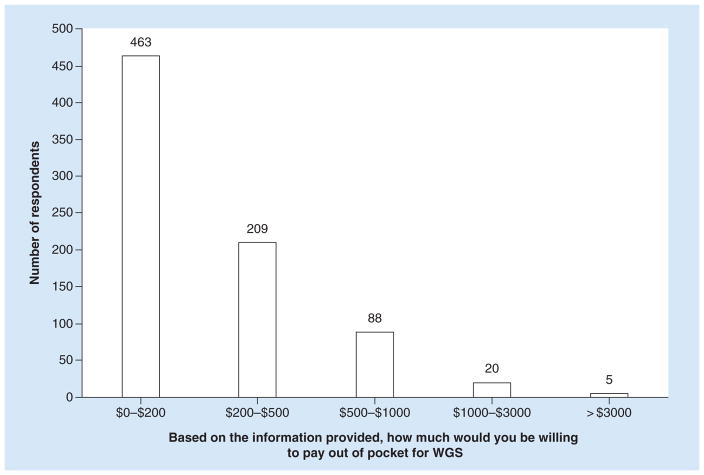
When asked about how much they would be willing to pay out of pocket for WGS, the majority of respondents (69%, n = 672) indicated they would not be willing to pay more than $500. Figure 2 summarizes the distribution of amounts that college seniors were willing to pay for WGS (n = 785). We found that sex, annual family household income and knowing someone who had genetic testing or having had genetic testing done personally were associated with significantly higher WTP (Table 2). Males were willing to pay more than females (p = 0.039), individuals who reported a family household income of US$100,000 or more were willing to pay more than those who reported a family household income of less than US$100,000 (p < 0.001), and those who knew someone who had genetic testing or had it done personally were willing to pay more than those who did or had not (p = 0.019). The college that respondents attended was not a significant predictor of WTP (p = 0.65).
Figure 2. Distribution of willingness to pay for college seniors (n = 785).
WGS: Whole-genome sequencing.
Discussion
On average, MedSeq Project participants were willing to pay US$1035 for WGS and in contrast, the majority of college seniors were not willing to pay more than US$500 for WGS. We found that various demographic characteristics including male, older age, higher education, higher family household income, better genomic knowledge and knowing someone who had genetic testing or having had genetic testing done personally were associated with a significantly higher WTP for WGS. Furthermore, even after controlling for education, income, age, genetic knowledge or experience with testing, males were willing to pay significantly more for WGS than females. Our observations around demographics are consistent with associations that have been observed with WTP around other health interventions [15]. However, in addition to the demographic factors, we found that better genomic knowledge and knowing someone who had genetic testing or having had genetic testing done personally is independently associated with significantly higher WTP. Our study explores differences in WTP for WGS between two populations that differed by age (college seniors population mostly younger than 29 years, compared with MedSeq Project population with a mean age of 55 years), life stage and life experience, which to our knowledge has not been explored in other research (Supplementary Online Table). This makes it challenging to directly compare these two populations, but also reflects a wide range of circumstances in the populations we have examined.
Other surveys in general population samples have explored individual knowledge, attitudes towards genetic testing and preferences for the receipt of genetic information, finding that men and women may differ in their views. One study of a representative sample of US adults revealed that males were more likely to have heard of personalized medicine [16]. In contrast, a Dutch survey showed that women had significantly more knowledge about genetic tests and were more likely to have had an assessment of family history to identify risks associated with hereditary conditions and risk assessment practices [17]. A recent systematic review showed mixed evidence for sex differences in attitudes and preferences toward genetic testing in relation to obesity, diabetes and heart disease [18]. Results from another systematic review suggested that women viewed genomic tests less positively than men and were less likely to pursue direct-to-consumer testing [19].
In summarizing key studies that estimated WTP for genetic/genomic testing (see Supplementary Online Table, which takes data from [20–32]), we found that individuals from the general population would be interested in genetic testing if free of charge [20,25–26], and most studies have found that few people are willing to pay more than $500 [20,25–28,32]. WTP is influenced by age, with younger individuals willing to pay more than older individuals [25]. These findings are consistent with the WTP estimates from our college seniors participants; however, our MedSeq Project participants were willing to pay more than US$500. The higher WTP estimates of MedSeq Project participants were made prior to learning their randomization status and probably reflects a combination of their older age, high income, high level of genetic knowledge and the enthusiasm for genetic testing that prompted them to voluntarily enroll in this trial of WGS.
In contrast to surveys within the general population, surveys of individuals with specific diseases (or of those at risk for specific diseases) revealed that these individuals are willing to pay for genetic testing that identifies benefit from treatment, changes in treatment or level of risk and is influenced by household income (those with higher income willing to pay more) and understanding of genetic information (Supplementary Online Table) [27,29–31]. Research by Eden et al. and Cuffe et al. found that prior knowledge or a better understanding of the condition or genetic testing led to higher WTP [27,29]. Concern about disease may also play an important role. Graves et al. reported that women with less understanding of genetic testing but more cancer worry were willing to pay more [30]. Similar to the findings with the general population, there is a wide range of estimates of what patients are willing to pay for genetic testing (US$100–13,000 but most less than US$2000). It is important to note that these studies tend to explore how genetic testing can inform or change treatment. These findings align more closely with the WTP estimate from our MedSeq Project participants.
Our results must be considered in light of certain limitations. One potential study limitation is that our populations are not representative of the general population in terms of sociodemographic factors. Another limitation is participation bias whereby those who were more interested in genetic testing might have been more likely to participate in each study than those who were not interested in the topic. The MedSeq Project population is highly knowledgeable about genomic concepts, well-educated and relatively affluent, and was also highly self-selected in that they had each agreed to participate in a sequencing study prior to being queried. The college seniors population included two different public university student populations, with an unequal number of participants from each. The San Francisco State University and UC Berkeley student populations in the current study shared important demographic similarities including sex distribution, average age, life stage and average personal education level; some differences were present including family household income and parental education level (data not shown). This combined population may not reflect the opinions of college students in general or the general population; however, it is important to note each student sample was similar to the overall demographic distribution of students from which it was drawn (data not shown).
Another potential limitation specific to the MedSeq Project population is that WTP was assessed prior to learning their randomization status or receiving WGS results, for those randomized to receive it. Receiving WGS information could impact one’s WTP. Further, both populations were assessing hypothetical WTP, which does not always reflect actual practice.
Additional research in larger, more diverse populations will be necessary to inform the future of genomics practice. Findings about WTP such as ours could be used to estimate the potential expected uptake of these personalized medicine technologies, and also to compare with observed rates of uptake in routine clinical practice over time.
Conclusion
As WGS becomes more available, these findings raise questions about access, coverage, affordability and the adoption of this personalized medicine technology into routine clinical practice. Differences in WTP in subgroups of individuals suggests that some individuals may end up with limitations on their access to WGS, which could lead to potential disparities. This type of heterogeneity, particularly by age, sex, life stage and life experience, is also important since the younger, more tech-savvy individuals are likely to be using these technologies in the future.
Our findings suggest WTP between US$0 and US$1000, coming close to the lower current cost of clinical WGS, which can range from US$1000 to US$15,000 [33]. Given the variation in coverage depending on insurance company, policies and type of testing [33], this raises concerns about access to WGS, especially for those who may not have insurance coverage. Policies on coverage and reimbursement for WGS, and personalized medicine more broadly, should be informed by public dialog and population preferences that consider the implications of access and equity.
Supplementary Material
Executive summary.
As whole genome-sequencing (WGS) techniques continue to improve and the associated costs decline, the use of WGS may become more widely adopted and integrated into routine clinical practice.
The volume and complexity of WGS information has both positive and negative consequences depending on individual preferences and values.
It is important to consider how the younger generation values these technologies as they are the future generation who are likely to be using them, which raises questions about how differences in age, life stage and life experience impact value and access.
Whether WGS can achieve its potential to improve patient outcomes will depend on what information is given to patients, and how patients and providers respond to and value the information provided.
Various demographic characteristics including male, older age, higher education, higher family household income, better genomic knowledge and knowing someone who had genetic testing or having had genetic testing done personally were associated with a significantly higher willingness to pay for WGS.
Our findings suggest willingness to pay between US$0 and US$1000, which is approaching the current cost of clinical WGS, which can range from US$1000 to US$15,000.
Policies on coverage and reimbursement for WGS, and personalized medicine more broadly, should be informed by public dialog and population preferences that consider the implications of access and equity.
Acknowledgments
The authors would like to thank J Rine (University of California, Berkeley) for his contributions to the College Seniors study.
Footnotes
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/full/10.2217/pme-2016-0075
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
The Partners Human Research Committee (Institutional Review Board for Brigham and Women’s Hospital at Harvard Medical School) and the Baylor College of Medicine Institutional Review Board approved the MedSeq Project study protocol. Institutional Review Boards at both UC Berkeley and San Francisco State University approved the study protocol.
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
KA Phillips was funded by NIH Grant (R01 HG007063) and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSFCTSI Grant Number UL1 TR000004. DA Marshall is supported by a Canada Research Chair, Health Services and Systems Research and the Arthur J.E. Child Chair in Rheumatology Outcomes Research. DA Marshall reports ad hoc consulting for Optum Insight for health economics and outcomes research. RC Green, A McGuire and JO Robinson are supported by NIH Grants U01 HG006500 and U19 HD077671. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15(4):258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl Res. 2009;154(6):277–287. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Bick D, Dimmock D. Whole exome and whole genome sequencing. Curr Opin Pediatr. 2011;23(6):594–600. doi: 10.1097/MOP.0b013e32834b20ec. [DOI] [PubMed] [Google Scholar]
- 4.Green RC, Rehm HL, Kohane IS. Chapter 9 – clinical genome sequencing. In: Ginsburg GS, Willard HF, editors. Genomic and Personalized Medicine. 2. Academic Press; London, UK: 2013. pp. 102–122. [Google Scholar]
- 5.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370(25):2418–2425. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 6.Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA. 2014;311(10):1035–1045. doi: 10.1001/jama.2014.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: Meeting the challenge one bin at a time. Genet Med. 2011;13(6):499–504. doi: 10.1097/GIM.0b013e318220aaba. Highlights key concepts of whole genome sequencing and definitions for types of results (clinically actionable, clinically valid but not directly actionable and unknown or no clinical significance) [DOI] [PubMed] [Google Scholar]
- 8.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neumann PJ, Cohen JT, Hammitt JK, et al. Willingness-to-pay for predictive tests with no immediate treatment implications: a survey of US residents. Health Econ. 2012;21(3):238–251. doi: 10.1002/hec.1704. [DOI] [PubMed] [Google Scholar]
- 10••.Marshall DA, Gonzalez JM, Johnson FR, et al. What are people willing to pay for whole-genome sequencing information, and who decides what they receive? Genet Med. 2016;18(12):1295–1302. doi: 10.1038/gim.2016.61. A recent publication of willingness to pay for whole genome sequencing in a general population sample. It explores willingness to pay for actionable and nonactionable findings. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Vassy J, Lautenbach D, Mclaughlin H, et al. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials. 2014;15(1):85. doi: 10.1186/1745-6215-15-85. Provides more details about the MedSeq Project randomized trial. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaphingst KA, Facio FM, Cheng MR, et al. Effects of informed consent for individual genome sequencing on relevant knowledge. Clin Genet. 2012;82(5):408–415. doi: 10.1111/j.1399-0004.2012.01909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helm M. Genetics and personalized medicine from the perspective of college students. Masters Sci Genet Counsel. 2015 [Google Scholar]
- 14.Haga SB, Barry WT, Mills R, et al. Public knowledge of and attitudes toward genetics and genetic testing. Genet Test Mol Biomarkers. 2013;17(4):327–335. doi: 10.1089/gtmb.2012.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noor Aizuddin A, Sulong S, Aljunid SM. Factors influencing willingness to pay for healthcare. BMC Public Health. 2012;12(2):1–1. [Google Scholar]
- 16.Garfield S, Douglas MP, MacDonald KV, Marshall DA, Phillips KA. Consumer familiarity, perspectives and expected value of personalized medicine with a focus on applications in oncology. Per Med. 2015;12(1):13–22. doi: 10.2217/pme.14.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vermeulen E, Henneman L, Van El CG, Cornel MC. Public attitudes towards preventive genomics and personal interest in genetic testing to prevent disease: a survey study. Eur J Public Health. 2014;24(5):768–775. doi: 10.1093/eurpub/ckt143. [DOI] [PubMed] [Google Scholar]
- 18.Collins J, Ryan L, Truby H. A systematic review of the factors associated with interest in predictive genetic testing for obesity, Type II diabetes and heart disease. J Hum Nutr Diet. 2014;27(5):479–488. doi: 10.1111/jhn.12179. [DOI] [PubMed] [Google Scholar]
- 19.Goldsmith L, Jackson L, O’connor A, Skirton H. Direct-to-consumer genomic testing: systematic review of the literature on user perspectives. Eur J Hum Genet. 2012;20(8):811–816. doi: 10.1038/ejhg.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosompra K, Ashikaga T, Flynn BS, Worden JK, Solomon LJ. Psychosocial factors associated with the public’s willingness to pay for genetic testing for cancer risk: a structural equations model. Health Educ Res. 2001;16(2):157–172. doi: 10.1093/her/16.2.157. [DOI] [PubMed] [Google Scholar]
- 21.Herbild L, Bech M, Gyrd-Hansen D. Estimating the Danish populations’ preferences for pharmacogenetic testing using a discrete choice experiment. The case of treating depression. Value Health. 2009;12(4):560–567. doi: 10.1111/j.1524-4733.2008.00465.x. [DOI] [PubMed] [Google Scholar]
- 22.Najafzadeh M, Johnston KM, Peacock SJ, et al. Genomic testing to determine drug response: measuring preferences of the public and patients using Discrete Choice Experiment (DCE) BMC Health Serv Res. 2013;13:454. doi: 10.1186/1472-6963-13-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geelhoed EA, Harrison K, Davey A, Walpole IR. Parental perspective of the benefits of genetic testing in children with congenital deafness. Public Health Genomics. 2009;12(4):245–250. doi: 10.1159/000203780. [DOI] [PubMed] [Google Scholar]
- 24.Regier DA, Ryan M, Phimister E, Marra CA. Bayesian and classical estimation of mixed logit: an application to genetic testing. J Health Econ. 2009;28(3):598–610. doi: 10.1016/j.jhealeco.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Ries NM, Hyde-Lay R, Caulfield T. Willingness to pay for genetic testing: a study of attitudes in a Canadian population. Public Health Genomics. 2010;13(5):292–300. doi: 10.1159/000253120. [DOI] [PubMed] [Google Scholar]
- 26.Cherkas LF, Harris JM, Levinson E, Spector TD, Prainsack B. A survey of UK public interest in internet-based personal genome testing. PLoS ONE. 2010;5(10):e13473. doi: 10.1371/journal.pone.0013473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eden M, Payne K, Combs RM, Hall G, Mcallister M, Black GC. Valuing the benefits of genetic testing for retinitis pigmentosa: a pilot application of the contingent valuation method. Br J Ophthalmol. 2013;97(8):1051–1056. doi: 10.1136/bjophthalmol-2012-303020. [DOI] [PubMed] [Google Scholar]
- 28••.Regier DA, Peacock SJ, Pataky R, et al. Societal preferences for the return of incidental findings from clinical genomic sequencing: a discrete-choice experiment. CMAJ. 2015;187(6):E190–E197. doi: 10.1503/cmaj.140697. This reference is a recent publication of willingness to pay for incidental findings from clinical genome sequencing in a general population sample. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuffe S, Hon H, Qiu X, et al. Cancer patients acceptance, understanding, and willingness-to-pay for pharmacogenomic testing. Pharmacogenet Genomics. 2014;24(7):348–355. doi: 10.1097/FPC.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 30.Graves KD, Peshkin BN, Luta G, Tuong W, Schwartz MD. Interest in genetic testing for modest changes in breast cancer risk: implications for SNP testing. Public Health Genomics. 2011;14(3):178–189. doi: 10.1159/000324703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopits IM, Chen C, Roberts JS, Uhlmann W, Green RC. Willingness to pay for genetic testing for Alzheimer’s disease: a measure of personal utility. Genet Test Mol Biomarkers. 2011;15(12):871–875. doi: 10.1089/gtmb.2011.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levitt DM. Let the consumer decide? The regulation of commercial genetic testing. J Med Ethics. 2001;27(6):398–403. doi: 10.1136/jme.27.6.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakradhar S. Insurance companies are slow to cover next-generation sequencing. Nat Med. 2015;21(3):204–205. doi: 10.1038/nm0315-204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.