Key Points
KMT2C mutations occur in 15% and 25% of patients with cHCL and vHCL, respectively, along with CCND3 and U2AF1 mutations each in 13% of vHCLs.
NF1, NF2, N/KRAS, and IRS1 alterations contribute to clinical resistance to vemurafenib treatment in patients with cHCL.
Abstract
Classical hairy cell leukemia (cHCL) is characterized by a near 100% frequency of the BRAFV600E mutation, whereas ∼30% of variant HCLs (vHCLs) have MAP2K1 mutations. However, recurrent genetic alterations cooperating with BRAFV600E or MAP2K1 mutations in HCL, as well as those in MAP2K1 wild-type vHCL, are not well defined. We therefore performed deep targeted mutational and copy number analysis of cHCL (n = 53) and vHCL (n = 8). The most common genetic alteration in cHCL apart from BRAFV600E was heterozygous loss of chromosome 7q, the minimally deleted region of which targeted wild-type BRAF, subdividing cHCL into those hemizygous versus heterozygous for the BRAFV600E mutation. In addition to CDKN1B mutations in cHCL, recurrent inactivating mutations in KMT2C (MLL3) were identified in 15% and 25% of cHCLs and vHCLs, respectively. Moreover, 13% of vHCLs harbored predicted activating mutations in CCND3. A change-of-function mutation in the splicing factor U2AF1 was also present in 13% of vHCLs. Genomic analysis of de novo vemurafenib-resistant cHCL identified a novel gain-of-function mutation in IRS1 and losses of NF1 and NF2, each of which contributed to resistance. These data provide further insight into the genetic bases of cHCL and vHCL and mechanisms of RAF inhibitor resistance encountered clinically.
Introduction
Hairy cell leukemia (HCL) comprises the clonal hematologic malignancies of classical (cHCL) and variant (vHCL). Although cHCL and vHCL share expression of CD11c and CD103, only cHCL expresses CD25, CD123, CD200, and annexin A1. Furthermore, cHCL and vHCL differ in therapeutic response and prognosis, with vHCL responding poorly to purine analogs (PAs), with a median survival less than half that of cHCL.1-5 In addition, the mutations present in each HCL subtype are distinct, with BRAFV600E mutations in ∼100% of cHCLs,6,7 whereas ∼30% of vHCLs harbor activating mutations in MAP2K1,8,9 encoding the MEK1 kinase just downstream of BRAF. These data have furthered our understanding of and therapeutic approaches for HCL; however, studies of diverse cancers marked by the BRAFV600E mutation suggest that additional alterations are frequently required for tumor initiation and/or progression in BRAFV600E-mutant cells.9-13 To this end, we recently detected coexistence of CDKN1B and BRAFV600E mutations in 16% of cHCLs.14 Currently, however, additional examples of recurrent genomic alterations that coexist with BRAFV600E or MAP2K1 mutations in HCL are not known, nor have recurrent mutations in MEK1 wild-type vHCL been identified. We therefore performed deep sequencing and copy number (CN) analysis to gain additional insights into the pathogenesis and mechanisms of therapeutic resistance in HCL.
Methods
Diagnostic bone marrow or peripheral blood mononuclear cells (MNCs) were obtained from 53 patients with cHCL (22 treatment naïve, 4 relapsed/refractory and PA refractory, and 27 relapsed/refractory) and 8 with vHCL (all relapsed/refractory) from Memorial Sloan-Kettering Cancer Center, University Hospital Heidelberg, and the Munich Leukemia Laboratory. Twenty-six of these patient cases were previously sequenced for CDKN1B alone.14 Treatment groups were defined as treatment naïve (those patients receiving no prior treatment), relapsed/refractory (patients with HCL who relapsed after therapy using a PA ≥1 year but ≤2 years after the first course or ≤4 years after a second or later course of a PA), or relapsed/refractory and PA refractory (patients with no response to PAs or any relapse ≤1 year after the initiation of PAs).15 Patient samples were collected after obtaining written informed consent. The use of human materials was approved by the institutional review boards of each institution in accordance with the Declaration of Helsinki.
Analysis of MNCs, fluorescence-activated cell sorter–purified HCL cells, and, for some cases, granulocytes (11 cHCL and 3 vHCL patient cases) was performed using the MSK-IMPACT targeted next-generation sequencing assay, which sequences all coding regions of 585 genes recurrently mutated in leukemias, lymphomas, and solid tumors (supplemental Table 1, available on the Blood Web site). Sequencing and analysis were performed as previously described.16-18 The FACETS algorithm was used to estimate tumor purity, ploidy, and allele-specific CN from sequencing data of tumor-normal pairs as previously described.18,19 Additional experimental details are described in the supplemental Methods.
Results and discussion
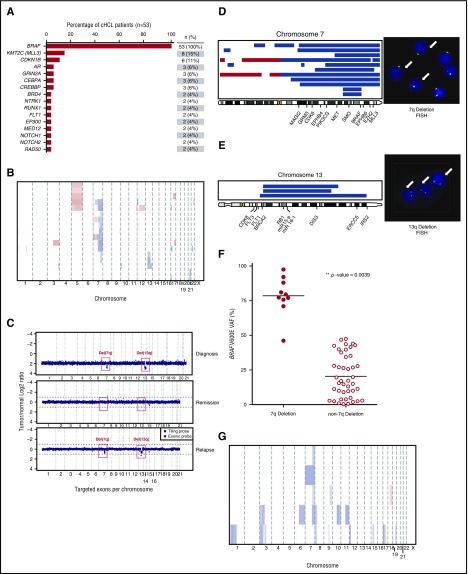
We performed targeted sequencing of 585 genes recurrently mutated in hematologic and solid tumors across 53 patients with cHCL. BRAFV600E mutation was present in 100% of patients with cHCL (Figure 1A; supplemental Figures 1 and 2; supplemental Table 2), and the next most commonly mutated genes in cHCL were the histone methyltransferase KMT2C (MLL3) and CDKN1B, occurring in 15% (8 of 53) and 11% (6 of 53) of patients, respectively. KMT2C was affected by predicted loss-of-function mutations throughout the coding region (supplemental Figure 2B). Other recurrent mutations in cHCL affected genes involved in transcriptional regulation (BRD4, CEBPA, CREBBP, RUNX1, EP300, and MED12), Notch signaling (NOTCH1 and NOTCH2), and DNA repair (RAD50; Figure 1A; supplemental Figure 1; supplemental Table 2).
Figure 1.
Genomic alterations in cHCL and vHCL. (A) Histogram of mutations in cHCL cohort (n = 53 patients) present in ≥2 patients. (B) CN analysis of the cHCL cases. Curated segmentation data for 53 cHCL samples. In the red-blue scale, white corresponds to a normal (diploid) CN log ratio, blue is a deletion, and red is a gain. (C) CN variation plots of peripheral blood MNCs from a single patient with cHCL at initiation, remission, and relapse from BRAF inhibitor treatment illustrating deletion of 7q and 13q [del(7q) and del(13q), respectively] regions at times of treatment initiation and relapse but not in disease remission. Genes mapped in the region of del(7q) with representative fluorescence in situ hybridization (FISH) with 7q deletion (white arrows; probes: red = 7q31; green = centromeric probe chromosome 7 [CEP7]) (D) and del(13q) with representative FISH with 13q14 deletion (white arrows; probes: red = 13q14; green = 13q34) (E). (F) BRAFV600E variant allele frequency (VAF) in patient cases with or without del(7q). (G) CN analysis of 8 cases of vHCL.
The most recurrent CN alterations in cHCL were deletions of chromosomes 7q and 13q and gains of chromosome 5. Chromosome 7q and 13q deletions were confirmed by fluorescence in situ hybridization (Figure 1B-E; supplemental Table 3). These CN changes were evident in peripheral blood MNCs at disease initiation and relapse but not during remission, consistent with their somatic nature in HCL cells (Figure 1C). Although recurrent 7q deletions have previously been reported in cHCL,2-4,14 additional genes in the minimally deleted region of 7q beyond BRAF included SMO (7q32) and BRAF (7q34) itself. As a result, 7q deletion resulted in loss of heterozygosity of the BRAFV600E allele (Figure 1D,F), as evidenced by the higher BRAFV600E variant allele frequency of 7q-deleted versus 7q-diploid cHCL (79% vs 22%, respectively; Wilcoxon matched-pairs signed rank test P = .0039). Recurrent 13q deletions in cHCL included the tumor suppressor RB1 and the miR-15a and miR-16-1 microRNA cluster at 13q14.3, which have been well studied in chronic lymphocytic leukemia20 (Figure 1E).
Prior genomic analyses identified MAP2K1 mutations in ∼30% of patients with vHCL, as well as individuals with mutations in CCND3, TP53, U2AF1, and ARID1A.8 Sequencing across 8 additional patients with vHCL identified change-of-function mutations in both CCND3 and U2AF1, each occurring in 13% (1 of 8) of patients with vHCL, and mutations in TP53 (38%; 3 of 8 patients; supplemental Figure 3A-B; supplemental Table 4). Of note, the reported TP53 mutations occurred at well-known hotspots, and 1 TP53 mutation (TP53 P301fs*44) was homozygous. The CCND3 and U2AF1 mutations were absent in cHCL, suggesting additional genetic differences between cHCL and vHCL. CCND3 mutations (p.R271fs; p.D286fs) are predicted to lead to loss of the PEST domain and increased expression of CCND321,22 (supplemental Figure 3C). The U2AF1 mutation identified occurred at a previously described hotspot region23 and altered the RNA splicing preferences of the protein distinct from loss of function24 (supplemental Figure 3D). These findings may have therapeutic relevance, because it is speculated that CCND3 mutations confer sensitivity to CDK4/CDK6 inhibitors,22 whereas those in U2AF1 confer sensitivity to spliceosome inhibitors.25 Other mutations affecting genes involved in transcriptional regulation (CEBPA, CREBBP, DDX3X, and PBRM1) and chromatin remodeling (KMT2C [MLL3], KDM6A, and KDM5C) were also identified (supplemental Figure 3A-B).
As with cHCL, chromosome 7q deletions were also present in vHCL (consistent with previous reports of 7q deletions in 20% of vHCLs3,4; (Figure 1G; supplemental Figure 3A; supplemental Table 5). In addition, we also identified recurrent 3p deletions in vHCL. This region includes a critical tumor suppressor locus encoding VHL, SETD2, BAP1, and PBRM1 and is commonly deleted in renal and lung carcinomas.26
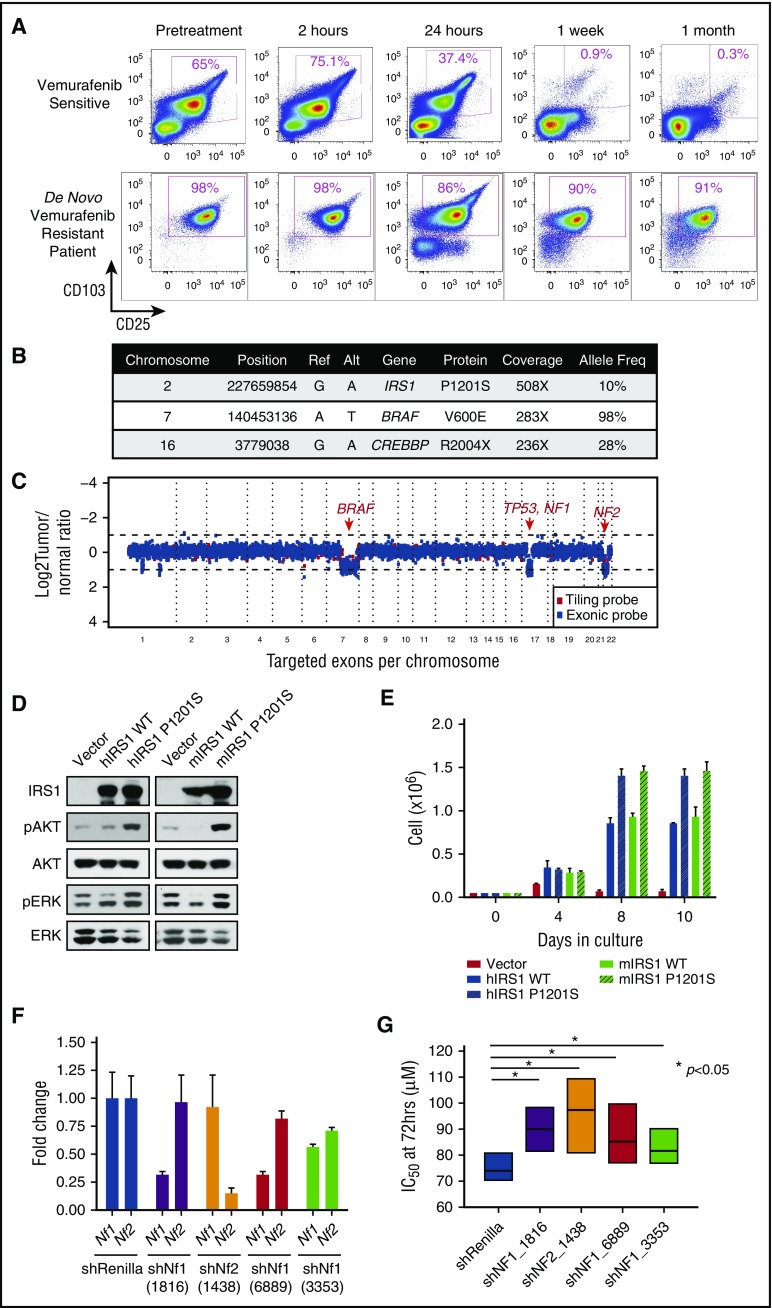
In addition to providing knowledge of disease biology and novel therapeutic targets, we also sought to understand if any of the mutations identified might affect response to therapy. For example, we previously described KRAS mutations in a patient with cHCL who developed vemurafenib resistance.15 Interestingly, here we identified an activating mutation in NRAS in a treatment-naïve patient with cHCL (supplemental Figure 2A; supplemental Table 2). Moreover, we also detected mutations in the kinases NTRK1 and FLT1 in BRAFV600E-mutant cHCL, the functional impact of which is not known. In addition, 1 patient experienced complete de novo vemurafenib resistance when treated in the phase 2 clinical trial of vemurafenib for relapsed/refractory cHCL (Figure 2A). Genomic analysis of the pretreatment sample uncovered a clonal hemizygous BRAFV600E mutation, as well as heterozygous deletions of BRAF, NF1, NF2, and TP53 and subclonal mutations in CREBBP and IRS1 (Figure 2A-C). IRS1 encodes a signaling adaptor protein that relays signals from IGF-1R to the MAPK and PI3K-AKT pathways. Stable expression of mutant IRS1P1201S activated PI3K-AKT signaling and phosphorylated ERK1/2, leading to cytokine-independent growth of Ba/F3 cells (Figure 2D-E). Both NF1 and NF2 encode tumor suppressors that have been experimentally implicated in RAF inhibitor resistance in epithelial cancer cells27,28 but have not been described in the context of cHCL or clinical RAF inhibitor resistance in hematologic malignancies. Consistent with the heterozygous deletion of NF1 and NF2, the de novo vemurafenib-resistant patient showed decreased expression of both NF1 and NF2, whereas patients without NF1 or NF2 copy loss who were responsive to vemurafenib as part of the same clinical trial did not demonstrate decreased expression of NF1/NF2 (supplemental Figure 4A-B). To understand the functional role of Nf1 and Nf2 loss and the potential contribution to RAF inhibitor resistance, we performed short hairpin RNA–mediated downregulation of Nf1 or Nf2 in Ba/F3 cells stably expressing BRAFV600E. Silencing of either Nf1 or Nf2 alone (Figure 2F-G) or concomitant downregulation of Nf1 and Nf2 simultaneously (supplemental Figure 4C-D) conferred vemurafenib resistance in vitro.
Figure 2.
Gain-of-function mutation in IRS1 and NF1/2 loss result in de novo vemurafenib resistance in cHCL. (A) Serial flow cytometric analyses of cHCL cells (CD103+/CD25+) in the peripheral blood of a patient with cHCL before vemurafenib initiation and during the first month of treatment (top), as well as a different patient before vemurafenib initiation until time of death (bottom). Mutations (B) and CN alterations (C) detected in pretreatment cHCL sample from the vemurafenib-resistant patient. (D) Western blot analysis of human (hIRS1) and mouse IRS1 (mIRS1) complementary DNA constructs in wild-type (WT) or mutant forms on AKT and ERK phosphorylation related to empty vector. (E) Cell growth of IRS1-expressing Ba/F3 cells from (D) after interleukin-3 withdrawal. (F) Quantitative reverse transcription polymerase chain reaction of Nf1 and Nf2 expression after anti-Nf1 or -Nf2 short hairpin (shRNA) knockdown. The numbers below each shRNA indicate the shRNA oligonucleotide sequence (as shown in supplemental Methods). (G) IC50, 50% inhibitory concentration (IC50) of BRAFV600E-expressing Ba/F3 cells to vemurafenib with or without knockdown of mNf1 or mNf2.
Combined, these data identify numerous novel drivers of HCL. Although activating MAPK mutations are critical for both cHCL and vHCL, our data suggest additional shared cooperating alterations, as well as disease-specific alterations targeting BRAF, KMT2C, and CDKN1B in cHCL and MAP2K1, CCND3, U2AF1, TP53, and KMT2C in vHCL. Finally, these data nominate several novel potential therapeutic approaches for vHCL and identify diverse causes for RAF inhibitor resistance through activation of signaling pathways parallel to the MAPK pathway.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
This work was supported by the American Society of Hematology Senior Research Training Award for Fellows and the New York State Council on Graduate Medical Education Empire Clinical Research Investigator Program Fellowship (B.H.D., J.T.); grants from the Hairy Cell Leukemia Foundation (O.A.-W., J.H.P.); a Mildred Scheel Professorship (Dt Krebshilfe) (T.Z.); and grants from the Edward P. Evans Foundation, the Department of Defense Bone Marrow Failure Research Program (BM150092 and W81XWH-12-1-0041), National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (R01 HL128239), an NIH National Cancer Institute K08 Clinical Investigator Award (1K08CA160647-01), the Josie Robertson Investigator Program, a Damon Runyon Clinical Investigator Award, an award from the Starr Foundation (I8-A8-075), the Leukemia and Lymphoma Society, the Pershing Square Sohn Cancer Research Alliance, and a Memorial Sloan-Kettering Cancer Center Core Grant (P30 CA008748) (O.A.-W.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: B.H.D., J.T., H.W., J.M.B., S.S., E.K., Y.R.C., S.S.C., L.W., S.X.L., C.C.O., B.S.T., M.F.B., and O.A.-W. performed the experiments and analyzed data; B.G., S.D., J.M.B., J.H., T.W., C.C.O., R.T., T.H., M.S.T., J.H.P., T.Z., and O.A.-W. collected patient material and clinical data; and B.H.D., T.Z., and O.A.-W. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thorsten Zenz, Im Neuenheimer Feld 410, 69120 Heidelberg, Germany; e-mail: thorsten.zenz@nct-heidelberg.de; and Omar Abdel-Wahab, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: abdelwao@mskcc.org.
References
- 1.Grever MR, Abdel-Wahab O, Andritsos LA, et al. Consensus guidelines for the diagnosis and management of patients with classic hairy cell leukemia. Blood. 2017;129(5):553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falini B, Martelli MP, Tiacci E. BRAF V600E mutation in hairy cell leukemia: from bench to bedside. Blood. 2016;128(15):1918-1927. [DOI] [PubMed] [Google Scholar]
- 3.Matutes E, Martínez-Trillos A, Campo E. Hairy cell leukaemia-variant: disease features and treatment. Best Pract Res Clin Haematol. 2015;28(4):253-263. [DOI] [PubMed] [Google Scholar]
- 4.Hockley SL, Morgan GJ, Leone PE, et al. High-resolution genomic profiling in hairy cell leukemia-variant compared with typical hairy cell leukemia. Leukemia. 2011;25(7):1189-1192. [DOI] [PubMed] [Google Scholar]
- 5.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Vol 2 4th ed Geneva, Switzerland: WHO Press; 2008. [Google Scholar]
- 6.Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364(24):2305-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arcaini L, Zibellini S, Boveri E, et al. The BRAF V600E mutation in hairy cell leukemia and other mature B-cell neoplasms. Blood. 2012;119(1):188-191. [DOI] [PubMed] [Google Scholar]
- 8.Waterfall JJ, Arons E, Walker RL, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46(1):8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mason EF, Brown RD, Szeto DP, et al. Detection of activating MAP2K1 mutations in atypical hairy cell leukemia and hairy cell leukemia variant. Leuk Lymphoma. 2017;58(1):233-236. [DOI] [PubMed] [Google Scholar]
- 10.Michaloglou C, Vredeveld LC, Soengas MS, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720-724. [DOI] [PubMed] [Google Scholar]
- 11.Sviderskaya EV, Hill SP, Evans-Whipp TJ, et al. p16(Ink4a) in melanocyte senescence and differentiation. J Natl Cancer Inst. 2002;94(6):446-454. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich S, Hüllein J, Lee SC, et al. Recurrent CDKN1B (p27) mutations in hairy cell leukemia. Blood. 2015;126(8):1005-1008. [DOI] [PubMed] [Google Scholar]
- 15.Tiacci E, Park JH, De Carolis L, et al. Targeting mutant BRAF in relapsed or refractory hairy-cell leukemia. N Engl J Med. 2015;373(18):1733-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients [published correction appears in Nat Med. 2017;23(8):1004]. Nat Med. 2017;23(6):703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross DS, Zehir A, Cheng DT, et al. Next-generation assessment of human epidermal growth factor receptor 2 (ERBB2) amplification status: clinical validation in the context of a hybrid capture-based, comprehensive solid tumor genomic profiling assay. J Mol Diagn. 2017;19(2):244-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cutrona G, Matis S, Colombo M, et al. Effects of miRNA-15 and miRNA-16 expression replacement in chronic lymphocytic leukemia: implication for therapy [published online ahead of print 3 February 2017]. Leukemia. doi:10.1038/leu.2016.394. [DOI] [PubMed] [Google Scholar]
- 21.Curiel-Olmo S, Mondéjar R, Almaraz C, et al. Splenic diffuse red pulp small B-cell lymphoma displays increased expression of cyclin D3 and recurrent CCND3 mutations. Blood. 2017;129(8):1042-1045. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. [DOI] [PubMed] [Google Scholar]
- 24.Ilagan JO, Ramakrishnan A, Hayes B, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25(1):14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirai CL, White BS, Tripathi M, et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat Commun. 2017;8:14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagenkord JM, Gatalica Z, Jonasch E, Monzon FA. Clinical genomics of renal epithelial tumors. Cancer Genet. 2011;204(6):285-297. [DOI] [PubMed] [Google Scholar]
- 27.Whittaker SR, Theurillat JP, Van Allen E, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3(3):350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.