Abstract
The tumor suppressor TP53 is the most frequently mutated gene product in human cancer. Close to half of all solid tumors carry inactivating mutations in the TP53 gene, while in the remaining cases, TP53 activity is abrogated by other oncogenic events, such as hyperactivation of its endogenous repressors MDM2 or MDM4. Despite identification of hundreds of genes regulated by this transcription factor, it remains unclear which direct target genes and downstream pathways are essential for the tumor suppressive function of TP53. We set out to address this problem by generating multiple genomic data sets for three different cancer cell lines, allowing the identification of distinct sets of TP53-regulated genes, from early transcriptional targets through to late targets controlled at the translational level. We found that although TP53 elicits vastly divergent signaling cascades across cell lines, it directly activates a core transcriptional program of ∼100 genes with diverse biological functions, regardless of cell type or cellular response to TP53 activation. This core program is associated with high-occupancy TP53 enhancers, high levels of paused RNA polymerases, and accessible chromatin. Interestingly, two different shRNA screens failed to identify a single TP53 target gene required for the anti-proliferative effects of TP53 during pharmacological activation in vitro. Furthermore, bioinformatics analysis of thousands of cancer genomes revealed that none of these core target genes are frequently inactivated in tumors expressing wild-type TP53. These results support the hypothesis that TP53 activates a genetically robust transcriptional program with highly distributed tumor suppressive functions acting in diverse cellular contexts.
The transcription factor TP53 is the most commonly inactivated gene product in cancer (Lawrence et al. 2014). Point mutations in the TP53 gene inactivate its tumor suppressor function and often confer the mutant protein with oncogenic properties (Freed-Pastor and Prives 2012; Hainaut and Pfeifer 2016). In the remainder of tumors that express the wild-type protein, TP53 activity is repressed by alternative means, such as hyperactivation of its repressors MDM2 and MDM4 (Leach et al. 1993; Riemenschneider et al. 1999). In the absence of cellular stress, basal TP53 activity is repressed by several mechanisms, including masking of its transactivation domains, proteasomal degradation, and reduced mRNA translation (Leach et al. 1993; Kubbutat et al. 1997; Takagi et al. 2005). In response to myriad stress stimuli, these repressive mechanisms are relieved, leading to unmasking of the TP53 transactivation domains, increased TP53 protein levels, and subsequent transactivation of TP53 target genes (Vousden and Prives 2009).
Despite intensive research efforts, the exact mechanisms by which TP53 prevents cancer development remain unclear. Over more than three decades of research, TP53 was shown to elicit multiple cellular responses that could prevent tumor progression, including cell cycle arrest, senescence, apoptosis, and DNA repair (Vousden and Prives 2009). However, recent investigations using mouse models concluded that major effector pathways, such as cell cycle arrest and apoptosis, are dispensable for TP53 tumor suppression under some conditions (Brady et al. 2011; Li et al. 2012b; Valente et al. 2013). These observations triggered a flurry of activity to identify new TP53 target genes involved in tumor suppression. Several studies employed a combination of chromatin occupancy assays and steady-state RNA expression measurements to generate lists of genes that are bound by TP53 within arbitrary distances from their promoters and that show changes in mRNA expression at various times after TP53 activation (Wei et al. 2006; Li et al. 2012a; Nikulenkov et al. 2012; Kenzelmann Broz et al. 2013; Menendez et al. 2013; Schlereth et al. 2013; Wang et al. 2014). However, independent meta-analyses revealed very little overlap among the catalogs of TP53 target genes obtained by these different research teams (Allen et al. 2014; Verfaillie et al. 2016; Fischer 2017). Given the fact that these investigations employed different cell types, it is possible that the lack of conservation is due to cell-type–specific configurations of the TP53 transcriptional program. Therefore, there is a clear need in the field to define the true extent to which the TP53 signaling cascade varies across different cell types and to define the contribution of both core and cell-type–specific gene expression programs to tumor suppression.
Here, we report a comprehensive characterization of the TP53 signaling cascade in three cancer cell types of different origins in response to TP53 activation upon pharmacological inhibition of MDM2 with Nutlin-3. We measured TP53 chromatin binding by ChIP-seq, activity of RNA polymerases by GRO-seq, and both total and polysome-associated mRNAs by RNA-seq. Using these data, we identified distinct sets of genes whose expression is affected by TP53 activation, including early direct transcriptional targets, late targets with and without proximal TP53 binding, and genes subject to translational control upon TP53 activation. Our analyses revealed massive differences in the TP53 signaling cascade across cell types at multiple regulatory steps along the central dogma. However, we also uncovered a conserved core program composed of ∼100 direct target genes involved in diverse cellular pathways with potential tumor suppressive activities. Using shRNA screens to measure effects on cell proliferation and survival, with either a library targeting direct TP53 targets or a genome-wide library, we found that TP53 itself was the only gene whose inactivation confers a significant selective advantage during Nutlin-3 treatment. Furthermore, these in vitro screening experiments led to the rapid selection for mutations in the TP53 gene itself. Genomic analysis of thousands of patient samples across different cancer types revealed that none of the core target genes are individually inactivated at a high frequency in TP53 wild-type tumors. Altogether, our results demonstrate the existence of a core set of genes, directly activated by TP53 in widely different cellular contexts, with highly distributed tumor suppressive functions.
Results
Multi-omics analysis of the TP53 signaling cascade
To comprehensively define the signaling cascade initiated by TP53, we employed a multi-omics experimental pipeline to identify genes whose expression is regulated upon TP53 activation at various steps along the central dogma, from direct transcriptional targets to genes regulated at the translational level. We activated TP53 specifically with Nutlin-3, a small molecule inhibitor of the TP53-MDM2 interaction (Vassilev et al. 2004), which induces a global pattern of TP53 binding to chromatin similar to that induced by DNA damage (Verfaillie et al. 2016). We then measured genome-wide patterns of TP53 chromatin binding via ChIP-seq, direct TP53 transactivation of RNA polymerases via GRO-seq, and global impacts of TP53 activation on both mRNA abundance and translation by sequencing of total and polysomal RNA, respectively (Fig. 1A; Supplemental Files S1–S4).
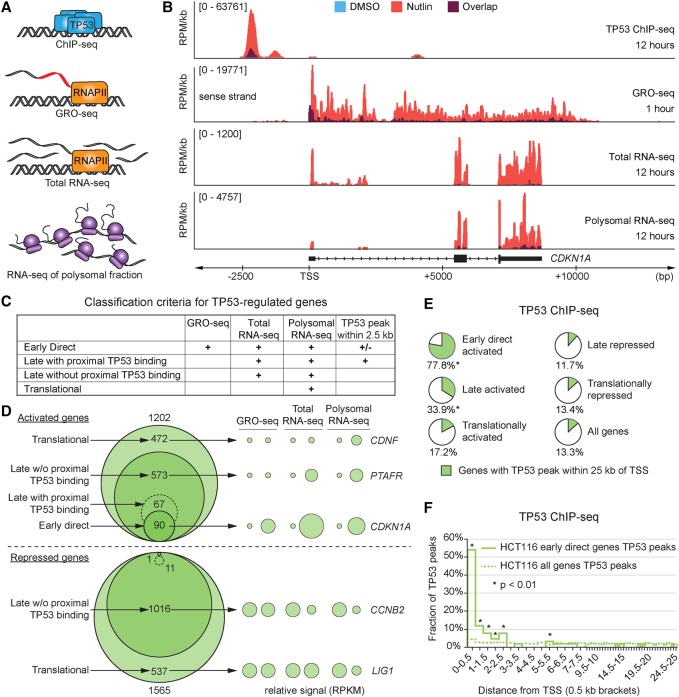
Figure 1.
Multidimensional analysis of the TP53 network. (A) Schematic of the four data sets generated for identification of different classes of TP53-regulated genes. (B) Genome browser snapshots of the CDKN1A locus. HCT116 cells were treated with 10 µM Nutlin-3 or 0.2% DMSO (vehicle) for indicated times. (RPM/kb) Reads per million per kilobase, (TSS) transcription start site. (C) Summary of criteria used for classification of genes within the TP53 network. (D) Venn diagrams display the number of genes in each class following TP53 activation in HCT116 cells. Bubble plots show relative RPKM signals derived from GRO-seq and RNA-seq experiments for example genes regulated at different steps of the central dogma. See Supplemental Methods for details. (E) Pie charts show fractions of genes with a TP53 binding peak within 25 kb of the TSS in HCT116 cells. Asterisks indicate fractions where P < 0.01, using a χ2 test with Yates’ correction, compared to the “All genes” group. (F) Distribution of TP53 peaks within 25 kb of the TSS in 0.5-kb bins. Statistics: χ2 with Yates’ correction, P < 0.01. See also Supplemental Figure S1 and Supplemental Files S1–S5.
We first employed this approach in HCT116 cells, a colorectal carcinoma cell line with a well-characterized wild-type TP53 response. The quality of these data sets is illustrated for the CDKN1A locus that encodes the cell cycle inhibitor CDKN1A (p21) (Fig. 1B), a canonical TP53 target gene (el-Deiry et al. 1993). ChIP-seq effectively identified TP53 binding at known upstream enhancers at −2.4 and −1.3 kb (el-Deiry et al. 1993); GRO-seq detected rapid up-regulation of RNA polymerase activity 1 h after Nutlin-3 treatment, even before obvious TP53 protein accumulation (Supplemental Fig. S1A); and RNA-seq detected increased levels of total and polysome-bound mRNA at 12 h (Fig. 1B), by which time TP53 and CDKN1A protein accumulation is apparent by Western blot (Supplemental Fig. S1A). The specificity of Nutlin-3 for TP53 activation is demonstrated by the negligible changes in the global transcriptome detected in HCT116 TP53−/− cells (Supplemental Fig. S1B).
Using these data sets, we next defined four classes of genes regulated by TP53 (Fig. 1C; Supplemental File S5). We focused on genes corresponding to mRNAs with differential enrichment in polysomal fractions, as these are most likely to be translated and thus functionally relevant to TP53-dependent tumor suppression. First, among the 1202 mRNAs with increased levels at polysomes, 472 did not show a concurrent significant increase in total steady-state mRNA levels. We refer to these genes as “translational targets” (e.g., CDNF) (Fig. 1D). Second, of the remaining 730 mRNAs showing up-regulation in both total and polysome-associated mRNA levels at 12 h, 90 were transcriptionally induced at the 1-h time point as seen by GRO-seq. We refer to these as “early direct targets” (e.g., CDKN1A) (Fig. 1D). ChIP-seq confirmed that most early direct targets (77.8%) have a TP53 binding event within 25 kb of their transcriptional start site (TSS), a much greater fraction than for all genes (13.3%) (Fig. 1E). Third, we noted that the frequency of TP53 binding within 25 kb was also higher for many other genes up-regulated at the total and polysomal levels (33.9%), suggesting that GRO-seq analysis at 1 h may have excluded some direct targets activated at a later time point. Alternatively, differences in the sensitivity and statistical analysis of GRO-seq versus RNA-seq could explain why GRO-seq would not detect all direct targets. To partly address this issue, we analyzed the spatial distribution of TP53 binding events near the early direct targets, which showed that the frequency of TP53 peaks is significantly higher relative to the genome average only within 2.5 kb of the TSS (Fig. 1F). We therefore employed this 2.5-kb window to define “late targets with proximal TP53 binding,” which are likely enriched for additional direct transcriptional targets (63 genes) (Fig. 1D), and “late targets without proximal TP53 binding,” which are likely enriched for indirect transcriptional targets (573 genes, e.g., PTAFR) (Fig. 1D).
A similar exercise was performed for down-regulated RNAs, which led to identification of 537 “translationally repressed” genes and 1027 “late repression targets” (Fig. 1D). Analysis of TP53 binding revealed that neither of these two classes display a significantly higher fraction of TP53 occupancy within 25 kb of the TSS relative to all genes (Fig. 1E), with only 11 repressed genes having a TP53 binding peak within 2.5 kb of their TSS. This indicates that gene repression downstream from TP53 activation is almost exclusively indirect. Furthermore, GRO-seq identified only one gene (MCM5) whose lower RNA levels could be explained by decreased transcription.
Together, these results indicate that TP53 activation by Nutlin-3 leads to early direct transactivation of a relatively small number of genes, followed by a later wave of direct transactivation, and subsequent indirect effects across the transcriptome including activation, repression, and translational control.
Cell-type–specific differences expand along the TP53 signaling cascade
Next, we investigated the extent of conservation and cell-type specificity of the TP53 signaling cascade by analyzing two additional cancer cell lines expressing wild-type TP53: MCF7 breast carcinoma cells and SJSA osteosarcoma cells. To provide a stringent test of TP53 network conservation and to maximize our ability to identify cell-type–specific TP53 programs, we chose cell types of different origins from HCT116, with different mutational spectra (Supplemental Table S1 in Supplemental Material) and distinct responses to Nutlin-3 (Fig. 2A,B). Whereas all three cell types undergo cell cycle arrest at early time points of Nutlin-3 treatment (Supplemental Fig. S2A), they display varying degrees of apoptosis upon prolonged treatment, with SJSA cells showing the greatest loss of viability and highest apoptotic index (Fig. 2A). Furthermore, although the three cell types show equivalent TP53 stabilization and CDKN1A accumulation at 48 h, only SJSA cells show CASP3 activation, a measure of commitment to apoptosis (Fig. 2B). Note that SJSA cells harbor an MDM2 amplification, which results in lower basal TP53 protein levels relative to HCT116 (high basal TP53) and MCF7 (intermediate basal TP53) cells (Supplemental Fig. S2B).
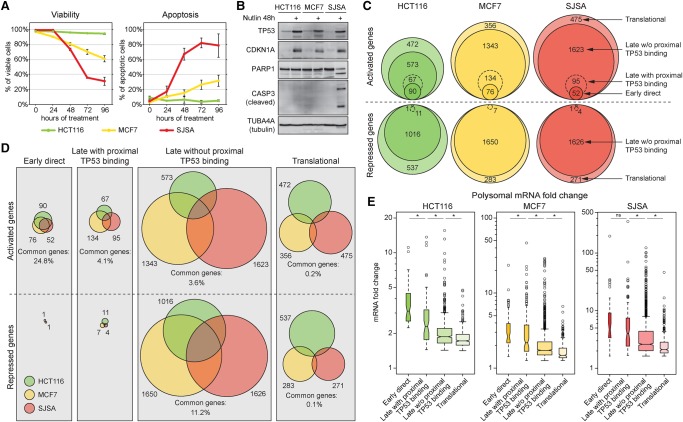
Figure 2.
Cell-type–specific configurations of the TP53 signaling cascade. (A) Flow cytometric analysis of cellular viability (propidium iodide exclusion assay) and apoptosis (Annexin-V staining). Data points represent mean ± SD of three independent experiments. (B) Western blots showing TP53 and CDKN1A accumulation in HCT116, MCF7, and SJSA cells treated with 10 µM Nutlin-3 for 48 h. Only SJSA cells show cleavage of PARP and CASP3. (C,D) Venn diagrams illustrate fractions of genes up- or down-regulated in response to TP53 activation in each cell type. (E) Box plots showing fold change distributions of polysome-associated mRNAs across each cell line for the indicated categories. Statistics: Mann-Whitney U test, P < 0.01. See also Supplemental Figures S2 and S3, and Supplemental Files S1–S5.
Our ChIP-seq and RNA-seq data are in agreement with recent meta-analyses of genomics data sets showing that TP53 binding is more conserved than mRNA signatures across different cell types (Supplemental Fig. S2C,D; Verfaillie et al. 2016; Fischer 2017), which indicates that much of the cell-type–specific regulation observed is not defined by TP53 binding. For example, whereas BBC3 and MDM2 are common direct targets associated with TP53 binding events in all three cell lines (Supplemental Fig. S3A), early direct targets such as DDIT4, YBX3, NYNRIN, ZNF488, TRIM22, and PRKY show cell-type–specific transactivation that cannot be explained by differential TP53 binding (Supplemental Fig. S3B–D). Our GRO-seq and polysomal RNA-seq data show that signaling differences expand along the central dogma (Supplemental Fig. S2E,F). Because our data sets were generated and analyzed in parallel under identical conditions, they reveal the massive impact of cell-type–specific regulatory events acting subsequent to TP53 binding to chromatin. To confirm that these differences were not due to our choice of statistical and fold change cut-offs, we generated heat maps ranking each set of cell-type–specific ChIP peaks or genes according to the FDR-adjusted P-value, which showed that there are clear cell-type–specific differences in mean fold change across the three lines (Supplemental Fig. S2G). Furthermore, Spearman rank correlation analysis shows that the highest correlations are among the peaks or genes that are common across all three cell lines, while each of the cell-type–specific sets generally have lower correlations between cell lines (Supplemental Fig. S2H). This confirms that our intersection-based comparison of peaks and genes across cell lines reveals true qualitative and quantitative cell-type–specific differences in TP53 signaling.
Next, we classified genes regulated by TP53 in MCF7 and SJSA cells using the same criteria as for HCT116 cells. Interestingly, all three cell lines display similar overall configurations of the TP53 signaling cascade: a small number of early direct targets; larger numbers of late activated and repressed targets; and an intermediate number of translationally regulated genes (Fig. 2C). The patterns of TP53 binding to each gene class are conserved among cell types, with ∼80% of early direct targets having a binding event within 25 kb of the TSS (Supplemental Fig. S2I) and the highest fraction within 2.5 kb of the TSS (Supplemental Fig. S2J). The hallmarks of indirect gene repression are also conserved in MCF7 and SJSA cells, where a nonsignificant fraction of down-regulated genes are proximally bound by TP53 (Fig. 2C,D; Supplemental Fig. S2I). Of note, relative to HCT116, the number of early direct genes is lower in MCF7 and SJSA cells (76 and 52 genes, respectively) (Fig. 2C), which could be explained by the varying degree of basal TP53 available for immediate transactivation upon MDM2 inhibition (Supplemental Fig. S2B).
Analysis of the degree of cell-type specificity at each stage of the TP53 signaling cascade revealed that conservation is greatest among the early direct genes, with increasingly larger differences among the late and translationally regulated targets (Fig. 2D). For example, the three-way overlap in early direct targets is much greater (24.8%) than the overlap in the translationally activated targets (0.2%) (Fig. 2D). Thus, as the TP53 signaling cascade propagates from the nucleus to the ribosomes, the modifying effects of cell-type–specific regulators become increasingly evident. Of note, the late repression programs show more conservation (11.2%) than the late activation programs (3.6%–4.1%) (Fig. 2D), which could be explained by the fact that the repression programs are dominated by mRNAs associated with cell proliferation (Supplemental Fig. S2K; Allen et al. 2014; Fischer et al. 2014; Fischer 2017). We also observed a high degree of conservation among long noncoding RNAs (lncRNAs) regulated by TP53 in each cell line (Supplemental Fig. S2L; Supplemental File S1). Finally, we noted that the impacts of the early direct, late, and translational programs are quantitatively different at the polysomal RNA level, whereby early direct targets display significantly greater Nutlin-3-induced changes than the late activated and translationally activated genes (Fig. 2E).
In sum, these results reveal a common pattern of the TP53 signaling cascade across cell types, defined by a small number of direct transactivation effects leading to a much greater number of indirect effects, with massive expansion of cell-type–specific regulation at each subsequent step of the central dogma.
Early direct and late targets display differences in TP53 binding, RNA polymerase activity, and chromatin environment
Next, we sought to identify potential mechanisms that might explain differences in temporal activation between early direct targets and late targets with proximal binding, which are likely enriched for additional direct targets. First, we hypothesized that early direct targets are regulated by enhancers with greater TP53 occupancy. Indeed, in all three cell types, TP53 peaks associated with early direct targets display stronger enrichment relative to late targets with proximal binding, which in turn have stronger signals relative to all other TP53 peaks across the genome (Fig. 3A). However, TP53 enrichment is not an absolute discriminator, with many early targets being associated with low occupancy sites and vice versa (see also box and whisker plots in Supplemental Fig. S4A). Interestingly, TP53 peaks associated with the early direct targets are significantly closer to the TSS than those associated with late targets with proximal binding (Supplemental Fig. S4B). Next, we investigated if these enhancers exhibit differences in transcriptional activity, as defined by the production of enhancer-derived RNAs (eRNAs). Indeed, GRO-seq revealed that the TP53-responsive enhancers associated with early targets produce more eRNAs, both before and after Nutlin-3 treatment (Fig. 3B; Supplemental Fig. S4C). In turn, late targets with proximal binding produce more eRNAs on average than all other TP53 binding events across the genome.
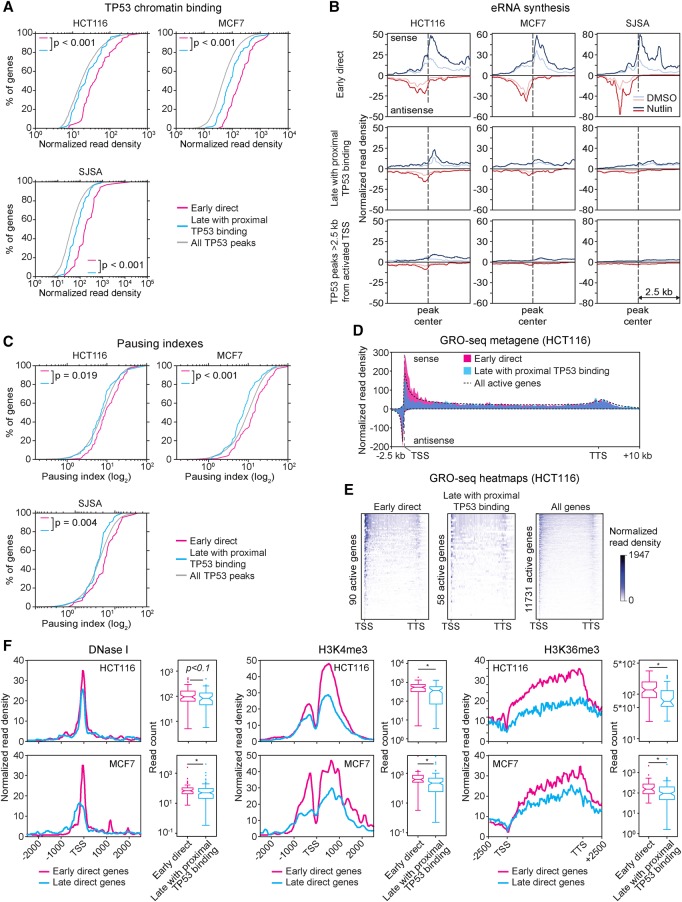
Figure 3.
Early and late TP53 target genes display distinct regulatory features. (A) Cumulative distribution plots of TP53 ChIP-seq peak read densities for TP53 binding events within 25 kb of the transcription start site of early direct targets (magenta), late targets with proximal binding (turquoise), and all TP53 peaks across the genome (gray). Statistics: Mann-Whitney U test. (B) Metaprofiles of GRO-seq signals within 2.5 kb from the TP53 ChIP-seq peaks associated with genes in the indicated categories. Normalized read density represents GRO-seq reads per bp per 10 million mapped reads. (C) Cumulative distribution plots of RNA polymerase pausing indexes for transcriptionally active genes in the indicated categories in HCT116 cells. Statistics: Mann-Whitney U test. (D) Metagene profiles showing GRO-seq signal distribution at direct early versus late targets with proximal binding, in comparison to all transcriptionally active genes in vehicle (DMSO)-treated HCT116 cells. Normalized read density represents GRO-seq reads per bp per 10 million mapped reads, adjusted for gene length. (E) Heat maps displaying GRO-seq signals (sense strand only) at active early direct targets, late targets with proximal binding, and all active genes in vehicle-treated HCT116 cells. Normalized read density represents GRO-seq reads per bp per 10 million mapped reads, adjusted for gene length. (F) Metagene profiles and box and whisker plots of DNase I sensitivity, H3K4 trimethylation, and H3K36 trimethylation at early direct targets and late targets with proximal TP53 binding. Data obtained from the ENCODE project. Statistics: Mann-Whitney U test, P < 0.01. See also Supplemental Figure S4.
Although the idea of RNA polymerase II holoenzyme pausing as a prerequisite for rapid gene activation is a subject of debate (Adelman et al. 2009; Lin et al. 2011), we tested for differences in pausing between early direct and late TP53 targets with proximal binding. Early direct targets tend to display higher “pausing indexes,” indicative of greater occupancy by paused RNA polymerases at their promoters (Fig. 3C–E). Furthermore, analysis of ENCODE data for HCT116 and MCF7 cells shows that the TSSs of early direct genes are more sensitive to DNase I digestion and more strongly decorated with histone H3 lysine 4 trimethylation (H3K4me3) (Fig. 3F), two chromatin features that have been associated with rapid activation upon transcription factor binding (Guertin and Lis 2013). We also observed high levels of H3K36me3 within the gene body of early targets, a sign of higher basal transcriptional activity (Fig. 3F).
Altogether, these results indicate that early direct genes reside in chromatin environments that are more permissive to TP53 binding and/or transcriptional activation and suggest the existence of regulatory mechanisms that could influence the timing of activation among TP53 target genes in a cell-type–specific fashion.
TP53 induces a core program of functionally diverse target genes across cell types
We reasoned that because TP53 exerts tumor suppressive activity in a wide variety of tissues, this important biological function could be mediated by a shared core set of TP53 target genes induced regardless of cellular context. We define this core program as genes that are early direct targets or late targets with proximal binding in at least one cell line and induced by Nutlin-3 (total RNA-seq FDR adjusted P-value < 0.05) in all three cell lines (103 genes in total) (Fig. 4A; Supplemental File S6). Of note, 75 of the 103 genes were detected by GRO-seq in at least one cell line, ∼20% were classified as early direct in one cell type but late target with proximal binding in another cell type (23 genes), and another ∼20% fell within the late target without a proximal binding category in one cell type (24 genes). This last group would include direct targets regulated by distal (>2.5 kb) enhancers.
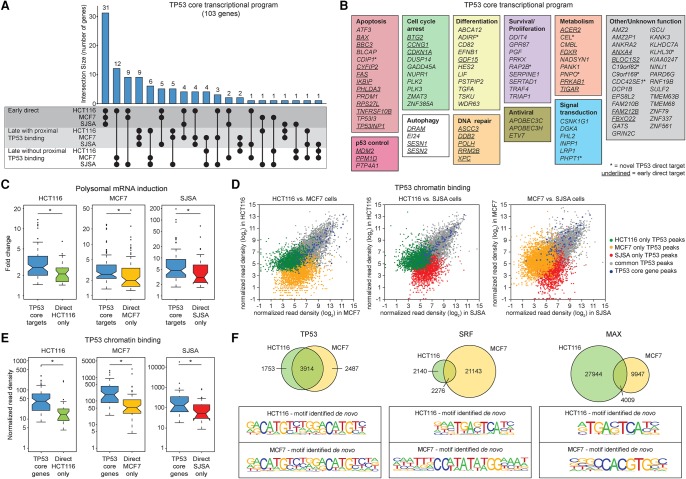
Figure 4.
Identification of a core TP53 transcriptional program. (A) Upset plot showing conserved core TP53 target genes derived from the early direct and late target genes with and without proximal binding identified in HCT116, MCF7, and SJSA cells. (B) The core TP53 transcriptional program is composed of genes functioning in multiple effector pathways and genes of unknown function. Early direct target genes are underlined, previously unidentified targets are indicated with asterisks. (C) mRNA fold changes of core TP53 target genes versus cell-type–specific direct TP53 target genes (early direct and late targets with proximal binding combined). Statistics: Mann-Whitney U test, P < 0.01. (D) Scatter plots displaying normalized read densities for each TP53 ChIP-seq peak detected, for each pairwise comparison between cell lines. (E) Box and whisker plots showing normalized ChIP-seq read densities for TP53 peaks within 25 kb of the transcription start site of core TP53 target genes, compared to TP53 peaks associated with cell line-specific direct target genes. Statistics: Mann-Whitney U test, P < 0.01. (F) Venn diagrams comparing unique and overlapping (within a 100-bp window) transcription factor binding events in HCT116 versus MCF7 cells for TP53, SRF, and MAX. Bottom panels show the position weight matrices identified during de novo motif discovery for each cell line. See also Supplemental Figure S5 and Supplemental Files S6 and S7.
Importantly, the core TP53 program includes genes involved in all major known effector pathways including cell cycle arrest, senescence, differentiation, DNA repair, metabolic control, and autophagy (Fig. 4B). Although the three cell types exhibit varying degrees of apoptosis in response to Nutlin-3 treatment, the core program includes many prominent pro-apoptotic genes (e.g., BAX, BBC3). This indicates that the core program alone is insufficient to specify cell fate choice in response to Nutlin-3. Even the core early direct program is multifunctional (Fig. 4B, underlined), suggesting that cell fate choice is not simply defined by temporal differences in the activation of cell cycle arrest versus apoptotic genes. Of note, the core program contains multiple genes per effector pathway (e.g., 10 genes involved in cell cycle arrest, 14 genes involved in apoptosis), which may allow for cooperative, distributed, and redundant actions of these genes, as well as 10 genes not previously identified as direct TP53 targets (Fig. 4B, asterisks; Supplemental File S6). Analysis of previously published data sets revealed that most genes in the core program are also induced by TP53 in additional cancer cell lines using different TP53 activating stimuli (Supplemental File S7), as well as in untransformed human cell lines (Supplemental Fig. S5B).
We next asked if there are any differences between core and cell-type–specific genes that might reflect or influence their regulation by TP53 across cell types. We noticed that the core program displays stronger induction of polysome-associated mRNAs relative to cell-type–specific direct targets (Fig. 4C), suggesting that the core program may have a greater impact on the proteome than cell-type–specific targets. Comparison of TP53 binding sites across the three cell types revealed conserved binding to high occupancy sites, while cell-type–specific peaks largely display lower occupancy (Fig. 4D). The low occupancy peaks are not likely to be ChIP-seq artifacts, as essentially all show increases in TP53 binding upon Nutlin-3 treatment (Supplemental Fig. S5C) and >80% contain known TP53 binding motifs under the peak (Supplemental Table S2 in Supplemental Material). Within this context, peaks associated with genes in the core program display stronger TP53 binding in all cell types, relative to those associated with cell-type–specific direct targets (Fig. 4E). This indicates that TP53 can bind to a subset of specific enhancers across vastly different cell types, regardless of potential differences in chromatin landscapes, to activate a core multifunctional program. Analysis of ENCODE data for multiple transcription factors in HCT116 and MCF7 cell lines reveals that TP53 is unique in terms of shared binding sites and conservation of the underlying DNA sequence (Fig. 4F; Supplemental Fig. S5D,E). Unlike TP53, many transcription factors show distinct binding patterns and less apparent sequence specificity across cell lines. For example, both Serum Response Factor (SRF) and MAX display higher ratios of unique to shared peaks and differing underlying motifs in each cell line (Fig. 4F). Furthermore, TP53 binds to a much greater fraction of the total occurrences of the enriched motif across the genome than the other transcription factors (Supplemental Fig. S5E). Collectively, these results indicate that sequence specificity is a key determinant of TP53 binding across cell-type–specific chromatin landscapes, whereas other transcription factors may be more sensitive to availability of auxiliary factors and chromatin architecture.
Altogether, these results demonstrate the existence of a core TP53 transcriptional program comprising multiple effector pathways, driven by strong TP53 binding sites bound across different cell types, and which is activated regardless of cell fate choice.
The anti-proliferative activity of TP53 is highly distributed among its target genes
Having identified TP53-regulated genes, both core and cell-type–specific, we embarked on a functional test of their contributions to TP53-dependent suppression of cell survival and proliferation upon Nutlin-3 treatment. We employed an RNAi-based screening strategy that allowed us to compare the effects of gene products relative to that of TP53 itself (Fig. 5A). First, we designed a custom library containing 3100 shRNAs targeting 331 TP53 target genes (p53TARGET library) (see Supplemental Material). This focused library contains shRNAs targeting genes directly regulated by TP53 identified in this study and additional known and putative direct targets compiled through literature searches. Second, we used a genome-wide library with ∼165,000 shRNAs targeting ∼20,000 genes, enabling us to test not only the contribution of indirect targets but also any other gene products outside the TP53 network (TRC library) (see Supplemental Material). We used both libraries to carry out positive selection screens in SJSA cells. Amplification of the MDM2 locus in SJSA cells likely minimizes selection for other inactivating mutations within the TP53 program. Thus, the tumor suppressive activity of TP53 is mostly intact as evidenced by the fact that Nutlin-3 induces complete tumor regression in SJSA xenografts (Vassilev et al. 2004; Tovar et al. 2006).
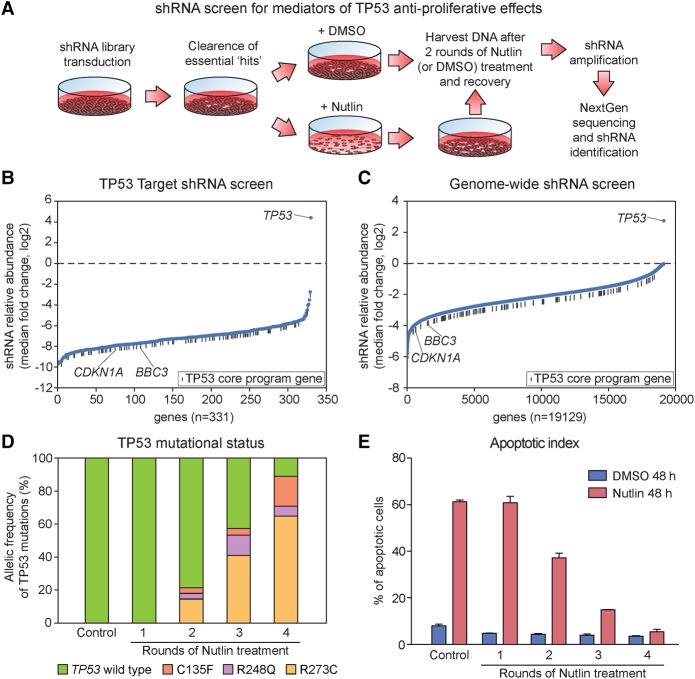
Figure 5.
shRNA screens reveal that the anti-proliferative activity of TP53 is highly distributed among its target genes. (A) Schematic of experimental design. After library transduction, SJSA cells were propagated in culture to deplete cells carrying shRNAs targeting essential genes. SJSA cultures were then treated with Nutlin-3 or vehicle (DMSO) for 48 h, and surviving cells were then allowed to recover and propagate after drug removal. Two rounds of treatment and recovery were carried out before analysis of shRNA abundance. See Supplemental Methods for details. (B,C) Ranking of all genes targeted in each screen by median fold change (Nutlin-3/DMSO) of all shRNAs targeting each gene, from those showing strongest shRNA depletion to strongest shRNA enrichment. (D) Analysis of TP53 mutation status after multiple rounds of Nutlin-3 treatment and recovery as in A reveals the rapid appearance of inactivating mutations in the TP53 locus. (E) SJSA cells treated as in A become resistant to the apoptotic effects of Nutlin-3 after four rounds of treatment. After each round of recovery, cells were exposed to Nutlin-3 for 48 h and the fraction of apoptotic cells was measured by Annexin-V staining. See also Supplemental File S8.
Knockdown of genes required for the anti-proliferative activity of TP53 would be expected to block cell cycle arrest and/or apoptosis, leading to enrichment of the corresponding shRNA in the Nutlin-3-treated cultures. After two rounds of treatment and recovery, we determined changes in shRNA abundance via next-generation sequencing (Fig. 5B,C; Supplemental File S8). Both screens revealed that TP53 is the only gene for which multiple shRNAs were enriched in the Nutlin-3-treated populations. Surprisingly, even shRNAs targeting CDKN1A or BBC3, known mediators of cell cycle arrest and apoptosis, respectively, did not confer significant selective advantage in this setting. The lack of consistent enrichment of shRNAs targeting any gene other than TP53 itself was not the result of a lack of coverage, because 97% of the shRNAs in our target library, and 90% of those in our genome-wide library, were present in the DMSO control samples. We considered increasing the number of treatment and recovery cycles to maximize our chances of identifying anti-proliferative genes. However, additional rounds of Nutlin-3 treatment led to the rapid enrichment of cells carrying mutant TP53 alleles (Fig. 5D), concomitant with loss of the apoptotic phenotype (Fig. 5E). Many of these nonsynonymous mutations were at “hot-spot” mutation sites for which oncogenic “gain-of-function” has been described (C135, R248, R273), which could explain their rapid selection during multiple rounds of Nutlin-3 treatment.
Altogether, these results demonstrate that, in the context of these in vitro evolution experiments, the anti-proliferative effects of TP53 are highly distributed among its target genes.
Core TP53 target genes are not frequently inactivated in wild-type TP53 tumors
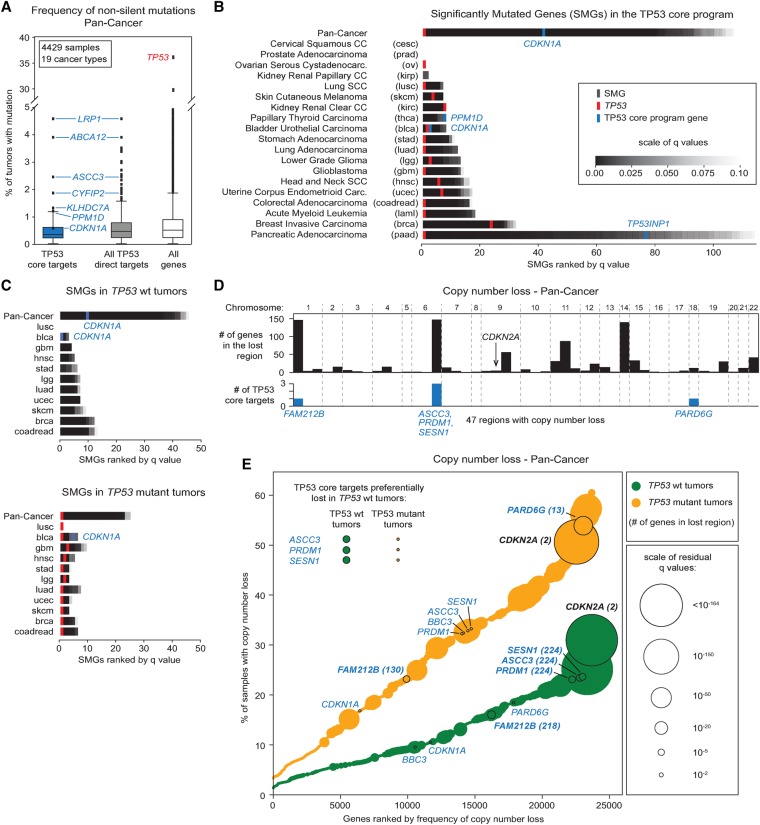
Although our screens did not identify individual genes that are crucial for TP53-dependent anti-proliferative activity in vitro, this could be due to differences from the physiological context in which TP53 normally exerts its tumor-suppressive functions in the human body. We hypothesized that core TP53 target genes with consistent and essential functions in tumor suppression would be significantly inactivated in a variety of cancer types, with a tendency toward mutual exclusivity with TP53 mutations. To test these ideas, we analyzed genomic data for 4429 samples from 19 different cancer types available through The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) (Lawrence et al. 2014). We found that frequencies of nonsynonymous mutations for core TP53 target genes, as well as all direct TP53 target genes (defined here as all early direct targets plus late targets with proximal binding), are well below the mutation frequency for TP53 itself, which was mutated in 36% of samples (Fig. 6A; Supplemental File S9). We sought to identify genes with mutation rates significantly (q < 0.1) above background, accounting also for factors such as gene length, chromatin state, and transcription rate, using MutSigCV (Lawrence et al. 2013). Expectedly, TP53 was significantly mutated across all cancers and in most individual tumor types (Fig. 6B). Of the core genes, only CDKN1A had a mutation rate considered significant in a pan-cancer analysis (Fig. 6B; Supplemental File S9). In individual cancer types, only CDKN1A, PPM1D, and TP53INP1 scored as significantly mutated in bladder urothelial carcinoma, papillary thyroid carcinoma, and pancreatic adenocarcinoma, respectively (Fig. 6B). None of the other core genes had mutation rates that were statistically significant, even those most frequently mutated (e.g., LRP1, ABCA12, ASCC3, CYFIP2) (Fig. 6A,B). Next, we asked whether mutations in any of the core genes tend to be mutually exclusive with TP53 mutations in cancer types with sufficient sample numbers for wild-type and mutant TP53. Note that, consistent with the role of TP53 in DNA repair, TP53 mutations correlate with higher overall mutation frequency and, consequently, a lower number of “significantly mutated genes” (SMGs) as detected by MutSigCV (Supplemental Fig. S6A). This exercise revealed that CDKN1A was the only core gene significantly mutated when segregating TP53 wild-type from mutant tumors (Fig. 6C). In the pan-cancer analysis, the CDKN1A mutation rate was significant only in wild-type tumors and, while it was considered significant in bladder cancer regardless of TP53 status, it ranked more highly in the tumors bearing wild-type TP53 alleles (Fig. 6C; Supplemental Fig. S6B). This suggests that in some specific settings, CDKN1A mutations can decrease, in small measure, the selective pressure to inactivate TP53. The small number of TP53 mutant samples for the tumor types in which PPM1D and TP53INP1 are significantly mutated prevented a similar mutual exclusivity analysis. Altogether, our analyses of nonsilent mutations failed to identify a core target gene whose inactivation could consistently bypass the need for TP53 mutation during cancer progression across diverse tissue types.
Figure 6.
Select TP53 target genes are inactivated at low rates in human cancers expressing wild-type TP53. (A) Frequency of nonsilent mutations for TP53 core target genes, all direct TP53 target genes (early direct plus late targets with proximal binding), and all genes in the genome across 4429 tumor samples. (B) Significantly mutated genes (SMGs) in the TP53 core program identified by MutSigCV in pan-cancer and cancer-type–specific analyses. SMGs are ranked based first on q-values and then by mutation frequency. For the full list of SMGs, see Supplemental File S9. (C) SMG identification as in B dividing tumors based on their TP53 mutation status. Only tumor types with sufficient numbers in each group were analyzed. (D) Genome regions showing significant copy number losses identified with GISTIC 2.0 (q < 0.01) in a pan-cancer analysis. Lost TP53 core target genes are highlighted in blue. The complete list of significantly lost regions can be found in Supplemental File S10. (E) Cumulative distribution plots for copy number loss frequencies identified with GISTIC 2.0 in a pan-cancer analysis. The size of each point (gene) reflects the significance of the loss (q-value). TP53 wild-type (green) and mutant (orange) tumors were analyzed and plotted separately. Genes highlighted in bold show a significant copy number loss (GISTIC 2.0, q < 0.01). See also Supplemental Figure S6 and Supplemental Files S9, S10.
Next, we asked if core TP53 target genes are frequently inactivated by genomic deletion during tumor evolution. We analyzed TCGA samples with available copy-number variation (CNV) data using the GISTIC 2.0 software (Mermel et al. 2011). First, we carried out a pan-cancer analysis and found only five core target genes to be lost at a significant rate (FAM212B, ASCC3, PRDM1, SESN1, and PARD6G) (Fig. 6D; Supplemental File S10). However, these genes all reside within large, frequently deleted chromosomal regions containing many additional genes (Fig. 6D; Supplemental Fig. S6C), which contrasts with the deletion pattern of CDKN2A, a tumor suppressor locus located in a small region of Chromosome 9 that is frequently deleted (Fig. 6D,E). Despite this caveat, we repeated the CNV analysis, accounting for TP53 status, to examine whether deletion of these core TP53 target loci was mutually exclusive with TP53 mutation. Once again, we observed signs of greater genetic instability in TP53 mutant tumors, which display more CNV across the genome (Supplemental Fig. S6D). Interestingly, we did find that ASCC3, PRDM1, and SESN1 are more significantly and frequently deleted in TP53 wild-type tumors (Fig. 6E; Supplemental Fig. S6D). This was not evident for FAM212B and PARD6G, or for CDKN1A and BBC3, which are not significantly lost, regardless of TP53 status (Fig. 6E). Although it is possible to speculate that the loss of the region of Chromosome 6 containing ASCC3, PRDM1, and SESN1 could contribute to tumor development, potentially bypassing—in a small measure—the need for TP53 mutation during cancer evolution, the fact that this region contains 145 other genes prevents any definitive conclusion.
Finally, we performed the CNV analysis for individual tumor types, which revealed that several core target genes are significantly deleted in specific cancer types. For example, TNFRSF10B was significantly lost only in prostate adenocarcinomas and HES2 only in kidney renal clear cell carcinoma (Supplemental Fig. S6E). When accounting for TP53 status, we found additional core genes significantly deleted in both TP53 wild-type and mutant tumors; however, this is always in large chromosomal regions with many other genes. For example, five core genes are located on a large region of Chromosome 11, containing >1200 genes, that is preferentially lost in TP53 wild-type head and neck squamous cell carcinoma (HNSCC) (Supplemental Fig. S6F–I).
Altogether, our analyses of cancer genome data suggest that different core TP53 target genes make context-dependent, small fractional contributions to the overall tumor suppressive activity of TP53.
Discussion
Despite decades of research, the identity of the tumor suppressive pathways governed by TP53, the most commonly inactivated gene in human cancer, remains a matter of debate. The transcriptional activator function of TP53 is clearly required for tumor suppression. Experimentally, point mutations inactivating the two N-terminal activation domains disrupt both target gene transactivation and tumor suppression (Brady et al. 2011), while in human cancers TP53 hotspot mutations impair DNA binding activity (Soussi et al. 2005). Therefore, much research has been devoted to the identification of direct TP53 target genes and downstream effector pathways mediating tumor suppression. To date, the overall result of these efforts is a vast collection of literature with many seemingly contradictory claims, whereby many genes or effector pathways have been reported as either critical or dispensable for tumor suppression. For example, in murine MYC-driven lymphoma models, depletion of the TP53 target gene BBC3/PUMA inhibits apoptosis and bypasses the requirement for TP53 mutation during the development of the disease (Hemann et al. 2004). In contrast, BBC3/PUMA is not required for tumor suppression in the context of murine thymic lymphomas (Valente et al. 2013). Similarly, two different reports employing TP53 alleles defective for transactivation of pro-apoptotic genes concluded that TP53-dependent apoptosis is either required (Timofeev et al. 2013), or dispensable (Li et al. 2012b) for tumor suppression.
At least three scenarios could help explain these apparently contradictory findings. First, the most important TP53 target genes mediating tumor suppression may not have been identified yet. This potential gap in our knowledge has triggered many efforts to identify novel TP53-regulated genes and pathways, with various teams pointing to autophagy, ferroptosis, or metabolism as the key to understanding tumor suppression by TP53 (Vousden and Ryan 2009; Kenzelmann Broz et al. 2013; Jiang et al. 2015). Second, it is possible that different TP53 target genes and effector pathways mediate tumor suppression in differing contexts. Third, the tumor suppressive activity of TP53 may be genetically robust, as a result of functions that are distributed and/or redundant across its transcriptional program. Our results here support the latter two scenarios, which are not mutually exclusive. Our genomics data demonstrate that TP53 consistently activates a multifunctional gene expression program in cancer cell lines of different origin, engaging multiple effector pathways, regardless of context or cell fate choice. This gene expression program consists of a conserved set of core direct target genes embedded within a larger gene network that is regulated in a context-dependent fashion and that likely modulates the ultimate outcome of TP53 activation. Taken together, our data indicate that the contribution of individual TP53 target genes is highly distributed across a robust tumor suppressive network, a scenario where inactivating mutations in the TP53 locus itself confers the strongest selective advantage during tumor evolution (Wagner 2005).
Due to the small sample size, our integrated analyses of multiple genomics data sets from three cancer cell lines may not be sufficient to distinguish between variation in gene expression due to cell and tumor type or heterogeneous cell lines. Nonetheless, our approach was sufficient to identify a core set of ∼100 shared TP53 target genes, most of which are also activated by other TP53-activating stimuli and in untransformed cells. Our ChIP-seq data confirm previous studies indicating that TP53 binds to a common set of sites across cell types, regardless of chromatin context, and that most TP53 binding events do not result in transactivation of the nearest gene (Verfaillie et al. 2016; Fischer 2017). Furthermore, our results strongly support the notion that TP53 only directly activates transcription and that all repression downstream from TP53 activation is indirect, as suggested previously (Allen et al. 2014; Verfaillie et al. 2016; Fischer 2017; Tonelli et al. 2017). During the preparation of this manuscript, a meta-analysis of numerous TP53-related ChIP-seq and RNA-seq data sets was published (Fischer 2017) that agrees with our findings that TP53 is solely an activator and that the core TP53 response contains genes involved in multiple pathways, including apoptosis and cell cycle arrest. In addition, our multi-omics approach revealed thousands of gene expression changes caused by TP53 activation, including the identification of many previously uncharacterized direct targets, some of which have undefined biological functions. Moreover, our results pave the way for future studies to define how TP53 signaling is expanded—in a cell-type–specific fashion—at the indirect and translational levels.
Our results also contribute to our understanding of the mechanisms of cell fate choice upon TP53 activation. It is well established that most cell types undergo cell cycle arrest in response to Nutlin-3 treatment (Tovar et al. 2006). In our experimental system, all three cell lines undergo cell cycle arrest, yet only SJSA cells exhibit a strong apoptotic response. Importantly, the core program contains many potent pro-apoptotic genes, whose induction is clearly insufficient to trigger apoptosis in HCT116 and MCF7 cells. This implies that indirect targets or factors outside the TP53 program play an important role in cell fate choice. For example, we found that factors outside the TP53 program in HCT116 cells can suppress apoptosis (Sullivan et al. 2012) and that engagement of the extrinsic apoptotic pathway is required to elicit cell death during TP53 activation with genotoxic agents (Henry et al. 2012). Our shRNA screens in SJSA cells indicate that no single gene, other than TP53 itself, is required for a robust apoptotic response in vitro. Although it remains possible that the situation differs in vivo, we found scant evidence of significant rates of mutation for any core TP53 target gene in TCGA data. Our results agree with a similar screen in which TP53 was the top hit and all core genes failed to score as required for Nutlin-3-induced cytotoxicity in MCF7 cells (Brummelkamp et al. 2006).
Finally, the multi-omics approach described here should be useful for understanding cell-type–specific signaling cascades governed by other master transcription factors. To what degree are the features revealed for the TP53 signaling cascade applicable to other ubiquitous transcription factors? In this regard, it is our sincere hope that our work will inspire similar analyses for other transcriptional regulators of biomedical importance, to advance our understanding of context-dependent gene expression control mechanisms, and eventually enable strategies for therapeutic manipulation of their activity in the clinical setting.
Methods
Cell culture
HCT116, MCF7, and SJSA cells were cultivated in McCoy's, DMEM, or RPMI media, respectively (Gibco, Thermo Fisher Scientific). Media were supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1% antibiotic-antimycotic solution (Life Technologies, Thermo Fisher Scientific). All cells were plated at indicated densities 24 h before treatment and cultivated under a humidified atmosphere with 5% CO2 at 37°C. Nutlin-3 was purchased from Cayman Chemical.
Cell viability, apoptosis, and BrdU incorporation assays
Viability of cells was assessed by propidium iodide (PI) exclusion assay. Briefly, cells were plated at a uniform density (2 × 104 cells/cm2) and after 24 h exposed to either DMSO (0.2%) or 10 µM Nutlin-3. Cells were harvested at indicated times by trypsinization and labeled with PI (1 µg/mL) for flow cytometric analysis (Accuri C6, Becton Dickinson). Collected samples contained at least 105 cells and PI-negative cells were identified as viable. The fraction of apoptotic cells was determined by Annexin-V binding assay as previously described (Gomes et al. 2006). Cells cultivated and treated as described above were harvested by trypsinization and resuspended in Annexin-V binding buffer (10 mM HEPES pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) and labeled with Annexin-V-FITC (Invitrogen) and PI (10 µg/mL) for 15 min in the dark, before analysis by flow cytometry. Cellular proliferation was measured indirectly by BrdU incorporation rate. One hour prior to harvest, 1 µg/mL of BrdU was added to the culture medium. Trypsinized cells were fixed in 70% ethanol overnight. DNA denaturation was performed in 2 M HCl with 0.5% Triton X-100 at 37°C for 10 min. After neutralization with 0.1 M NaBO4, cells were labeled with anti-BrdU antibody (sc-51514, Santa Cruz Biotechnology), stained with Alexa-488-labeled secondary antibody (A21202, Life Technologies/Invitrogen), and analyzed by flow cytometry. Data represent average numbers of BrdU positive cells from three independent experiments (each run in duplicate).
Western blotting
Cells were washed twice with cold PBS and lysed in SDS buffer (1% SDS, 0.1 M Tris pH 7.4, 10% glycerol) supplemented with protease inhibitors. Lysates were sonicated and heated at 95°C for 5 min. The total protein content was normalized, and samples were resolved by standard SDS-PAGE. After Western transfer, PVDF membranes were incubated with the following antibodies: TP53 (OP43 – DO1, Calbiochem), CDKN1A (sc-817, Santa Cruz Biotechnology), PARP (SA250, Enzo), CASP3 (#9661, Cell Signaling Technology), and TUBA4/tubulin (T9026, Sigma-Aldrich). After incubation with HRP-labeled secondary antibodies (Santa Cruz Biotechnology), proteins were visualized by enhanced chemiluminescence using an LAS4000 digital imaging system (GE Healthcare).
ChIP-seq
ChIP was performed as described before (Galbraith et al. 2013) and described in detail in the Supplemental Methods. Briefly, after drug or vehicle treatment, cells were cross-linked using 1% formaldehyde, washed twice with cold PBS, lysed in RIPA buffer, and cell lysates sonicated. Following immunoprecipitation with anti-TP53 antibody, immune complexes were washed, bound proteins eluted, and cross-linking reversed prior to DNA purification. Sequencing libraries were prepared using Illumina's TruSeq ChIP Library Preparation kit, and barcoded libraries sequenced on an Illumina HiSeq2000 instrument.
GRO-seq
Both nuclear run-on and sequencing library preparation were carried out using a modified protocol described by Wang et al. (2011) and are described in detail in the Supplemental Methods. Briefly, after drug or vehicle treatment, cells were harvested in hypotonic lysis buffer before nuclei preparation. Nuclear run-on was performed in the presence of Br-UTP for 5 min before RNA extraction and base hydrolysis. BrU-containing RNA was purified using anti-BrdU agarose beads, phosphorylated with T4 PNK, and subjected to polyadenylation and reverse transcription reactions. cDNA was subjected to circularization by ssDNA circular ligase. ssDNA was then used for preparation of next generation sequencing libraries and sequencing using an Illumina HiSeq2000 instrument.
RNA-seq
Polysomal fractions were prepared as described previously (Zaccara et al. 2014). Total RNA was extracted using TRI Reagent, according to the manufacturer's instructions. PolyA+ RNA was purified using the Dynabeads mRNA DIRECT Micro Purification Kit (Life Technologies). Libraries for Ion Torrent sequencing were prepared using the Ion Total RNAseq v2 kit (Life Technologies). Ion Torrent template preparation was carried out using the Ion PI Template OT2 200 kit v2 and sequenced in an Ion Torrent Proton instrument. See Supplemental Methods for details.
shRNA screens
shRNA libraries were acquired from the Functional Genomics Facility at the University of Colorado (see Supplemental Methods for details). Libraries were packaged into lentiviral particles using HEK293FT cells, and target cells transduced and selected with puromycin. After 1 wk, transduced cells were treated with either 0.2% DMSO or 10 µM Nutlin-3 for 48 h and allowed to recover. After a second round of treatment and recovery, genomic DNA was extracted and used in PCR reactions to amplify shRNA cassettes. shRNA libraries were prepared as described in the Supplemental Methods before sequencing using an Illumina HiSeq 2000 instrument.
Data access
All sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE86222. For previously published data sets, see Supplemental Table S3 in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Mary Allen, Robin Dowell, and W. Lee Kraus for initial assistance with the GRO-seq protocol and data analysis, Claire Bensard and Rose Byrne for preparation of the p53TARGET shRNA library, Jean-Luc Fraikin for early work with SJSA cells, Eliana Gomez for troubleshooting PCR amplification of shRNA cassettes, Steve Harris for helpful discussions, and Ryan Henry and Molishree Joshi at the Functional Genomics Facility for support with shRNA experiments. This study was supported primarily by National Institutes of Health (NIH) grant R01 CA117907 to J.M.E. Additional support was provided by National Science Foundation (NSF) grants MCB-1243522/MCB-1627615 to J.M.E., NIH grant 1F32CA199716-01 to A.L.G., and the Mary Miller & Charlotte Fonfara-LaRose Down Syndrome & Leukemia Research Fund. The Howard Hughes Medical Institute supported this work through an Early Career Award to J.M.E. between 2009 and 2015. The Functional Genomics Facility at the University of Colorado Cancer Center is supported by the NIH, National Cancer Institute (NCI) grant P30 CA046934.
Author contributions: Z.A. designed and performed experiments, analyzed data, and cowrote the manuscript. M.D.G. designed and performed RNA-seq experiments, interpreted and analyzed data, and cowrote the manuscript. A.L.G. performed shRNA screens. S.Z. performed polysomal RNA purification and RNA-seq. A.P. performed most bioinformatics analyses. M.M. performed quantitative analysis of TP53 expression. K.D.S. developed protocols for shRNA screens and data analysis. A.I. contributed to experimental design and data interpretation. J.M.E. conceived and designed experiments, interpreted data, and cowrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.220533.117.
Freely available online through the Genome Research Open Access option.
References
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. 2009. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci 106: 18207–18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL, et al. 2014. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife 3: e02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, et al. 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. 2006. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol 2: 202–206. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825. [DOI] [PubMed] [Google Scholar]
- Fischer M. 2017. Census and evaluation of p53 target genes. Oncogene 36: 3943–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Steiner L, Engeland K. 2014. The transcription factor p53: not a repressor, solely an activator. Cell Cycle 13: 3037–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. 2012. Mutant p53: one name, many proteins. Genes Dev 26: 1268–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, et al. 2013. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 153: 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 20: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin MJ, Lis JT. 2013. Mechanisms by which transcription factors gain access to target sequence elements in chromatin. Curr Opin Genet Dev 23: 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P, Pfeifer GP. 2016. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb Perspect Med 6: a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Zilfou JT, Zhao Z, Burgess DJ, Hannon GJ, Lowe SW. 2004. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci 101: 9333–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RE, Andrysik Z, Paris R, Galbraith MD, Espinosa JM. 2012. A DR4:tBID axis drives the p53 apoptotic response by promoting oligomerization of poised BAX. EMBO J 31: 1266–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. 2015. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD. 2013. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev 27: 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach FS, Tokino T, Meltzer P, Burrell M, Oliner JD, Smith S, Hill DE, Sidransky D, Kinzler KW, Vogelstein B. 1993. p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res 53: 2231–2234. [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. 2012a. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell 46: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. 2012b. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149: 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. 2011. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev 25: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez D, Nguyen TA, Freudenberg JM, Mathew VJ, Anderson CW, Jothi R, Resnick MA. 2013. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res 41: 7286–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. 2011. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, Kivioja T, Ignatiev I, Kel A, Taipale J, et al. 2012. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ 19: 1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemenschneider MJ, Buschges R, Wolter M, Reifenberger J, Bostrom J, Kraus JA, Schlegel U, Reifenberger G. 1999. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res 59: 6091–6096. [PubMed] [Google Scholar]
- Schlereth K, Heyl C, Krampitz AM, Mernberger M, Finkernagel F, Scharfe M, Jarek M, Leich E, Rosenwald A, Stiewe T. 2013. Characterization of the p53 cistrome—DNA binding cooperativity dissects p53's tumor suppressor functions. PLoS Genet 9: e1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T, Kato S, Levy PP, Ishioka C. 2005. Reassessment of the TP53 mutation database in human disease by data mining with a library of TP53 missense mutations. Hum Mutat 25: 6–17. [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Padilla-Just N, Henry RE, Porter CC, Kim J, Tentler JJ, Eckhardt SG, Tan AC, DeGregori J, Espinosa JM. 2012. ATM and MET kinases are synthetic lethal with nongenotoxic activation of p53. Nat Chem Biol 8: 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. 2005. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123: 49–63. [DOI] [PubMed] [Google Scholar]
- Timofeev O, Schlereth K, Wanzel M, Braun A, Nieswandt B, Pagenstecher A, Rosenwald A, Elsasser HP, Stiewe T. 2013. p53 DNA binding cooperativity is essential for apoptosis and tumor suppression in vivo. Cell Rep 3: 1512–1525. [DOI] [PubMed] [Google Scholar]
- Tonelli C, Morelli MJ, Sabo A, Verrecchia A, Rotta L, Capra T, Bianchi S, Campaner S, Amati B. 2017. Genome-wide analysis of p53-regulated transcription in Myc-driven lymphomas. Oncogene 36: 2921–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, et al. 2006. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci 103: 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente LJ, Gray DH, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, Janic A, Strasser A. 2013. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep 3: 1339–1345. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848. [DOI] [PubMed] [Google Scholar]
- Verfaillie A, Svetlichnyy D, Imrichova H, Davie K, Fiers M, Kalender Atak Z, Hulselmans G, Christiaens V, Aerts S. 2016. Multiplex enhancer-reporter assays uncover unsophisticated TP53 enhancer logic. Genome Res 26: 882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell 137: 413–431. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. 2009. p53 and metabolism. Nat Rev Cancer 9: 691–700. [DOI] [PubMed] [Google Scholar]
- Wagner A. 2005. Distributed robustness versus redundancy as causes of mutational robustness. Bioessays 27: 176–188. [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Niu D, Lam TH, Xiao Z, Ren EC. 2014. Mapping the p53 transcriptome universe using p53 natural polymorphs. Cell Death Differ 21: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219. [DOI] [PubMed] [Google Scholar]
- Zaccara S, Tebaldi T, Pederiva C, Ciribilli Y, Bisio A, Inga A. 2014. p53-directed translational control can shape and expand the universe of p53 target genes. Cell Death Differ 21: 1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.