Abstract
Adenosine (A) to inosine (I) RNA editing is widespread in eukaryotes. In prokaryotes, however, A-to-I RNA editing was only reported to occur in tRNAs but not in protein-coding genes. By comparing DNA and RNA sequences of Escherichia coli, we show for the first time that A-to-I editing occurs also in prokaryotic mRNAs and has the potential to affect the translated proteins and cell physiology. We found 15 novel A-to-I editing events, of which 12 occurred within known protein-coding genes where they always recode a tyrosine (TAC) into a cysteine (TGC) codon. Furthermore, we identified the tRNA-specific adenosine deaminase (tadA) as the editing enzyme of all these editing sites, thus making it the first identified RNA editing enzyme that modifies both tRNAs and mRNAs. Interestingly, several of the editing targets are self-killing toxins that belong to evolutionarily conserved toxin-antitoxin pairs. We focused on hokB, a toxin that confers antibiotic tolerance by growth inhibition, as it demonstrated the highest level of such mRNA editing. We identified a correlated mutation pattern between the edited and a DNA hard-coded Cys residue positions in the toxin and demonstrated that RNA editing occurs in hokB in two additional bacterial species. Thus, not only the toxin is evolutionarily conserved but also the editing itself within the toxin is. Finally, we found that RNA editing in hokB increases as a function of cell density and enhances its toxicity. Our work thus demonstrates the occurrence, regulation, and functional consequences of RNA editing in bacteria.
RNA editing is a post-transcriptional process in which RNA bases are being altered (Knoop 2011). Adenosine (A) to inosine (I) RNA editing is the most prevalent form of editing in metazoans (Bazak et al. 2014). Inosine in turn can be identified by the translational or genetic machinery (e.g., reverse transcriptase) as a guanosine (G). A-to-I editing can recode proteins in eukaryotes (for example, humans and fungi) (Knoop 2011; Liu et al. 2016; Wang et al. 2016). The majority of editing events found in humans occur in untranslated regions, while only a small fraction of editing events are found in coding regions, of which only a few lead to nonsynonymous recoding (Ramaswami and Li 2014). All A-to-I editing events in mRNA are mediated by enzymes belonging to the ADAR (adenosine deaminase, RNA specific) family that was suggested to constitute a metazoan innovation (Grice and Degnan 2015). In bacteria, however, RNA editing was only reported in a single nucleotide site, within a tRNA for arginine, and it is mediated by the enzyme tRNA-specific adenosine deaminase (tadA) (Wolf et al. 2002).
Recent advances in sequencing technologies have facilitated the discovery of RNA modifications and edited sites in an unprecedented level both in the nucleus (Ramaswami et al. 2013; Bazak et al. 2014; Schwartz et al. 2014; Liu et al. 2016; Wang et al. 2016) and within organelles (Bar-Yaacov et al. 2013; Bentolila et al. 2013; Oldenkott et al. 2014). However, editing events in mRNA were so far not reported in bacteria.
Results
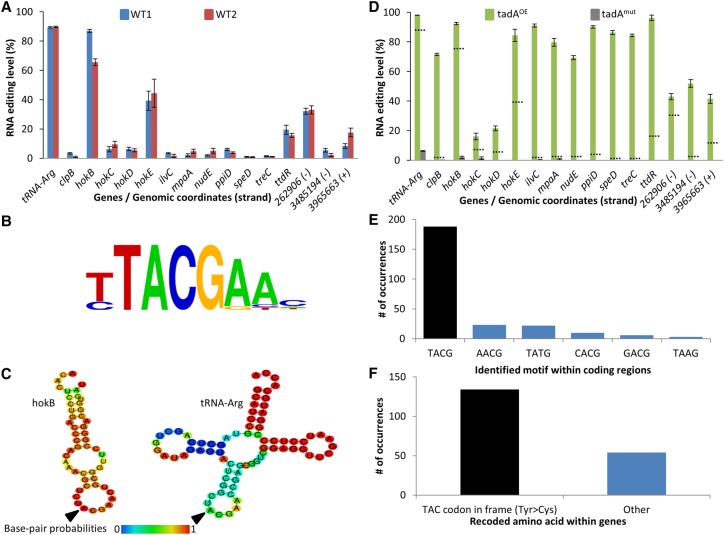
In order to identify novel editing events, we deep sequenced in parallel the RNA and DNA from two Escherichia coli strains (Fig. 1A). We used stringent parameters (Supplemental Fig. S1; Methods) to identify editing events that can manifest themselves as base differences between the DNA and RNA sequences. We identified 15 novel A-to-G RNA editing events (12 within known ORFs) in addition to the known editing site in tRNA-Arg (Fig. 1A; Supplemental Table S1). Strikingly, examining all 12 sites in which we detected editing within ORFs revealed that they are all predicted to recode a tyrosine (Tyr) encoded by the TAC codon into a cysteine (Cys) encoded by the TGC codon. While the majority of editing events were A-to-G, we also detected one additional genomic site which constituted a C-to-U substitution (which results in a synonymous substitution at the protein level) (Supplemental Table S1). All A-to-G editing events were embedded within a four-base-long motif TACG, with the edited A on the second position (Fig. 1B). Interestingly, this motif is completely identical to the known tadA recognition motif (Wolf et al. 2002) present on tRNA-Arg. In addition, tadA was previously shown to require for its activity a specific RNA secondary structure loop conformation around the edited site (Wolf et al. 2002). Indeed, RNA secondary structure modeling (Gruber et al. 2008) predicts that the edited base is also embedded within a loop in most of the newly identified sites (Fig. 1C; Supplemental Fig. S2). This raised the suspicion that tadA, which was so far believed to exclusively edit the anticodon of the tRNA-Arg, might be responsible for the editing of the aforementioned positions. Therefore, we performed RNA-seq on two additional strains, one overexpressing tadA from a plasmid and another harboring a tadA mutation (Supplemental Fig. S3) reported to completely abolish its activity in vitro and slightly reduce it in vivo in the NWL37 strain (Poulsen et al. 1992; Wolf et al. 2002). Since this strain was generated through an experimental lab evolution technique (interestingly, by exposing it to constant expression of the toxin hokC), we reasoned that it is possible that the evolved strain might also contain additional mutations. Therefore, rather than using this evolved strain, we have “surgically” introduced only the inactivating mutation into tadA’s genome version in the background of our strain (see Methods). Consistent with our hypothesis, overexpressing tadA dramatically increased the editing levels in all A-to-G sites, while in the tadA mutant, editing levels were abolished or dramatically reduced in all sites, including in the tRNA-Arg (Fig. 1D; Supplemental Fig. S4; Supplemental Table S1). Thus, all A-to-G substitutions in our RNA-seq data are likely to represent an adenosine to inosine editing event (hereafter A-to-I). Notably, editing levels of the C-to-U event (which did not harbor the tadA motif) were unaffected upon overexpression of tadA as well as in the tadA mutant (Supplemental Table S1). Overexpressing tadA revealed 252 additional A-to-I sites in coding regions that demonstrate RNA editing levels of at least 10% (Fig. 1E; Supplemental Table S2). Of these, 188 (75%) are embedded within the TACG recognition motif and show a significant enrichment for recoding a Tyr into a Cys codon (χ12 = 1.35 × 10−12) (Fig. 1E,F; Supplemental Tables S2, S3), raising the hypothesis that they might represent additional targets for this enzyme. Thus, we showed that A-to-I RNA editing in protein-coding genes occurs in bacteria, recodes Tyr into a Cys codon, and is mediated by tadA, an enzyme previously thought to be a tRNA-specific deaminase. Notably, there is no correlation between RNA editing levels and mRNA expression levels of the 12 genes that are edited by tadA in wild-type strains (Fig. 1A; Supplemental Fig. S5).
Figure 1.
RNA editing occurs in E. coli and it is mediated by tadA. (A) RNA-seq (and DNA-seq) from two WT E. coli strains (Top10 and MG1655-EcM2.1, blue and red, respectively) reveals 15 novel A-to-G(I) RNA editing sites in E. coli in addition to the known editing site in tRNA-Arg. Notably, all sites found in known genes (12 out of the 15 sites) recode a tyrosine (TAC) into a cysteine (TGC) codon. The three RNA editing sites that do not occur in known ORFs are denoted by their genomic coordinates and genomic strand (+ or −). RNA editing levels are defined here as the number of reads with a G at the position out of all reads that cover the position. RNA samples were extracted in mid-log phase at OD600 ∼ 0.7. (B) All sites share a common four-base DNA motif which is identical to tadA's recognition motif. (C) RNA secondary structure modeling predicts that edited sites are embedded within a loop. Here, the secondary structure of hokB (as well as tRNA-Arg) is presented (the RNA secondary structure modeling of all other targets found in this work is shown in Supplemental Fig. S2). (D) Overexpressing (green) or mutating (gray) tadA increases or reduces the editing level, respectively. Dotted lines represent the average editing levels measured for each gene in the two WT strains. RNA samples were extracted in mid-log phase at OD600 ∼ 0.5–0.6. (E) Overexpression of tadA reveals additional putative editing sites, of which 75% are embedded within the canonical motif (TACG, black bar), while the rest deviate by one base from the canonical motif. (F) Out of 188 editing sites which occur within genes, 134 (black bar) recode a Tyr into a Cys codon (71%). Error bars in parts A and D represent standard errors of measuring editing level in a given coverage. Exact values can be found in Supplemental Tables S1 and S2.
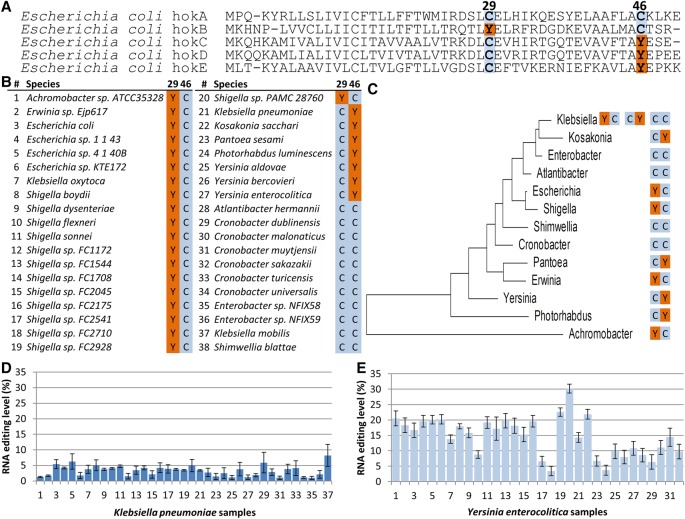
Interestingly, we found that four of the A-to-I editing sites observed in wild-type cells occur within the ORF of genes belonging to the hok family of host-killing toxins (Fig. 1A). Proteins encoded by this family belong to the hok-sok toxin-antitoxin module that confer membrane de-polarization, which results in growth inhibition and potentially cell death (Pedersen and Gerdes 1999; Verstraeten et al. 2015). Multiple sequence alignment of the toxins belonging to the hok family revealed that the editing event which recodes a Tyr (TAC) into a Cys (TGC) codon at position 29 of hokB aligns against a conserved genome-encoded Cys residue in the other hok members. Remarkably, in the other toxins, hokC, hokD, and hokE, editing recodes another position in the peptide, 46, there too converting a Tyr (TAC) into a Cys (TGC) codon (Fig. 2A). hokB in turn harbors a DNA-encoded Cys at position 46. Thus, across the hok family of E. coli toxins, there are two positions, of which one is always hard-coded with Cys in the genome and the second contains a hard-code Tyr that can be converted into a second Cys upon editing of the RNA (except in hokA). The conserved Tyr is always encoded in these positions through the TAC codon which is contained in the editing motif TACG, and never with the synonymous codon for this amino acid, TAT, which does not confer to the editing motif consensus. The five genome-encoded hok genes share about a third of their sequence (i.e., conserved positions). At the RNA level, in all of them the edited site resides within a predicted secondary structure loop; however, hokB’s sequence around the edited site is the only one with complete identity to the loop of tRNA-Arg, which is the known substrate of tadA (Fig. 1C; Supplemental Fig. S2). All five Hok genes encode a short peptide (∼50 amino acids) with an N-terminus that is embedded within the membrane, while the C-terminus is located within the periplasm (Poulsen et al. 1991). We therefore modeled the 3D structure of all five hok peptides which displayed a conserved 3D structure of an alpha helix at the N-terminus and two beta strands at the C-terminus (Supplemental Fig. S6). Notably, the residues at position 29 and 46 of hokB reside each in one of the two beta strands and are predicted here to be in close proximity to each other. Four of the toxins (hokA, hokC, hokD, and hokE) were reported to be inactive in E. coli (Pedersen and Gerdes 1999), thus raising the interesting possibility that high levels of RNA editing can be found in functional, rather than nonfunctional, hok members. Nevertheless, our results suggest that these genes are at least transcribed and that hokC, hokD, and hokE are edited (Fig. 1A; Supplemental Fig. S5; Supplemental Table S1). We therefore focused further on hokB that was shown to be active (Verstraeten et al. 2015) and demonstrated the highest level of RNA editing. When we analyzed multiple sequence alignments of annotated hokB’s orthologs from different bacterial species, we found that most orthologs either have a Tyr encoded by the editable TAC codon (embedded within the TACG motif) at position 29 and a Cys at position 46, or, in other orthologs, a Cys at position 29 and an editable Tyr codon at position 46. Note that some species in this sample have Cys at both positions (Fig. 2B,C; Supplemental Table S4). This remarkable correlated pattern raised the question whether hokB mRNA editing can occur and can be detected in other species. Indeed, by analyzing multiple publicly available RNA-seq data sets (Leskinen et al. 2015), we observed A-to-I mRNA editing that recodes position 46 in two pathogenic bacteria, Klebsiella pneumonia and Yersinia enterocolitica (Fig. 2D,E; Supplemental Tables S5, S6). It is possible that hokB RNA is edited in additional species which currently are lacking publicly available RNA-seq data sets or only have an insufficient number of RNA reads (>10×) that cover hokB. Thus, not only is hokB evolutionarily conserved, but also RNA editing within it can be identified in species other than E. coli.
Figure 2.
Evolutionary analyses suggest an interplay between the recoded and a hard-coded cysteines in hokB. (A) Multiple sequence alignment of five hok proteins encoded by the E. coli genome (NC_000913.3). The hokB edited version recapitulates the cysteine at position 29 which is hard-coded in the genome of all other hok protein family members. Symmetrically, hokC, hokD, and hokE editing sites (position 46) recapitulate the cysteine at the same position of hokB. (B) Multiple sequence alignment of hokB of a representative nonredundant set of orthologs from bacterial species harboring an annotated hokB gene suggests interplay between peptide residues at positions 29 and 46. Notably, all the Tyr codons at position 29 or 46 are encoded by the editable codon (TAC, embedded within the TACG motif). The complete alignment can be found in Supplemental Table S4. (C) A maximum likelihood phylogenetic tree based on the 16S rRNA gene, showing the amino acid composition at hokB's positions 29 and 46 in each bacterial genus with species harboring an annotated hokB. (D,E) RNA editing in hokB was identified in publicly available Klebsiella pneumoniae (37) and Yersinia enterocolitica (32) samples with sufficient coverage (≥51×) of RNA reads and at least two reads supporting an editing event. This editing event is predicted to recode position 46 (Tyr>Cys) in hokB. SRA accession numbers can be found in Supplemental Tables S5 and S6. Error bars represent standard errors of measuring editing level in a given coverage.
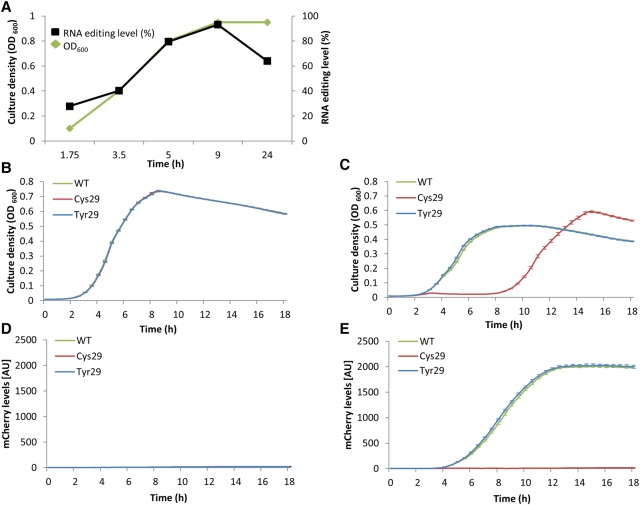
hokB was implicated in arresting cellular growth via membrane depolarization, and by doing so, it was found to mediate antibiotic tolerance through a mechanism of persistence (Verstraeten et al. 2015). Additionally, it was demonstrated that expression of hokB is elevated in response to starvation (Verstraeten et al. 2015). Since one of the characteristics of reaching culture stationary phase is a lack of nutrients, we aimed to examine if the editing levels of hokB change as a function of cellular density of the bacterial culture. Indeed, editing levels of this toxin's mRNA site were found to increase from ∼28% at early logarithmic phase to ∼93% when the culture enters stationary phase (Fig. 3A; Supplemental Table S7). Thus, in addition to elevation in toxin expression, hokB’s RNA editing levels are elevated during culture growth. We further asked how the predicted change in amino acid, from Tyr to Cys at position 29, can affect hokB's activity. To answer this question, we first mutated the genomic hokB gene. The first version was a positive control, as it contained the WT version of the toxin (hokB-WT) with the codon TAC coding for Tyr; the second version mimicked constitutive editing, with the codon TGC encoding for a Cys yet hard-coded into the DNA (hokB-Cys29); the third version of the mutated toxin had the Tyr at the edited position, yet with the synonymous codon TAT that is noneditable (hokB-Tyr29) (Supplemental Table S7). No observable difference in growth was detected between the three strains (Supplemental Fig. S7). This lack of observable phenotype could be expected given that hokB’s expression is governed by high levels of the alarmone (p)ppGpp, a condition that is observed in only 1/10,000 cells (Gerdes and Maisonneuve 2015) during logarithmic phase. Therefore, we utilized a previously established strategy (Verstraeten et al. 2015) of overexpressing hokB from a plasmid to acquire a detectable signal in order to facilitate the study of the activity of the three versions of the toxin we created. In order to detect potential functional differences between hokB-WT, hokB-Cys29, and hokB-Tyr29, we expressed the three versions of the toxin from an inducible plasmid in an E. coli strain that lacks its genome-encoded hokB. When each of the hokB versions is induced, we observed reduction in growth rate and yield compared to cells not expressing hokB (Fig. 3B,C). Yet, while the toxicity observed in the hokB-WT and hokB-Tyr29 versions is relatively mild, it is very dramatic in the hokB-Cys29 version (Fig. 3B,C). In fact, the Cys29 strain shows signs of growth only 9 h after induction of the toxin. Note that the growth observed in the Cys29 strain after 9 h is probably a result of some (potentially genetic) adaptation that allows the cells not to express the toxin and therefore support growth, while the two other strains still express it (Fig. 3D,E). In other words, the Cys29 version is so toxic that it only allows a small fraction of cells in the population that do not express hokB to grow. Since the frequency of such cells is very low (below the technical detection), growth of Cys29 cells is only visible after 9 h. Nonetheless, if we allow all three strains to grow to mid-logarithmic phase without expression of hokB and only then inducing hokB expression, we observe a clear mCherry signal, including in the Cys29 strain, indicating that hokB gene is intact and can be induced in all three variants (Supplemental Figs. S8–S10). This result further supports our conclusion that the lack of growth observed upon induction of the hokB-Cys29 version is due to hokB toxicity rather than lack of hokB expression from the plasmid. Thus, RNA editing that converts Tyr to Cys at position 29 of hokB enhances its toxicity.
Figure 3.
hokB mRNA editing increases with cell density and enhances its toxicity. (A) hokB mRNA editing levels (black) in E. coli MG1655-EcM2.1 WT strain as measured in different culture densities (green). Notably, the standard error of measuring editing levels in a given coverage in all samples was smaller than 0.00012%. (B–E) The E. coli Top10-ΔhokB strain was transformed with inducible plasmids harboring the WT (green), constitutively edited (Cys29, red), and noneditable (Tyr29, blue) hokB versions fused to mCherry reporter protein (N-terminus). (B) Growth analysis without induction of hokB (0% arabinose). (C) Growth analysis with induction of hokB (0.2% arabinose). (D) mCherry levels without induction (0% arabinose). (E) mCherry levels with induction (0.2% arabinose). Error bars represent standard error for 14 replicates (B–E).
Discussion
Why do bacteria exercise RNA editing? In mammalian cells, editing occurs mostly in noncoding regions (Bazak et al. 2014). In bacteria, however, as we have shown, out of the 15 novel A-to-I sites, 12 occur within known protein-coding genes and recode a Tyr into a Cys codon. RNA editing in bacterial coding regions could provide another layer of post-transcriptional regulation, and it can contribute to proteome diversity, as was recently suggested in cephalopods (e.g., octopus and squids) (Liscovitch-Brauer et al. 2017). In hokB’s case, RNA editing appears to provide another layer of regulation of toxicity. This editing-induced increase in toxicity of hokB could either represent a change in toxicity per protein molecule or an increase in its amount (e.g., by a stabilizing effect).
Turning editing on or off can affect the RNA and even the protein sequence within relatively short physiological time scales. This is demonstrated here in our observation that RNA editing levels in hokB increase with culture density. In addition, editing may allow cells to obtain both the edited and unedited versions of hokB, and even “play” with the ratio between them, generating phenotypic heterogeneity between genetically identical cells. Such cell-to-cell variability, when exercised in the activation pattern of host-killing toxins, can potentially affect the antibiotic persistence they confer and thus might form an even more complex bet-hedging mechanism than was previously suggested (Verstraeten et al. 2015).
Why do we observe different editing levels among the 15 newly discovered mRNA editing sites? It was shown that RNA secondary structure is important for tRNA-Arg editing (Wolf et al. 2002). Therefore, difference in secondary structure and/or additional sequence features might affect editing levels by affecting tadA-RNA interaction. Indeed, tadA's structure in complex with a tRNA-Arg loop (Losey et al. 2006) demonstrates that the enzyme interacts with seven of the tRNA substrate nucleotides that constitute the entire hepta-loop. Thus, loop size and additional sequence features could affect enzyme-substrate interaction and hence editing level. Indeed, the only newly discovered RNA target that has complete sequence identity to the tRNA seven loop nucleotides is hokB, which reassuringly shows the highest editing level, second only to the tRNA. All other targets either differ in their sequence (surrounding the core TACG motif) or loop size. Future studies are needed to examine if all detected mRNA editing events have functional consequences or whether some of them represent an “accidental” activity due to sequence/structure similarity to tadA's substrates. tadA is found in most bacterial species (Yokobori et al. 2013). Therefore, our work sets the stage for investigating RNA editing in other bacterial species that harbor this enzyme. Moreover, tadA’s orthologs (such as Tad1p and ADAT) are found in eukaryotes (yeast [Wang et al. 2016] and human [Grice and Degnan 2015], for example). Since we now implicated tadA in mRNA editing, in addition to its established role in tRNA editing, future studies should examine whether its orthologs are involved in mRNA editing in other organisms too. In conclusion, RNA editing occurs in bacteria and can recode protein sequences, potentially affecting their function as well as cell physiology, at least in hokB’s case. Thus, sequence variation among bacteria should also be examined at the RNA level.
Methods
RNA and DNA purification
RNA and DNA were purified using the GeneJET RNA Purification kit (Thermo Fisher Scientific) and Wizard Genomic DNA Purification kit (Promega), respectively, according to the manufacturer's protocol. Cultures were grown on LB supplemented with ampicillin (100 µg/mL). RNA was purified from a culture at the middle of logarithmic phase (OD600 in a 1-cm cuvette ∼0.8) for whole transcriptome sequencing and hokB MAGE (multiplex automated genome engineering) strains; different ODs as specified in Figure 3A. DNA was extracted and purified at stationary phase.
cDNA synthesis
One microgram of total RNA was subjected to cDNA synthesis using either the M-MLV cDNA Synthesis kit (Promega) or SuperScript II (Thermo Fisher Scientific), following the manufacturer's protocol.
PCR reaction mix, primers, and conditions
All data regarding PCR reaction mix, conditions, and primers can be found in Supplemental Tables S8 and S9. PCR products were visualized by an EtBr-stained 1% agarose gel. PCR fragments were purified using a Wizard SV Gel and PCR Clean-Up System (Promega), following the manufacturer's protocol.
Massively parallel deep sequencing
RNA was treated with a Ribo-Zero rRNA Removal kit (Illumina). Libraries for sequencing RNA to examine RNA editing levels in different optical densities of microbial cultures (OD600) were constructed by designing PCR primers targeting the hokB gene with tails that match Illumina adapters (PCR1). A second PCR (PCR2) was carried out to attach the adapters for the Illumina run. Total DNA and RNA libraries of wild-type and of hokB versions that are expressed from plasmids were sequenced using 151-nt or 75-nt paired-end reads, respectively, on the NextSeq platform (Illumina). Bacterial strains that were sequenced were: RNA-seq – WT1 (Top10), WT2 (MG1655-EcM2.1), tadA overexpression (Top10 harboring a pBAD plasmid overexpressing tadA), and tadA mutant (MG1655-EcM2.1 strain with an introduced tadA mutation); and DNA-seq – WT1 (Top10) and WT2 (MG1655-EcM2.1).
Analysis of massively parallel sequencing data
E. coli sequencing reads were aligned against the MG1655 Genome (NC_000913.3). For multiple sequence alignment, we utilized BWA-MEM (Li and Durbin 2009) with default parameters. Only reads that were aligned to the corresponding genome were used for further analyses. SAMtools (Li et al. 2009) was used to convert the SAM to the BAM sequence format. MitoBam Annotator (Zhidkov et al. 2011) transformed the BAM files into tables containing all parameters (e.g., base composition, coverage per base, etc.) for each position in the E. coli genome. These tables were used to identify sites that differ between RNA samples and their corresponding DNA base. Initial RNA editing sites were considered high quality only if identified in at least 30 sequence reads that contain a high-quality base call (≥30 Phred quality score); if their minimal read fraction was at least 3%; if each site contained at least five forward and reverse reads; they presented a mixture of only 2 nucleotides; and after manual inspection using the Integrative Genomics Viewer (Robinson et al. 2011) to exclude signal stemming from the edges of reads or low-complexity regions (mononucleotide regions). We also aimed to identify edited sites below 3% (but more than 1%). Therefore, after establishing that tadA was the mediating enzyme, we searched for sites shared between the two WT strains and the tadA overexpressing strain that display an A-to-I editing level of at least 1% in the WT and harbor a tadA recognition motif. Three additional sites were detected, demonstrating a dramatic increase in editing levels when tadA was overexpressed (Fig. 1A,D; Supplemental Table S1). In addition, BLAST analysis excluded that the RNA editing signal (a mixture of nucleotides at the identified edited sites) stems from paralogous regions in the E. coli genome (Supplemental Table S10). BLAST was performed by searching against the E. coli genome with a 101-bp fragment encompassing the edited site plus 50 bases upstream of and downstream from it. Finally, we examined and compared the final editing sites between all four RNA-seq and two DNA-seq data sets we obtained (some did not pass the initial thresholds). RNA expression analysis was performed by using python/2.7.6 HTSeq (Anders et al. 2015) for read count per gene. FPKM values were calculated manually.
Multiplex automated genome engineering
In order to create a mutation in tadA (T2697699G, D64E) that was previously shown to reduce its activity (Wolf et al. 2002), we used the E. coli strain MG1655-EcM2.1 (a specially designed strain for high MAGE efficiency) to carry out one successive MAGE cycle as previously described (Wang et al. 2009; Bar-Yaacov et al. 2016). We used 77-bp single-strand oligonucleotides to target the lagging strand in the tadA gene: G*A*TAATTTTGCATCACC AGACCACCCTGCCGCAGGGCCATGATTTCTGCATGTGCGGTGGGCTCATGGCGACCAA*T*C. Similarly, we used MAGE to recode hokB gene sequence once into a TGC Cys codon (T1491986C) mimicking constitutive editing and once into a noneditable TAT Tyr codon (C1491985T, synonymous Tyr mutation). We used the following 90-bp single-strand oligonucleotides to target the lagging strand in the hokB gene: A*T*CTGCATTACGATTCTGACATTCACACTCCTGACCCGACAAACGCTCTGCGAACTGCGGTTCCGGGACGGTGATAAGGAGGTTGCTG*C*G (for T1491986C) and A*T*CTGCATTACGATTCTGACATTCACACTCCTGACCCGACAAACGCTCTATGAACTGCGGTTCCGGGACGGTGATAAGGAGGTTGCTG*C*G (for C1491985T). The mutated base is underlined, and asterisks represent phosphorothioate bonds. Briefly, cells were grown overnight at 30°C. Then, 30 µL of the saturated culture were transferred into fresh 3 mL of LBL (10 g of tryptone, 5 g of NaCl, and 5 g of yeast extract per liter) medium until reaching an OD = 0.4 (measured in a 1-cm cuvette in this section) and then moved to a shaking water bath (350 RPM) at 42°C for 15 min, after which it was moved immediately to ice. Next, 1 mL was transferred to an Eppendorf tube and cells were washed twice with double-distilled water (DDW) at a centrifuge speed of 13,000g for 30 sec. Next, the bacterial pellet was dissolved in 50 µL of DDW containing 2 µM of SS-DNA oligo and transferred into a cuvette. Electroporation was performed at 1.78 kV, 200 ohms, 25 µF. After electroporation, the bacteria were transferred into 2 mL of fresh LBL and incubated at 34°C until reaching an OD = 0.8, diluted in 1:10−4 and 1:10−5 ratios, and seeded on LB+ampicillin agar plates (100 µg/mL). To identify positive MAGE colonies (referred to as bacterial strains throughout the text), we PCR-amplified fragments encompassing the E. coli genomic region with primers corresponding to the mutated and WT form (differing in one base in their 3′ end – PCR3). Successful PCR amplification implies successful MAGE mutagenesis. To verify this interpretation, we amplified a second fragment encompassing the mutated position in tadA (PCR4) and Sanger sequenced it. Similarly, we used PCR5 (to identify colonies) and PCR6 (to validate the colonies using Sanger sequencing) to validate MAGE mutagenesis in hokB. The sequences were aligned and visualized using SnapGene Viewer 3.1.2 (GSL Biotech LLC).
Plasmid construction and transformation
In order to examine the functional role of RNA editing in hokB, we utilized the plasmid previously used to examine hokB's activity (Verstraeten et al. 2015) and constructed two additional plasmids (also harboring an ampicillin resistance cassette): pBAD-mCherry-linker-hokB(WT)—a generous gift from Prof. Jan Michiels from KU Leuven–University of Leuven—pBAD-mCherry-linker-hokB(Cys29), and pBAD-mCherry-linker-hokB(Tyr29). By using PCR7 and PCR8, we mutated the TAC codon corresponding to position 29 in hokB into a TGC (Tyr>Cys) and TAT (editable Tyr>noneditable Tyr) codons, respectively. All three plasmids were transformed into a Top10-ΔhokB strain (another generous gift from Prof. Jan Michiels) (Verstraeten et al. 2015).
We also constructed a pBAD-mCherry-linker-tadA(WT) plasmid. Specifically, we amplified the tadA gene and the plasmid backbone with overlapping (∼20 nt) tails using PCR9 and PCR10. These fragments were subjected for NEB-assembly (New England Biolabs) according to the manufacturer's protocol. The plasmid was transformed into a Top10 WT strain, and single colonies were isolated, grown, and frozen (−80°C) for future assays.
PCR and Sanger sequence of hokB
PCR11 was performed to sequence the hokB gene/transcript from corresponding DNA and RNA samples from the WT strain (MG1655-EcM2.1) as well as RNA from the tadA mutant.
Liquid growth measurements
Cultures were grown at 30°C and 37°C for the genomic (MG1655-EcM2.1)- and plasmid (Top10-ΔhokB)-encoded hokB versions for 48 h in LB medium, back diluted in a 1:100 ratio, and dispensed on 96-well plates containing LB medium supplemented with 100 µg/mL ampicillin (150 µL per well) and either with arabinose (final concentration of 0.2% (Fig. 3C,E) or without arabinose (no hokB expression; control) (Fig. 3B,D). Wells were measured for optical density at OD600 and mCherry fluorescence levels at 575 nm (excitation) and 620 nm (emission) wavelengths. Measurements were taken at 15-min intervals. Growth comparisons were performed using 96-well plates (Thermo Scientific). For each strain harboring a different version of hokB, a growth curve was obtained by averaging over well-dispersed 14 wells. The 96-well plate was divided as following: 12 wells are blank control and the remaining 84 wells were divided between the bacterial stains harboring the plasmid encoding for the three different hokB versions, with and without induction; thus, 84/6 = 14. Measurements for Figure 3A were conducted in a 1-cm cuvette, and values were divided by 2 for qualitative presentation purposes and comparison to the measurements shown in the growth curves (Fig. 3B,C).
Detecting mRNA editing in hokB of Yersinia enterocolitica and Klebsiella pneumoniae
RNA-seq data sets were downloaded from the SRA database (https://www.be-md.ncbi.nlm.nih.gov/sra). Accession numbers and parameters (e.g., coverage per base) of samples with identified mRNA editing in hokB are found in Supplemental Table S5 (K. pneumoniae) and S6 (Y. enterocolitica). We analyzed 46 Y. enterocolitica and 338 K. pneuomoniae samples and detected editing in 32 and 37 samples, respectively. Alignment and file manipulation were performed as described above. Notably, the rest of the species in Figure 2B were not assessed for their editing level since they did not meet the criteria of having a sufficient number of RNA reads (>10×) that cover hokB or did not have publicly available RNA-seq data sets.
Identifying tadA's motif
We used weblogo at http://weblogo.berkeley.edu/logo.cgi to identify the conserved, four-base motif which is identical to the tadA recognition motif.
RNA secondary structure prediction
In order to examine the RNA secondary structure, we extracted the RNA sequence 25 bases upstream of and downstream from the edited site (inclusive). We then used this sequence in the Vienna RNA Websuite (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) to predict the RNA secondary structure with minimum free energy.
hok proteins 3D structure prediction
We used the RaptorX (Källberg et al. 2012) package at http://raptorx.uchicago.edu/StructurePrediction/predict/ to predict the 3D structure of hokA, B, C, D, and E (using default parameters).
Analysis of hokB orthologs in other bacterial species
In order to examine the amino acid composition at positions 29 and 46 of hokB, we downloaded hokB gene sequence from organisms with annotated hokB from the NCBI nucleotide website (https://www.ncbi.nlm.nih.gov/nuccore/). We constructed a nonredundant set of orthologs with one hokB sequence (gene and protein) per species (Supplemental Table S4). The sequence identities at positions 29 and 46 are presented in Figure 2B.
Multiple sequence alignment
MSA was performed by using ClustalW (default parameters) embedded in the MEGA5 package (Tamura et al. 2011) and the MAFFT server (http://mafft.cbrc.jp/alignment/server/).
Phylogenetic analyses
We used the 16S ribosomal RNA to build a genus phylogenetic tree to visualize the amino acid composition in hokB's positions 29 and 46 in an evolutionary context. We used the 16S ribosomal RNA from one representative from each genus (Supplemental Table S11). The evolutionary tree was inferred by using the maximum likelihood method based on the Tamura–Nei model (Tamura and Nei 1993). The tree with the highest log likelihood (−4907.0796) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Joining and BIONJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with superior log likelihood value (Saitou and Nei 1987; Gascuel 1997). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 13-nt sequences. All positions containing gaps and missing data were eliminated. There were a total of 1398 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (Tamura et al. 2011).
Statistical analysis
In order to examine whether the enrichment for recoding a Tyr into a Cys codon is significant, we performed a test for goodness of fit. Specifically, we counted how many times TACG occurs in coding regions and in which frame it occurs, and thus, what is the amino acid change predicted to occur upon RNA editing. We then compared it to the distribution we obtained from sites detected to be edited (>10%) after overexpressing tadA. See Supplemental Table S3 for numbers and calculations.
Confocal microscopy
Cells were grown for 3.5 h without arabinose until reaching mid-logarithmic phase and then induced with 0.2% arabinose (final concentration) for 1 h. Cells were visualized under a confocal microscope (LSM 780, Zeiss) to obtain high-resolution images (10 µm). As a control, we used uninduced cells that were taken from the same culture prior to adding arabinose. Image processing was performed using FIJI (Schindelin et al. 2012).
Data access
The RNA and DNA sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP103577. The Sanger sequences that correspond to the chromatograms in this study have been submitted to the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers MF554632–MF554636.
Supplementary Material
Acknowledgments
We thank the DIP foundation for grant support. We thank the Minerva Foundation for establishment of the Minerva Center for Live Emulation of Genome Evolution in the Lab. We thank Professor Jan Michiels and Dr. Natalie Verstraeten from KU Leuven–University of Leuven for providing strains and plasmids. We thank Dima Zabezhinsky for his help in the confocal microscopy analysis. We thank Shlomit Gilad and the INCPM center at the Weizmann Institute for their part in sequencing the total DNA and RNA samples of E. coli. We thank Nathalie Balaban, Erez Levanon, Avigdor Eldar, Rotem Sorek, Deborah Fass, and all the people in the Pilpel lab for useful discussion and comments.
Author contributions: D.B.Y. raised the original idea and performed all the experiments; E.M., R.T., C.S., and O.D. participated in experiments; D.B.Y., E.M., O.D., and Y.P. designed the experiments; D.B.Y., E.M., T.B., O.D., and Y.P. analyzed the data; Y.P. supervised the project; D.B.Y., E.M., S.S., O.D., and Y.P. interpreted the results; D.B.Y. and Y.P. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.222760.117.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yaacov D, Avital G, Levin L, Richards AL, Hachen N, Jaramillo BR, Nekrutenko A, Zarivach R, Mishmar D. 2013. RNA–DNA differences in human mitochondria restore ancestral form of 16S ribosomal RNA. Genome Res 23: 1789–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, Blumberg A, Schlesinger O, Bieri P, Greber B. 2016. Mitochondrial 16S rRNA is methylated by tRNA methyltransferase TRMT61B in all vertebrates. PLoS Biol 14: e1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 24: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Oh J, Hanson MR, Bukowski R. 2013. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet 9: e1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14: 685–695. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Maisonneuve E. 2015. Remarkable functional convergence: alarmone ppGpp mediates persistence by activating type I and II toxin-antitoxins. Mol Cell 59: 1–3. [DOI] [PubMed] [Google Scholar]
- Grice LF, Degnan BM. 2015. The origin of the ADAR gene family and animal RNA editing. BMC Evol Biol 15: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. 2008. The Vienna RNA Websuite. Nucleic Acids Res 36: W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. 2012. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7: 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V. 2011. When you can't trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci 68: 567–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskinen K, Varjosalo M, Skurnik M. 2015. Absence of YbeY RNase compromises the growth and enhances the virulence plasmid gene expression of Yersinia enterocolitica O:3. Microbiology 161: 285–299. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscovitch-Brauer N, Alon S, Porath HT, Elstein B, Unger R, Ziv T, Admon A, Levanon EY, Rosenthal JJ, Eisenberg E. 2017. Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell 169: 191–202.e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Q, He Y, Chen L, Hao C, Jiang C, Li Y, Dai Y, Kang Z, Xu J-R. 2016. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res 26: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losey HC, Ruthenburg AJ, Verdine GL. 2006. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat Struct Mol Biol 13: 153–159. [DOI] [PubMed] [Google Scholar]
- Oldenkott B, Yamaguchi K, Tsuji-Tsukinoki S, Knie N, Knoop V. 2014. Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 20: 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Gerdes K. 1999. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol 32: 1090–1102. [DOI] [PubMed] [Google Scholar]
- Poulsen L, Refn A, Molin S, Andersson P. 1991. Topographic analysis of the toxic Gef protein from Escherichia coli. Mol Microbiol 5: 1627–1637. [DOI] [PubMed] [Google Scholar]
- Poulsen LK, Larsen NW, Molin S, Andersson P. 1992. Analysis of an Escherichia coli mutant strain resistant to the cell-killing function encoded by the gef gene family. Mol Microbiol 6: 895–905. [DOI] [PubMed] [Google Scholar]
- Ramaswami G, Li JB. 2014. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res 42: D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O'Connell MA, Li JB. 2013. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods 10: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10: 512–526. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC. 2015. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 59: 9–21. [DOI] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. 2009. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460: 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IX, Grunseich C, Chung YG, Kwak H, Ramrattan G, Zhu Z, Cheung VG. 2016. RNA–DNA sequence differences in Saccharomyces cerevisiae. Genome Res 26: 1544–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J, Gerber AP, Keller W. 2002. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J 21: 3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S-i, Kitamura A, Grosjean H, Bessho Y. 2013. Life without tRNAArg–adenosine deaminase TadA: evolutionary consequences of decoding the four CGN codons as arginine in Mycoplasmas and other Mollicutes. Nucleic Acids Res 41: 6531–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhidkov I, Nagar T, Mishmar D, Rubin E. 2011. MitoBamAnnotator: a web-based tool for detecting and annotating heteroplasmy in human mitochondrial DNA sequences. Mitochondrion 11: 924–928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.