Abstract
In the era of genome-wide association studies (GWAS) and personalized medicine, predicting the impact of single nucleotide polymorphisms (SNPs) in regulatory elements is an important goal. Current approaches to determine the potential of regulatory SNPs depend on inadequate knowledge of cell-specific DNA binding motifs. Here, we present Sasquatch, a new computational approach that uses DNase footprint data to estimate and visualize the effects of noncoding variants on transcription factor binding. Sasquatch performs a comprehensive k-mer-based analysis of DNase footprints to determine any k-mer's potential for protein binding in a specific cell type and how this may be changed by sequence variants. Therefore, Sasquatch uses an unbiased approach, independent of known transcription factor binding sites and motifs. Sasquatch only requires a single DNase-seq data set per cell type, from any genotype, and produces consistent predictions from data generated by different experimental procedures and at different sequence depths. Here we demonstrate the effectiveness of Sasquatch using previously validated functional SNPs and benchmark its performance against existing approaches. Sasquatch is available as a versatile webtool incorporating publicly available data, including the human ENCODE collection. Thus, Sasquatch provides a powerful tool and repository for prioritizing likely regulatory SNPs in the noncoding genome.
Major efforts are being made to understand the links between genetic variation within human populations and predisposition to a wide range of common diseases and traits. Currently, more than 2000 genome-wide association studies (GWAS) have identified almost 17,000 single nucleotide polymorphisms (SNPs) (Welter et al. 2014), but as yet, their contribution to functional variation has been limited. This is mainly because most SNPs (∼94%) associated with common diseases and traits are in noncoding regions of the genome (Rockman and Kruglyak 2006; Maurano et al. 2015), where their functional role is unclear and the genes whose expression they affect are not known.
Regulation of gene expression occurs via the binding of tissue-specific and general transcription factors (TFs) to regulatory sequences (promoters, enhancers, and boundary elements). Whereas promoters can be easily linked to the genes they regulate, enhancers and boundary elements are widely dispersed, lying tens to thousands of kilobases (kb) upstream or downstream, within the genes they regulate or within the introns of unrelated genes (Natoli and Andrau 2012; Pennacchio et al. 2013). It has been suggested that many SNPs may influence gene expression by altering the binding of transcription factors to these regulatory elements (Bauer et al. 2013; Pasquali et al. 2014). Importantly, recent improvements in chromosome conformation capture (3C) techniques have enabled us to link remote regulatory elements to the genes they control; however, performing informative 3C depends on identifying the causal regulatory SNP (Edwards et al. 2013; Hughes et al. 2014; Davies et al. 2016) or a small set of prioritized variants.
Interpretation of GWAS data is frequently confounded by linkage disequilibrium (LD). Hence, causative SNPs within a group of linked polymorphisms are frequently prioritized by intersection with annotated regulatory elements or their potential to alter derived TF binding motifs. Major drawbacks of such approaches are their dependency on incomplete ChIP-seq data and associated transcription factor position weight matrices (PWMs) in specific cell types. Such analyses are entirely dependent on knowing which transcription factors bind which motifs and how binding is affected by sequence variation in the cell type under investigation, which is known for fewer than 100 of the 2000–3000 known transcription factors in a very limited number of cell types (Vaquerizas et al. 2009).
DNase I accessibility assays are unbiased and identify all common regulatory elements (Thurman et al. 2012). Regulatory SNPs that alter TF binding can be identified from altered chromatin accessibility at individual regulatory elements. At high sequence read depths, stable TF binding at such elements can be assessed at near base-pair resolution using DNase-seq digital footprints (Fig. 1A; Hesselberth et al. 2009; Neph et al. 2012). Therefore, in principle, analysis of digital footprints could reveal alterations in TF binding (Sung et al. 2016; Vierstra and Stamatoyannopoulos 2016) due to SNPs (Maurano et al. 2015; Moyerbrailean et al. 2016). In practice, this approach is limited by the necessity to generate DNase-seq data from appropriate cell types with and without the informative SNPs. Furthermore, depending on the level of DNase-seq signal at different genomic regions, digital footprints may be weak and consequently difficult to interpret.
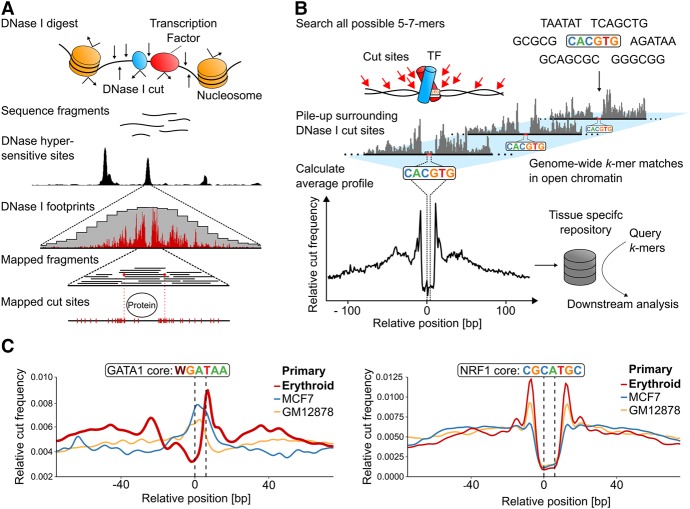
Figure 1.
Generation of k-mer-based, genome-wide average footprints. (A) The principle of generating the digital DNase I footprints occurring in a single genomic location. Exposure of nuclei to DNase I causes frequent double cuts in open chromatin regions, releasing small fragments that can be identified by sequencing. Deep sequencing allows for high-density mapping of the ends of these fragments, which represents the DNase I digestion points and reveals positions protected by TF binding. (B) Sasquatch overview. Genome-wide cut frequency profiles are piled up over all occurrences of all possible short sequences (k-mers) within DNase I hypersensitive sites. For k-mers associated with TF binding, the resulting average cut frequency profiles display average footprints characterized by a cut-depleted center flanked by shoulders of enriched cut frequency. (C) Average profiles reflect tissue specificity of TF footprints as demonstrated by an erythroid-specific TF GATA1 and by a ubiquitous TF NRF1. Average cut profiles were calculated from DNase-seq of primary erythroid, breast cancer (MCF-7), and B-lymphocyte cells (GM12878). The highlighted dotted black lines indicate the location of the motif within the footprint.
Meta-approaches that overcome these limitations use DNase footprints and allele-specific sensitive signals, piled-up over matches to known PWMs (Pique-Regi et al. 2011; Maurano et al. 2015; Moyerbrailean et al. 2016). They are able to make predictions of binding and how this may be affected by variants at bp resolution; however, again, such analyses depend on incomplete catalogs of PWMs, which limits their use. As yet, there is no clear consensus on how best to identify or predict regulatory SNPs using DNase-seq footprints (Sung et al. 2016; Vierstra and Stamatoyannopoulos 2016).
To address these limitations, we have developed Sasquatch, a comprehensive and unbiased approach to prioritize regulatory variants based on DNase footprinting.
Using both publicly available data and data generated in primary erythroid cells as part of this study, we illustrate the efficiency of Sasquatch in prioritizing SNPs associated with previously validated changes in TF binding, re-evaluate regulatory SNPs reported in the literature, and demonstrate its use for interpreting GWAS data.
Results
We have developed Sasquatch to assess the cell-type–specific TF binding potential based on DNase footprints. Sasquatch requires no prior knowledge of the underlying binding motifs. Additionally, it requires only a single DNase-seq track of any genotype, with only moderate sequence coverage. Sasquatch uses all possible variations of the four-letter DNA code to assess the potential of short sequences (k-mers) to be bound directly by proteins, in a specific cell type. Importantly this approach uses direct sampling of biological data rather than in silico prediction. Preanalysis of all potential 5-, 6-, and 7-mers means that any sequence can be rapidly interrogated for protein binding within a user-friendly webtool (http://apps.molbiol.ox.ac.uk/sasquatch/cgi-bin/foot.cgi). We preprocessed all human ENCODE DNase-seq data sets (Maher 2012; Romanoski et al. 2015) and other publicly available data to create a large repository of cell- and tissue-specific DNase footprints.
Using Sasquatch to identify cell-specific binding motifs within DNase I hypersensitive sites
Sasquatch searches for all 5-, 6-, and 7-mers that lie within the DNase I hypersensitive sites (DHSs) present in a specific cell type. It stores their occurrences, piles up the surrounding DNase I cut sites (Fig. 1B), and produces a footprint profile for each k-mer. Importantly, the footprints are normalized for DNase I sequence bias and strand specificity (see Supplemental Methods; Supplemental Fig. S1; Lazarovici et al. 2013; He et al. 2014; Sung et al. 2014; Yardimci et al. 2014).
Previously reported DNase footprints for known DNA binding proteins (Church and Gilbert 1984; Hesselberth et al. 2009; Holwerda and de Laat 2012; Sung et al. 2014) were faithfully reproduced in the Sasquatch analysis, with a depletion of DNase I cuts over known protein bound sequences (the footprint) flanked by an increased frequency of cutting (shoulders) at the edges of the bound protein (Fig. 1B,C). The depth of the footprint relative to the shoulders reflects occupancy of the bound site (Neph et al. 2012; Sung et al. 2014). Henceforth, we refer to the distribution of relative cut frequencies as the profile and the characteristic average footprint within a profile as the footprint.
To quantify footprints, we implemented an algorithm to detect the shoulders within a profile. We then calculated the shoulder-to-footprint ratio (SFR)—the ratio of the relative cut frequency of the shoulders to the footprint (Fig. 2A). Based on this method for calculating the SFR, profiles lacking a visible footprint score around 1.0–1.2, while distinct footprints score >1.4.
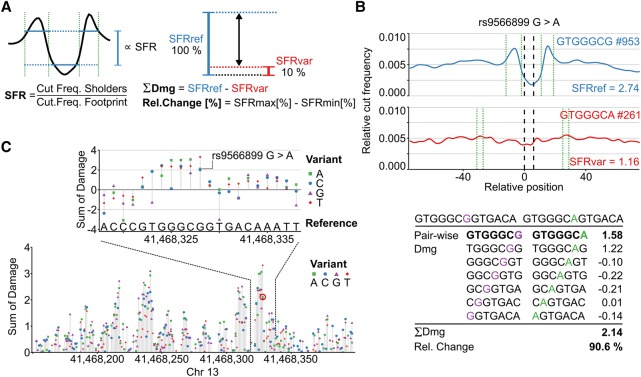
Figure 2.
Predicting the damage potential of single nucleotide variants. (A) The average footprint strength is quantified by calculating the shoulder-to-footprint ratio (SFR). The damaging potential (Dmg) is quantified as the difference between the reference and the variant SFR and also expressed as the relative change (Rel. Change) in footprint strength with respect to the higher SFR. (B) The damaging potential of a SNP, for example, rs9566899, is then assessed by summing the pairwise damage using a sliding window (shown below). The average profiles of the highest scoring k-mer pair are plotted (blue = reference, red = variant). Green dotted lines bracket the automatically detected shoulder regions. The number of occurrences of the respective k-mer within DHSs is indicated by #. (C) Illustration of extending the approach to perform in silico mutation by predicting the impact of every possible mutation at every position in a region of interest. The resulting in silico mutation plots highlight potentially severe mutations and reveal sequences associated with strong TF binding potential in an unbiased and tissue-specific manner. The rs9566899 A allele is indicated (red circle in lower plot).
Sasquatch allows informative analysis of a wide range of DNase-seq protocols sequenced to variable depths
To ensure usability across publicly available DNase-seq data, we confirmed that the number of DHSs and the sequencing depth do not affect the detection or the shape of footprints (Supplemental Figs. S2, S3). Different DNase-seq protocols only affect the shoulder shape (Supplemental Fig. S4). Sasquatch can even be applied to low-input DNase-seq protocols (Lu et al. 2016) (liDNase-seq, Supplemental Fig. S5), allowing evaluation of very low cell numbers. Although we found no general relationship between frequency of k-mer occurrence and quality of footprint, for k-mers with very low (<100) representation within DHSs, we observed a significant increase in background noise due to undersampling. Therefore, while care is required when interpreting average profiles below this frequency, their underrepresentation in active regions within a given cell type may suggest that such sequences may not be of functional importance in such cells. Finally, it is important to note that the SFR may be influenced by the nature of the TF tested. For example, we find the GATA1 core motif has a consistently lower SFR than the NRF1 core motif (Fig. 1C), probably reflecting the nature of their interactions with DNA and features of their structures. Considering the increased use of ATAC-seq (Buenrostro et al. 2013), we adapted Sasquatch for the use of ATAC-seq data. However, intrinsic confounding factors in ATAC-seq, such as Tn5 cutting preferences, are currently not well understood (Tsompana and Buck 2014; Madrigal 2015). Using ATAC-seq data (Supplemental Fig. S6) and matched background generated in primary erythroid cells, we observed that ATAC-seq recovers some but not all footprints that are detectable in DNase-seq data over known binding sites, and the footprint shapes differ strikingly (Supplemental Fig. S7). Furthermore, we have reason to believe that the transposase introduces different cutting biases (MC Suciu, R Schwessinger, M Kassouf, J Telenius, DR Higgs, JR Hughes, in prep.), suggesting there is a need for more research into the transposase-based cleavage before it can be modeled reliably for footprinting. We made the software adaption to ATAC-seq data available for further investigation of the underlying differences in footprinting but discourage its use for exhaustive variant screens at this stage.
Average profiles of protein–DNA interactions identify widely expressed and cell-restricted TFs
Transcription factors such as NRF1 (Chan et al. 1993) show clear, high-scoring footprints across all cell types analyzed (Supplemental Fig. S8), while the profiles for GATA1, a known regulator in erythroid tissue, form a distinctive profile with a high-scoring footprint only in erythroid cells (Fig. 1C). Based on such profiles, we can infer details about TF binding across cell types. For example, GATA1 binding is frequently associated with binding of its partner protein (TAL1) to a half E-box motif 8 base pairs (bp) 5′ of the GATA core motif (Kassouf et al. 2010). This is mirrored by the one-sided expansion of the average GATA footprint in the 5′ direction and consequently reduced prominence of the 5′ shoulder (Supplemental Fig. S9).
Assessing the potential effects of DNA variants on TF binding
By changing a specific consensus sequence scored as a footprint, Sasquatch can report how well a variant of this sequence would also act as a binding site for its cognate protein in that cell type. The difference between the reference and variant footprint is calculated as a “damage-score” (Dmg), which quantifies the relative ability of the variant sequence to form a footprint and so its effect on TF binding (Fig. 2A). A positive total damage score predicts that a variant will damage the average footprint, while a negative score predicts a positive influence on protein binding. To increase robustness, we consider each variant in its 13-bp context. Additionally, Sasquatch provides a visual assessment illustrating the potential of each nucleotide change to damage the predicted footprint. (Fig. 2B) and can quickly score 1000s of variants in batch mode as all k-mer data are precalculated.
Because sampling true neutral variants from binding site-rich open chromatin is difficult, we devised a simulation strategy to calibrate the damage score thresholds empirically at 0.5 and 1.0 for a relaxed and stringent cutoff, respectively (Supplemental Fig. S10; Supplemental Methods). The meta-footprinting approach can also be used within k-mers covering a known binding motif to dissect the relative importance of each base at each position by in silico mutating and predicting the severity of base changes (Supplemental Fig. S11). Furthermore, changes in the footprint shape can indicate differential factor binding (e.g., differential E-box usage). This approach can also be applied to longer genomic sequences (up to 1000s of bp) limited only by run time. For example, Sasquatch can analyze a complete DHS in the form of in silico mutation, predicting the impact of every possible base substitution at every genomic position. These in silico mutation plots (Fig. 2C) can reveal clusters of single positions within a DHS that are particularly sensitive to variation which, in turn, can reveal the positions of known TF binding sites. As a guide to which TFs might bind the k-mers identified, we integrated the JASPAR 2016 (Mathelier et al. 2016) motif database into the analysis.
Sasquatch validation in erythroid tissues
Many regulatory elements, as well as the TFs and cofactors binding them, have been well characterized in erythroid cells; we therefore initially tested Sasquatch using data generated from human and mouse erythroid cells. Interestingly, when we tested all k-mers perfectly matching the binding motifs of several TFs active in erythroid cells (GATA1, NRF1, E-box, and NFE2), we surprisingly found that we could detect strong evidence of footprints in the majority of occurrences in DHSs, suggesting that the presence of a perfect motif is sufficient to allow binding of that TF as long as the chromatin is accessible (Supplemental Fig. S12).
We then took advantage of the existence of deeply characterized regulatory elements within the erythroid system and the observation that the large majority of in silico mutations within open chromatin regions do not alter binding potential (Supplemental Fig. S13) to test the ability of Sasquatch to identify the positions of potential TF binding sites in regulatory regions.
The alpha-globin promoters interact with five distal regulatory elements with the signatures of enhancers and bind many of the known erythroid master transcription factors, of which two (called R1 and R2) act as strong enhancers in vivo (Hay et al. 2016). Sasquatch in silico mutation plots derived from mouse erythroid cells effectively identified the known conserved TF binding motifs within these elements with one notable exception.
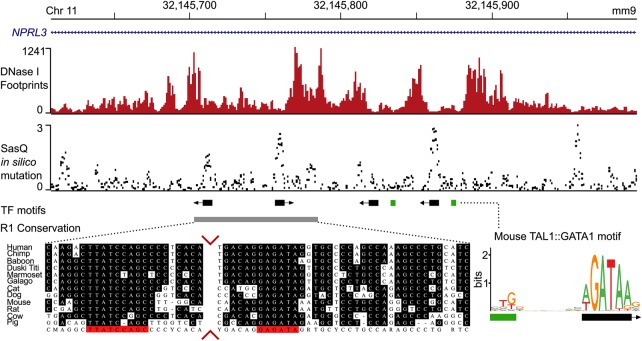
The R1 element was initially identified using sequence conservation analysis across 23 species (Hughes et al. 2005) and appeared to be a simple structure containing just two conserved GATA1 binding sites which were presumed to direct all of the GATA1 and TAL1 binding at this element (Fig. 3). However, analysis of DNase-seq data from mouse primary erythroid cells (Ter 119+) (Hosseini et al. 2013; Marques et al. 2013) shows that the DHS extends beyond these conserved GATA binding sites, incorporating two additional GATA sites. All four sites are associated with strong DNase footprints. Aligning the damage plot from Sasquatch with both the conservation analysis and DNase footprinting shows that it detects all four E-box-GATA1 binding motifs, two of which were initially discarded on the basis of their lack of conservation. These results emphasize the unique ability of high-resolution DNase-seq assays to reveal species-specific regulatory features. Our meta-analysis approach can detect the same functional elements where data of sufficient depth are not publicly available or difficult to generate due to cell number limitations.
Figure 3.
Overlay of DNase I footprints, Sasquatch in silico mutation predictions, and sequence conservation. The top panel shows deep DNase I footprints in mouse primary erythroid cells over the mouse alpha-globin locus R1 enhancer, which is present within an intron of NPRL3. Sasquatch's in silico mutation analysis using the same DNase-seq data reveals clusters of predicted high-damaging variants overlapping the footprints and known GATA1 binding motifs (middle panel). While sequence conservation analysis can identify the two leftmost binding sites, the in silico mutation analysis can also identify the murine-specific binding sites. Sequence conservation analysis adapted from Hughes et al. (2005). The conservation panel was trimmed to visualize both conserved sites (red arrows).
Analysis of sequence variation underlying loss or gain of TAL1 binding
To demonstrate the effectiveness of Sasquatch, we interrogated the effect of two well-characterized binding site mutations and SNPs within regions previously shown to variably bind the transcription factor TAL1 (Lower et al. 2013) using tissue-matched DNase-seq data generated from human primary erythroid cells.
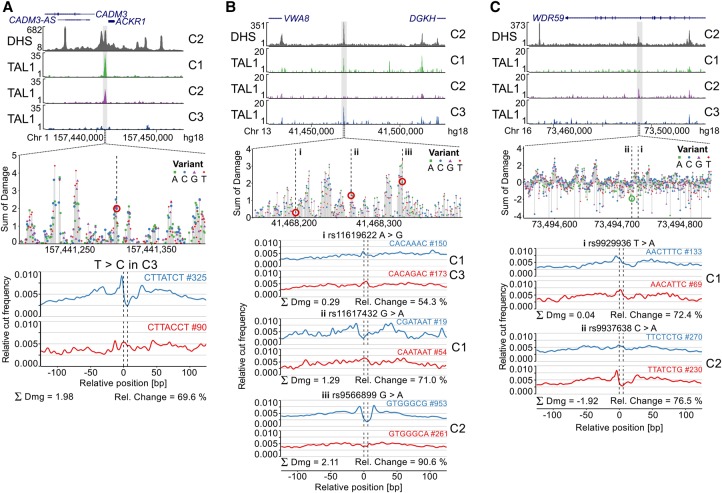
The expression of the Duffy blood group protein is known to be highly polymorphic due to the linked resistance to Plasmodium vivax (de Carvalho and de Carvalho 2011). The T-to-C substitution in the ACKR1 gene promoter abrogates its expression (Tournamille et al. 1995), which Sasquatch detects as a strong damage score of 1.98 (Fig. 4A). The block of potential damage overlies a known GATA1 binding site and causes complete loss of binding of the GATA1 cofactor TAL1 in a homozygous individual (Supplemental Fig. S6).
Figure 4.
Prioritizing SNPs associated with differential TAL1 binding at single base-pair resolution. Several SNPs associated with differential TAL1 ChIP-seq peaks between three individuals (Lower et al. 2013) (C1, C2 and C3) were analyzed using Sasquatch. (A) A known T-to-C substitution at position −33 in the promoter region of the ACKR1 gene that abrogates its expression causing a form of the Duffy-negative genotype (de Carvalho and de Carvalho 2011). Sasquatch identifies a distinct depletion of a GATA1 footprint in the variant sequence. The variant lies within a cluster of damaging variants, strongly indicating a TF binding site, and is associated with abolishment of TAL1 binding in C3. (B) Three SNPs (rs11619622, rs11617432, and rs9566899) have been identified within a DHS on Chromosome 13 overlapping with TAL1 binding in C1 and C3 but not C2. Analysis of the rs11619622 A>G SNP present in C1 and C3 did not show striking differences between the reference and variant profile. The rs11617432 G>A SNP present only in C1 appears to disrupt a less common GATA binding motif, is predicted to have an intermediate damaging potential, but appears insufficient to abolish TAL1 binding. In contrast, the rs9566899 G>A SNP, found within a potential C2H2 zinc finger motif and present only in C2, shows the strongest predicted damage and appears sufficient to abrogate TAL1 binding in C2. (C) Sasquatch is able to predict the introduction of potential novel binding sites. Two SNPs (rs9929936 and rs9937638) have been found in an intragenic DHS associated with TAL1 binding only in C2. The rs9929936 T allele present in C1 potentially shows weak binding potential, but no alteration is caused by the variant. In contrast, the rs9937638 C>A SNP present in C2 shows the potential to introduce a novel GATA site. The sliding window approach identifies rs9937638 C>A to have a strong negative damaging potential in line with the observed TAL1 binding. Interestingly, the in silico mutation plot identifies nearby peaks of damage, potentially indicating bound motifs within the DHS that may support TAL1 binding.
Similarly, we analyzed 16 SNPs previously shown to also be associated with the loss or gain of TAL1 binding in erythroid tissues (Lower et al. 2013). Twelve of the 16 variants show intermediate or high damage scores (five intermediate: absolute total damage >0.5; seven high: absolute total damage >1.0). These 12 SNPs were associated with altered footprints either in the reference or in the variant profiles (Fig. 4B,C; Supplemental Table S1). The disrupted footprints included GATA1 sites, a known partner of TAL1, but also additional erythroid TF binding sites such as KLF1 (Kassouf et al. 2010).
In one illustrative example, three variants lie within the same intergenic hypersensitive site upstream of the DGKH locus on Chr 13 (Fig. 4B), which shows polymorphic binding of TAL1. The rs11619622 variant is found in two individuals (C1 and C3) positive for TAL1 binding in this region; rs11617432 is also found in one of these individuals (C1), and a third SNP rs9566899 is found in an individual negative for TAL1 binding (C2). As these data stand, it is not possible to prioritize a mechanism where rs11619622 promotes binding of TAL1 over a mechanism where rs9566899 disrupts binding of TAL1. The lack of any negative damage scores for SNPs rs11619622 and rs11617432 (gain of binding potential) and the high damage score of rs9566899 (2.11) strongly suggest that loss of binding potential is due to rs9566899 (marked “iii” in Fig. 4B; Supplemental Fig. S14). In silico analysis shows rs9566899 to be in a region of high potential damage (Fig. 4B) indicative of a TF binding site, which is most similar to KLF1, a cofactor of TAL1 (Supplemental Fig. S9; Kassouf et al. 2010; Tallack et al. 2010), suggesting that TAL1 binding to this element is KLF1-dependent.
Formation of new elements can be as detrimental to gene regulation as their disruption (De Gobbi et al. 2006). However, without regulatory data from the correct cells and genotype, these events are undetectable and so of unknown frequency. Sasquatch can prioritize the investigation of such events by detecting an enhanced footprint (negative damage value) associated with a sequence variant.
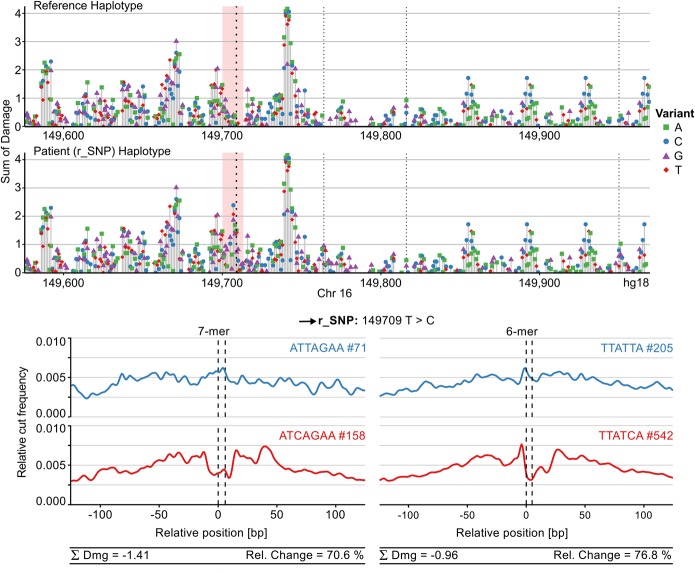
In a study by De Gobbi et al. (2006), the down-regulation of the alpha-globin genes in an alpha-thalassemia patient was caused by the formation of a cryptic promoter element in the alpha-globin locus. The position of this element between the alpha-globin genes and enhancers suggested that it was interfering with the interaction of the enhancers and promoters, leading to gene down-regulation. Of the seven variants associated with this new element, six could be discounted by analysis of unaffected family members to leave a single SNP 195 as the causative variant. Sasquatch analysis of these seven variants clearly identifies SNP 195 as the strongest change in footprint formation and, importantly, shows it to be a strong gain of function (Fig. 5; Supplemental Fig. S15), coherent with the genetics and proposed mechanism. Motif analysis in the original publication and in Sasquatch showed the 195 variant to form a new GATA1 site, and GATA1 and TAL1 were shown to be recruited only in the affected individual. The in silico analysis of this region in both the affected and reference haplotype showed the existence of clusters of potential binding sites around the novel GATA1 site (Fig. 5), providing insight into what other motifs may be required for the gain of function.
Figure 5.
Characterizing a gain-of-function SNP. Sasquatch is capable of identifying a gain-of-function SNP associated with down-regulation of the HBA genes (De Gobbi et al. 2006) and characterizing its regulatory context using in silico mutation analysis. The black arrow indicates the position of the regulatory SNP (r_SNP). The in silico mutation plot identifies an introduction of a novel TF binding site (red panel) in the patient haplotype. Furthermore, the novel peak appears to lie adjacent to other potentially active binding sites, possibly contributing to the rSNP's regulatory context. Weaker dotted lines indicate known SNPs in the patient, neither of which appears to be associated with a strong change in footprint potential (see Fig. 5; Supplemental Fig. S1). The gain of a novel GATA footprint is captured both in 7-mer and 6-mer analysis, while 6-mers excel at capturing the characteristic shape of the GATA footprint.
Prioritizing variants identified by GWAS for functional analysis
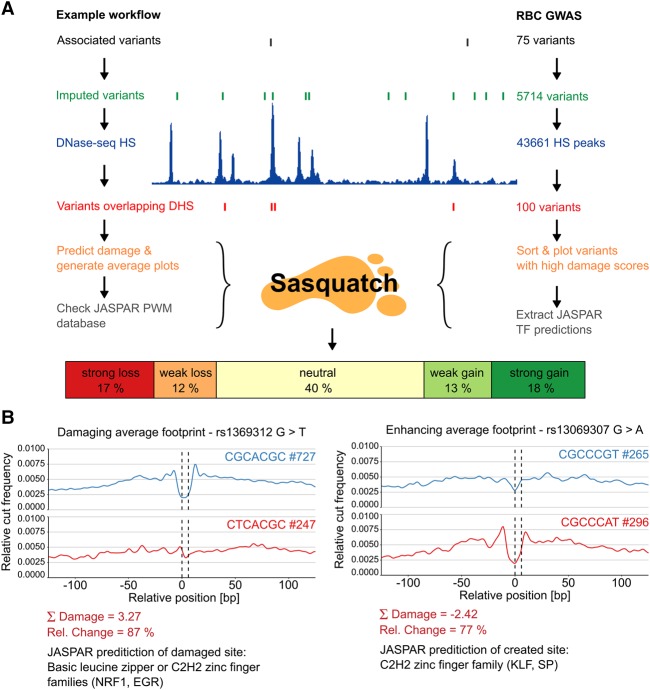
GWAS have identified large numbers of SNPs that are significantly associated with common disease. Considering the very large number of variants usually found in LD with single GWAS hits (Maurano et al. 2012), functional validation of causal changes is logistically and financially prohibitive. To address this, we applied Sasquatch to prioritize GWAS hits for multiple blood cell traits (van der Harst et al. 2012) to rank variants by impact on average TF footprints (Fig. 6).
Figure 6.
Prioritization of GWAS variants with Sasquatch. (A) The workflow for assessing and prioritizing GWAS variants according to their footprint-damaging potential is shown on the left. We employed Sasquatch using variants found to be significantly associated with red blood cell phenotypes (van der Harst et al. 2012). Starting from 75 GWAS-identified SNPs, variants were imputed and filtered for occurrence within genome-wide DHSs in primary erythroid tissue. For the 100 intersecting SNPs, damage scores were calculated and visualized using Sasquatch. (Note that variants not overlapping DHSs might still be interesting because of the potential introduction of novel TF-binding sites.) The top-ranked footprint-damaging (rs1369312 G>T) and enhancing variants (rs13069307 G>A) from this analysis are shown in B.
The GWAS hits (van der Harst et al. 2012) were further imputed to identify all variants in strong linkage disequilibrium using the latest release of 1000 Genomes data. A total of 5714 variants were in LD (r2 > 0.8, 500 kb) with the reported SNPs. All variants were filtered for their location in open chromatin (DHSs), and the damage scores were calculated for the resulting 100 candidate SNPs (Fig. 6A). The damage scores ranged from 4.14 to −3.11, suggesting some variants damage, while others enhance footprints. The variant footprints range from clear loss or gain (98% relative change) to weaker, quantitative changes in footprint strengths (25%). Using this workflow, we prioritized all the candidates into neutral and loss or gain of binding potential, both with two levels of confidence depending on the strength of the damage (Supplemental Table S2). Two of the strongest variants are illustrated in Figure 6B (variants rs1369312 G>T and rs13069307 G>A). The rs1369312 SNP shows a loss of an NRF1 or EGR family TF, with a relative change of 87%, one of the highest scores. At the opposite end of the spectrum, rs13069307 G>A appears to introduce a new binding site for KLF1, with a relative change of 77% in the negative damage score. The result of the Sasquatch workflow is a sorted list of candidate variants (100) with intuitive visualization providing fast and efficient prioritization of thousands of variants for functional analysis.
Using Sasquatch to analyze functionally validated variants from nonerythroid tissues
To demonstrate Sasquatch's utility in nonerythroid tissues, we assessed the impact of previously described functionally validated variants from either prostate (Huang et al. 2014) or breast cancer (Hsiung et al. 2014) GWAS where both publicly available DNase-seq data sets (He et al. 2014) and functional validation in matched cell types were available.
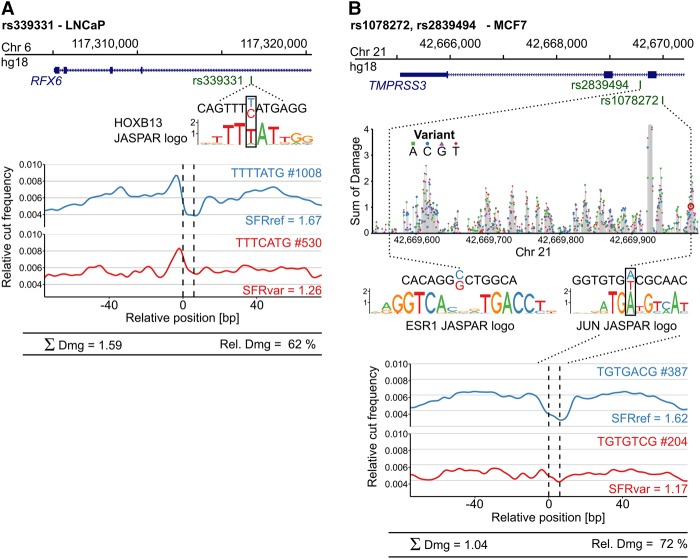
The common SNP rs339331 was identified by multiple GWAS as highly associated with a risk of prostate cancer. The T-to-C change disrupts a potential HOXB13 binding site in intron 4 of RFX6 (Huang et al. 2014). Using ChIP-seq and allele-specific expression analysis, Huang and coworkers functionally validated this variant and demonstrated that it affects HOXB13 binding and modulates RFX6 expression in an allele-specific manner. Sasquatch analysis using DNase-seq in a prostate cancer cell line (LNCaP) showed a severe reduction in the score of the variant footprint and a relative change of 62% (Fig. 7A). Two SNPs, rs2839494 and rs1078272, have been identified as significantly associated with higher survival rates in breast cancer patients (Hsiung et al. 2014). Hsiung and colleagues functionally validated the SNPs using ESR1-responsive luciferase assays and proposed rs2839494 C>G to alter ESR1 binding directly and rs1078272 A>T to alter binding of the cofactor RAD21. Sasquatch predicts only rs1078272 as having a damaging effect in MCF-7 cells (Fig. 7B), with a damage score of 1.04 (72% relative change) and a clear disruption of the footprint (rs2839494: Dmg = 0.039). In silico mutation over the locus places rs1078272 in a cluster of damaging variants in close proximity to other potentially strong binding sequences. The predicted disruption of a potential JUN binding site emphasizes rs1078272 A>T as a likely causal variant, as JUN overexpression has been linked with overall tumor cell aggressiveness in breast cancer cells (Smith et al. 1999).
Figure 7.
Assessing the footprint-damaging potential of functionally validated SNPs in nonerythroid tissues. Sasquatch-predicted damaging potential recapitulates functionally validated SNPs identified from GWAS studies in prostate and breast cancer. (A) SNP rs339331 T>C has been identified as significantly associated with prostate cancer risk and functionally validated to impair RFX6 expression by disrupting a HOXB13 binding site (Huang et al. 2014). Comparison of Sasquatch average profiles reflects the binding impairment and predicts a high damaging potential. (B) Two intronic SNPs (rs2839494 and rs1078272) found within an estrogen response element have been identified as significantly associated with survival in breast cancer patients and are functionally validated (Hsiung et al. 2014). Sasquatch predicts rs1078272 A>T to damage a potential JUN binding site, associated with a clear abolishment of a footprint in the average profile. In silico mutation identified rs1078272 to be located within a cluster of damaging variants. In contrast, rs2839494 C>G, proposed to directly alter ESR1 binding, is not detected. This is most likely because Sasquatch is limited in detecting large motifs spanning multiple noninformative bases, due to the use of short k-mers, which is also reflected in the lack of signal in the in silico mutation plot at that region. For the in silico mutation analysis, the T base at SNP rs107827 A>T retrieved from the hg18 reference genome has been changed from the minor to the major SNP allele.
We could not detect the proposed alteration of ESR1 binding of rs2839494 using Sasquatch and the in silico mutation plot shows no strikingly damaging variants over that specific region (Fig. 7B). This highlights a limitation of the k-mer based approach for detecting binding events over large motifs due to low numbers of matches for long sequences in DHS sites. Although such large TF binding motifs are uncommon, they represent a source of false negatives in Sasquatch and may be better addressed using alternative approaches.
Benchmarking Sasquatch against current state-of-the-art neuronal network approaches
Direct benchmarking for the identification of variants that alter TF binding is difficult, as no gold standard validated sets exist. Recent approaches for prioritizing causal variants have used: (1) expression or DNase I hypersensitive quantitative trait loci (eQTL, dsQTLs); (2) sets of regulatory element-associated variants or noncoding GWAS lead SNPs; (3) correlation with the output of other prediction tools; and (4) massively parallel reporter assays, all of which do not directly imply a change in TF binding associated with the SNP. Additionally, negative sets are commonly derived from random sampling and assumed to be true negatives while the positive sets are often used without consideration of the potentially causal SNPs being in high LD with the sentinel SNPs.
To this end, we have developed a novel benchmark data set that, as directly as possible, addresses the core functionality of Sasquatch, the alteration of TF binding, and yet provides a fair and appropriate comparison with existing relevant approaches.
This data set is based on a genome-wide study of TF binding QTLs (bQTLs) for five transcription factors in lymphoblastoid cell lines (Tehranchi et al. 2016). As these cell lines have been extensively studied in the ENCODE Consortium, DNase-seq data as well as a large number of histone modification and TF ChIP-seq data are publicly available. These data sets allow for a fair comparison of Sasquatch's performance against the neuronal network (NN) approaches (Basset and DeepSEA), whose performance is dependent on such matched deep data sets and include the lymphoblastoid GM12878 cell line in both their training regimes (Zhou and Troyanskaya 2015; Kelley et al. 2016).
For each factor addressed in the bQTL study (JUND, RELA, POU2F1, SPI1), we retrieved the 25 most significant bQTLs, and pooled and matched them against dbSNP. After imputation, we grouped the SNPs into high-confidence LD blocks and predicted the impact of each SNP with Sasquatch, Basset, and DeepSEA and measured the fraction of LD blocks that could be explained by one or more SNPs from each tool. For Sasquatch, we used the total damage, for Basset the SNP accessibility difference (SAD), and for DeepSEA we used the functional significance score (funsig), which combines sequence conservation with the NN output, GM12878 DNase I predictor alone, and the smallest E-value across all GM12878 predictors (TF, histone marks, and DNase I). We used a set of stringent and relaxed thresholds for comparison (Supplemental Fig. S16).
Compared to DeepSEA's funsig score, Sasquatch achieved more explainable blocks using stringent and slightly less using the relaxed thresholds, with Sasquatch showing comparable performance to DeepSEA's DNase I predictor. Interestingly, the majority of blocks that were flagged by Sasquatch but not by both of DeepSEA's DNase I and funsig score were, in fact, flagged up by one of the latter two.
Therefore, scanning exhaustively all cell-type–associated predictors for any change might be a valid approach for prioritizing causal SNPs and, in fact, taking the smallest E-value across all predictors flags up the largest proportion of LD blocks in our test set, although it is difficult to know the number of false positives without a concerted effort by the field to test this enriched data set with in-depth ChIP experiments.
Changes in TF binding may not necessarily always coincide with changes in DNase I hypersensitivity, and as Basset has been specifically trained to predict the latter, its poor performance in predicting changes in TF binding could be expected (Supplemental Fig. S16). However, as it is only trained on ENCODE DNase-seq, we included it in the comparison to demonstrate the capabilities of learning NNs on DNase-seq data alone and as a comparator for the two other approaches.
It is important to note that, while DeepSEA has been simultaneously trained on large collections of ENCODE data, Sasquatch requires only a single data set. Therefore, when diverse genomic data are available for a specific cell type, neuronal network approaches may represent the method of choice, which can, of course, be performed in conjunction with Sasquatch analysis. However, if such resources are limited, as is most often the case, Sasquatch can yield comparable results from a single DNase-seq data set.
Discussion
An important current challenge in molecular genetics is to understand how genome variation alters gene expression and how this defines genetic traits and predisposition to disease. However, most variants found in GWAS studies lie in noncoding DNA, and therefore, being able to discriminate between true regulatory SNPs and variants in LD with them is a major roadblock in interpreting GWAS.
Most regulatory elements can be identified by DNase-seq and/or ATAC-seq (Schaub et al. 2012; Levo and Segal 2014). The patterns of chromatin accessibility vary greatly between cell types and regulatory SNPs and thus need to be investigated in the appropriate cell type. Simple intersection with these data does not differentiate between causal SNPs and nonfunctional linked sequence variants within these regions. To address this, we have developed Sasquatch to rank and prioritize SNPs lying within regions of open chromatin for their potential to cause changes in gene expression based on their likelihood of causing a change in TF binding. Sasquatch combines the power of comprehensive and unbiased k-mer-based analysis with the single base-pair resolution of DNase digital footprinting.
Other approaches proposed to predict regulatory SNPs include (1) prioritization based on overlaps with functional annotations like ENCODE data (Kircher et al. 2014; Gulko et al. 2015); (2) changes in PWM matching scores (Coetzee et al. 2015; Schmidt et al. 2015); (3) TF-centric approaches, via DNase footprints or allelic skewed DNase-seq reads over precomputed PWMs (Pique-Regi et al. 2011; Maurano et al. 2015; Moyerbrailean et al. 2016); and (4) sequence-based machine learning approaches using defined training sets of TF ChIP-seq and/or chromatin accessibility as a proxy (Arvey et al. 2012; Alipanahi et al. 2015; Lee et al. 2015; Svetlichnyy et al. 2015; Zhou and Troyanskaya 2015; Kelley et al. 2016; Zeng et al. 2016). Specifically, the recent convolutional neuronal network-based approaches show promise for integrating a wider sequence context into their predictions (Alipanahi et al. 2015; Zhou and Troyanskaya 2015; Kelley et al. 2016). This would complement the Sasquatch's k-mer-based approach. Nevertheless, no clear consensus has been reached about which approaches are able to accurately and efficiently prioritize regulatory SNPs. Since the number of sufficiently validated regulatory variants is still limited and a gold standard set is missing, benchmarks vary largely between publications (Li et al. 2017) depending on how the test data sets have been constructed.
All of these methods have inherent limitations. ChIP-seq or chromatin accessibility peaks frequently contain common and presumably neutral polymorphisms, and so presence within a peak alone is not evidence of disruption. Additionally, the number of TFs currently amenable to ChIP-seq is only a fraction of the total number (2000–3000) and cannot cover the complete regulatory landscape of even a single cell type. Similarly, due to the current incomplete understanding of the sequences used by transcription factors across human development, any dependence on PWM collections will necessarily be skewed and contain many false negatives. Machine learning approaches do away with the limitation of PMWs, learning directly from the underlying sequences, but are highly dependent on the availability of extensive training sets, are complex, computationally demanding, and only indirectly predict the effect on protein binding.
Sasquatch provides a relatively simple and yet informative approach, requiring only a single DNase-seq data set from the appropriate cell type. Furthermore, it can use data from any genotype to assess variants that are appropriate to that cell type. Sasquatch can use publicly available data of any reasonable depth and quality, generated by any of the existing DNase-seq protocols, including low-input DNase-seq protocols (Lu et al. 2016), giving access to analysis of data from small cell numbers (Supplemental Fig. S5). The approach allows for the reuse of any existing data from the appropriate cell type, and the current implementation gives access to all of the current human ENCODE data that has been preprocessed and is available to the scientific community via a rapid user-friendly webtool. Within this platform, the average profiles of the footprints are automatically plotted and quantified relative to reference genome plots and within the context of the whole regulatory elements using in silico mutation plots. To enable custom analysis of unpublished data, all analysis tools have been made available via GitHub and as Supplemental Software.
One limitation of the k-mer-based approach is its inability to evaluate the influence of cobinding factors or the genomic context on the average footprint of the sequences interrogated. However, as our implementation of the algorithm is rapid and computationally efficient, this can potentially be overcome in the future by developments that involve longer (gapped) k-mers (Ghandi et al. 2014). Another limitation of Sasquatch arises from the inherited properties of DNase footprints. TFs that yield only weak footprints will result in weaker average footprints and TFs that do not have a DNase footprint in the first place will be impossible to detect. This consequently translates into the ability to infer potential binding changing variants and the current implementation might disfavor certain TFs with weak footprints. Furthermore, if a variant would exchange the TF bound to a sequence rather than abolishing binding, the damage-based prediction might not be able to pick this up. However, investigating the associated profiles with such changes can prove useful for probing differential factor occupancy (see E-box usage example, Supplemental Fig. S11D). It is important to note that some changes in transcription factor binding may not be associated with a change in an underlying binding site if the binding event is dependent on long-range interactions with other elements in the manner of promoters and regulatory elements, which would fit with observations that variations in ChIP-seq signal are frequently not explainable by very local SNPs (Karczewski et al. 2011). However, in this case, one would assume that any genetic association (such as in GWAS) would have highlighted this distal site and so be tractable by the Sasquatch approach.
In summary, driven by the current challenges in the GWAS field, we have developed an approach to quickly and effectively prioritize sequence changes based on their ability to bind protein in vivo using cell- and tissue-specific data. We have provided this as a public interface which removes any major computational barriers and gives access to a large and ever increasing repository of public data with the aim of improving our understanding of how noncoding variants affect gene expression and predisposition to human disease.
Methods
Cell source, culture, preparation, and DNase-seq protocol (tissue and background)
Human primary erythroid stem cell progenitors were isolated from peripheral blood, using CD34 coupled magnetic beads. Fifty million cells were used for the DNase-seq protocol.
The DNase-seq protocol was performed as previously published (Hosseini et al. 2013). For the background libraries, 100 ng of genomic DNA were incubated with DNase I for 3 min, and libraries were created as described in Supplemental Methods.
ATAC-seq protocol
ATAC-seq was performed as previously published (Buenrostro et al. 2013). For details, see also Supplemental Methods. For the ATAC-seq background, 100 ng of genomic DNA was incubated with the Tn5 transposase and the library was generated following the protocol described in Supplemental Methods.
Data preprocessing
Paired-end reads were processed using our in-house DNase-seq pipeline. In brief, reads were mapped using Bowtie (v0.12.8) (Langmead et al. 2009). Sequencing adapters of unmapped reads were trimmed using Trim Galore! (v0.3.1) (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and remapped. Duplicates were removed using SAMtools (v0.1.19) (Li et al. 2009), and regions blacklisted for mappability issues were removed (Source: ENCODE, http://hgdownload.cse.ucsc.edu/goldenpath/hg19/encodeDCC/wgEncodeMapability/wgEncodeDukeMapabilityRegionsExcludable.bed.gz).
DHSs were then called as peaks from the DNase-seq signal. Peak-calling was performed using a SeqMonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/)-like contig generator with manually optimized parameters according to visual inspection. Detailed information and parameters for mapping and peak-calling are listed per data set in Supplemental Table S3. For filtering GWAS variants, we used a slightly relaxed stringency peak call using MACS2 (Zhang et al. 2008) with default settings (v2.0.10.20131216, default options include -q 0.05). MACS2 peaks were also used for mouse data.
From the aligned reads, strand-specific DNase I cut sites were extracted by assigning only the 5′ end of each mapped read to the respective reference strand. Thus, each DNase I cut is attributed to the first base of the read, which is the first base after the expected cut site. Minus strand cut positions were corrected by subtracting 1 from each mapped 5′ end.
ATAC-seq data were processed as described for the DNase-seq data, but the mapped cut sites were corrected by offsetting plus strand reads by +4 and minus strand reads by −5 bp (Buenrostro et al. 2013). Henceforth, ATAC-seq footprints were processed and analyzed as described for DNase-seq below.
Retrieving and processing publicly available data
We retrieved DNase-seq data from three different publicly available sources (DFCI, Dana-Farber Cancer Institute [He et al. 2014], GSE51915; Duke, ENCODE, DNase I HS, Duke University; UW, ENCODE, DNase I HS, University of Washington). Details for preprocessing public data are listed in Supplemental Methods. When available, replicates were processed separately, and their cut profiles were merged afterwards by averaging the k-mer occurrences and DNase I cut profiles.
Previously published mouse Ter119+ DNase-seq data are available under accession number GSE49460.
Recording k-mer-based DNase I cut profiles
For every possible k-mer with (5-, 6-, 7-mers), the average DNase I cut profile of the 300-bp window surrounding the k-mer was calculated. Only k-mer occurrences within DHSs were considered. DHSs (extended by 150 bp to each side) were scanned with a sliding window of length 300 bp + k-mer length. At every position, the mapped DNase I cut sites within the window were recorded, multiplied with a sequence bias weighting factor, and attributed to the center k-mer. Sequence bias weight factors were calculated according to the 6-mer (Neph et al. 2012; Lazarovici et al. 2013; He et al. 2014; Sung et al. 2014; Yardimci et al. 2014)-based DNase I cutting preference on genomic DNA (see Supplemental Methods for details). The total number of k-mer occurrences and the summed up surrounding cut profiles were stored. The cut profiles were recorded in a strand-specific fashion.
Calculating relative cut probability profiles and estimating shoulders and SFR
For every selected tissue and k-mer, the number of occurrences and the associated cut profile can be retrieved from the stored files. For downstream analysis, only the 250-bp window surrounding the k-mer of interest was used. The relative DNase I cut probability in the 250-bp window was calculated by dividing the weighted cut sites per position by the sum of all weighted cuts within the window. The profiles were not explicitly normalized for k-mer occurrence as this is captured in the relative cut probability.
Strand-specific profiles were merged while accounting for the DNase-seq strand-imbalance (Piper et al. 2015). Relative cut probabilities upstream of the centric k-mer were retrieved solely from the plus-strand profile, while downstream probabilities were derived only from the minus strand. In the centric k-mer region, the profiles were averaged. ATAC-seq-derived profiles were equally merged from both strands. For downstream quantitative analysis, the profiles were smoothed using a normal kernel smoothing with a bandwidth of 5 bp (R ksmooth stats base package v3.2.3). To define average footprints in the profiles, footprint shoulders were estimated and the region in between was defined as the footprint. Therefore, edges within the cut profiles were estimated by filtering the profiles with a 1D Sobel operator to approximate the second derivative where zero-crossings correspond to turning points. Zero-crossings were detected and separated into upstream and downstream. To speed up the analysis of noisy profiles, only zero-crossings with a maximum distance of 50 bp to the k-mer center were considered. Optimal shoulder positions and shoulder sizes were estimated by maximizing the SFR. To speed up the analysis, only shoulder sizes of 4, 6, 8, or 10 bp were considered. To quantify the footprint strength, the SFR was calculated as the ratio of the average relative cut probability of both shoulder regions to the average probability in the footprint region. This measure is comparable to the footprint-occupancy score employed by ENCODE-related footprint analysis (Neph et al. 2012). However, the SFR is simplified, as no pseudocounts are required for average profiles and the SFR was chosen to maximize the score for strong footprints rather than minimizing it.
Calculating damage and comparing sequences
To compare footprint characteristics of two profiles, for example from a reference and a variant k-mer in the same tissue, the difference in their SFR was calculated (SFRvariant, SFRreference) as the damage in footprinting potential introduced by that variant. Thus, a positive damage reflects a weakened footprint and a negative damage an enhanced footprint. To ensure robustness over a larger (13-bp) sequence context, every single base-pair variant was analyzed as part of multiple 7-mers, occupying different positions (1–7) in a sliding window over the 13-bp sequences centered on the variant of interest. After calculating the reference, variant SFRs, and their pairwise damage scores for every sliding 7-mer window, the total damage was then calculated as the sum of all damage scores from the k-mer comparisons. Per default, a 7-mer scheme was employed, querying the 13-bp sequence window centered on the variant position. In addition, the pairwise relative change was calculated for the highest scoring k-mer pair as the percentage reduction in the SFR with respect to the highest SFR (reference or variant): 100% − [100%/(SFRmax – 1)] × (SFRmin − 1).
In silico mutation
In silico mutation of genomic sequences was performed by comparing at every genomic position, every possible variant against the reference base. For that, the respective 13-bp sequence windows centered on the variant were queried, dissected, compared, and summed up as described above. Output was a single summed-up damage score per possible mutation.
SNP prioritization workflow
For prioritizing SNPs associated with red blood cell phenotypes, we retrieved the list of 75 GWAS-identified SNPs from van der Harst et al. (2012). The SNPs were imputed using the rAggr proxy search online tool (http://raggr.usc.edu/) with default values and the 1000 Genomes (The 1000 Genomes Project Consortium 2015) pilot 1 database (r2 = 0.8, distance limit = 500 kb, population panels = CEU, SA). The resulting 5714 variants were intersected with DHSs from the human primary erythroid cells DNase-seq data called with MACS2 (default settings with relaxed q-value cut off 0.05). One hundred SNPs intersected with DHSs. Reference and variant sequences of the respective 13-bp windows centered on the SNP position were retrieved. The DHSs intersecting SNPs were than processed with the batch query mode of Sasquatch's sequence comparison, and the total damage per variant was estimated. Afterward, SNPs were sorted and classified into neutral or strong and weak gain or loss according to their total damage with cutoffs of 1.0, 0.5, −0.5, and −1.0.
Benchmarking against Basset and DeepSEA
The full model of DeepSEA (v0.94) was downloaded and run with default parameters. The Basset framework was cloned from GitHub (accessed on August 10, 2017) and the full training data set was downloaded and reconstructed according to the manual. The internal primary erythroid DNase-seq set was processed and added to the training set, and the network was trained from scratch following the online manual.
For benchmarking, binding QTLs of five distinct transcription factors identified from pooled Yoruban lymphoblastoid cell lines were downloaded from Tehranchi et al. (2016). For each factor, the 25 most significant SNPs were extracted and queried against dbSNP (Sherry et al. 2001) (build 149 - GRCh37p13). Only SNPs for which an rsID could be extracted were kept. These SNPs were imputed using plink (v1.9) against the YRI individuals from 1000 Genomes (v3 release 20130502). SNPs with an R2 ≥ 0.8 were queried against dbSNP, and all SNPs with rsID and the sentinel SNP were grouped into LD blocks and their reference and alternative allele extracted.
The impact of every SNP in GM12878 was predicted with all three tools. For each tool, a set of stringent and a set of relaxed thresholds was defined and the fraction of explainable LD blocks calculated. For Sasquatch, we used the absolute value of the total damaging score (stringent: 1.0, relaxed 0.5). For Basset, we used 0.1 SAD for both. For DeepSEA, we extracted the functional significance score, the E-value of hypersensitive predictor alone, and the minimum E-value across all GM12878 predictors (histone modification, TFs, hypersensitivity), and used 0.05 as relaxed and 0.01 as stringent cutoffs.
Additional R packages
Additional R (R Core Team 2016) packages used for analysis and visualization are the following:
Biostrings (v2.36.4, http://bioconductor.org/packages/release/bioc/html/Biostrings.html)
BSgenome (https://rdrr.io/bioc/BSgenome/)
cowplot (v0.6.2, https://github.com/wilkelab/cowplot)
ggplot2 (Wickham 2009)
JASPAR 2016 (Mathelier et al. 2016)
RColorBrewer (v1.1.1-2, https://CRAN.R-project.org/package=RColorBrewer)
reshape2 (v1.4.2, https://github.com/hadley/reshape)
rtracklayer (v1.30.4) (Lawrence et al. 2009)
TFBSTools (v1.6.1) (Tan and Lenhard 2016)
Software availability
The webtool is available under http://apps.molbiol.ox.ac.uk/sasquatch/cgi-bin/foot.cgi. The R implementation and source code for preprocessing novel data is available via GitHub (https://github.com/rschwess/sasquatch) and as Supplemental Software.
Data access
Raw data sets of deep DNase-seq and ATAC-seq from human erythroid cells and the appropriate backgrounds have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE86393. The repository of preprocessed publicly available DNase-seq data is available via the webtool.
Supplementary Material
Acknowledgments
J.R.H. would like to acknowledge the support of the Medical Research Council (reference MC_UU_12009/4), the Wellcome Trust via a Strategic Award (reference 106130/Z/14/Z), and the Institutional Strategic Support Fund (reference 105605/Z/14/Z). R.S. would like to acknowledge the Wellcome Trust supporting award (reference 203728/Z/16/Z). M.C.S. would like to acknowledge the Wellcome Trust supporting award (reference 097309/Z/11/Z).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.220202.117.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipanahi B, Delong A, Weirauch MT, Frey BJ. 2015. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol 33: 831–838. [DOI] [PubMed] [Google Scholar]
- Arvey A, Agius P, Noble WS, Leslie C. 2012. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res 22: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, et al. 2013. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Han XL, Kan YW. 1993. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci 90: 11371–11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. 1984. Genomic sequencing. Proc Natl Acad Sci 81: 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee SG, Coetzee GA, Hazelett DJ. 2015. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics 31: 3847–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JO, Telenius JM, McGowan SJ, Roberts NA, Taylor S, Higgs DR, Hughes JR. 2016. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods 13: 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho GB, de Carvalho GB. 2011. Duffy Blood Group System and the malaria adaptation process in humans. Rev Bras Hematol Hemoter 33: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de Jong P, et al. 2006. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science 312: 1215–1217. [DOI] [PubMed] [Google Scholar]
- Edwards SL, Beesley J, French JD, Dunning AM. 2013. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 93: 779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M, Lee D, Mohammad-Noori M, Beer MA. 2014. Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput Biol 10: e1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulko B, Hubisz MJ, Gronau I, Siepel A. 2015. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat Genet 47: 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D, Hughes JR, Babbs C, Davies JO, Graham BJ, Hanssen LL, Kassouf MT, Oudelaar AM, Sharpe JA, Suciu MC, et al. 2016. Genetic dissection of the α-globin super-enhancer in vivo. Nat Genet 48: 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, et al. 2014. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods 11: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. 2009. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holwerda S, de Laat W. 2012. Chromatin loops, gene positioning, and gene expression. Front Genet 3: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini M, Goodstadt L, Hughes JR, Kowalczyk MS, de Gobbi M, Otto GW, Copley RR, Mott R, Higgs DR, Flint J. 2013. Causes and consequences of chromatin variation between inbred mice. PLoS Genet 9: e1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung CN, Chu HW, Huang YL, Chou WC, Hu LY, Hsu HM, Wu PE, Hou MF, Yu JC, Shen CY. 2014. Functional variants at the 21q22.3 locus involved in breast cancer progression identified by screening of genome-wide estrogen response elements. Breast Cancer Res 16: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Whitington T, Gao P, Lindberg JF, Yang Y, Sun J, Vaisanen MR, Szulkin R, Annala M, Yan J, et al. 2014. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat Genet 46: 126–135. [DOI] [PubMed] [Google Scholar]
- Hughes JR, Cheng JF, Ventress N, Prabhakar S, Clark K, Anguita E, De Gobbi M, de Jong P, Rubin E, Higgs DR. 2005. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc Natl Acad Sci 102: 9830–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR. 2014. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet 46: 205–212. [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Tatonetti NP, Landt SG, Yang X, Slifer T, Altman RB, Snyder M. 2011. Cooperative transcription factor associations discovered using regulatory variation. Proc Natl Acad Sci 108: 13353–13358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassouf MT, Hughes JR, Taylor S, McGowan SJ, Soneji S, Green AL, Vyas P, Porcher C. 2010. Genome-wide identification of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res 20: 1064–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Snoek J, Rinn JL. 2016. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res 26: 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Gentleman R, Carey V. 2009. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25: 1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarovici A, Zhou T, Shafer A, Dantas Machado AC, Riley TR, Sandstrom R, Sabo PJ, Lu Y, Rohs R, Stamatoyannopoulos JA, et al. 2013. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc Natl Acad Sci 110: 6376–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Gorkin DU, Baker M, Strober BJ, Asoni AL, McCallion AS, Beer MA. 2015. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet 47: 955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M, Segal E. 2014. In pursuit of design principles of regulatory sequences. Nat Rev Genet 15: 453–468. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Alvarez RV, Sharan R, Landsman D, Ovcharenko I. 2017. Quantifying deleterious effects of regulatory variants. Nucleic Acids Res 45: 2307–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower KM, De Gobbi M, Hughes JR, Derry CJ, Ayyub H, Sloane-Stanley JA, Vernimmen D, Garrick D, Gibbons RJ, Higgs DR. 2013. Analysis of sequence variation underlying tissue-specific transcription factor binding and gene expression. Hum Mutat 34: 1140–1148. [DOI] [PubMed] [Google Scholar]
- Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. 2016. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165: 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigal P. 2015. On accounting for sequence-specific bias in genome-wide chromatin accessibility experiments: recent advances and contradictions. Front Bioeng Biotechnol 3: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. 2012. ENCODE: the human encyclopaedia. Nature 489: 46–48. [DOI] [PubMed] [Google Scholar]
- Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP. 2013. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol 14: R131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. 2016. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44: D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Haugen E, Sandstrom R, Vierstra J, Shafer A, Kaul R, Stamatoyannopoulos JA. 2015. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat Genet 47: 1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyerbrailean GA, Kalita CA, Harvey CT, Wen X, Luca F, Pique-Regi R. 2016. Which genetics variants in DNase-seq footprints are more likely to alter binding? PLoS Genet 12: e1005875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Andrau JC. 2012. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet 46: 1–19. [DOI] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. 2012. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L, Gaulton KJ, Rodriguez-Segui SA, Mularoni L, Miguel-Escalada I, Akerman I, Tena JJ, Moran I, Gomez-Marin C, van de Bunt M, et al. 2014. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 46: 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. 2013. Enhancers: five essential questions. Nat Rev Genet 14: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J, Assi SA, Cauchy P, Ladroue C, Cockerill PN, Bonifer C, Ott S. 2015. Wellington-bootstrap: differential DNase-seq footprinting identifies cell-type determining transcription factors. BMC Genomics 16: 1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. 2011. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 21: 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Rockman MV, Kruglyak L. 2006. Genetics of global gene expression. Nat Rev Genet 7: 862–872. [DOI] [PubMed] [Google Scholar]
- Romanoski CE, Glass CK, Stunnenberg HG, Wilson L, Almouzni G. 2015. Epigenomics: roadmap for regulation. Nature 518: 314–316. [DOI] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. 2012. Linking disease associations with regulatory information in the human genome. Genome Res 22: 1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EM, Zhang J, Zhou W, Chen J, Mohlke KL, Chen YE, Willer CJ. 2015. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics 31: 2601–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Wise SC, Hendricks DT, Sabichi AL, Bos T, Reddy P, Brown PH, Birrer MJ. 1999. cJun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive and hormone resistant phenotype. Oncogene 18: 6063–6070. [DOI] [PubMed] [Google Scholar]
- Sung MH, Guertin MJ, Baek S, Hager GL. 2014. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell 56: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MH, Baek S, Hager GL. 2016. Genome-wide footprinting: ready for prime time? Nat Methods 13: 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlichnyy D, Imrichova H, Fiers M, Kalender Atak Z, Aerts S. 2015. Identification of high-impact cis-regulatory mutations using transcription factor specific random forest models. PLoS Comput Biol 11: e1004590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallack MR, Whitington T, Yuen WS, Wainwright EN, Keys JR, Gardiner BB, Nourbakhsh E, Cloonan N, Grimmond SM, Bailey TL, et al. 2010. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res 20: 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G, Lenhard B. 2016. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32: 1555–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehranchi AK, Myrthil M, Martin T, Hie BL, Golan D, Fraser HB. 2016. Pooled ChIP-seq links variation in transcription factor binding to complex disease risk. Cell 165: 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournamille C, Colin Y, Cartron JP, Le Van Kim C. 1995. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet 10: 224–228. [DOI] [PubMed] [Google Scholar]
- Tsompana M, Buck MJ. 2014. Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Harst P, Zhang W, Mateo Leach I, Rendon A, Verweij N, Sehmi J, Paul DS, Elling U, Allayee H, Li X, et al. 2012. Seventy-five genetic loci influencing the human red blood cell. Nature 492: 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. 2009. A census of human transcription factors: function, expression and evolution. Nat Rev Genet 10: 252–263. [DOI] [PubMed] [Google Scholar]
- Vierstra J, Stamatoyannopoulos JA. 2016. Genomic footprinting. Nat Methods 13: 213–221. [DOI] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. 2014. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42: D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- Yardimci GG, Frank CL, Crawford GE, Ohler U. 2014. Explicit DNase sequence bias modeling enables high-resolution transcription factor footprint detection. Nucleic Acids Res 42: 11865–11878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Hashimoto T, Kang DD, Gifford DK. 2016. GERV: a statistical method for generative evaluation of regulatory variants for transcription factor binding. Bioinformatics 32: 490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Troyanskaya OG. 2015. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods 12: 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.