Abstract
Bromodomain PHD finger transcription factor (BPTF) is the largest subunit of nucleosome remodeling factor (NURF), a member of the ISWI chromatin-remodeling complex. However, the clinical consequences of disruption of this complex remain largely uncharacterized. BPTF is required for anterior-posterior axis formation of the mouse embryo and was shown to promote posterior neuroectodermal fate by enhancing Smad2-activated wnt8 expression in zebrafish. Here, we report eight loss-of-function and two missense variants (eight de novo and two of unknown origin) in BPTF on 17q24.2. The BPTF variants were found in unrelated individuals aged between 2.1 and 13 years, who manifest variable degrees of developmental delay/intellectual disability (10/10), speech delay (10/10), postnatal microcephaly (7/9), and dysmorphic features (9/10). Using CRISPR-Cas9 genome editing of bptf in zebrafish to induce a loss of gene function, we observed a significant reduction in head size of F0 mutants compared to control larvae. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) and phospho-histone H3 (PH3) staining to assess apoptosis and cell proliferation, respectively, showed a significant increase in cell death in F0 mutants compared to controls. Additionally, we observed a substantial increase of the ceratohyal angle of the craniofacial skeleton in bptf F0 mutants, indicating abnormal craniofacial patterning. Taken together, our data demonstrate the pathogenic role of BPTF haploinsufficiency in syndromic neurodevelopmental anomalies and extend the clinical spectrum of human disorders caused by ablation of chromatin remodeling complexes.
Keywords: 17q24.2 deletion, head size, intellectual disability, PSMD12, zebrafish
Introduction
Chromatin remodeling, an essential process regulating DNA accessibility and transcriptional activation, is controlled by covalent histone modifications and ATP-dependent nucleosome translocation which involves five conserved protein complexes: (1) SWI/SNF (a.k.a. BRG1-associated factors [BAF]), (2) ISWI (imitation switch), (3) CHD (chromatin helicase DNA-binding), (4) INO80/SWR1, and (5) ATRX.1 Thus far, pathogenic variants in 11 chromatin remodeling genes involving SWI/SNF (BAF), CHD, and ATRX have been implicated in congenital disorders as well as cancer development, i.e., ARID1A (Coffin-Siris syndrome 2 [MIM: 614607]), ARID1B (Coffin-Siris syndrome 1 [MIM: 135900]), SMARCA2 (Nicolaides-Baraitser syndrome [MIM: 601358]), SMARCA4 (Coffin-Siris syndrome 4 [MIM: 614609] and Rhabdoid tumors [MIM: 613325]), SMARCB1 (Coffin-Siris syndrome 3 [MIM: 614608], Rhabdoid tumors [MIM: 609322], and Schwannomatosis [MIM: 162091]), SMARCD2 (specific granule deficiency 2 [MIM: 617475]), SMARCE1 (Coffin-Siris syndrome 5 [MIM: 616938]), CHD2 (epileptic encephalopathy, childhood-onset [MIM: 615369]), CHD4 (Sifrim-Hitz-Weiss syndrome [MIM: 617159]), CHD7 (CHARGE syndrome [MIM: 214800] and hypogonadotropic hypogonadism 5 with or without anosmia [MIM: 612370]), and ATRX (alpha-thalassemia/mental retardation syndrome [MIM: 301040]). To date, neither INO80/SWR1 nor ISWI genes have been associated with disease in humans.
The ISWI family member NURF (nucleosome remodeling factor) is an evolutionarily conserved key transcriptional regulator of development2 in a locus-specific manner3, 4 by virtue of the complex’s chromatin remodeling activity.5, 6 In vertebrates, NURF consists of SNF2L (ISWI homolog encoded by SMARCA1), pRBAP46/48, and the largest subunit BPTF (bromodomain PHD finger transcription factor).7, 8, 9 Human BPTF contains two PHD finger domains followed proximally by a bromodomain (BRD). The second PHD finger of BPTF mediates binding of the NURF complex to chromatin with trimethylation of histone H3 lysine 4 (H3K4me3).10 In a combinatorial manner via multivalent interactions together with the PHD finger, the BRD binds to acetylated lysine 16 in histone H4 (H4K16ac), enabling the selective targeting of BPTF to chromatin that contains both histone marks, thereby increasing its selectivity.11, 12, 13, 14
Heterozygous Bptf mutant mice are apparently normal and fertile with no obvious phenotype.15 Analyses of homozygous mice have revealed that BPTF is essential for the formation of mesoderm, endoderm, and differentiated ectoderm lineages and is required for the establishment of the embryonal anterior-posterior axis during early development; homozygotes show lethality at embryonic day (E)7.5 to E8.5 with 100% penetrance.16 BPTF was also found to be important for trophoblast differentiation during early mouse development (E6.5).15 Additionally, depletion of Bptf has enhanced T cell-mediated antitumor immunity in two syngeneic mouse models of cancer.17
Consistent with murine mutant data, Bptf was shown previously to promote neuroectodermal posteriorization in zebrafish embryos. RNA in situ hybridization and protein analyses demonstrated that bptf is expressed ubiquitously during zebrafish early embryonic development, and it is expressed abundantly in the zebrafish head through 30 hr post fertilization (hpf).18 Morpholino-based suppression of bptf results in abnormal anterior patterning and defects in neural posteriorization; these phenotypes are attributed to misregulated TGF-β/Smad2 signaling and concomitant restriction of wnt8a expression.18 The TGF-β pathway is critical for cellular growth, differentiation, and apoptosis and was shown to be an important driver in neurogenesis and central nervous system development.19 Moreover, zebrafish wnt8a was shown to function in early-stage mesoderm patterning and posteriorization of the neuroectoderm.20
In humans, BPTF is ubiquitously expressed.10 Amplification and overexpression of BPTF were reported in a variety of cancers including breast, lung, and brain.21, 22, 23, 24, 25, 26 BPTF was shown as essential for T cell homeostasis and function.27 BPTF also inhibits fNK cell activity and the abundance of natural cytotoxicity receptor co-ligands.28 Lastly, BPTF maintains chromatin accessibility and the self-renewal capacity of mammary gland stem cells.29
Here, we describe phenotypic manifestations of germline loss-of-function (LoF) variants in BPTF in ten unrelated individuals with an autosomal-dominant neurodevelopmental disorder and show with an in vivo zebrafish model that bptf is relevant to brain development and craniofacial patterning.
Material and Methods
Subjects
The study cohort consists of ten unrelated case subjects. Individuals 1–3 were found in the exome database of 9,056 individuals referred for clinical whole-exome sequencing (WES) at Baylor Genetics (BG). Individuals 4 and 5 were identified in the clinical database of 75,795 subjects referred for clinical chromosomal microarray analysis (CMA) at BG. These five subjects were chosen through filtering for potentially LoF variants in previously unsolved case subjects with overlapping neurological phenotypes. Subsequently, we identified three additional individuals: via DECIPHER30 individuals 6 (DECIPHER 275557) and 7 (DECIPHER 264215) from the Deciphering Developmental Disorders (DDD) Study cohort31 and individual 8 from the University of Groningen, the Netherlands. Through the online matchmaker platform GeneMatcher,32 individuals 9 and 10 were identified from the Institute of Human Genetics, Friedrich-Alexander-Universität Erlangen-Nürnberg in Erlangen, Germany and Virginia Commonwealth University in Richmond, respectively. Written informed consent (for individuals 1–3, 6–10) was obtained in accordance with protocols approved by the appropriate human subject ethics committees; individuals 4 and 5 were covered by a protocol (with waiver of consent) approved by Baylor College of Medicine. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC).
Microarray and Molecular Analyses
Individuals 1–3 were analyzed at BG Laboratories by trio WES (individual 1) or proband-only WES (individuals 2 and 3) using the capture design based on VCRome by NimbleGen.33 The mean coverage of target bases was >120×, and >96% target bases were covered at >20×. PCR amplification and Sanger sequencing to verify all candidate variants were done according to standard procedures in the proband and the parents when available, and candidate variants were annotated using the BPTF RefSeq transcript GenBank: NM_004459.6. The two copy-number variant (CNV) deletions in individuals 4 and 5 were detected at BG using customized exon-targeted oligo arrays (OLIGO V8.1.1 and V11.2),34, 35 which cover more than 1,700 and 4,800 disease-associated genes, respectively, with exon-level resolution. CNV deletion junction fragments were amplified using long-range PCR with LA Taq DNA polymerase (TaKaRa Bio) and primers designed by Primer3 software. Individuals 6 and 7 were recruited and analyzed by WES in the DDD Study.31 The variant in individual 8 was detected in the Department of Human Genetics, Radboud University Medical Center, in Nijmegen, the Netherlands. The mutation in individual 9 was identified within an exome Pool-Seq screening approach which will be published elsewhere (B. Popp, unpublished data); validation and segregation analysis was performed by Sanger sequencing followed by genetic fingerprinting using the PowerPlex 21 system (Promega) to confirm de novo occurrence. For individual 10, parent-proband trio WES was performed at Ambry Genetics using the IDT xGen Exome Research Panel and analyzed as previously described.36, 37 On average, ∼96.6% of the target bases were covered at >20× for the trio. The de novo frameshift alteration identified in this patient by WES was confirmed by Sanger sequencing and interpreted as a (suspected) candidate gene finding.37
CRISPR-Cas9 Genome Editing
We identified a single zebrafish (Danio rerio) ortholog of BPTF using reciprocal BLAST (Ensembl ID: ENSDART00000109601; GRCz10; 51% similarity to human BPTF; Figure S1). The CRISPR single-guide (sgRNA) target was identified with ChopChop software38 and synthesized using the GeneArt Precision gRNA Synthesis Kit (Invitrogen) according to manufacturer’s instructions as described.39 To generate F0 mutants, 75 pg of sgRNA and 150 pg of CAS9 protein (PNA bio, CP01) were injected directly into the cell of 1-cell stage zebrafish embryos. The efficiency of the sgRNA was determined by extracting genomic DNA from F0 embryos at 2 dpf by proteinase K digestion (Life technologies, AM2548). The sgRNA genome editing site was PCR amplified and resulting PCR products were denatured and slowly reannealed (denaturing at 95°C for 5 min, ramped down to 85°C at −1°C/s and then to 25°C at −0.1°C/s) to facilitate heteroduplex formation. Heteroduplexes were detected by 15% polyacrylamide gel electrophoresis38 (n = 6 F0 embryos and 1 uninjected control embryo) followed by cloning and sequencing of PCR amplicons to estimate mosaicism.
Zebrafish Embryo Injections
Zebrafish embryos were collected from natural matings of -1.4col1a1:egfp40 heterozygous adults. For CRISPR experiments, one-cell stage embryos were injected with 1 nL, with the investigator masked to injection cocktail. Embryos were maintained in fresh embryo media (0.3 g/L NaCl, 75 mg/L CaSO4, 37.5 mg/L NaHCO3, 0.003% methylene blue) at 28°C until phenotypic endpoints. Larvae were phenotyped for cell death and cell proliferation at 2 days post fertilization (dpf); and for head size and craniofacial patterning at 3 dpf.
Zebrafish Phenotyping
Whole-Mount TUNEL Assay
Apoptotic cell death was detected by the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay as described.41 Briefly, 2 dpf embryos were dechorionated and fixed in 4% paraformaldehyde (PFA) at 4°C overnight and then in 100% methanol at −20°C for 2 hr. After rehydration in PBS, embryos were permeabilized with proteinase K (10 μg/mL) and postfixed with 4% PFA. Embryos were incubated in equilibration buffer and then with TdT enzyme followed by anti-digoxigenin provided in ApopTag Red In Situ Apoptosis Detection Kit (Millipore) as suggested by the manufacturer. We imaged the dorsal anterior aspect of whole-mounts with Z stack image capture using a Nikon AZ100 fluorescent microscope. TUNEL stain was quantified by counting positive cells in defined regions of the head with ImageJ software.
Cell Proliferation Assay
At 2 dpf, embryos were dechorionated and fixed in Dent’s solution overnight at 4°C. Embryos were rehydrated in PBS with a stepwise reduced concentration of methanol, and then bleached, postfixed with 4% PFA, and permeabilized using proteinase-K. Embryos were then washed twice in IF buffer (1% BSA, 0.1% Tween-20 in 1× PBS) and incubated overnight with anti-p-histone H3 (PH3; 1:500, Santa Cruz Biotechnology, sc-8656-R). Following two washes in IF buffer, embryos were placed in secondary antibody solution containing Alexa Fluor 488 goat anti-rabbit IgG (1:500; Invitrogen) in blocking solution for 1 hr at room temperature. Z stacked images were captured using a Nikon AZ100 fluorescent microscope. Immunostained cells in defined regions of the dorsal aspect of the head were counted with ImageJ software.
Automated Live Phenotyping of Larval Head Size and Craniofacial Patterning
We positioned and imaged live 3 dpf larvae with the Vertebrate Automated Screening Technology (VAST; software v.1.2.3.6) to capture dorsal (head size; bright field) or ventral (craniofacial skeleton; fluorescent excitation at 470 nm) images. Larvae were anesthetized, pattern recognition templates were created, and all VAST operational settings were similar to those described.42 Once recognized inside the 600 μm borosilicate capillary, each larva was rotated to capture images using a 5× fluar objective and Axiocam 503 monochromatic camera (Zen Pro software; Zeiss). Head size and ceratohyal cartilage angle were measured in respective images using ImageJ software and pairwise comparisons to determine statistical significance were made via a Student’s t test.
Results
Clinical Findings
We identified 10 unrelated individuals with unique apparent non-mosaic variants in BPTF. The affected individuals included six males and four females, aged 2.1 to 13 years at the last clinical assessment (Table 1, Figure 1A). Common features included developmental delay (DD)/intellectual disability (ID) (10/10), speech delay (10/10), microcephaly (7/9), motor delay (8/10), hypotonia (5/10), and dysmorphic features (9/10), which included prominent nose (7/10), up-slanting or short palpebral fissures (4/10), and flaring of eyebrows (2/10), 5th finger clinodactyly (3/10), and bulbous halluces/broad great toes/sandal foot (5/10) (Table 1). Of note, individual 3 had clinical WES as part of an NIH-supported pediatric cancer study due to a diagnosis of pheochromocytoma, and WES resulted in a VHL pathogenic heterozygous variant c.499C>T (p.Arg167Trp) (GenBank: NM_000551.3) (Table S1).43 The medical record and WES requisition form had also noted mild developmental delay and speech delay.
Table 1.
Clinical and Genetic Findings in Individuals with BPTF Variants
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | Subject 5 | Subject 6 | Subject 7 | Subject 8 | Subject 9 | Subject 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Variant (GRCh37; GeneBank NM_004459.6) | chr17: g.65890220dup | chr17: g.65908838_65908839del | chr17: g.65972049A>T | chr17: g.65700188-65896330del (196 kb) | chr17: g.65898399-65986981del (89 kb) | chr17: g.65905867_65905878del | chr17: g.65914918G>A | chr17: g.65971957T>G | chr17: g.65850431del | chr17: g.65889796del |
| c.2860dup | c.5216_5217del | c.8650A>T | c.-121653_2922-3575del | c.2922-1506_∗8577del | c.3360_3370+1del | c.5770G>A | c.8558T>G | c.989del | c.2744del | |
| p.Glu954Glyfs∗5 | p.Val1739Glyfs∗96 | p.Lys2884∗ | CNV deletion | CNV deletion | r.spl? | p.Ala1924Thr | p.Met2853Arg | p.Leu330Argfs∗28 | p.Asn915Thrfs∗36 | |
| Exon | 9 | 13 | 29 | 1–9 | 10–30 | 12 | 14 | 29 | 2 | 8 |
| Variant type | frameshift | frameshift | nonsense | CNV deletion | CNV deletion | splicing & frameshift | missense | missense | frameshift | frameshift |
| Variant inheritance | de novo | mother negative, father unavailable | de novo | de novo | unknown | de novo | de novo | de novo | de novo | de novo |
| Ethnicity | white | Latino | Latino | Latino | Latino | white | white | white | white | white |
| Gender | M | M | M | F | F | M | M | F | M | F |
| Age at last assessment (years) | 2.1 | 7.9 | 10.9 | 10 | 4.3 | 13 | 11 | 11 | 7.11 | 12 |
| Weight at birth (kg) (Z score) | 2.7 (Z = −1.42) | 2.63 (Z = −0.8) | ND | 3.4 (Z = 0.00) | 2.4 (Z = −3.6) | 4.39 (Z = 0.84) | 1.89 (Z = −3.44) | 2.27 (Z = −3.5) | 3.46 (Z = −0.06) | 2.69 (Z = −1.38) |
| Length at birth (cm) (Z score) | ND | 47 (Z = −0.7) | ND | 50 (Z = 0.28) | 48 (Z = −3.82) | ND | ND | 45 (Z = −3.8) | 51 (Z = −0.39) | 48.3 (Z = 0.46) |
| Head circumference at birth (cm) (Z score) | 33 (Z = −1.13) | 33 (Z = −0.3) | ND | 33 (Z = −1.11) | ND | 36 (Z = 0.11) | 33 (Z = −1.17) | ND | 34.5 (Z = −0.62) | 33 (Z = −0.74) |
| Weight at assessment (kg) (Z score) | 10.1 (Z = −2.29) | 20.6 (Z = −1.58) | 30.1 (Z = −0.98) | 37.8 (Z = 0.65) | 17 (Z = 0,13) | 22.3 (Z = −1.83) | 23.2 (Z = −2.85) | 25.5 (Z = −2.68) | 19.5 (Z = −2.09) | 26.05 (Z = −3.53) |
| Height at assessment (cm) (Z score) | 84.5 (Z = −0.85) | 117.1 (Z = −1.85) | 142.2 (Z = −0.15) | 143 (Z = 0.70) | 88 (Z = −3.93) | 134 (Z = 0.01) | 126 (Z = −2.83) | 137 (Z = −2.30) | 120 (Z = −1.35) | 134.4 (Z = −3.03) |
| Head circumference at assessment (cm) (Z score) | 45 (Z = −2.05) | 49.2 (Z = −2.1) | ND | 51 (Z = −1.04) | 46 (Z = −2.48) | 50.2 (Z = −2.52) | 54.2 (Z = −0.35) | 49 (Z = −2.68) | 48.3 (Z = −2.90) | 49.5 (Z = −2.05) |
| Developmental delay/intellectual disability | mild | severe | mild | moderate | mild autistic spectrum disorder | moderate autistic spectrum disorder | severe | mild | moderate aggression and distractedness, disturbed sleep rhythm and sleeping problems | mild |
| Speech/language delay | + | + | + | + | + | + | + | + | + | + |
| Motor delay | + | + | + | + | − | − | + | + | + | − |
| Hypotonia | + | − | − | + | − | − | + | − | + | + |
| Microcephaly | + | + | ND | − | + | + | − | + | + | + |
| Brain anomalies | small anterior pituitary | ND | ND | brain MRI: bilateral nonspecific multifocal white matter lesions | − | ND | pituitary hypoplasia | brain MRI: reduced signal intensity of frontal and temporal white matter | MRI at age 4 years 8 months: periventricular white matter lesions | MRI brain: borderline low positioned cerebellar tonsil on right; no true Chiari |
| Dysmorphism | − | multiple lateral flaring of eyebrows, bilateral occipital protuberances, long nasal bridge with mildly hypolastic alae nasi | large helices of both ears (similar to father) | ocular hypertelorism, epicantal folds, up-slanting palpebral fissures, prominent nose | up-slanting palpebral fissures, hypertelorism, mediofacial hypoplasia | high palate, prominent nose and columella, thin upper lip | lateral flaring of eyebrows, prominent supraorbital ridges, short palpebral fissures, broad nasal tip, prominent gum line, Peg like, disorganized teeth | short palpebral fissures, long nose, mildly hypolastic alae nasi, full lower lip, prominent ears, micrognathia | long nasal bridge, small mouth and micrognathia | telecanthus, prominent nasal root with mildly bulbous nasal tip, micrognathia |
| Ophthalmological anomalies | − | outward deviation of one eye | − | − | − | − | severe myopia and convergent squint | − | cataract in right eye, hyperopia | myopia |
| Skeletal abnormalities | − | pes planus, 5th digit clinodactyly, windswept 2nd toe with lateral deviation, broad short great toes | − | advanced bone age | 5th digit clinodactyly | slender fingers and toes, bulbous halluces | wrinkly hands, flexed fingers, 5th finger clinodactyly, bulbous halluces, overlapping toes | broad halluces, premature eruption of permanent mandibular central incisors | sandal gap of both feet | congenital hip dysplasia, small hands |
Abbreviations: MRI, magnetic resonance imaging; ND, not determined because of non-availability or non-applicability
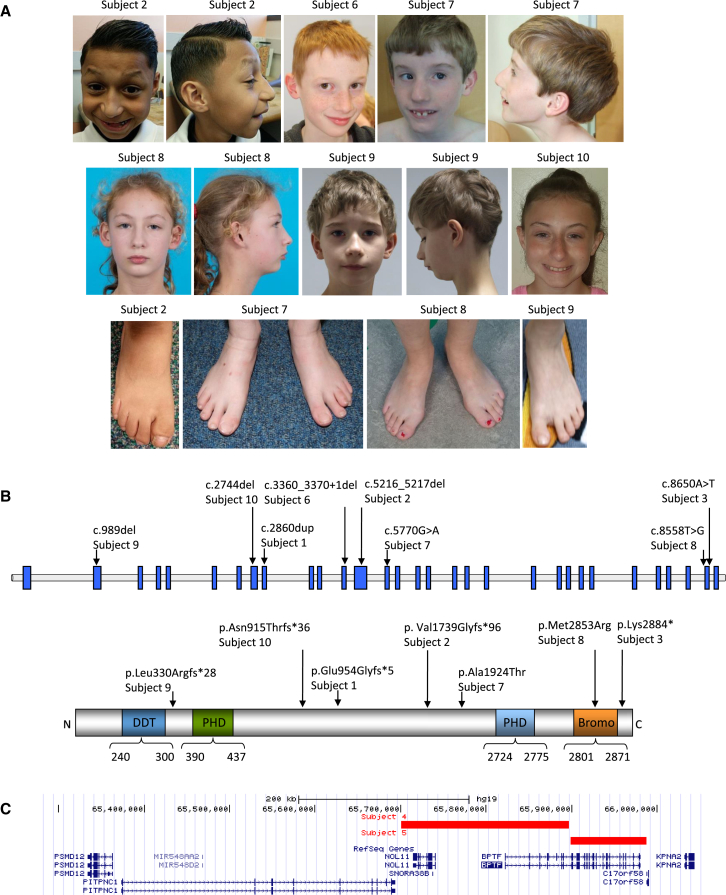
Figure 1.
Clinical and Genetic Findings of Individuals with BPTF Variants
(A) Clinical features of subjects 2 and 6–10. Note the lateral flaring of eyebrows, long nasal bridge, windswept 2nd toe with lateral deviation, and broad short great toe in subject 2; prominent nose and columella and thin upper lip and in subject 6; prominent supraorbital ridges, lateral flaring of eyebrows, short palpebral fissures, convergent squint, broad nasal tip, disorganized teeth, bulbous halluces, and overlapping toes in subject 7; short palpebral fissures, long nose, prominent ears, mildly hypolastic alae nasi full lower lip, micrognathia, and broad halluces in subject 8; long nasal bridge, small mouth, micrognathia, and sandal gap in subject 9; and telecanthus, prominent nasal root with mildly bulbous nasal tip, and micrognathia in subject 10.
(B) A schematic of BPTF, the protein, and the effects of variants identified in this study. Domains represented in colors are: different transcription factors domain (DDT), PHD finger domain (PHD), and bromodomain (Bromo). The domain structure and amino acid numbering is based on the NCBI reference sequences GenBank: NP_003482.3 and NP_00450.3.
(C) A schematic of chromosomal region 17q24.2 showing the deletions affecting BPTF in the UCSC browser.
Clinical CMA and WES
Exome sequencing identified four frameshifting indels affecting the coding exons (c.989del [p.Leu330Argfs∗28], c.2744del [p.Asn915Thrfs∗36], c.2860dup [p.Glu954Glyfs∗5], and c.5216_5217del [p.Val1739Glyfs∗96]); one splicing/frameshifting indel (c.3360_3370+1del), which is predicted to eliminate 11 nucleotides of the coding region plus the first nucleotide of the invariant splice donor site sequence (GT); one nonsense variant (c.8650A>T [p.Lys2884∗]); and two missense variants (c.5770G>A [p.Ala1924Thr] and c.8558T>G [p.Met2853Arg]) (GenBank: NM_004459.6) (Figure 1B). In eight subjects, the variants arose de novo; the origin of the other two changes could not be determined (Table 1). The eight truncating variants are predicted to result in haploinsufficiency of the gene. The two de novo missense variants, c.5770G>A (p.Ala1924Thr) and c.8558T>G (p.Met2853Arg), are predicted to be likely damaging by most of the in silico prediction programs, including PolyPhen-2 and MutationTaster (Table S2). Human splicing finder also reported the generation of a cryptic acceptor site for the variant c.8558T>G, although MaxEntScan and SPIDEX did not find a significant splice alteration. The two variants are not present in the Genome Aggregation Database (gnomAD) database (>122,000 individuals; accessed on 6/18/2017). In addition, Met2853 is located in the bromodomain; 3D simulation based on crystal structure of PHD finger-linker-bromodomain fragment of human BPTF (PDB: 3UV2) indicates that the substitution of Met with the positively charged Arg in the hydrophobic core may destabilize the 3D conformation of the BRD (Figure S2). Our collective data indicate that the missense variants likely disrupt the protein function.
CMA using V11.2 array in subject 4 revealed an ∼157 kb CNV deletion on chromosome 17q24.2 (65,720,270–65,877,348, GRCh37/hg19), involving BPTF, and a ∼1 kb CNV deletion of unknown significance on chromosome 14q11.2 (21,681,066-21,682,235), involving a non-disease-associated gene HNRNPC. CMA of the parental samples showed no evidence of these deletions. CMA using V8.1.1 array in subject 5 showed an ∼63 kb CNV deletion on chromosome 17q24.2 (65,909,023–65,972,166), involving BPTF and a likely benign ∼703 kb CNV duplication on chromosome 2q12.3 (108,403,193–109,106,402). Parental samples were not available. DNA sequencing of the junction fragment in subject 4 mapped the proximal breakpoint within AluSx between chr17: 65,700,189 and 65,700,200 and the distal breakpoint within AluSz6 between chr17: 65,896,331 and 65,896,342 with 12 bp microhomology, eliminating the 5′ end of the gene through exon 9. PCR analyses showed no evidence of low-level somatic mosaicism for the copy-number loss on 17q24.2 in either parent. In subject 5, the proximal deletion breakpoint was mapped within a unique sequence between chr17: 65,898,399 and 65,898,404 and the distal breakpoint within AluSp between 65,986,981 and 65,986,986 with a 4 bp GTGA microhomology, eliminating exon 10 through the 3′ end of the gene (Figures 1C and S3).
CRISPR/CAS9 Genome Editing of bptf in Zebrafish Results in Head Size Reduction Likely Caused by an Increase in Neuronal Cell Death
Although suppression of bptf implicated discrete morphogenetic pathways in aberrant neural patterning,18 the eventual effects on zebrafish head size or craniofacial structures were unexplored previously in transient bptf knockdown models. To determine the relevance of BPTF disruption to patient phenotype, we used CRISPR-CAS9 to generate a bptf LoF model. We and others have shown previously that neuroanatomical defects can be modeled in zebrafish as a direct readout for analogous defects observed in humans.31, 44, 45, 46, 47 We designed a CRISPR single-guide RNA (sgRNA) targeting exon 9 of bptf, injected 75 pg sgRNA and 150 pg of CAS9 into the cell of 1-cell-stage zebrafish embryos, and detected efficient disruption of the locus through the introduction of small insertions or deletions (90% mosaicism; Figure S1, Table S3). First, we asked whether we could detect any differences in the head size of 3 dpf F0 mutant larvae. We observed a significant reduction in a defined region of the head of bptf CRISPR F0 mutants compared to either control larvae or compared to larval batches injected with sgRNA alone (without CAS9); (p ≤ 0.0001, n = 26–52 larvae/batch, repeated, Figure 2). To determine the cellular basis of this head size reduction in bptf F0 mutants, we performed TUNEL staining and PH3 staining to assess cell death and cell proliferation, respectively. We observed a significant increase in cell death in F0 mutants compared to control larvae and larvae injected with sgRNA alone (p = 0.0004 versus control, p = 0.0003 versus sgRNA alone, n = 20 larvae per condition, repeated). However, we did not detect any significant differences in cell proliferation between bptf F0 CRISPR mutants and controls (Figure 2).
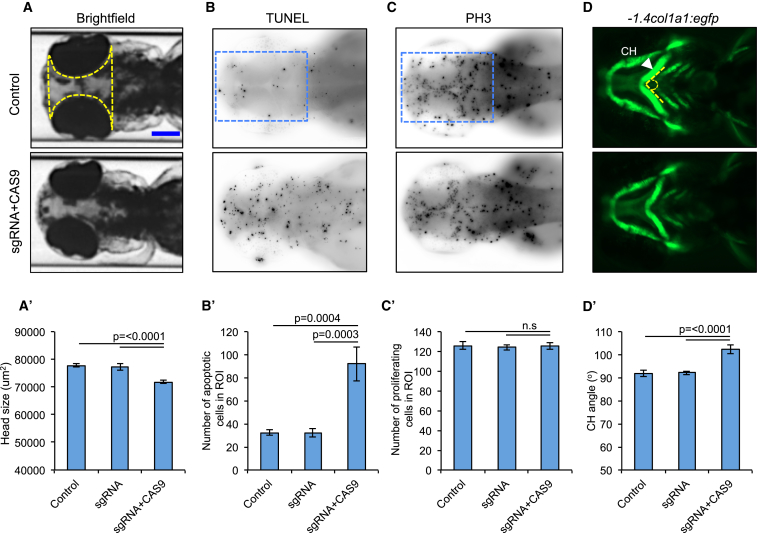
Figure 2.
bptf CRISPR F0 Mutants Exhibit Microcephaly, Apoptosis in Anterior Structures, and Aberrant Patterning of the Branchial Arches
(A) Representative dorsal images of 3 dpf zebrafish larvae show a reduction in head size (yellow dotted area) in F0 mutants. Scale bar (blue), 150 μm.
(A′) Quantification of head size area in bptf F0 mutants (p ≤ 0.0001 versus control, n = 26–52 larvae/batch, repeated). Solid yellow line indicates scale bar.
(B) Representative dorsal images of 2 dpf zebrafish larvae display increased apoptosis in the forebrain-midbrain region (blue dotted rectangle).
(B′) Apoptotic cell count in bptf CRISPR F0 mutants (p = 0.0004 versus control, n = 20 larvae/condition per batch, repeated).
(C) Representative dorsal images of 2 dpf zebrafish larvae do not show cell proliferation defects in anterior structures (blue dotted rectangle).
(C′) Cell proliferation count in bptf CRISPR F0 mutants (n.s., no statistical significance, n = 20 larvae/condition per batch).
(D) Representative ventral views of -1.4col1a1:egfp larvae imaged live at 3 dpf show an increase in the ceratohyal cartilage (CH; arrowhead) angle (dotted line) in F0 mutants.
(D′) Quantification of the ceratohyal angle in bptf F0 CRISPR mutants (p ≤ 0.0001, n = 47–59 larvae/batch, repeated). Error bars indicate standard error of the mean (SEM).
bptf F0 Mutants Display Abnormal Craniofacial Patterning
To determine the relevance of BPTF ablation to the dysmorphic features observed in our cohort and to assess the role of BPTF in craniofacial patterning, we injected bptf sgRNA and CAS9 in -1.4col1a1:egfp transgenic embryos and assessed cartilage structures. We and others have shown that zebrafish can serve as robust direct models for craniofacial abnormalities observed in humans.39, 42, 48, 49, 50, 51, 52 We assessed the craniofacial patterning by measuring the angle of ceratohyal (CH) cartilage in ventral images acquired from 3 dpf larvae. We observed a significant increase of the CH angle in bptf CRISPR F0 mutants (102° versus 91° for F0 mutants versus controls; p ≤ 0.0001, n = 47–59 larvae/batch, repeated; Figure 2). Importantly, injection of bptf sgRNA alone was indistinguishable from that of controls, excluding the possibility of sgRNA toxicity inducing phenotypic differences.
Discussion
Here, we describe syndromic anomalies in ten individuals with potentially deleterious variants in BPTF, which encodes the largest subunit of the ATP-dependent chromatin-remodeling ISWI family member NURF. We identified five small frameshift indels, one nonsense variant, two missense variants, and two CNV deletions. For all affected individuals (n = 8) whose parents were both studied, the BPTF variant occurred de novo. BPTF is predicted to be highly intolerant to LoF mutations (probability of LoF intolerance, pLI = 1.0 in ExAC, accessed 6/18/2017).53 Our data indicate that the most likely pathomechanism of BPTF is haploinsufficiency. All individuals in this study had variants that affect BPTF only, with the exception of individual 4, who carried a deletion that affects BPTF and NOL11. However, the low pLI score for NOL11 (pLI = 0.03) suggests that disruptions of this gene are unlikely to cause an autosomal-dominant disorder (Figure 3). These data are supported further by the observation that zebrafish F0 mutants with depleted bptf display relevant neuroanatomical and craniofacial defects. However, further studies in stable bptf mutants will be required to correlate precisely gene dosage with phenotype in this model system.
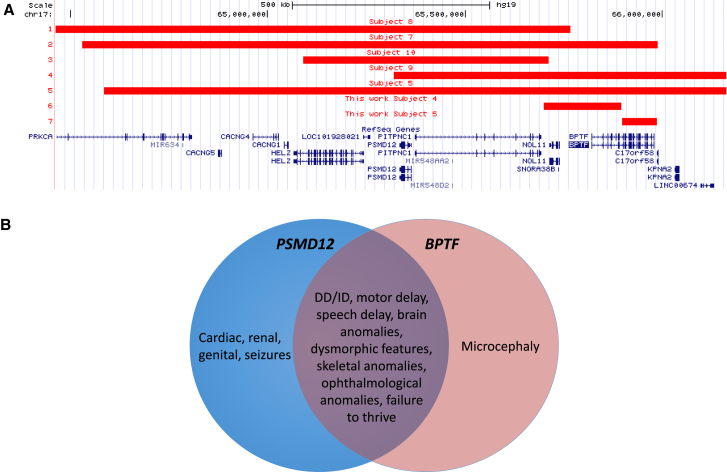
Figure 3.
Analysis of the 17q24.2 Microdeletion Region
(A) Deletions in affected individuals reported previously39 and in this study.
(B) Phenotype comparison of PSMD12-39 and BPTF-related disorders. Individuals with deletions affecting both genes are expected to have blended phenotypes.
In addition to the variable degree of DD/ID and speech delay present in all reported subjects studied, the most consistent finding is postnatal microcephaly,54 observed in 7/9 subjects. Interestingly, dysfunction of five genes reported previously to cause chromatin remodeling disorders are associated with microcephaly (SMARCA2, SMARCA4, SMARCB1, SMARCE1, and CHD7) and one is associated with macrocephaly (CHD4), suggesting that disruption of chromatin remodeling can have global consequences on brain development likely by deregulating multiple transcription factors. Unlike primary microcephaly, usually caused by a decrease in the number of neurons generated during neurogenesis, secondary microcephaly is thought to result from the postnatal reduction of dendritic processes, myelination, and synaptic connections.55 Corroboratively, our studies of bptf deficiency in zebrafish showed a significant increase in neuronal cell death.
Reduction in brain volume found in microcephalic individuals is often associated with brain anomalies, DD/ID, motor disabilities, epilepsy, and ophthalmological disorders.56 No epilepsy was observed in our cohort. Brain anomalies detected in MRI include small anterior pituitary in subject 1, bilateral nonspecific multifocal white matter lesions in subject 2, pituitary hypoplasia in subject 7, reduced signal intensity of frontal and temporal white matter in subject 8, periventricular white matter lesions in subject 9, and borderline low-lying cerebellar tonsil on the right in subject 10. Ophthalmological abnormalities include: outward deviation of the eye (individual 1); left colobomatous (iris and disk) microphthalmia; slight inferior lens subluxation; telecanthus (individual 10); myopia (individuals 7 and 10); up-slanting or short palpebral fissures (individuals 4, 5 and 7, 8, respectively); and cataract and hyperopia (individual 9). The most characteristic facial feature observed is prominent nose (observed in subjects 2, 4, 6, 7–10). Other commonly shared features include toe/foot (subjects 2, 6, 7–9) and finger (subjects 2, 5–7, and 10) anomalies (Figure 1A, Table 1).
BPTF is located within the 17q24.2 microdeletion region, which encompasses multiple genes including BPTF and PSMD12. Recently, we have reported de novo disruption of the proteasome regulatory subunit PSMD12 (MIM: 604450).39 Reported subjects included four (individuals 1–4) with single-nucleotide variants in PSMD12, three (individuals 5, 7, and 9) with CNV deletions on 17q24.2 affecting PSMD12, BPTF, and other genes, and two (individuals 8 and 10) with deletions on 17q24.2 encompassing PSMD12 and neighboring genes but leaving BPTF intact.39 We hypothesize that the clinical phenotype of 17q24.2 microdeletion is associated with haploinsufficiency of the affected PSMD12 and/or BPTF, and possibly additional genes. Within the reported deletion region on 17q24.2, five genes have pLI scores greater than 0.95 and are predicted to be highly intolerant of LoF mutations, including PSMD12, BPTF, and three other genes that are currently not implicated in disease: HELZ, PITPNC1, and PRKCA (Figure 3). The remaining protein-coding genes in this microdeletion region are predicted to be LoF tolerant. Comparison of the phenotypic features indicate that PSMD12- and BPTF-related disorders share clinical phenotypes including DD/ID, dysmorphisms, skeletal anomalies, and failure to thrive (Figure 3). The presence of cardiac, renal, genital, and ophthalmological defects and seizures were more commonly seen in individuals with PSMD12 mutations while microcephaly was more commonly associated with a BPTF disruption. Notably, Küry et al.39 also reported that individuals with 17q24.2 deletions involving PSMD12 and BPTF had more prominent microcephaly than those with isolated PSMD12 deficiency, supporting further the causative role of BPTF in microcephaly.
In addition to BPTF, there are more than 40 diverse proteins containing BRDs;57 more than 20 of them display linked PHD fingers and BRDs.58 Importantly, BRD-containing proteins have been implicated in numerous developmental disorders,59 e.g., EP300 in Rubinstein-Taybi syndrome (RSTS2 [MIM: 613684]),60 BRWD3 (MRX93 [MIM: 300659]),61 WDR11 (HH14 [MIM: 614858]),62 and BRPF1 (IDDDFP [MIM: 617333]).63 Moreover, genes encoding BRD-containing proteins have been also found mutated in multiple cancers.57, 64 For example, a number of them have been found at chromosomal translocation breakpoint fusions mapping at the BRDs and affecting their function,57 e.g., BRD4-NUT fusion is detected in NUT midline carcinomas.65 Notably, based on promising results in murine models, BRD proteins have become targets for drug development and translation.66, 67
In aggregate, we propose that haploinsufficiency of BPTF results in an augmented neuronal death, which likely occurs mainly during the postnatal period and manifests with acquired microcephaly and neurodevelopmental abnormalities. Characteristic facial features involving nose and eyes as well as anomalies involving fingers and toes, resembling those reported in patients with 17q24.2 deletions,68, 69, 70, 71, 72 enabled us to define a novel syndromic disorder. Genes encoding BRD and/or PHD represent strong candidates that could possibly contribute to human genetic disorders that involve a constellation of ID/DD, microcephaly, and dysmorphic features.
Acknowledgments
We thank all families for participating in this study. We thank Mr. Z. Kupchinsky for zebrafish husbandry and automated imaging support. Evaluation of subject 3 was through support from U01HG006485 to S.E.P. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from https://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust. E.E.D. is supported by US NIH grant R01 MH106826. The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. S.E.P. is a member of the Scientific Advisory Board (SAB) of Baylor Genetics Laboratories. A.R. is supported by CHROMATIN-Net funded by German Federal Ministry of Education and Research (BMBF, grant number 01GM1520A). N.K. is a paid consultant for and holds significant stock of Rescindo Therapeutics, Inc. Y.Y. is a member of the Scientific Advisory Board (SAB) of Veritas Genetics China. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered by Baylor Genetics.
Published: September 21, 2017
Footnotes
Supplemental Data include Supplemental Note, 3 figures, and 3 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.08.014.
Contributor Information
Paweł Stankiewicz, Email: pawels@bcm.edu.
Yaping Yang, Email: yapingy@bcm.edu.
Accession Numbers
The accession numbers for the data reported in this paper are Leiden OpenVariation Database (LOVD) #00106517–00106527 and ClinVar SCV000584200–SCV000584209.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
Baylor Genetics Laboratory, http://bmgl.com/
CHOPCHOP, http://chopchop.cbu.uib.no/
Database of Genomic Variants (DGV), http://dgv.tcag.ca/dgv/app/home
DECIPHER, https://decipher.sanger.ac.uk/
ExAC Browser, http://exac.broadinstitute.org/
GeneMatcher, https://genematcher.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
Human Splicing Finder, http://www.umd.be/HSF3/
MaxEntScan, http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Primer3, http://bioinfo.ut.ee/primer3
SPIDEX, http://tools.genes.toronto.edu/
SWISS-MODEL, http://swissmodel.expasy.org/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu. Rev. Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badenhorst P., Voas M., Rebay I., Wu C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002;16:3186–3198. doi: 10.1101/gad.1032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai X., Larschan E., Kwon S.Y., Badenhorst P., Kuroda M.I. Regional control of chromatin organization by noncoding roX RNAs and the NURF remodeling complex in Drosophila melanogaster. Genetics. 2007;176:1491–1499. doi: 10.1534/genetics.107.071571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon S.Y., Xiao H., Glover B.P., Tjian R., Wu C., Badenhorst P. The nucleosome remodeling factor (NURF) regulates genes involved in Drosophila innate immunity. Dev. Biol. 2008;316:538–547. doi: 10.1016/j.ydbio.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 5.Hamiche A., Sandaltzopoulos R., Gdula D.A., Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 6.Tsukiyama T., Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 7.Alkhatib S.G., Landry J.W. The nucleosome remodeling factor. FEBS Lett. 2011;585:3197–3207. doi: 10.1016/j.febslet.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barak O., Lazzaro M.A., Lane W.S., Speicher D.W., Picketts D.J., Shiekhattar R. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 2003;22:6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao H., Sandaltzopoulos R., Wang H.M., Hamiche A., Ranallo R., Lee K.M., Fu D., Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 10.Jones M.H., Hamana N., Shimane M. Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics. 2000;63:35–39. doi: 10.1006/geno.1999.6070. [DOI] [PubMed] [Google Scholar]
- 11.Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Müller S., Pawson T. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Ilin S., Wang W., Duncan E.M., Wysocka J., Allis C.D., Patel D.J. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruthenburg A.J., Li H., Milne T.A., Dewell S., McGinty R.K., Yuen M., Ueberheide B., Dou Y., Muir T.W., Patel D.J., Allis C.D. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 15.Goller T., Vauti F., Ramasamy S., Arnold H.H. Transcriptional regulator BPTF/FAC1 is essential for trophoblast differentiation during early mouse development. Mol. Cell. Biol. 2008;28:6819–6827. doi: 10.1128/MCB.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landry J., Sharov A.A., Piao Y., Sharova L.V., Xiao H., Southon E., Matta J., Tessarollo L., Zhang Y.E., Ko M.S. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayes K., Alkhatib S.G., Peterson K., Alhazmi A., Song C., Chan V., Blevins T., Roberts M., Dumur C.I., Wang X.Y., Landry J.W. BPTF depletion enhances T-cell-mediated antitumor immunity. Cancer Res. 2016;76:6183–6192. doi: 10.1158/0008-5472.CAN-15-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Y., Liu X., Liu Z., Wei S., Shang H., Xue Y., Cao Y., Meng A., Wang Q. The chromatin remodeling protein Bptf promotes posterior neuroectodermal fate by enhancing Smad2-activated wnt8a expression. J. Neurosci. 2015;35:8493–8506. doi: 10.1523/JNEUROSCI.0377-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobolyi A., Vincze C., Pál G., Lovas G. The neuroprotective functions of transforming growth factor beta proteins. Int. J. Mol. Sci. 2012;13:8219–8258. doi: 10.3390/ijms13078219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erter C.E., Wilm T.P., Basler N., Wright C.V., Solnica-Krezel L. Wnt8 is required in lateral mesendodermal precursors for neural posteriorization in vivo. Development. 2001;128:3571–3583. doi: 10.1242/dev.128.18.3571. [DOI] [PubMed] [Google Scholar]
- 21.Buganim Y., Goldstein I., Lipson D., Milyavsky M., Polak-Charcon S., Mardoukh C., Solomon H., Kalo E., Madar S., Brosh R. A novel translocation breakpoint within the BPTF gene is associated with a pre-malignant phenotype. PLoS ONE. 2010;5:e9657. doi: 10.1371/journal.pone.0009657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai M., Lu J.J., Guo W., Yu W., Wang Q., Tang R., Tang Z., Xiao Y., Li Z., Sun W. BPTF promotes tumor growth and predicts poor prognosis in lung adenocarcinomas. Oncotarget. 2015;6:33878–33892. doi: 10.18632/oncotarget.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong Y.C., Liu D.C., Li X.P., Dai S.P. BPTF biomarker correlates with poor survival in human NSCLC. Eur. Rev. Med. Pharmacol. Sci. 2017;21:102–107. [PubMed] [Google Scholar]
- 24.Lee J.H., Kim M.S., Yoo N.J., Lee S.H. BPTF, a chromatin remodeling-related gene, exhibits frameshift mutations in gastric and colorectal cancers. APMIS. 2016;124:425–427. doi: 10.1111/apm.12512. [DOI] [PubMed] [Google Scholar]
- 25.Richart L., Carrillo-de Santa Pau E., Río-Machín A., de Andrés M.P., Cigudosa J.C., Lobo V.J., Real F.X. BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nat. Commun. 2016;7:10153. doi: 10.1038/ncomms10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao S., Liu L., Fang M., Zhou X., Peng X., Long J., Lu X. BPTF associated with EMT indicates negative prognosis in patients with hepatocellular carcinoma. Dig. Dis. Sci. 2015;60:910–918. doi: 10.1007/s10620-014-3411-0. [DOI] [PubMed] [Google Scholar]
- 27.Wu B., Wang Y., Wang C., Wang G.G., Wu J., Wan Y.Y. BPTF is essential for T cell homeostasis and function. J. Immunol. 2016;197:4325–4333. doi: 10.4049/jimmunol.1600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayes K., Elsayed Z., Alhazmi A., Waters M., Alkhatib S.G., Roberts M., Song C., Peterson K., Chan V., Ailaney N. BPTF inhibits NK cell activity and the abundance of natural cytotoxicity receptor co-ligands. Oncotarget. 2017 doi: 10.18632/oncotarget.17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frey W.D., Chaudhry A., Slepicka P.F., Ouellette A.M., Kirberger S.E., Pomerantz W.C.K., Hannon G.J., Dos Santos C.O. BPTF maintains chromatin accessibility and the self-renewal capacity of mammary gland stem cells. Stem Cell Reports. 2017;9:23–31. doi: 10.1016/j.stemcr.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boone P.M., Bacino C.A., Shaw C.A., Eng P.A., Hixson P.M., Pursley A.N., Kang S.H., Yang Y., Wiszniewska J., Nowakowska B.A. Detection of clinically relevant exonic copy-number changes by array CGH. Hum. Mutat. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiszniewska J., Bi W., Shaw C., Stankiewicz P., Kang S.H., Pursley A.N., Lalani S., Hixson P., Gambin T., Tsai C.H. Combined array CGH plus SNP genome analyses in a single assay for optimized clinical testing. Eur. J. Hum. Genet. 2014;22:79–87. doi: 10.1038/ejhg.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 37.Farwell Hagman K.D., Shinde D.N., Mroske C., Smith E., Radtke K., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Alcaraz W.A. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet. Med. 2017;19:224–235. doi: 10.1038/gim.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Küry S., Besnard T., Ebstein F., Khan T.N., Gambin T., Douglas J., Bacino C.A., Craigen W.J., Sanders S.J., Lehmann A. De novo disruption of the proteasome regulatory subunit PSMD12 causes a syndromic neurodevelopmental disorder. Am. J. Hum. Genet. 2017;100:352–363. doi: 10.1016/j.ajhg.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu X., Xu Y., Yu S., Lu L., Ding M., Cheng J., Song G., Gao X., Yao L., Fan D. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep. 2014;4:6420. doi: 10.1038/srep06420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kague E., Gallagher M., Burke S., Parsons M., Franz-Odendaal T., Fisher S. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE. 2012;7:e47394. doi: 10.1371/journal.pone.0047394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isrie M., Breuss M., Tian G., Hansen A.H., Cristofoli F., Morandell J., Kupchinsky Z.A., Sifrim A., Rodriguez-Rodriguez C.M., Dapena E.P. Mutations in either TUBB or MAPRE2 cause circumferential skin creases Kunze type. Am. J. Hum. Genet. 2015;97:790–800. doi: 10.1016/j.ajhg.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golzio C., Willer J., Talkowski M.E., Oh E.C., Taniguchi Y., Jacquemont S., Reymond A., Sun M., Sawa A., Gusella J.F. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parsons D.W., Roy A., Yang Y., Wang T., Scollon S., Bergstrom K., Kerstein R.A., Gutierrez S., Petersen A.K., Bavle A. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2015.5699. Published online January 28, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H.T., Lee M.S., Choi J.H., Jung J.Y., Ahn D.G., Yeo S.Y., Choi D.K., Kim C.H. The microcephaly gene aspm is involved in brain development in zebrafish. Biochem. Biophys. Res. Commun. 2011;409:640–644. doi: 10.1016/j.bbrc.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama T., Al-Maawali A., El-Quessny M., Rajab A., Khalil S., Stoler J.M., Tan W.H., Nasir R., Schmitz-Abe K., Hill R.S. Mutations in PYCR2, encoding pyrroline-5-carboxylate reductase 2, cause microcephaly and hypomyelination. Am. J. Hum. Genet. 2015;96:709–719. doi: 10.1016/j.ajhg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ta-Shma A., Khan T.N., Vivante A., Willer J.R., Matak P., Jalas C., Pode-Shakked B., Salem Y., Anikster Y., Hildebrandt F. Mutations in TMEM260 cause a pediatric neurodevelopmental, cardiac, and renal syndrome. Am. J. Hum. Genet. 2017;100:666–675. doi: 10.1016/j.ajhg.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bohnsack B.L., Kasprick D.S., Kish P.E., Goldman D., Kahana A. A zebrafish model of axenfeld-rieger syndrome reveals that pitx2 regulation by retinoic acid is essential for ocular and craniofacial development. Invest. Ophthalmol. Vis. Sci. 2012;53:7–22. doi: 10.1167/iovs.11-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dauber A., Golzio C., Guenot C., Jodelka F.M., Kibaek M., Kjaergaard S., Leheup B., Martinet D., Nowaczyk M.J., Rosenfeld J.A. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am. J. Hum. Genet. 2013;93:798–811. doi: 10.1016/j.ajhg.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frosk P., Arts H.H., Philippe J., Gunn C.S., Brown E.L., Chodirker B., Simard L., Majewski J., Fahiminiya S., Russell C., FORGE Canada Consortium. Canadian Rare Diseases: Models & Mechanisms Network A truncating mutation in CEP55 is the likely cause of MARCH, a novel syndrome affecting neuronal mitosis. J. Med. Genet. 2017;54:490–501. doi: 10.1136/jmedgenet-2016-104296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon C.T., Weaver K.N., Zechi-Ceide R.M., Madsen E.C., Tavares A.L., Oufadem M., Kurihara Y., Adameyko I., Picard A., Breton S. Mutations in the endothelin receptor type A cause mandibulofacial dysostosis with alopecia. Am. J. Hum. Genet. 2015;96:519–531. doi: 10.1016/j.ajhg.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw N.D., Brand H., Kupchinsky Z.A., Bengani H., Plummer L., Jones T.I., Erdin S., Williamson K.A., Rainger J., Stortchevoi A. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet. 2017;49:238–248. doi: 10.1038/ng.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seltzer L.E., Paciorkowski A.R. Genetic disorders associated with postnatal microcephaly. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:140–155. doi: 10.1002/ajmg.c.31400. [DOI] [PubMed] [Google Scholar]
- 55.Woods C.G. Human microcephaly. Curr. Opin. Neurobiol. 2004;14:112–117. doi: 10.1016/j.conb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 56.von der Hagen M., Pivarcsi M., Liebe J., von Bernuth H., Didonato N., Hennermann J.B., Bührer C., Wieczorek D., Kaindl A.M. Diagnostic approach to microcephaly in childhood: a two-center study and review of the literature. Dev. Med. Child Neurol. 2014;56:732–741. doi: 10.1111/dmcn.12425. [DOI] [PubMed] [Google Scholar]
- 57.Fujisawa T., Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 2017;18:246–262. doi: 10.1038/nrm.2016.143. [DOI] [PubMed] [Google Scholar]
- 58.Ruthenburg A.J., Li H., Patel D.J., Allis C.D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J., Zhao G., Gao X. Development of neurodevelopmental disorders: a regulatory mechanism involving bromodomain-containing proteins. J. Neurodev. Disord. 2013;5:4. doi: 10.1186/1866-1955-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roelfsema J.H., White S.J., Ariyürek Y., Bartholdi D., Niedrist D., Papadia F., Bacino C.A., den Dunnen J.T., van Ommen G.J., Breuning M.H. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Field M., Tarpey P.S., Smith R., Edkins S., O’Meara S., Stevens C., Tofts C., Teague J., Butler A., Dicks E. Mutations in the BRWD3 gene cause X-linked mental retardation associated with macrocephaly. Am. J. Hum. Genet. 2007;81:367–374. doi: 10.1086/520677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim H.G., Ahn J.W., Kurth I., Ullmann R., Kim H.T., Kulharya A., Ha K.S., Itokawa Y., Meliciani I., Wenzel W. WDR11, a WD protein that interacts with transcription factor EMX1, is mutated in idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am. J. Hum. Genet. 2010;87:465–479. doi: 10.1016/j.ajhg.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan K., Rousseau J., Littlejohn R.O., Kiss C., Lehman A., Rosenfeld J.A., Stumpel C.T., Stegmann A.P., Robak L., Scaglia F., DDD Study. CAUSES Study Mutations in the chromatin regulator gene BRPF1 cause syndromic intellectual disability and deficient histone acetylation. Am. J. Hum. Genet. 2017;100:91–104. doi: 10.1016/j.ajhg.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huether R., Dong L., Chen X., Wu G., Parker M., Wei L., Ma J., Edmonson M.N., Hedlund E.K., Rusch M.C. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat. Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.French C.A., Miyoshi I., Kubonishi I., Grier H.E., Perez-Atayde A.R., Fletcher J.A. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–307. [PubMed] [Google Scholar]
- 66.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang C.Y., Filippakopoulos P. Beating the odds: BETs in disease. Trends Biochem. Sci. 2015;40:468–479. doi: 10.1016/j.tibs.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 68.Naud M.E., Tosca L., Martinovic J., Saada J., Métay C., Drévillon L., Benoit V., Brisset S., Tachdjian G. Prenatal diagnosis of a 2.5 Mb de novo 17q24.1q24.2 deletion encompassing KPNA2 and PSMD12 genes in a fetus with craniofacial dysmorphism, equinovarus feet, and syndactyly. Case Rep. Genet. 2017;2017:7803136. doi: 10.1155/2017/7803136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stewart D.R., Pemov A., Johnston J.J., Sapp J.C., Yeager M., He J., Boland J.F., Burdett L., Brown C., Gatti R.A. Dubowitz syndrome is a complex comprised of multiple, genetically distinct and phenotypically overlapping disorders. PLoS ONE. 2014;9:e98686. doi: 10.1371/journal.pone.0098686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bartnik M., Nowakowska B., Derwińska K., Wiśniowiecka-Kowalnik B., Kędzior M., Bernaciak J., Ziemkiewicz K., Gambin T., Sykulski M., Bezniakow N. Application of array comparative genomic hybridization in 256 patients with developmental delay or intellectual disability. J. Appl. Genet. 2014;55:125–144. doi: 10.1007/s13353-013-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vergult S., Dauber A., Delle Chiaie B., Van Oudenhove E., Simon M., Rihani A., Loeys B., Hirschhorn J., Pfotenhauer J., Phillips J.A., 3rd 17q24.2 microdeletions: a new syndromal entity with intellectual disability, truncal obesity, mood swings and hallucinations. Eur. J. Hum. Genet. 2012;20:534–539. doi: 10.1038/ejhg.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blyth M., Huang S., Maloney V., Crolla J.A., Karen Temple I. A 2.3 Mb deletion of 17q24.2-q24.3 associated with ‘Carney Complex plus’. Eur. J. Med. Genet. 2008;51:672–678. doi: 10.1016/j.ejmg.2008.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.