Abstract
Amyloids are traditionally observed in the context of disease. However, there is growing momentum that these structures can serve a beneficial role where the amyloid carries out a specific function. These so called ‘functional amyloids’ have all the structural hallmarks of disease-associated amyloids, raising the question as to what differentiates a well-behaved benign amyloid from a lethally destructive one. Here, we review our work on the repeat domain (RPT) from Pmel17, an important functional amyloid involved in melanin biosynthesis. Particularly, we focused our attention on the unique reversible aggregation-disaggregation process of RPT that is controlled strictly by solution pH. This pH dependence of RPT amyloid formation functions as a switch to control fibril assembly and maintains the benign nature that is associated with functional amyloids.
Keywords: melanosomes, melanin, transmission electron microscopy, pH, kinetics
1 Introduction to Amyloids
Amyloids are highly-ordered protein aggregates that have a filamentous morphology.[1] These structures are un-branched with indefinite length and diameters between 2–20 nm. They are rich in β-sheet secondary structure and are formed by non-covalent polymerization of protein monomers that assemble with their β-strands running approximately perpendicular to the fibril axis. This distinctive protein assembly fold is also termed as cross-β structure, where the distances between inter β-strands and β-sheets is 4.7 and 10 Å as determined by x-ray diffraction.[2]
Biochemical and physical properties of amyloids include resistance to proteolysis (e.g. proteinase-K), chemical denaturation (molar concentrations of guanidinium-HCl and urea), and moderate detergent extraction such as Triton X-100 and sodium dodecyl sulfate.[3] Staining amyloid with dyes such as Congo red gives a yellow-green birefringence under polarized light,[4] while an intense fluorescence signal is observed upon binding to thioflavin-T (ThT).[5] Transmission electron microscopy (TEM) and atomic force microscopy (AFM) have been instrumental in gross structural characterization of amyloid fibrils.[6] In fact, TEM has been used to identify amyloid polymorphism, a phenomenon where molecular structures of fibrils formed by a given polypeptide can vary, depending on the precise details of growth conditions.[7] Such changes can result in different fibril morphologies, e.g. ribbons or twists.[7b, 8] Electron microscopy can also measure the mass per length of fibrils, which gives the number of protein monomers (or fraction of a monomer) that compose a unit length of the fibril, determining important constraints for structural model building.[9]
While amyloids are generally referred to in a negative context, since their presence is associated with neurodegenerative diseases such as Alzheimer’s,[10] Parkinson’s,[11] and Huntington’s,[12] some fulfill a beneficial role where the amyloid carries out a particular function. Examples have been identified in bacteria,[13] fungi,[14] and humans,[15] offering molecular and cellular insights of using amyloids to carry out a function.
In this review, we first discuss two hypotheses that may differentiate pathological from functional amyloids. This is followed by reviewing our work on a functional amyloid involving the repeat domain (RPT) of Pmel17 that forms amyloid only under acidic melanosomal pH.[16] This work shows that unlike pathological amyloids, these RPT fibrils rapidly dissolve to monomeric species at neutral pH. From a biological perspective, we hypothesize that if RPT fibrils were to escape from melanosomes, they would be exposed to a more neutral environment, and eventually dissolve and maintain a soluble benign form. This highly pH dependent mechanism ensures that RPT only aggregates in the melanosome.
1.1 Structural Differences between Pathological vs. Functional Amyloid
Although amyloids are generally non-crystalline, short synthetic peptides (4–10 residues) derived from amyloid-forming proteins have been crystallized and atomic structure determination reveals β-strands with an interlocking of self-complementary surfaces of adjacent β-strands, a structure termed “steric zipper”.[17] These structures are proposed to be a characteristic hallmark of amyloid structures. Some of the most informative structural information on fibrils from full-length proteins have come from solid-state NMR (ssNMR)[2,18] and electron paramagnetic resonance (EPR) spectroscopy.[19] For ssNMR uniformly 13C,15N-labeled samples offer the most valuable information, but fibril polymorphism still makes structural determination challenging.[20] Despite this limitation, high resolution atomic models have been obtained for amyloid structures. [7b, 21]
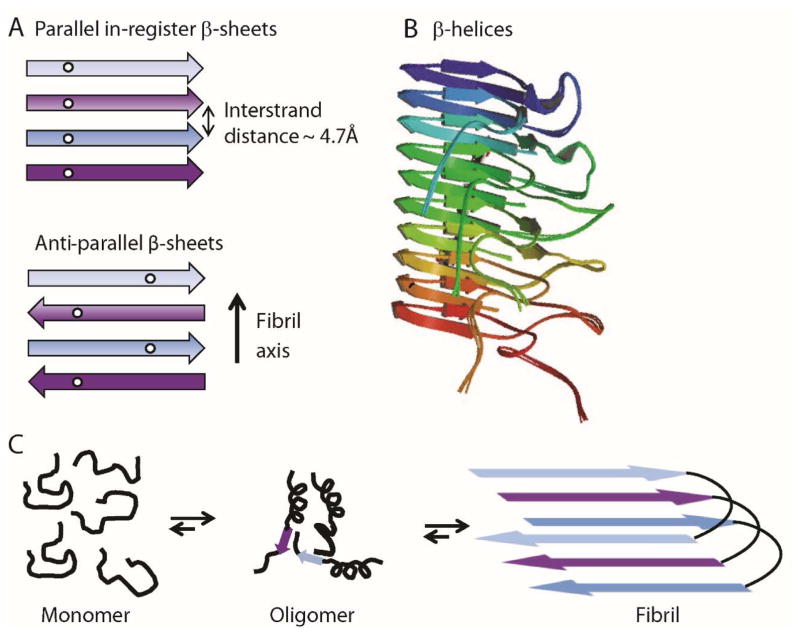
Parallel in-register β-sheet structures appear to be the most common underlying architecture observed for pathological amyloids (Figure 1A). [1b, 22] Here, the β-strands lie perpendicular to the fibril axis and form hydrogen bonds running parallel with the fibril. Each residue within a single polypeptide forms an aligned row along the long axis of the fibril. The best characterized pathological amyloid structure is that of Aβ1–40, a protein aggregate associated with Alzheimer’s disease.[7a, 23] A sole exception to parallel in-register β-sheet structure is a mutant form of Aβ1–40 associated with early-onset familial Alzheimer’s disease that was found to form fibrils with antiparallel strands (Figure 1A).[24]
Figure 1.
(A) Schematic showing parallel in-register β-sheet structures composed of individual polypeptides stacking in-register every ~4.7 Å along the fibril axis (top). Antiparallel β-sheet structures are also composed of polypeptides stacking every ~4.7 Å but β-strands alternately run in opposite directions (bottom). (B) β-Helices of Het-S are composed of a single polypeptide wrapping around an axis, forming intramolecular parallel β-sheets (PDB accession no. 2LBU). (C) Schematic diagram depicting fibril formation via oligomeric intermediates associated with pathological amyloids.
For functional amyloids, structural information is generally lacking; however, a full atomic model of the HET-s prion protein of Podospora anserina was revealed by ssNMR to be a β-helix (or solenoid) model (Figure 1B) containing parallel β-sheets, with the strands wrapping around an axis in a helical arrangement.[21a] Unlike parallel in-register β-sheets, parallel strands in these helices had intramolecular backbone hydrogen bonds. Furthermore, no fibril polymorphism was observed suggesting a single structure is formed. The Curli protein from E.coli which is a filamentous homopolymer that functions in extracellular biofilm formation[25] is suggested to also form a β-helical structure.[26] Based on these two structures, a β-helical model has been suggested to be more common amongst functional rather than pathological amyloids, possibly providing the molecular basis for their disparate biological behavior.
1.2 Cellular Control During Amyloid Formation
There is gaining momentum that oligomers, increasingly pinpointed as the pathogenic agents,[27] offer clues as to what differentiates a well-behaved amyloid, from a lethally destructive amyloid. Studies now target amyloid toxicity to oligomeric intermediates that build-up during amyloid formation (Figure 1C).[27–28] For example, it has been observed that soluble oligomers of Aβ correlate better with disease severity than the insoluble fibrillar deposits that are present in amyloid plaques, suggesting that oligomeric forms are the toxic species.[29]
For functional amyloids, a possible solution to avoid toxicity would be tight kinetic control during amyloid formation. Insights from the functional bacterial amyloid ‘Curli’ show cell regulation at several steps by other proteins that facilitate localization and nucleate rapid polymerization of the protein CsgA.[13] Thus, multiple cellular factors in E. coli have evolved to promote efficient amyloid assembly and ensure that amyloid fibrillization is spatially sequestered in the extracellular matrix. Another example comes from the ingenious method of storing peptide hormones in secretory granules in a concentrated amyloid-state, and rapidly releasing non-toxic functional monomers, providing an elegant mechanism for bypassing stable oligomeric species.[15b, 30]
Newly discovered functional amyloids from RNA proteins have also been shown to reversibly transform from a soluble state into polymeric, amyloid-like fibrils. While the biological function for this phenomenon is unknown,[31] rapid aggregation kinetics that circumvent oligomeric intermediates have been suggested, which again could solely explain their non-toxic nature.[31a]
2 Pmel17 and Its Role in Melanin Synthesis
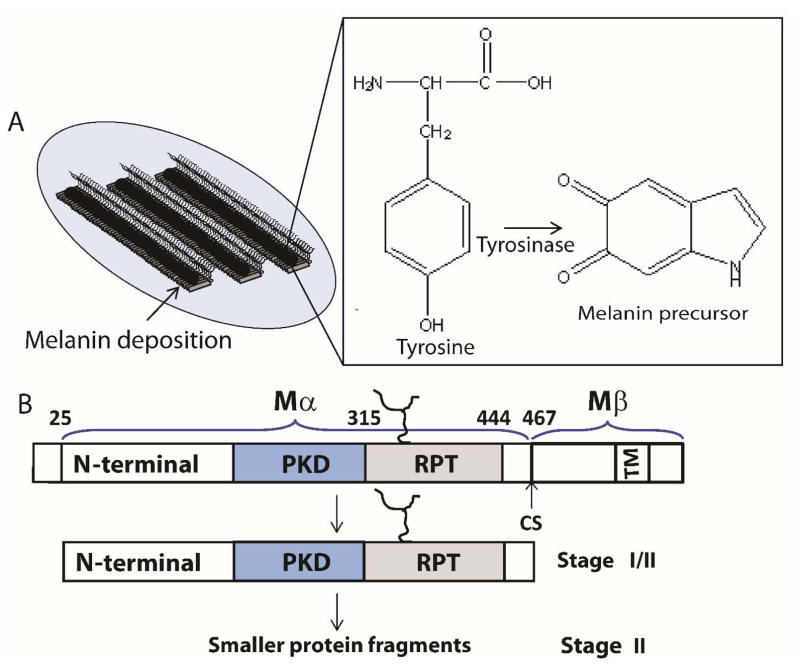
Pmel17 is a transmembrane protein that is associated with intralumenal fibrous striations in melanosomes, organelles where melanin is formed and stored.[32] Melanin synthesis involves a multitude of oxidation steps initiated by the action of tyrosinase on the substrate tyrosine (Figure 2A). Specifically, tyrosine is converted to L-DOPA (L-3,4-dihydroxyphenylalanine), which is readily oxidized to the melanin precursor, indole-5,6-quinone. The subsequent polymerization of these precursors to form melanin is suggested to occur on these filamentous striations, which acts as a scaffold for depositing melanin.[33] Since melanin intermediates are cytotoxic, sequestration of these molecules provide a detoxifying environment that permits safety during melanin biosynthesis (Figure 2A).
Figure 2.
(A) Schematic representation showing stage III/IV melanosomes. Melanin is depicted as black polymer deposits on preformed fibrils (left). Chemical transformation of L-tyrosine by tyrosinase to the melanin precursor, indole-5,6-quinone (right). (B) Stage I contains full-length Pmel17, which undergoes membrane cleavage, liberating the Mα from the Mβ transmembrane fragment. Further proteolytic processing of Mα (Stage II) generates smaller fragments containing at least two domains, including RPT, which contributes to the fibrillar matrix.
Melanosome maturation consists of four morphological stages at the ultrastructural level that is defined based on Pmel17 fibril formation and melanin synthesis.[33] Stages I and II are defined by formation of fibrils which initiates during stage I and is completed by stage II. Ellipsoidal shaped stage II melanosomes harbor mature fibrils that are found in parallel arrays, spanning the length of the organelle. These fibrils are approximately 6–10 nm in diameter. Once mature fibrils are formed, melanin synthesis begins with enrichment of the enzyme, tyrosinase, catalyzing melanin formation during stages III/IV. TEM images reveal that these fibrils grow 2-fold in diameter reaching 14–20 nm upon melanin deposition.[33] In highly pigmented cells, fibrils are completely masked with melanin.
Pmel17 was first identified genetically in 1930, as the ‘silver gene’ in mice, whose mutation resulted in hypopigmentation.[34] However, it took more than 60 years for the gene to be mapped to the silver locus and cloned.[35] Transient expression of Pmel17 in nonmelanocytic cell lines (e.g. HeLa cells) induced fibril formation, reminiscent to those seen in melanosomes.[36] Pmel17 is synthesized as a 668-residue protein with a single transmembrane domain, a short C-terminal cytoplasmic domain and a large luminal domain exposed within the lumen of the endoplasmic reticulum (Figure 2B). After a series of N- and O-glycosylation steps, Pmel17 is delivered to endosomes where it associates internally with intraluminal vesicles (IVLs). Cleavage by a protease activity between residues 467 and 468 produces a C-terminal fragment Mβ (residues 468-668) including the transmembrane and cytoplasmic domains and an N-terminal fragment Mα (residues 25–467) with most of the luminal domain (Figure 2B).[32, 37] Within Mα, several distinct regions have been ascribed based primarily on primary amino acid sequence. An N-terminal region with a lack of sequence homology to other proteins aside from the closely related homolog gpNmb[38] contains two N-glycosylation sites and cysteine residues that likely participate in disulfide bond formation. Currently, no structural information is available for this region. The immediately adjacent region is referred to as the polycystic kidney disease (PKD)-like domain due to its sequence homology to a short peptide consensus sequence in the protein polycystin-1.[39] It is suggested to contain a β-sandwich domain like PKD but no structural information is available for this domain. Finally, there is a region called the repeat domain (RPT) due to its series of 10 imperfect repeats of 13 residues each rich in proline, serine, threonine, and glutamic acid residues (Figure 3A).[36] Deletion of any of these regions was shown in vivo to either disrupt Pmel17 trafficking within endosomes or ablate fibril formation.[36, 40]
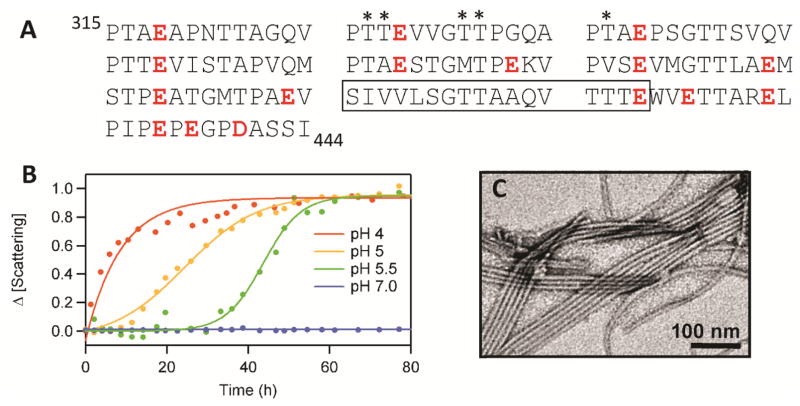
Figure 3.
(A) The RPT sequence is composed of 10 imperfect repeats rich in Pro, Ser, Thr, and Glu (in red). The conserved amyloid core region determined by solid-state NMR is boxed. Putative O-glycosylation sites are marked with an asterisk. (B) pH dependent aggregation of RPT monitored by light scattering.[50] (C) Representative TEM image of RPT fibrils formed at pH 5.[49]
3 Identifying the Amyloid-forming Region of Pmel17
Pmel17 derived fibrils were first shown to be amyloidogenic using purified melanosome fractions from bovine eyes that were positively stained with thioflavin S (ThS) and Congo red, two known amyloidogenic dyes.[15a] A Triton X-100 insoluble melanosomal extract showed an almost exclusive overlap between ThS stained particles and Pmel17 immunofluorescence. Supporting the involvement of Pmel17 in amyloid formation, a recombinant fragment Mα (residues 25-467) expressed in E. coli showed rapid aggregation upon dilution out of guanidinium-HCl.[15a] These structures had all the hallmarks of amyloid with its rapid aggregation suggestive to be biologically relevant for a functional amyloid.
Our attempts to replicate this rapid or even gradual fibril formation with an identical Mα construct using the reported conditions were unsuccessful.[41] While it was observed that Mα aggregated upon dilution out of denaturant, in our hands the morphology of these aggregates visualized by TEM looked more amorphous than filamentous. Supporting this observation, Mel220 cells overexpressed with Pmel17 were shown to generate Mα that was found in the Triton-X100 soluble fraction, implying Mα is not in an insoluble amyloid state.[42]
To define the likely amyloidogenic region within Pmel17, several fragments from the Mα region (residues 25–467) were expressed and purified from E. coli.[41] Of all the fragments tested, only the protein fragment corresponding to the RPT domain (residues 315–444) (Figure 2B)[36] formed amyloid. Starting as an unstructured protein at pH 5, RPT then aggregated with typical sigmoidal kinetics, consisting of a lag, growth and stationary phase (Figure 3B). TEM images taken post aggregation at pH 5 showed straight amyloid fibrils with a diameter of ~10 nm (Figure 3C). Amyloid presence was also confirmed by Congo red birefringence, proteinase-K resistance as well as being β-sheet rich using circular dichroism (CD) spectroscopy, electron diffraction and ssNMR.[41] Deletion of RPT in vivo ablates fibril formation.[36, 40] Also, antibodies directed against the only known O-glycosylation site in RPT (between residues 328 to 344) (Figure 3A) decorates ex vivo fibrils,[36, 43] further validating RPT as a constituent of melanosomal fibrils.
While our work highlights the requirement of RPT in fibril formation, the groups of Marks[38, 44] and Cresswell[42, 45] offer a different perspective as to what composes the amyloid-forming region in melanosomes. Rather, they consider the PKD domain as the fibril core with the RPT domain likely serving a regulatory rather than a structural role. Our reasons for believing that the RPT domain constitutes the amyloid-forming region in melanosomes are further discussed in more detail in section 10.
4 RPT Fibrils Enhance Melanin Synthesis
Using a Pmel17 knockout mutation in mice, a 40–50% reduction in melanin content in hair was demonstrated, suggesting melanosomal fibrils contribute to overall melanin content.[46] To recapitulate fibrils enhance melanin formation, an in vitro assay was performed using RPT fibrils, tyrosinase, and L-3,4-dihydroxyphenylalanine at pH 5. In the presence of RPT fibrils, a 6-fold increase in the yield of melanin was observed when compared to a control reaction containing only soluble RPT.[41] This observation is consistent with in vivo data showing a strong correlation between the presence of fibrils and total melanin production.[46] Interestingly, other amyloids such as the yeast prion Sup35 and the fungal prion Het-s also enhanced melanin formation,[41] suggesting a specific amyloid structure is not critical to enhancing melanin synthesis.
5 Structural Features of RPT Fibrils
Using ssNMR, RPT fibrils formed at pH 5 adopt the parallel in-register β-sheet architecture that is generally observed in human pathogenic amyloids.[47] By labelling at least one or a few residues with13C-labelled amino acid (Ala, Val, or Met) in the carbonyl position, the nearest intra/intermolecular 13C distance between β-strands in a β-sheet was 0.5 nm, indicative of parallel in-register structure. This observation is inconsistent with the prevailing notion that all functional amyloids adopt a β-solenoid structure.[1b]
ssNMR measurements of three independently prepared 13C15N-labeled RPT fibril samples at pH 5 revealed fibril polymorphism.[47] Here, subtle variations in aggregating conditions led to fibrils that were readily distinguishable in ssNMR spectra. Sequence assignment showed only a subset (~30%) of the full amino acid sequence contributes to the immobilized fibril core. A term ‘segmental polymorphism’[48] was used to describe these polymorphic fibrils that differ in the identity of the protein segments that form the fibril core. While these RPT polymorphs had distinct amyloid core-forming segments, the C-terminal residues 406–422 were always shown to have β-sheet backbone assignments indicating a key role for the C-terminus in fibril assembly. Additional supporting evidence comes from proteinase K resistance,[49] a lack of prolines between residues 403-431, W423 fluorescence[50] as well as computational methods that predict the amyloid-forming segments between residues 403–413.[47] Corroborating these data, a peptide segment 405VSIVVLSGT413 was shown to self-aggregate into amyloid affirming this region as the most aggregation prone.[51]
6 The RPT Domains of Pmel17 Orthologs Form Amyloid
To observe if fibril formation is conserved amongst other repeat regions from various Pmel17 orthologs, repeat sequences from mouse (mRPT) and zebrafish (zRPT) Pmel17 were studied.[52] Sequence comparison between human and mouse repeat regions share only 50% sequence identity, while those of zebrafish are completely unrelated. In addition to these, a known splice variant of human Pmel17 (sRPT) where 42 amino acids is deleted from repeats 5 to 8 was also investigated.[53]
Upon incubation at pH 5 with gentle agitation, all three RPT orthologs generated fibrils but with differing fibril morphology. For mRPT, twisted fibrils formed at pH 5 with an average diameter of 14 nm. Mass per unit length measurements of unstained mRPT fibrils gave a mean mass/length 2.12 ± 0.3 monomers/4.7 Å, suggesting paired filaments. In the case of zRPT, TEM images showed these fibrils to be thinner (~4 nm) and more prone to bundling into higher formed aggregates. The sRPT fibrils formed bundles of twisted fibrils, which are clearly different from the straight, single fibril morphology observed for RPT. ssNMR measurements from labeled mRPT, zRPT, and sRPT fibrils revealed all three samples adopt a parallel in-register structure.[52]
7 Mechanism of pH Dependent RPT Amyloid Formation
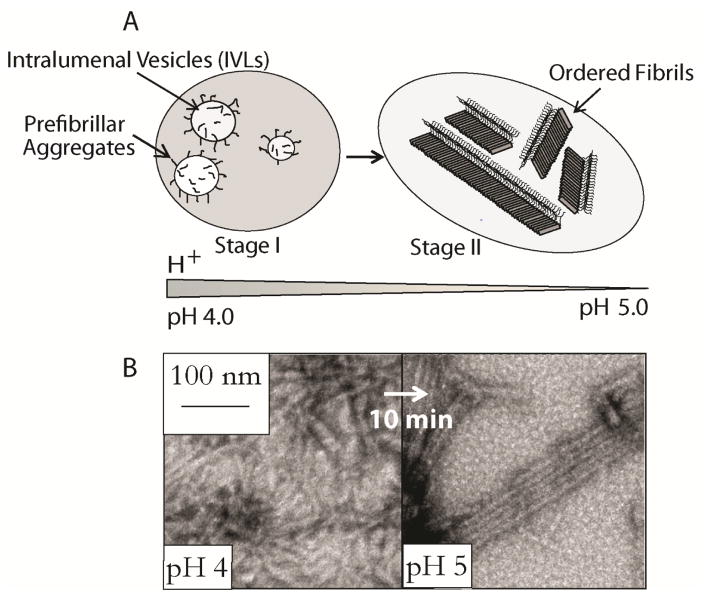
Melanosome maturation is accompanied by a change in pH starting more acidic (pH ~4.0) during stages I and II and likely approaching near neutral pH in fully melanized melanosomes (Figure 4A).[54] Our observation that RPT forms amyloid at pH 5 suggests that intramelanosomal pH could influence RPT amyloid formation. A combination of techniques including ThT and Trp fluorescence, light scattering and TEM were used to observe RPT structural changes as a function of pH (Figure 3B).[50] At low pH (~4.0), aggregation of RPT was rapid with a moderate ThT response and formation of small pre-fibrillar structures that have an amorphous appearance.[50] Between pH 5–5.5, aggregation was initiated within hours with TEM images revealing long un-branched fibrils, a typical morphology associated with amyloid and in vivo melanosomal fibrils [1b, 33]. At pH > 6.0, RPT remained soluble, showing no signs of secondary structure by CD spectroscopy and the presence of aggregates by TEM.[50]
Figure 4.
Schematic representation showing the morphological stages of melanosome maturation. Stage I contains prefibrillar aggregates generated from Pmel17 processing and their association with intraluminal vesicles (IVLs). Stage II consists of long fibrillar striations that run in parallel arrays spanning the length of the melanosome. (B) pH induced RPT fibril morphological conversion. TEM images (left) of aggregated RPT at pH 4, after adjusting the pH to 5 (right).[50]
To mimic RPT amyloid formation as a function of pH during melanosome maturation, an in vitro assay was performed where preformed RPT aggregates at pH 4.0 were first titrated to pH 5.0. Rapid (< 10 min) morphological changes occurred with these aggregates changing from small squiggles into long unbranched fibrils (Figure 4B). This structural change is reminiscent of the transition observed in stages I and II melanosomes, where organized arrays of intralumenal fibrils are shown by Stage II.[33] The biological time frame is also relevant since incorporation of Mα into insoluble fibrils occurs within minutes or at most a few hours, as judged by metabolic pulse/chase assays.[32, 37]
This pH dependent aggregation process would imply that protonation sites likely from specific carboxylic acids modulate amyloid kinetics. A total of 15 Glu and 1 Asp are present throughout the RPT sequence with a bias more towards the C-terminus. Since the C-terminus is likely the amyloid core region, we investigated the role of specific Glu residues: E404, E422, E425 and E430.[49] To assess the importance of protonation as well as hydrogen bonding at these sites, both Ala and Gln mutations were generated as well as single, double, and quadruple mutants to evaluate potential allosteric interactions.
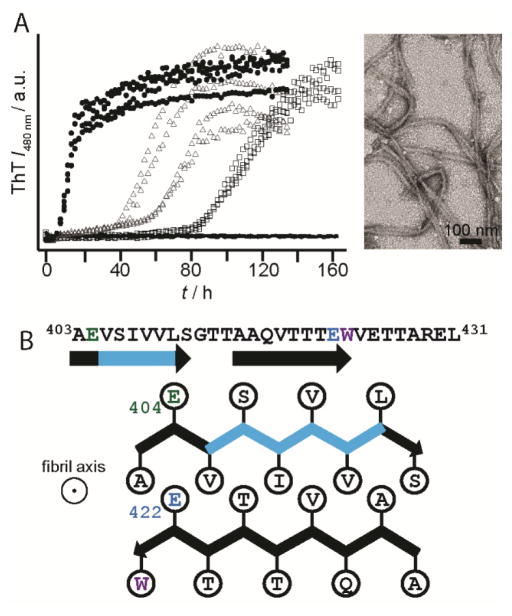
Aggregation kinetics revealed residue E422 as the critical side chain in controlling the pH sensitivity of RPT amyloid. Here, E422 mutants had a substantial impact, shifting the pH dependence more than one pH unit, with fibrils now forming at pH 6.5 (Figure 5A). Mutation at E404, E425, or E430 had little to no effect. We suggest that protonation at E422 is essential for initiating amyloid formation, while other Glu residues play a lesser role in this process. From a structural perspective, both E404 and E422 reside within the amyloid-forming region of RPT. A schematic representation for one possible conformation is shown in Figure 5B with two β-strands (indicated by arrows) formed by residues 403–411 and 415–423 aligned perpendicular to the filament axis.
Figure 5.
(A) Effect of pH on the aggregation kinetics of E422Q RPT monitored by ThT fluorescence at pH 5.5 (circle), 6.0 (triangle), 6.5 (square), and 7 (line). Representative TEM images E422Q fibrils at pH 6.5.[49] (B) Schematic diagram showing a possible conformation of the central core of RPT filaments; it is composed of two β-strands (403–411) and (415–423) and depicts the side-chain orientations for E404 (green) and E422 (blue) as being outside and inside the β-strands, respectively. The fibril axis is coming out of the page. Removal of the critical amyloidogenic region, VSIVVL (cyan), completely abrogates fibril formation.[49]
8 Lysolipids Modulate RPT Fibril Formation
The association of Pmel17 processing and fibril formation with IVLs during early stage melanosome development (Figure 4A) suggests lipids might influence RPT fibrilogenesis. Since melanosome membranes contain a high content of lysophospholipids (lysolipids), their effects on RPT fibril formation were studied.[55] Vesicles containing either negatively charged lysophosphatidylglycerol (LPG) or zwitterionic lysophosphatidylcholine (LPC) stimulated RPT fibril formation, with LPG exerting the greater effect by reducing the apparent lag phase during aggregation.
Since fibril oligomers are likely to be the cytotoxic agent, enhanced fibril aggregation due to association with IVLs would circumvent the over-population of these intermediates. Additionally, sequestration of melanin precursors on fibrils would be promoted, suggesting a dual role for lipid involvement during fibril formation.
9 Biophysical Characterization of RPT Fibril Disassembly
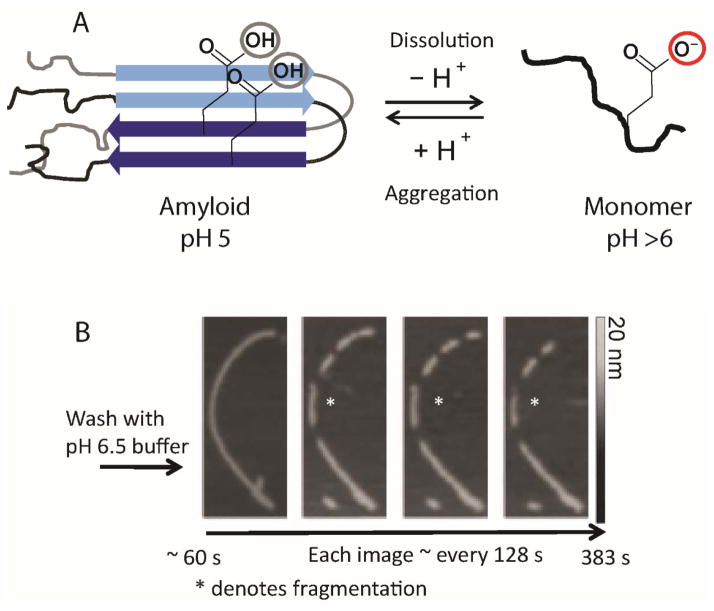
The unique pH dependence of RPT amyloid formation was shown to be reversible, where pre-formed fibrils at pH 5 rapidly disassembled upon exposure to neutral pH (Figure 6A).[41, 49–50, 56] This reversible polymerization behavior is different from those exhibited by pathological amyloids, which only behave similarly when subjected to the harshest of treatments, for example, chemical denaturants and non-physiological pH. However, functional amyloids formed from peptide hormones[15b] and RNA proteins[31c] also show a reversible aggregation/disaggregation processes, which fit well with that observed for RPT.
Figure 6.
(A) Schematic diagram showing the reversible pH disassembly of RPT fibrils. The critical deprotonation/protonation event of E422 residue that is responsible for controlling the pH dependence of amyloid fibril formation is illustrated. (B) Real-time AFM measurements of RPT fibril dissolution. Fibrils formed at pH 5.0 were washed with pH 6.5 buffer and monitored over time (an asterisk denotes one disassembly event). Gray scale indicates fibril height.[56]
Using AFM the fibril disassembly of RPT was studied at the ultrastructural level.[56] Upon washing preformed RPT fibrils with pH 6.5 buffer, fragmentation of larger fibrils followed by the complete disappearance of the smaller fragments on the order of minutes was visualized by AFM (Figure 6B). A technique known as scanning force kymography was used to examine the kinetics of fibril disassembly, which revealed asymmetric dissolution rates at both fibril ends. This observation could result from fibril polymorphism where different fibril conformations have varying sensitivity towards pH.
At the residue level, our data revealed fibrillar aggregates incubated at near neutral pH and monitored by solution-state NMR revealed all Glu residues became deprotonated upon fibril disassembly.[49] An absence of populated oligomers with only monomer formation was observed by both solution-state NMR and size exclusion chromatography. The fact that (toxic) intermediate states are not populated during RPT fibril disassembly at near neutral pH provides mechanistic insight into the benign nature of these fibrils.
10 The Dark Side of RPT
While it is universally agreed that the RPT domain is a feature of melanosome fibrils, others pinpoint the PKD domain as the amyloid-forming region and suggest RPT likely serves a regulatory rather than a structural role. While we cannot preclude the involvement of the PKD domain,[44] we do not believe the prevailing reasons against RPT forming amyloid in vivo are sufficient and conclusive.
First, the fact that the RPT domain is thought to be charged with many O-linked, sialylated glycans in vivo has been over-stated. In fact, the only known glycosylation sites are located in the region between residues 328–344, [36] which is located far away from the C-terminal RPT amyloid core that we have characterized.[47, 49–50, 55–56] Specifically, in our work, we determined by deleting residues, 405–410, where there is no glycosylation, amyloid formation is ablated.[49] In addition, the notion that a glycosylated RPT would be unable to fibrillate in vivo is questioned by the fact that a short C-terminal segment of RPT composed of 405VSIVVLSGT413 alone was capable of self-assembling into amyloid fibrils.[51] This implies that short small peptides might serve as the minimum requirement needed to form in vivo melanosome fibrils. Most importantly, there are two studies that directly show that deletion of RPT results in melansomes devoid of fibrils while processing and trafficking of Pmel17 are unaffected[36, 40] and immunohistochemistry (HMB45 antibody) shows that O-glycosylated RPT is a component of fibrils.[44]
Second, since the RPT domain is the most variable part in the sequence of Pmel17, it was suggested that the diversity in the number and sequence of repeats in RPT makes it an improbable candidate for the amyloid core for an evolutionarily conserved functional amyloid.[44] However, our data reveal very divergent mouse and zebrafish RPT domains form amyloid, in a similar pH-dependent way as human RPT, indicating that the variation of RPT sequence should not discount its ability to be amyloidogenic.
Third, the contradictory work on a different recombinant RPT fragment (residues 303–467) which suggested that RPT does not form fibrils in vitro[44] should be taken with caution, since it contains extra residues on both the N- and C-termini that would change the properties of the protein. For example, constructs slightly expanded from our RPT region such as residues 293–456 did not exhibit filamentous morphology by TEM.[41]
Lastly, the unique reversible pH dependence of RPT is questioned because there is no current evidence of fibril dissolution in mature melanosomes, which are thought to approach neutral pH. We believe that upon melanosome maturation, the fibrils are fully coated by melanin and hence would be (1) difficult to assess whether fibril dissolution has indeed occurred and (2) the deposition of melanin in mature melanosomes would likely protect fibrils from disassembly at any pH. We also note that it remains controversial as to the exact intramelanosomal pH during melanin synthesis.[57] In our view, since both domains are shown to be essential for melanosome fibril formation, we believe that only by isolating ex vivo fibrils would their structural properties unequivocally be revealed. Thus, the identities of the fibrillar components in melanosomes remain to be validated.
11 Summary and Outlook
In this review, we highlighted our work on the repeat domain of Pmel17 that forms amyloid fibrils only at the physiological pH (4–5.5) of melanosomes. Collectively, we have revealed an elegant mechanism that shows a reversible aggregation/disaggregation process of RPT that is strictly controlled by pH. We have identified the molecular origin of this pH dependence to the charge neutralization of E422. Remarkably, the removal of a single negative charge at E422 out of a total of 16 carboxylic acids shifted the pH dependence by a full pH unit. Fibril dissolution studies have indicated that the disassembly process is fully reversible with no detectable oligomers by AFM, solution-state NMR spectroscopy, and size-exclusion chromatography. We have also shown that RPT aggregation is strongly influenced by the negatively-charged lysophosphatidylglycerol, an abundant melanosomal lipid, whereas its two-chained phospholipid counterpart, phosphatidylglycerol, had no effects. As melanosomes are enriched in lysolipids, it is proposed that membrane lipids contribute to the regulation of Pmel17 amyloid formation in vivo, consistent with the observation of fibril association with IVLs.
This unique pH dependence to control fibril assembly has strong biological implications, where fibril formation is formed under its acidic melanosomal environment. While RPT amyloid is sequestered, in an event of releasing RPT amyloid into the cytosol, the ability for RPT to rapidly disaggregate at neutral pH provides an essential response to avoid cytotoxicity associated with amyloid structures. This reversible aggregation/disaggregation process is analogous to amyloids from peptide hormones and RNA proteins, suggesting a common mechanism may be shared amongst human functional amyloids. The emerging concept of “functional” amyloids is challenging the way we view amyloids, especially regarding how cells can deal with and manipulate amyloids. We foresee that more discoveries will be forthcoming involving dynamic assemblies of functional organized amyloid structures.
Acknowledgments
This work is supported by the Intramural Research Program at the National Institutes of Health, National Heart, Lung, and Blood Institute.
References
- 1.a) Greenwald J, Riek R. Structure. 2010;18:1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]; b) Shewmaker F, McGlinchey RP, Wickner RB. J Biol Chem. 2011;286:16533–16540. doi: 10.1074/jbc.R111.227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tycko R. Ann Rev Phys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. Annual Reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Mitsui K, Doi H, Nukina N. Amyloid, Prions, and Other Protein Aggregates. 2006;412:63–76. doi: 10.1016/S0076-6879(06)12005-4. [DOI] [PubMed] [Google Scholar]; b) Peretz D, Supattapone S, Giles K, Vergara J, Freyman Y, Lessard P, Safar JG, Glidden DV, McCulloch C, Nguyen HOB, Scott M, DeArmond SJ, Prusiner SB. J Virol. 2006;80:322–331. doi: 10.1128/JVI.80.1.322-331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Divry P, Florkin M. Comptes Rendus Seances Soc Biol Fil. 1927;97:1808–1810. [Google Scholar]
- 5.Vassar PS, Culling CFA, Taylor HE. Am J Pathol. 1959;35:718–718. [Google Scholar]
- 6.a) Cohen AS, Calkins E. Nature. 1959;183:1202–1203. doi: 10.1038/1831202a0. [DOI] [PubMed] [Google Scholar]; b) Stine WB, Snyder SW, Ladror US, Wade WS, Miller MF, Perun TJ, Holzman TF, Krafft GA. J Protein Chem. 1996;15:193–203. doi: 10.1007/BF01887400. [DOI] [PubMed] [Google Scholar]
- 7.a) Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]; b) Paravastu AK, Leapman RD, Yau WM, Tycko R. Proc Natl Acad Sci USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Petkova AT, Yau WM, Tycko R. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Proc Natl Acad Sci USA. 2009;106:14339–14344. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy MP, LeVine H. J Alzheimers Dis. 2010;19:311–323. [Google Scholar]
- 11.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, DiIorio G, Golbe LI, Nussbaum RL. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 12.Ross CA, Tabrizi SJ. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 13.Barnhart MM, Chapman MR. Annu Rev Microbiol. Vol. 60. Palo Alto: 2006. pp. 131–147. Annual Reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Proc Natl Acad Sci USA. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Plos Biology. 2006;4:100–107. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KPR, Simon R, Schubert D, Eisenberg D, Rivier J, Sawchenko P, Vale W, Riek R. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGlinchey RP, Yap TL, Lee JC. Phys Chem Chem Phys. 2011;13:20066–20075. doi: 10.1039/c1cp21376h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]; b) Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bockmann A, Meier BH. Prion. 2010;4:72–79. doi: 10.4161/pri.4.2.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margittai M, Langen R. Q Rev Biophys. 2008;41:265–297. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- 20.Tycko R. Neuron. 2015;86:632–645. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a) Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]; b) Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, Wan W, Stubbs G, Schwieters CD, Lee VMY, George JM, Rienstra CM. Nat Struct Mol Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benzinger TLS, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Proc Natl Acad Sci USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Proc Natl Acad Sci USA. 2009;106:7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Qiang W, Yau WM, Luo YQ, Mattson MP, Tycko R. Proc Natl Acad Sci USA. 2012;109:4443–4448. doi: 10.1073/pnas.1111305109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tycko R, Sciarretta KL, Orgel J, Meredith SC. Biochemistry. 2009;48:6072–6084. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shewmaker F, McGlinchey RP, Thurber KR, McPhie P, Dyda F, Tycko R, Wickner RB. J Biol Chem. 2009;284:25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glabe CG. J Biol Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fandrich M. J Mol Biol. 2012;421:427–440. doi: 10.1016/j.jmb.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 30.Nespovitaya N, Gath J, Barylyuk K, Seuring C, Meier BH, Riek R. J Am Chem Soc. 2016;138:846–856. doi: 10.1021/jacs.5b08694. [DOI] [PubMed] [Google Scholar]
- 31.a) Hervas R, Li LY, Majumdar A, Fernandez-Ramirez MD, Unruh JR, Slaughter BD, Galera-Prat A, Santana E, Suzuki M, Nagai Y, Bruix M, Casas-Tinto S, Menendez M, Laurents DV, Si K, Carrion-Vazquez M. Plos Biology. 2016;14 doi: 10.1371/journal.pbio.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Majumdar A, Cesario WC, White-Grindley E, Jiang HQ, Ren FZ, Khan M, Li LY, Choi EML, Kannan K, Guo FL, Unruh J, Slaughter B, Si K. Cell. 2012;148:515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]; c) Kato M, Han TNW, Xie SH, Shi K, Du XL, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei JM, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berson JF, Theos AC, Harper DC, Tenza D, Raposo G, Marks MS. J Cell Biol. 2003;161:521–533. doi: 10.1083/jcb.200302072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurbain I, Geerts WJC, Boudier T, Marco S, Verkleij AJ, Marks MS, Raposo G. Proc Natl Acad Sci USA. 2008;105:19726–19731. doi: 10.1073/pnas.0803488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn LC, Thigpen LW. J Hered. 1930;21:495–498. [Google Scholar]
- 35.Kwon BS, Chintamaneni C, Kozak CA, Copeland NG, Gilbert DJ, Jenkins N, Barton D, Francke U, Kobayashi Y, Kim KK. Proc Natl Acad Sci U S A. 1991;88:9228–9232. doi: 10.1073/pnas.88.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoashi T, Muller J, Vieira WD, Rouzaud F, Kikuchi K, Tamaki K, Hearing VJ. J Biol Chem. 2006;281:21198–21208. doi: 10.1074/jbc.M601643200. [DOI] [PubMed] [Google Scholar]
- 37.Berson JF, Harper DC, Tenza D, Raposo G, Marks MS. Mol Biol Cell. 2001;12:3451–3464. doi: 10.1091/mbc.12.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theos AC, Watt B, Harper DC, Janczura KJ, Theos SC, Herman KE, Marks MS. Pigment Cell Melanoma Res. 2013:26. doi: 10.1111/pcmr.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bycroft M, Bateman A, Clarke J, Hamill SJ, Sandford R, Thomas RL, Chothia C. Embo J. 1999;18:297–305. doi: 10.1093/emboj/18.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theos AC, Truschel ST, Tenza D, Hurbain I, Harper DC, Berson JF, Thomas PC, Raposo G, Marks MS. Dev Cell. 2006;10:343–354. doi: 10.1016/j.devcel.2006.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGlinchey RP, Shewmaker F, McPhie P, Monterroso B, Thurber K, Wickner RB. Proc Natl Acad Sci USA. 2009;106:13731–13736. doi: 10.1073/pnas.0906509106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leonhardt RM, Vigneron N, Hee JS, Graham M, Cresswell P. Mol Biol Cell. 2013;24:964–981. doi: 10.1091/mbc.E12-10-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valencia JC, Rouzaud F, Julien S, Chen KG, Passeron T, Yamaguchi Y, Abu-Asab M, Tsokos M, Costin GE, Yamaguchi H, Jenkins LMM, Nagashima K, Appella E, Hearing VJ. J Biol Chem. 2007;282:11266–11280. doi: 10.1074/jbc.M608449200. [DOI] [PubMed] [Google Scholar]
- 44.Watt B, van Niel G, Fowler DM, Hurbain I, Luk KC, Stayrook SE, Lemmon MA, Raposo G, Shorter J, Kelly JW, Marks MS. J Biol Chem. 2009;284:35543–35555. doi: 10.1074/jbc.M109.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.a Leonhardt RM, Vigneron N, Rahner C, van den Eynde BJ, Cresswell P. J Biol Chem. 2010;285:16166–16183. doi: 10.1074/jbc.M109.097725. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Leonhardt RM, Vigneron N, Rahner C, Cresswell P. J Biol Chem. 2011;286:9321–9337. doi: 10.1074/jbc.M110.168088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hellstrom AR, Watt B, Fard SS, Tenza D, Mannstrom P, Narfstrom K, Ekesten B, Ito S, Wakamatsu K, Larsson J, Ulfendahl M, Kullander K, Raposo G, Kerje S, Hallbook F, Marks MS, Andersson L. PLoS Genet. 2011:7. doi: 10.1371/journal.pgen.1002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu KN, McGlinchey RP, Wickner RB, Tycko R. Biophys J. 2011;101:2242–2250. doi: 10.1016/j.bpj.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiltzius JJW, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, Soriaga AB, Cascio D, Rajashankar K, Eisenberg D. Nat Struct Mol Biol. 2009;16:973–U998. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGlinchey RP, Jiang ZP, Lee JC. Chembiochem. 2014;15:1569–1572. doi: 10.1002/cbic.201402074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfefferkorn CM, McGlinchey RP, Lee JC. Proc Natl Acad Sci USA. 2010;107:21447–21452. doi: 10.1073/pnas.1006424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Louros NN, Iconomidou VA. Biopolymers. 2016;106:133–139. doi: 10.1002/bip.22746. [DOI] [PubMed] [Google Scholar]
- 52.McGlinchey RP, Shewmaker F, Hu KN, McPhie P, Tycko R, Wickner RB. J Biol Chem. 2011;286:8385–8393. doi: 10.1074/jbc.M110.197152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nichols SE, Harper DC, Berson JF, Marks MS. J Invest Dermatol. 2003;121:821–830. doi: 10.1046/j.1523-1747.2003.12474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raposo G, Tenza D, Murphy DM, Berson JF, Marks MS. J Cell Biol. 2001;152:809–823. doi: 10.1083/jcb.152.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang ZP, Lee JC. J Mol Biol. 2014;426:4074–4086. doi: 10.1016/j.jmb.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGlinchey RP, Gruschus JM, Nagy A, Lee JC. Biochemistry. 2011;50:10567–10569. doi: 10.1021/bi201578h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhatnagar V, Anjaiah S, Puri N, Darshanam BNA, Ramaiah A. Arch Biochem Biophys. 1993;307:183–192. doi: 10.1006/abbi.1993.1577. [DOI] [PubMed] [Google Scholar]